Abstract
Glycosylation is a ubiquitous reaction controlling the bioactivity and storage of plant natural products. Glycosylation of small molecules is catalyzed by a superfamily of glycosyltransferases (GTs) in most plant species studied to date. We present crystal structures of the UDP flavonoid/triterpene GT UGT71G1 from Medicago truncatula bound to UDP or UDP-glucose. The structures reveal the key residues involved in the recognition of donor substrate and, by comparison with other GT structures, suggest His-22 as the catalytic base and Asp-121 as a key residue that may assist deprotonation of the acceptor by forming an electron transfer chain with the catalytic base. Mutagenesis confirmed the roles of these key residues in donor substrate binding and enzyme activity. Our results provide an initial structural basis for understanding the complex substrate specificity and regiospecificity underlying the glycosylation of plant natural products and other small molecules. This information will direct future attempts to engineer bioactive compounds in crop plants to improve plant, animal, and human health and to facilitate the rational design of GTs to improve the storage and stability of novel engineered bioactive compounds.
INTRODUCTION
Glycosylation is quantitatively the most significant reaction on earth. It is catalyzed by a superfamily of enzymes, the glycosyltransferases (GTs), which have been classified into >70 families (Campbell et al., 1997) (http://afmb.cnrs-mrs.fr/CAZY/index.html). Family 1 GT enzymes, the UDP glycosyltransferases (UGTs), transfer UDP-activated sugar moieties to specific acceptor molecules. Twenty-seven UGTs have been identified in the human genome (de Wildt et al., 1999; Li et al., 2001); these play central roles in the metabolism and detoxification of foreign chemicals such as carcinogens and hydrophobic drugs. By contrast, plants contain a very large number of UGTs that are involved in the glycosylation of natural products (Vogt and Jones, 2000; Jones and Vogt, 2001) and contain a highly conserved consensus signature sequence called the PSPG motif (Hughes and Hughes, 1994; Paquette et al., 2003). Plants collectively synthesize >200,000 natural products, and a significant proportion of them are glycosylated. Consistent with this chemical complexity, 117 putative UGT genes have been identified in the genome of Arabidopsis thaliana (Ross et al., 2001). Likewise, >100 UGTs are predicted in the model legume Medicago truncatula based on an analysis of currently available EST and genomic sequences (Achnine et al., 2005).
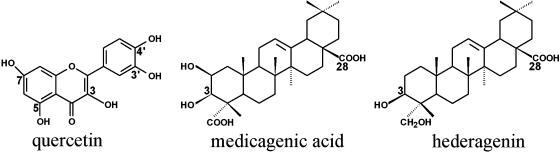
Glycosylation is generally seen as a mechanism for the detoxification of xenobiotics or for facilitating the storage of bioactive molecules in plant vacuoles (Sandermann, 1992; Paquette et al., 2003). However, in some cases, glycosylated plant natural products exhibit higher bioactivity than their corresponding aglycones, as in the case of the triterpene saponins (Osbourn, 2003). UGT71G1 was recently identified in M. truncatula as an enzyme functionally involved in the biosynthesis of saponins, although the enzyme exhibits more favorable kinetics with the flavonoid quercetin (Achnine et al., 2005). Triterpene glycosides have allelopathic, antimicrobial, insecticidal, and antiherbivory activities for plant protection (Osbourn et al., 1996; Small, 1996; Oleszek et al., 1999; Papadopoulou et al., 1999). They also have pharmacological, anticholesterolemic, anticancer, adjuvant, and hemolytic activities (Behboudi et al., 1999; Oh et al., 2000; Haridas et al., 2001), act on the cardiovascular, nervous, and digestive systems, and are used in sneezing powders, emetics, and cough syrups to facilitate expectoration (Dennehy, 2001). The saponins in M. truncatula are glycosides of the five triterpene aglycones soyasapogenol B, soyasapogenol E, medicagenic acid, hederagenin, and bayogenin (Huhman and Sumner, 2002). These aglycones share similar structures, and UGT71G1 recognizes both medicagenic acid and hederagenin as acceptors (Figure 1) for the sugar moiety delivered from UDP-glucose. It is not yet clear which positions on the triterpene scaffold are glycosylated by UGT71G1; O-glycosylation at positions 3 and 28 is most common for this class of compound, but C-glycosylation of plant natural products has also been observed, and these products are of particular pharmaceutical interest (Clemens et al., 2004). Understanding the structural basis for the glycosylation of triterpene natural products will facilitate future metabolic engineering of these compounds to improve the health of plants (resistance to disease and herbivory), animals (via improved forage quality), and humans (through drug discovery).
Figure 1.
Structures of Three Substrates of M. truncatula UGT71G1.
The known or predicted glycosylation sites are labeled with numbers: quercetin (3, 5, 7, 3′, and 4′), medicagenic acid (3 or 28), and hederagenin (3 or 28).
To date, 17 members of the GT superfamily (but none from plants) have been analyzed structurally (Goldsmith et al., 1989; Vrielink et al., 1994; Charnock and Davies, 1999; Gastinel et al., 1999, 2001; Ha et al., 2000; Pedersen et al., 2000, 2003; Unligil et al., 2000; Persson et al., 2001; Gibbons et al., 2002; Gibson et al., 2002; Buschiazzo et al., 2004; Fritz et al., 2004; Lobsanov et al., 2004; Mulichak et al., 2004), and these structures fall into two different topologies/folds, GT-A and GT-B (Breton et al., 2002; Hu and Walker, 2002; Coutinho et al., 2003). Plant UGTs have been predicted to be similar to the bacterial GT family 1 TDP-epi-vancosaminyltransferases GtfA, GtfB, and GtfD from Amycolatopsis orientalis (Mulichak et al., 2001, 2003, 2004; Hans et al., 2004; Bowles et al., 2005), which belong to the GT-B fold comprising two Rossmann-like domains. However, bacterial Gtfs share only ∼10% sequence identity with plant UGTs, making it very difficult to model plant glycosylation processes without knowledge of plant UGT crystal structures.
Here, we report the structures of UGT71G1 from M. truncatula bound to UDP or UDP-glucose. These plant UGT crystal structures reveal the detailed interactions between the enzyme and its donor substrate as well as the roles of the UGT signature motif in substrate recognition and the catalytic activity of the enzyme. Further mutational and structural analyses, and comparisons with other GT structures, will provide a basis for understanding the complex and often overlapping substrate specificity and regiospecificity within the plant UGT superfamily.
RESULTS
Overall Structure
The three-dimensional structure of UGT71G1 from M. truncatula bound with a UDP molecule was determined using the multiwavelength anomalous dispersion (MAD) method with a crystal of Se-Met–substituted enzyme and refined to 2.0-Å resolution with a native data set. Data collection, phasing, and refinement statistics are presented in Table 1.
Table 1.
Data Collection, Phasing, and Refinement Statistics
| Data Collection and Phasing Statistics | |||||
|---|---|---|---|---|---|
| Native (+ UDP) | Se peak (+ UDP) | Se inflection (+ UDP) | Se remote (+ UDP) | Complex (+ UPG) | |
| Resolution (Å) | 2.0 | 2.4 | 2.4 | 2.4 | 2.6 |
| Wavelength (Å) | 1.5418 | 0.9797 | 0.98 | 0.9393 | 1.5418 |
| Unique reflections | 62,825 | 36,121 | 35,553 | 34,461 | 29,545 |
| Completeness (%) | 98.2 (97.7) | 94.4 (81.8) | 97.0 (85.0) | 92.4 (78.4) | 99.8 (99.7) |
| Rsyma(%) | 4.1 (27.7) | 8.2 (26.0) | 5.7 (25.2) | 6.0 (28.6) | 5.9 (44.2) |
| I/σ (I) | 13.7 (2.7) | 16.5 (2.7) | 16 (2.6) | 16.1 (3.1) | 12.1 (2.3) |
| Figure of merit | 0.44 | ||||
| Refinement Statistics | ||
|---|---|---|
| Enzyme-UDP | Enzyme-UDP glucose (UPG) | |
| R factor (%) | 18.7 | 20.5 |
| Rfree(%) | 22.9 | 28.0 |
| Number of protein atoms | 7204 | 7204 |
| Number of solvent atoms | 728 | 17 |
| Number of ligand atoms | 50 (two UDP molecules) | 72 (two UDP-glucose molecules) |
| Average B factors (Å2) | 31.8 | 56.3 |
| RMSD from ideal values | ||
| Bond length (Å) | 0.0057 | 0.0070 |
| Bond angle (°) | 1.27 | 1.29 |
Numbers in parentheses are for the highest-resolution shell.
Rsym = Σhkl |I − <I>|/ΣI, where I is the observed intensity and <I> is the average intensity from observations of symmetry-related reflections. A subset of the data (8%) was excluded from the refinement and used to calculate the free R value (Rfree). R factor = Σ‖Fo| − |Fc‖/Σ|Fo|.
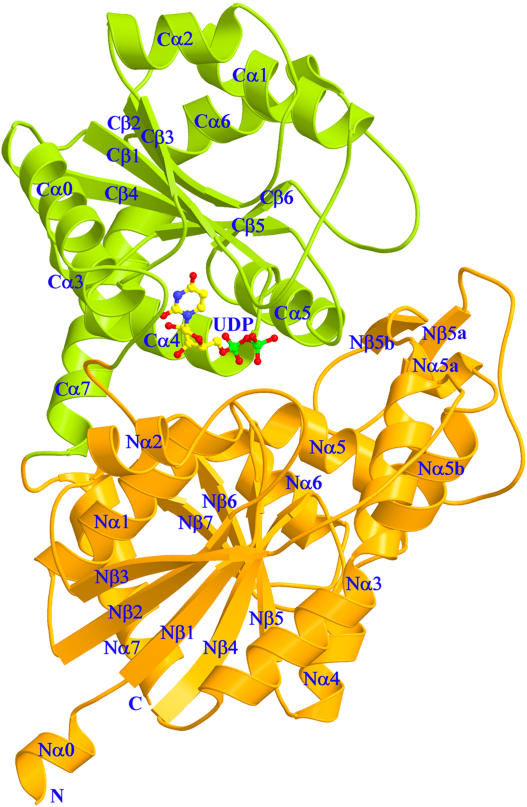
The structure of UGT71G1 consists of two N- and C-terminal domains with similar Rossmann-type folds and, as predicted, belongs to the GT-B fold (Figures 2 and 3). The N-terminal domain contains a central seven-stranded parallel β sheet flanked by eight α helices on both sides and a small two-stranded β sheet. The C-terminal domain contains a six-stranded β sheet flanked by eight α helices. The two domains pack very tightly and form a deep cleft with a UDP molecule bound.
Figure 2.
Ribbon Diagram of the Structure of UGT71G1 with Bound UDP.
The N- and C-terminal domains are shown in orange and green, with the secondary structures and the N and C termini labeled. The α helices and β strands in the N- and C-terminal domains are numbered separately. The UDP molecule is shown as a ball-and-stick model colored by atom type (nitrogen, blue; carbon, yellow; oxygen, red; phosphorus, green). Figures 2, 3B, 4, 5, and 6 were prepared with MOLSCRIPT (Kraulis, 1991) and RASTER3D (Merritt and Bacon, 1997).
Figure 3.
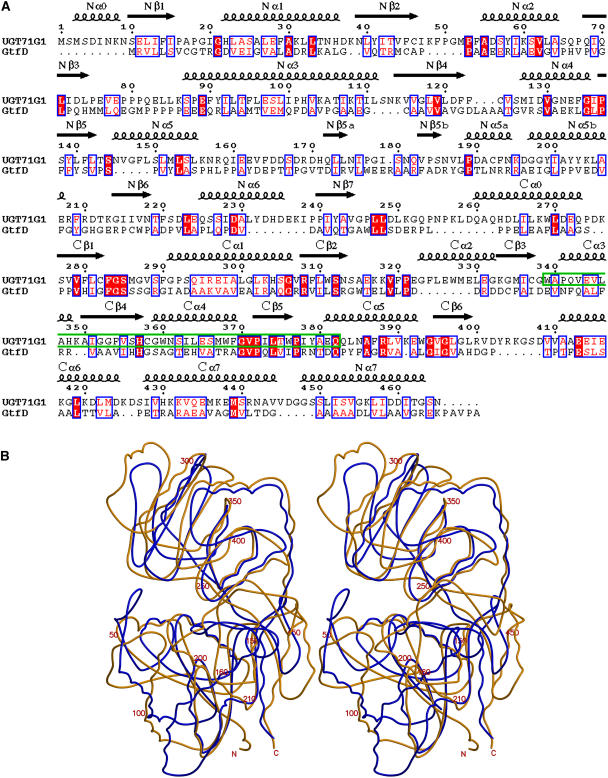
Comparison of M. truncatula UGT71G1 and A. orientalis GtfD.
(A) Structure-based sequence alignment of M. truncatula UGT71G1 and A. orientalis GtfD. The secondary structure elements observed in the UGT71G1 structure are shown above the alignment. The UGT signature motifs are enclosed in a green box. Conserved residues are highlighted. This figure was produced with ENDscript (Gouet and Courcelle, 2002).
(B) Stereo diagram showing the superimposition of the structures of UGT71G1 (molecule A; brown) and GtfD (blue; Protein Data Bank [PDB] code 1RRV).
The asymmetric unit contains two independent UGT71G1 molecules related by a noncrystallographic twofold axis. These two molecules are very similar to each other, with root mean square deviations (RMSD) of 1.04 and 1.52 Å for 456 Cα atoms and all atoms (residues 8 to 463), respectively. However, small differences were observed between the two molecules, especially in the region near the UDP binding site in the cleft between the N- and C-terminal domains, which included the C-terminal part of helix Nα1 (around residue Ile-20); loop Nβ2-Nα2 (around residue Pro-50), which is highly mobile with high B factors (∼80 Å2) and poorly defined electron density in both molecules A and B; and the loop linker between the N- and C-terminal domains (around residue Asn-255), the electron density of which was poorly defined in molecule B. The helix Nα1 (P18 to H36) and loop Nβ2-Nα2 and the following helix Nα2 (K48 to Q66) in molecule B shift away slightly from the C-terminal domain, with the largest movement, 2.6 Å, for the Cα atom of Leu-23, which is adjacent to the key catalytic residue His-22 (see below). This suggests that the flexibility in the cleft region between the two subunits allows for movement to occur during substrate binding and catalysis.
Comparison with Other Members of the GT Superfamily
There are three known structures of family 1 GTs, all for bacterial vancomycin UDP glucosyltransferase Gtfs. Superimposing the structures of plant UGT71G1 and bacterial GtfD revealed strong structural similarity, with an RMSD of 2.23 Å for 242 Cα atoms, although the sequence identity is very low (∼10%) (Figure 3). The largest difference between these two enzymes was observed in the region between the helix Nα5 and the strand Nβ6 (∼155 to 210) in their N-terminal domains. A two-stranded β sheet is present in this region of the UGT71G1 structure, resembling an arm flipped to the opposite side near the helix Cα5 of the C-terminal domain, compared with the corresponding region in the GtfD structure. This region forms one side of the substrate (acceptor molecule) binding pocket, and this large difference in conformation/folding changes the shape and size of the binding pocket. The comparison also shows that the similarity between the C-terminal domains of UGT71G1 and GtfD (RMSD = 2.13 Å for 137 Cα atoms) is higher than that between their N-terminal domains (RMSD = 2.34 Å for 120 Cα atoms), presumably because the C-terminal domains recognize similar donor molecules but the N-terminal domains recognize quite different acceptor molecules. The UGT71G1 structure is more compact in its N-terminal domain, presumably because its substrates, such as medicagenic acid and quercetin, are considerably smaller than the acceptor substrate desvancosaminyl vancomycin (DVV) of GtfD.
Comparison was also performed between the UGT71G1 structure and those of other GTs with the GT-B fold. Escherichia coli MurG (Coutinho et al., 2003) is closely related to UGT71G1 in the overall GT classification. Superimpositions between portions of the structures of UGT71G1 and MurG gave RMSD values of 2.27 Å (199 Cα atoms of the overall structures), 2.23 Å (95 Cα atoms in the N-terminal domains), and 1.95 Å (110 Cα atoms in the C-terminal domains).
Trehalose-6-phosphate synthetase OtsA, T4 bacteriophage β-glucosyltransferase (BGT), and glycogen synthase are relatively distant from UGT71G1 in classification, although they use the same UDP-sugar or, in the case of glycogen synthase, the related ADP-sugar. Superimposing the overall structures of UGT71G1 and OtsA, which have very little sequence identity, gave an RMSD of 2.21 Å (158 Cα atoms). Superimposing their N- and C-terminal domains gave RMSD values of 2.17 Å (96 Cα atoms) and 2.27 Å (70 Cα atoms), respectively; the N-terminal rather than the C-terminal domain is more similar to the corresponding domain of UGT71G1, in contrast with the situation with UGT71G1 and GtfD or MurG. Similarly, superimposing the N- and C-terminal domains of UGT71G1 and BGT gave RMSD values of 2.28 Å (79 Cα atoms) and 2.29 Å (70 Cα atoms), respectively.
Interactions between Enzyme and Donor Ligands
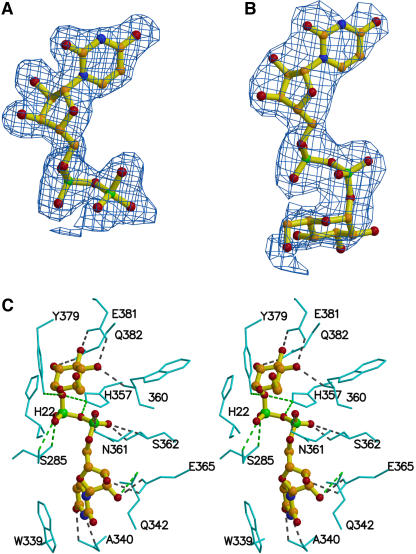
Although UDP-galactose (a nondonor) and quercetin were included for cocrystallization with UGT71G1, the only substrate electron density observed was for the UDP moiety of UDP-galactose (Figure 4A) as a result of possible hydrolysis. This is similar to the situation with the bacterial GtfD structure, in which only TDP was observed, although GtfD was cocrystallized with TDP-glucose (Mulichak et al., 2004). Soaking crystals of UGT71G1 with UDP-glucose resulted in a weak electron density map for the glucose portion of the molecule in the enzyme structure at 2.6 Å (Figure 4B). The B factor of the glucose moiety is higher than that of the UDP moiety, indicating either flexibility or low occupancy of the sugar attributable to the possible hydrolysis of UDP-glucose.
Figure 4.
Donor Molecules and Their Interactions with UGT71G1.
(A) A |Fobs| − |Fcalc| electron density omit map of bound UDP contoured at 1.5 σ is superimposed on a ball-and-stick model of the UDP molecule.
(B) A |Fobs| − |Fcalc| electron density omit map of bound UDP-glucose contoured at 1.5 σ is superimposed on a ball-and-stick model of the UDP-glucose molecule.
(C) Stereo diagram showing interactions between bound UDP-glucose and UGT71G1 side chains. The structure of UDP-glucose is shown as a ball-and-stick model. Hydrogen bonding interactions are indicated by dashed lines. Interactions observed only between UDP(-galactose) and UGT71G1 in the 2.0-Å structure are indicated by green dashed lines.
UDP or UDP-glucose is almost completely buried in a long, narrow channel mainly within the C-terminal domain of the enzyme. In the 2.0-Å structure of UGT71G1 complexed with UDP, the enzyme interacts with UDP mainly through residues in the UGT signature PSPG motif (residues 339 to 382) (Figure 4C, Table 2). The uracil ring of UDP interacts through hydrogen bonds between the N3 and O4 atoms of the ring and the main chain oxygen and nitrogen atoms of Ala-340 and also forms parallel π-stacking interactions with the indole ring of Trp-339. The ribose ring of UDP makes contacts with the enzyme through hydrogen bonds between its O2* and O3* atoms and the OE1 and OE2 atoms of Glu-365 and between its O2* atom and the NE2 atom of Gln-342. The α-phosphate forms hydrogen bonds through its O1A atom and the ND2 atom of Asn-361, its O2A atom and the OG and N atoms of Ser-362, and its O3A atom and the NE2 atom of His-357. The β-phosphate forms hydrogen bonds through its O3B oxygen and the OH atom of Tyr-379 and the NE2 atom of His-357 and between its O2B oxygen and the OG and N atoms of Ser-285, the only interacting residue outside of the UGT signature motif region.
Table 2.
Interactions between Enzyme Molecules (A or B) and Donors (UDP or UPG)
| Distance (Å)
|
|||||
|---|---|---|---|---|---|
| Protein Residue: Atom | Donor Atom | Molecule A, UDP | Molecule B, UDP | Molecule A, UPG | Molecule B, UPG |
| A340: N | O4 | 3.0 | 3.1 | 2.8 | 3.3a |
| O | N3, O2 | 2.7, 3.3 | 2.6, 3.3 | 2.6, 3.1 | 2.8, 2.9 |
| Q342: NE2 | O2* | 3.0 | 3.0 | 3.4 | 3.3 |
| E365: OE1 | O2* | 2.7 | 2.6 | 2.7 | 2.7 |
| OE2 | O2*, O3* | 3.2, 2.8 | 3.2, 2.8 | 3.1, 2.9 | 3.0, 2.6 |
| N361: ND2 | O1A | 3.1 | 3.2 | 3.1 | 3.2 |
| S362: N | O2A | 2.8 | 2.9 | 3.1 | 2.9 |
| OG | O2A | 2.5 | 2.8 | 2.5 | 2.5 |
| H357: NE2 | O3A | 2.9 | 2.9 | 3.4 | 3.2 |
| O3B | 3.0 | 3.2 | 3.4 | 3.4 | |
| Y379: OH | O3B | 2.4 | 2.6 | 3.3 (w. O2B) | 3.2(w. O2B) |
| S285: N | O2B | 2.7 | 2.9 | 3.4 | 3.3 |
| OG | O2B | 2.7 | 2.7 | 3.5 | 3.3 |
| W360: N | O4′ | 2.9 | 2.6 | ||
| E381: OE1 | O3′ | 2.4 | 2.4 | ||
| OE2 | O4′ | 2.6 | 3.0 | ||
| Q382: NE2 | O2′ | 2.7 | 2.9 | ||
Numbers for distances >3.3 Å are underlined.
Interactions between the enzyme and the glucose moiety of the donor are observed in the 2.6-Å structure of UGT71G1 in its complex with UDP-glucose (Figure 4C, Table 2). The O2′ and O3′ atoms of the glucose ring form hydrogen bonds with the NE2 atom of Gln-382 and the OE1 atom of Glu-381. The O4′ atom of the glucose ring contacts both the OE2 atom of Glu-381 and the N atom of Trp-360. On the other hand, several interactions observed between UDP and the enzyme are not present in the model of UDP-glucose bound to the enzyme. A small rotation and shift in the position of the β-phosphate of UDP-glucose weakens the interaction of the O2B and O3B atoms of β-phosphate and the O3A atom of α-phosphate with protein residues.
In UGT71G1 binding UDP-glucose, GtfD binding TDP, trehalose-6-phosphate synthetase OtsA binding UDP, MurG binding UDP-GlcNAc, and T4 bacteriophage BGT and its mutant D100A binding UDP or UDP-glucose, the sugar donor molecules are observed in a similar region of the enzymes (Gibson et al., 2002; Hu et al., 2003; Lariviere et al., 2003; Mulichak et al., 2004). The PSPG motif of UGT71G1 and the corresponding sugar binding regions in these different GTs share a very similar conformation, α/β/α/β, even though the sequence identities are very low and the detailed interactions with the sugar donors also vary. Superimpositions show that the UGT signature motif of UGT71G1 and the corresponding regions of GtfD and MurG share higher structural similarity, indicative of similar binding modes with sugar donors, and differ from those for OtsA and BGT, in which the first β strands of their α/β/α/β motifs are long and the second β strands cannot interact with sugar donors (Figure 5).
Figure 5.
Comparison of Donor Binding Regions of UGT71G1 and Other GT-B Fold Enzymes.
Stereo diagram showing superimposition of the UGT signature motif regions of UGT71G1 (yellow), GtfD (red), MurG (green; PDB code 1F0K), OtsA (cyan; PDB code 1GZ5), and BGT (black; PDB code 1J39).
The Acceptor Binding Site
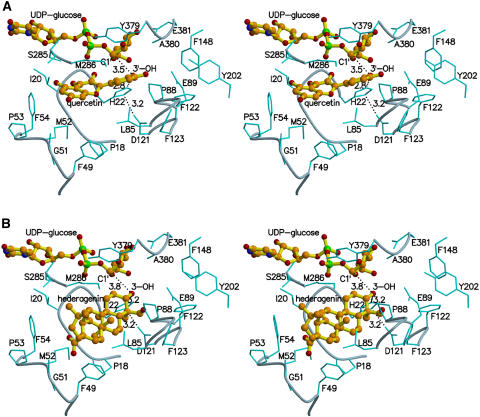
A very narrow, deep pocket is observed adjacent to the UDP binding site, mainly consisting of residues from the N-terminal domain (Figure 6), which would appear to be the binding pocket for acceptor molecules. The location of this pocket is similar to that of the substrate binding site observed in the structure of GtfD bound with DVV and TDP (Mulichak et al., 2004). In the GtfD structure, Asp-13 is close to both DVV and TDP and has been proposed as the general base for the catalysis of glycosyl transfer. His-22 occupies the corresponding position in UGT71G1 and is 4.7 Å from the C1′ atom of UDP-glucose, as shown in the structure of UGT71G1 (molecule A) complexed with UDP-glucose. Close to it (∼3 Å), there is an acidic residue (Asp-121) that may form an electron transfer chain to His-22 to help deprotonate acceptor molecules. The sequence alignment of multiple plant UGTs (Figure 7) shows that corresponding His and Asp residues are conserved in most plant UGTs.
Figure 6.
The Putative Acceptor Binding Pocket.
Stereo diagram showing quercetin (A) and hederagenin (B) docked into the proposed binding pockets. Quercetin, hederagenin, and UDP-glucose are shown as ball-and-stick models. Some protein residues in the acceptor binding pocket are labeled and shown in cyan as bond models. Distances (Å) between the OH group of acceptors and the atom NE2 of His-22 or the atom C1′ of UDP-glucose are labeled and indicated with dashed lines.
Figure 7.
Comparison of the Donor and Acceptor Regions of Functionally Characterized Plant UGTs.
Sequence alignment of 39 plant UGTs, showing the acceptor binding region (first three fragments: 1 to 55, 81 to 95, and 117 to 151) and the donor binding region (last two fragments: 282 to 286 and 339 to 382). Identical residues are highlighted, and similar residues are enclosed in boxes. Residues His-22 and Asp-121 are marked with asterisks. The alignment was performed using ClustalX (Thompson et al., 1997). The plant UGTs include Medicago truncatula UGT71G1 and UGT73K1; Allium cepa UGT73G1 and UGT73J1; Aralia cordata GaT; Bellis perennis UGAT; Brassica napus SGT1; Catharanthus roseus UGT2; Citrus maxima 1,2RhaT; Crocus sativus UGTCs2; Dorotheanthus bellidiformis UGT73A5; Gentiana triflora 3′GT; Glycyrrhiza echinata UGT73F1; Nicotiana tabacum GT1a, GT2, and GT3; Phaseolus lunatus ZOG1; Phaseolus vulgaris ZOX1; Sorghum bicolor UGT85B1; Stevia rebaudiana UGT74G1, UGT76G1, and UGT85C2; and Zea mays cisZOC1 and cisZOG2. All others are from Arabidopsis thaliana.
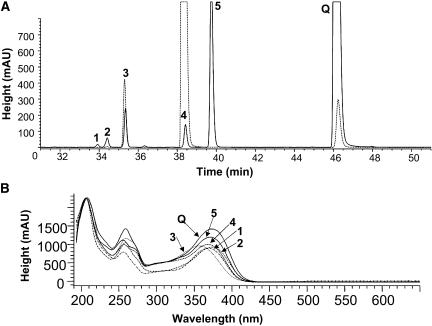
There are several potential O-glycosylation sites (five hydroxyl groups) on the quercetin molecule, and multiple products are formed after incubation of quercetin and UDP-glucose with UGT71G1 (Figure 8A). All five products from the in vitro incubation of recombinant UGT71G1 with UDP-glucose and quercetin exhibited a similar UV spectrum to that of quercetin (Figure 8B), had identical molecular ion masses (462.5 to 462.9 D) consistent with quercetin monoglucoside structures, and disappeared after incubation with β-glucosidase (data not shown). This indicates that, in spite of the production of one favored product (peak 5 in Figure 8A, identified as quercetin-3′-O-glucoside), UGT71G1 is capable of glucose transfer to each of the five hydroxyl groups on the quercetin molecule. To probe the structural basis of the apparently promiscuous acceptor binding to UGT71G1, molecular docking was performed using the software GOLD. Consistent with the multiple product specificity with quercetin and the fact that quercetin was not observed in the structure when included in the crystallization liquid or soaked into crystals, all five hydroxyl groups of quercetin could be docked close to His-22, which lies in the middle of the potential acceptor channel. Three aromatic residues, Phe-148, Phe-122, and Tyr-202, and an acidic residue, Asp-190, are located at one end of the channel; the other end is relatively larger, containing two aromatic residues, Phe-49 and Phe-54, and two Mets, Met-52 and Met-286. An aromatic residue, Phe-123, and an acidic residue, Glu-89, are in the middle of the channel, opposite UDP-glucose. The pocket is much longer and larger than quercetin, and quercetin may be fit in easily, with any of its hydroxyl groups located ∼3 Å from the NE2 atom of His-22.
Figure 8.
Glucosylation of Quercetin by Recombinant UGT71G1.
(A) HPLC trace showing the substrate quercetin (Q) and the five products (numbered 1 to 5). Solid line, complete reaction mixture; dashed line, standards for quercetin (Q), quercetin-3-O-glucoside (peak 3), and quercetin-4′-O-glucoside (peak 4). mAU, milli-absorption unit.
(B) UV absorption spectra of quercetin (Q) and products 1 to 5.
The docking in the active site of the hydroxyl groups at positions 3 or 3′ of quercetin gives fitness scores slightly higher than for the other three hydroxyls, and also more potential interactions between quercetin and the protein and donor. In the model with the 3′-OH docked in the active site (Figure 6A), hydrogen bonds may form between the 3-OH of quercetin and the O1B atom of UDP-glucose and between the 5-OH of quercetin and the OG atom of Ser-285.
The triterpene substrates hederagenin and medicagenic acid are more hydrophobic and significantly larger than quercetin. Compared with the docking results with quercetin, docking with medicagenic acid and hederagenin gave lower fitness scores and weaker interaction with the enzyme, consistent with previously reported kinetic data (Achnine et al., 2005). The molecular docking study for hederagenin and medicagenic acid also suggested that O-glycosylation at position 3 would be preferred over other positions. In the model docked with hederagenin (Figure 6B) or medicagenic acid, the OH group at position 3 points to the NE2 atom of His-22 (∼3.2 Å) and is located just above the C1′ atom of UDP-glucose (∼3.8 Å). Similar to the proposed general mechanism of other inverting GTs (Unligil and Rini, 2000), His-22 would act as an effective general base to deprotonate the OH group of the acceptor molecule, then the newly formed nucleophilic oxyanion would attack the C1′ carbon of the UDP-glucose directly to displace the UDP moiety. The carboxyl groups at position 28 of the triterpene acceptors may hydrogen bond with the main chain oxygen atom of Gly-51. The residues Pro-18, Phe-49, Pro-53, Phe-54, Leu-85, Pro-88, Ile-92, and Phe-123 provide a hydrophobic environment for the acceptor substrates. A solution of the model for docking the carboxyl oxygen atom at position 28 of hederagenin to the active site had a slightly lower fitness score than that for the 3-hydroxyl. In the former case, the hydroxyl group at position 3 may interact with the main chain oxygen atom of Gly-51.
Mutagenesis of UGT71G1
To further explore the roles of the potential key residues identified above, structure-directed point mutants of UGT71G1 were constructed, targeting residues involved in direct contact with the sugar donor, the proposed general base His-22, and its partner, Asp-121. Substitution of His-22 with Ala (H22A) led to the complete loss of enzyme activity, which is consistent with the proposed role of His-22 as the general base. Substitution of Asp-121 with Ala or Asn resulted in no detectable enzyme activity. This is similar to the situation with trypsin, for which the mutant D102N (in the catalytic triad) showed a large loss in activity (Sprang et al., 1987). Also similar to the case of trypsin, Asp-121 in UGT71G1 is nearly buried and forms a hydrogen bond with a catalytic His residue. Our results demonstrate that Asp-121 and its proper interaction with His-22 are essential for enzyme activity. In the model docked with acceptors, Asp-121 is not able to contact the acceptor molecules directly. This finding suggests that Asp-121 may play an important role in assisting His-22 by initialization or activation of the reaction through electron transfer. The E381A mutant also showed no detectable activity, demonstrating the key role of Glu-381 in recognition of the sugar donor substrate because the side chain of Glu-381 forms hydrogen bonds with the O3′ and O4′ atoms of the glucose ring of the donor (Figure 4C). By contrast, mutant A340F retained activity similar to that of the wild-type enzyme reported previously (Achnine et al., 2005). Ala-340 forms hydrogen bond interactions with the uracil ring of the donor through its main chain oxygen and nitrogen atoms; therefore, such bonds will still be supplied by Phe.
DISCUSSION
Comparisons between GT-B and GT-A Fold GTs
Comparison of the three-dimensional structures of UGT71G1 and several other GT-B fold enzymes revealed high similarity despite the very low sequence identity. Gtfs and MurG, with a similar donor binding motif, share higher similarity in their C-terminal domains than in their N-terminal domains, suggesting a similar sugar donor binding mode and probably a common evolutionary origin. OtsA and BGT, with a slightly different donor binding motif, share higher similarity in their N-terminal domains with UGT71G1. Glycogen synthase (PDB code 1RZV) shares higher similarity with UGT71G1 in the C-terminal domain, but its donor binding region is closer to that of OtsA or BGT (data not shown). This suggests that these enzymes have different sugar donor binding modes and evolutionary origins.
GT-A fold enzymes contain only a single extended domain (or two tightly associated and abutting domains) that recognizes both donor and acceptor (Davies et al., 2001; Coutinho et al., 2003). The structures of GT-A fold enzymes and the N- and C-terminal domains of GT-B fold enzymes are similar to each other. Superimposition between a GT-A enzyme, human β1,3-glucuronyltransferase I (family 43; PDB code 1FGG) (Pedersen et al., 2000), and each domain of UGT71G1 gave RMSD values of 2.3 Å for 80 Cα atoms in the N-terminal domain and 1.96 Å for 40 Cα atoms in the C-terminal domain. This finding suggests that GT-A and GT-B enzymes are evolutionarily related. During evolution, two domain enzymes were probably produced, with one domain recognizing similar donors and another adapting to divergent acceptors in the natural environment.
Substrate Binding and Catalysis by UGT71G1
Kubo et al. (2004) reported that a single mutation, H374Q, in Aralia cordata UDP-galactose:anthocyanin galactosyltransferase (corresponding to Gln-382 in UGT71G1) switched the preferred donor from UDP-galactose to UDP-glucose and suggested that the last residue, Gln or His, in this signature motif would define the sugar donor specificity. However, the Scutellaria baicalensis UDP-glucose:flavonoid glucosyltransferase mutant Q382H does not acquire galactosyltransferase activity, suggesting that other residues might also play roles in sugar recognition and specificity in plant UGTs. In the structure of UGT71G1 bound with UDP-glucose, the O2′ and O3′ atoms of the glucose ring of the donor form hydrogen bonds with the NE2 atom of Gln-382 and the OE1 atom of Glu-381, respectively. The O4′ atom of the glucose ring contacts both the OE2 atom of Glu-381 and the N atom of Trp-360. If a UDP-galactose is placed in the position of the UDP-glucose in the structure of UGT71G1, the O4′ atom of galactose points away from the OE2 atom of Glu-381 and can no longer form a hydrogen bond. This suggests that Glu-381 may be a key residue for sugar recognition and specificity.
A structural model for catalysis by plant UGTs has been proposed based on a modeling and mutagenesis study of UGT73A5, a betanidin glucosyltransferase from Dorotheanthus bellidiformis (Hans et al., 2004). However, the low identity (∼14%) between UGT73A5 and the bacterial GtfB used as the basis for the homology modeling made it difficult to model regions with insertion or deletion of amino acid residues. Nevertheless, His-22 was identified as a key residue for enzyme activity, although it was oriented differently from that shown in the crystal structure of UGT71G1, such that the acceptor binding pocket was predicted to be in a different location from that suggested by our docking results. The position of the binding pocket for the sugar donor was predicted correctly, but the sugar was predicted to lie in the opposite orientation to that revealed from our crystal structure, and His-370 (His-357 in UGT71G1) interacted with the phosphates rather than the uracil ring of the donor molecule. Based on the modeling study, Glu-394 was predicted to be part of a catalytic triad. However, as revealed in the crystal structure of UGT71G1, Glu-381 (corresponding to Glu-394 in UGT73A5) contacts the glucose moiety of the donor and plays an important role in sugar recognition and specificity. Rather than Glu-381, the highly conserved Asp residue (Asp-121) located in the acceptor binding pocket of UGT71G1 may be essential for enzyme catalysis.
In the model revealed by our crystal structures, Asp-121 interacts with His-22 at a distance of ∼3 Å. His-22 is close to the glucose ring of the sugar donor and the docked acceptor. The imidazole group of His-22 acts as a general base to deprotonate an OH group on the acceptor molecule and transfers the proton to Asp-121. The deprotonated acceptor, as a nucleophilic oxyanion, can then attack at the C1′ carbon center of the UDP-glucose from the opposite side of the sugar ring, with displacement of the UDP moiety. The crystal structure of UGT71G1 presented here thus provides a basis for understanding the catalytic mechanism for glycosyl transfer and also provides a starting point for homology modeling of diverse plant UGTs.
Substrate Specificity in the Plant UGT Superfamily
Most of the residues in direct contact with donor molecules, such as Trp-339, Gln-342, His-357, and Glu-365, are conserved in all 39 plant UGTs functionally characterized to date (Figure 7). In the position corresponding to Glu-381 of UGT71G1, Asp is frequently observed, which may enter into a similar interaction with the donor. At the position corresponding to Gln-382 of UGT71G1, Gln is conserved in glucosyltransferases, His is present and conserved in galactosyltransferases, and an Asn residue is observed in Arabidopsis UGT78D1, which uses UDP-rhamnose as the sugar donor. Residues at these two positions interact with the sugar moiety of the donor and therefore would be predicted to be critical for sugar specificity.
The apparent lack of stringent structural requirements for acceptor binding and catalysis is consistent with the evolution of varied and broad substrate specificities of many plant GTs (Vogt and Jones, 2000; Bowles et al., 2005). Of five UGTs that we have characterized recently from M. truncatula (Achnine et al., 2005), all have multiple in vitro substrates, suggesting multiple potential in vivo substrates. UGT71G1 can catalyze the glucosylation of two triterpenes (medicagenic acid and hederagenin), the flavonol quercetin, and the isoflavones genistein and biochanin A. For assessment of in vivo function, therefore, it is necessary to understand which potential substrates are present in the cell types in which the enzyme is expressed. In the case of methyl jasmonate–induced Medicago cell suspension cultures, such analysis points to UGT71G1 glycosylating medicagenic acid rather than the catalytically preferred substrates biochanin A or quercetin (Achnine et al., 2005).
When considered in terms of the multiple products formed from each potential substrate, such apparent promiscuity raises two important questions: why do higher plants possess so many different UGTs, and how are their activities regulated in vivo to allow for the production of a more limited number of products than suggested by their in vitro substrate/product specificities? The importance of UGTs for the detoxification of xenobiotics (allelochemicals before the human introduction of synthetic herbicides and pesticides) may have driven the evolution of new forms of the enzyme differing in residues around the acceptor binding pocket.
It has been suggested that UGTs may be associated in enzyme complexes with the early enzymes of the pathway in which they are involved, and such metabolic channeling would essentially limit a particular GT to a specific pathway irrespective of the enzyme's in vitro substrate specificity (Jorgensen et al., 2005). The presence and/or nature of physical interactions between any UGT and a related biosynthetic enzyme remain unknown. The structures described in this work provide a basis for the exploration of such interactions.
METHODS
Expression and Purification of UGT71G1
The UGT71G1 cDNA from Medicago truncatula (Achnine et al., 2005) was cloned into the pET28a expression vector (Novagen) with a hexahistidine tag and a thrombin cleavage site. Escherichia coli BL21(DE3) cells transformed with the plasmid were grown at 37°C in Luria-Bertani medium containing 50 μg/mL kanamycin until A600 = 0.6 to 0.8. Cultures were induced with 0.5 mM isopropyl 1-thio-β-galactopyranoside and grown overnight at 16°C. Cells were pelleted and resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10 mM imidazole, and 10 mM β-mercaptoethanol). After lysis with a French press and centrifugation at 12,000 rpm at 4°C for 20 min, Ni2+-nitrilotriacetic acid agarose was added to the supernatant containing the target proteins. After incubation for 40 to 60 min, the mixture was transferred into a disposable column and washed extensively with lysis buffer (∼50 column volumes). The His-tagged proteins were eluted with elution buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 250 mM imidazole, and 10 mM β-mercaptoethanol). Incubation with biotinylated thrombin during overnight dialysis at 4°C against 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 10 mM β-mercaptoethanol cleaved the N-terminal His tag. Dialyzed proteins were incubated with streptavidin agarose and Ni2+-nitrilotriacetic acid agarose to remove thrombin and the cleaved His tag, respectively. The proteins were further purified on Resource Q and Superdex-200 gel filtration columns (Amersham Pharmacia Biotech) and concentrated to 5 mg/mL in 10 mM NaCl, 10 mM Tris-HCl, pH 7.5, and 5 mM β-mercaptoethanol.
Se-Met–substituted UGT71G1 was prepared by expressing the recombinant protein in E. coli B834(DE3) cells (Novagen) grown in M9 minimal medium supplemented with Se-Met (Doublie, 1997). The procedure for expression and purification was similar to the procedure described above for the native protein.
Crystallization and Data Collection
Crystals of UGT71G1 were grown from hanging drops by the vapor diffusion method. UGT71G1 protein at a concentration of 5 mg/mL was mixed with 5 mM UDP-galactose and 5 mM quercetin at a 2:1 (v/v) ratio, then mixed with an equal volume of reservoir solution containing 40% (w/v) polyethylene glycol 3350, 0.2 M ammonium acetate, and 0.1 M sodium citrate, pH 5.6. The mixture was equilibrated over the reservoir solution at 20°C. Crystals typically grew over 2 to 5 d with dimensions of 0.1 × 0.1 × 0.2 mm.
Before data collection, the crystals were flash-cooled to −180°C. Data from a native protein crystal were measured to 2.0 Å with a Raxis IV++ image plate detector and a RU3H x-ray source. The crystal belonged to space group P21 (a = 53.7 Å, b = 90.6 Å, c = 101.8 Å, β = 102.6°). There were two molecules per crystallographic asymmetric unit, with 47% solvent content and a Matthews coefficient VM of 2.6 Å3/D. A 2.4-Å MAD data set from a Se-Met derivative crystal, using three wavelengths, was collected at the Gulf Coast Protein Crystallography Consortium beamline using a MAR CCD detector.
Crystals of UGT71G1 complexed with UDP-glucose were obtained by soaking the crystals described above with mother liquid containing saturated UDP-glucose. Five changes of this solution were made during the 3-d soaking period, the last being a few hours before freezing the crystal for data collection. A 2.6-Å diffraction data set was collected with a Raxis IV++ image plate detector and a RU3H x-ray source.
All data sets were processed using denzo and scalepack or HKL2000 (Otwinowski and Minor, 1997).
Structure Determination and Refinement
The structure of UGT71G1 was determined using the MAD method. The MAD data (40 to 2.4 Å) were analyzed with SOLVE (Terwilliger and Berendzen, 1999), and 20 of 24 Se sites were located, yielding an overall figure of merit of 0.44. The program RESOLVE (Terwilliger, 2000) was used for electron density modification, phase extension to 2.0 Å by combining with the native data, and automated model building. Interactive model building and crystallographic refinement were performed using the programs O (Jones et al., 1991) and CNS (Brünger et al., 1998), respectively. A bulk solvent correction was applied. B factors were refined individually for the structure of UGT71G1 complexed with UDP at 2.0 Å, and grouped B factor refinement was performed for the structure of UGT71G1 complexed with UDP-glucose at 2.6 Å. Water molecules were added with Arp/wArp (Lamzin et al., 2001) and checked manually for inclusion. In the models, the first two and seven amino acid residues in the N terminus were not observed in molecules A and B, respectively, and the last two residues in the C terminus were disordered in both molecules.
The program PROCHECK (Laskowski et al., 1993) was used to check the models. All backbone φ-ψ torsion angles are within allowed regions of the Ramachandran plot.
Molecular Docking
The automated docking program GOLD was used to dock acceptors into the UGT71G1 active site (Jones et al., 1997). Default genetic algorithm parameters for controlling the operation of the docking process were used. All docking calculations were restricted to the predicted binding pocket by defining the active site with residue His-22. For quercetin, medicagenic acid, and hederagenin, there are several potential glycosylation sites, and a distance constraint was used to define each potential site for calculation. GOLDscore was used to identify the lowest energy docking results. The hydrogen bonds and van der Waals contacts between ligands and enzyme were analyzed to identify the optimal binding mode.
Mutagenesis and Enzyme Assay
Site-directed mutants of UGT71G1 were constructed using the QuikChange strategy (Stratagene). The mutant proteins were expressed and purified using the same procedures described above for native protein except for the thrombin cleavage. Enzyme assay was performed essentially according to a reported method (Lim et al., 2003). Each GT activity assay mix (200 μL) contains 2 μg of recombinant protein, 50 mM Tris-HCl, pH 7.0, 14 mM β-mercaptoethanol, 5 mM UDP-glucose, and 1 mM quercetin. For mutants, increasing concentrations of substrates (2 mM quercetin and 10 mM UDP-glucose, or 5 mM quercetin and 25 mM UDP-glucose) and proteins (4 μg) were also used. The reaction was performed at 30°C for 1 h and stopped by the addition of 20 μL of trichloroacetic acid, quick-frozen, and stored at −20°C before HPLC analysis with a Waters Spherisorb 5u ODS2 C18 reverse-phase column (250 × 4.6 mm) on an Agilent HP1100 HPLC device equipped with an autosampler, a quaternary pump, and a diode array detector. Elution was as described previously (Achnine et al., 2005).
Accession Numbers
The coordinates and structure factors for the structures of UGT71G1 in complexes with UDP and UDP-glucose have been deposited to the PDB with codes 2ACV and 2ACW, respectively. The plant UGTs included in the alignment (Figure 7) and their accession numbers are as follows: Medicago truncatula UGT71G1 (accession number AAW56092) and UGT73K1 (AAW56091); Allium cepa UGT73G1 (AAP88406) and UGT73J1 (AAP88407); Aralia cordata GaT (BAD06514); Bellis perennis UGAT (BAD77944); Brassica napus SGT1 (AAF98390); Catharanthus roseus UGT2 (BAD29722); Citrus maxima 1,2RhaT (AAL06646); Crocus sativus UGTCs2 (AAP94878); Dorotheanthus bellidiformis UGT73A5 (CAB56231); Gentiana triflora 3′GT (BAC54092); Glycyrrhiza echinata UGT73F1 (BAC78438); Nicotiana tabacum GT1a (BAB60721), GT2 (BAB88935), and GT3 (BAB88934); Phaseolus lunatus ZOG1 (AAD04166); Phaseolus vulgaris ZOX1 (AAD51778); Sorghum bicolor UGT85B1 (AAF17077); Stevia rebaudiana UGT74G1 (AAR06920), UGT76G1 (AAR06912), and UGT85C2 (AAR06916); Zea mays cisZOC1 (AAK53551) and cisZOG2 (AAL92460); and Arabidopsis thaliana UGT71C1 (AAC35226), UGT72B1 (AAB61023), UGT72E2 (BAA97275), UGT73C5 (AAD20156), UGT73C6 (AAD20155), UGT74B1 (AAC00570), UGT74F2 (AAB64024), UGT84A2 (BAB02351), UGT84A1 (CAB10326), UGT84B1 (AAB87119), UGT76C1 (BAB10792), UGT76C2 (AAN28835), UGT78D1 (AAF19756), UGT85A1 (AAF18537), and UGT91B1 (BAA98174).
Acknowledgments
We thank S. Liu for initial protein purification and crystallization screening, J. Lin for technical assistance, L.W. Sumner, Z. Lei, and D. Huhman for mass spectrometric analysis, N. Duke (Structural Biology Center) and I. Koshelev (Industrial Macromolecular Crystallography Association–Collaborative Access Team) at the Advanced Photon Source (Argonne National Laboratory) for assistance with data collection of initial crystals, H. Bellamy and D. Neau of the Gulf Coast Protein Crystallography Consortium beamline of the Center for Advanced Microstructures and Devices (Louisiana State University) for help with Se-Met MAD data collection, and T.M.T. Hall and P. Xu for critical reading of the manuscript. This work was supported by National Science Foundation Grant 0416883 and the Samuel Roberts Noble Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Xiaoqiang Wang (xwang@noble.org).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.035055.
References
- Achnine, L., Huhman, D.V., Farag, M.A., Sumner, L.W., Blount, J.W., and Dixon, R.A. (2005). Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J. 41, 875–887. [DOI] [PubMed] [Google Scholar]
- Behboudi, S., Morein, B., and Villacres-Eriksson, M.C. (1999). Quillaja saponin formulations that stimulate proinflammatory cytokines elicit a potent acquired cell-mediated immunity. Scand. J. Immunol. 50, 371–377. [DOI] [PubMed] [Google Scholar]
- Bowles, D., Isayenkova, J., Lim, E., and Poppenberger, B. (2005). Glycosyltransferases: Managers of small molecules. Curr. Opin. Plant Biol. 8, 254–263. [DOI] [PubMed] [Google Scholar]
- Breton, C., Heissigerova, H., Jeanneau, C., Moravcova, J., and Imberty, A. (2002). Comparative aspects of glycosyltransferases. Biochem. Soc. Symp. 69, 23–32. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., et al. (1998). Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Buschiazzo, A., Ugalde, J.E., Guerin, M.E., Shepard, W., Ugalde, R.A., and Alzari, P.M. (2004). Crystal structure of glycogen synthase: Homologous enzymes catalyze glycogen synthesis and degradation. EMBO J. 23, 3196–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, J.A., Davies, G.J., Bulone, V., and Henrissat, B. (1997). A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 326, 929–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnock, S.J., and Davies, G.J. (1999). Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry 38, 6380–6385. [DOI] [PubMed] [Google Scholar]
- Clemens, D., Dirk, H., Sven-Eric, W., Koji, I., Monika, W., Ursula, V.M., Jon, S.T., and Andreas, B. (2004). The glycosyltransferase UrdGT2 catalyzes both C- and O-glycosidic sugar transfers. Angew. Chem. Int. Ed. Engl. 43, 2962–2965. [DOI] [PubMed] [Google Scholar]
- Coutinho, P.M., Deleury, E., Davies, G.J., and Henrissat, B. (2003). An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328, 307–317. [DOI] [PubMed] [Google Scholar]
- Davies, G.J., Charnock, S.J., and Henrissat, B. (2001). The enzymatic synthesis of glycosidic bonds: “Glycosynthases” and glycosyltransferases. Trends Glycosci. Glycotechnol. 13, 105–120. [Google Scholar]
- Dennehy, C. (2001). Botanicals in cardiovascular health. Clin. Obstet. Gynecol. 44, 814–823. [DOI] [PubMed] [Google Scholar]
- de Wildt, S.N., Kearns, G.L., Leeder, J.S., and van den Anker, J.N. (1999). Glucuronidation in humans. Pharmacogenetic and developmental aspects. Clin. Pharmacokinet. 36, 439–452. [DOI] [PubMed] [Google Scholar]
- Doublie, S. (1997). Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530. [PubMed] [Google Scholar]
- Fritz, T.A., Hurley, J.H., Trinh, L.B., Shiloach, J., and Tabak, L.A. (2004). The beginnings of mucin biosynthesis: The crystal structure of UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferase-T1. Proc. Natl. Acad. Sci. USA 101, 15307–15312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastinel, L.N., Bignon, C., Misra, A.K., Hindsgaul, O., Shaper, J.H., and Joziasse, D.H. (2001). Bovine alpha1,3-galactosyltransferase catalytic domain structure and its relationship with ABO histo-blood group and glycosphingolipid glycosyltransferases. EMBO J. 20, 638–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastinel, L.N., Cambillau, C., and Bourne, Y. (1999). Crystal structures of the bovine beta4-galactosyltransferase catalytic domain and its complex with uridine diphosphogalactose. EMBO J. 18, 3546–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons, B.J., Roach, P.J., and Hurley, T.D. (2002). Crystal structure of the autocatalytic initiator of glycogen biosynthesis, glycogenin. J. Mol. Biol. 319, 463–477. [DOI] [PubMed] [Google Scholar]
- Gibson, R.P., Turkenburg, J.P., Charnock, S.J., Lloyd, R., and Davies, G.J. (2002). Insights into trehalose synthesis provided by the structure of the retaining glucosyltransferase OtsA. Chem. Biol. 9, 1337–1346. [DOI] [PubMed] [Google Scholar]
- Goldsmith, E.J., Sprang, S.R., Hamlin, R., Xuong, N.H., and Fletterick, R.J. (1989). Domain separation in the activation of glycogen phosphorylase a. Science 245, 528–532. [DOI] [PubMed] [Google Scholar]
- Gouet, P., and Courcelle, E. (2002). ENDscript: A workflow with web interface to display sequence and structure information. Bioinformatics 18, 767–768. [DOI] [PubMed] [Google Scholar]
- Ha, S., Walker, D., Shi, Y., and Walker, S. (2000). The 1.9 Å crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis. Protein Sci. 9, 1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans, J., Brandt, W., and Vogt, T. (2004). Site-directed mutagenesis and protein 3D-homology modelling suggest a catalytic mechanism for UDP-glucose-dependent betanidin 5-O-glucosyltransferase from Dorotheanthus bellidiformis. Plant J. 39, 319–333. [DOI] [PubMed] [Google Scholar]
- Haridas, V., Higuchi, M., Jayatilake, G.S., Bailey, D., Mujoo, K., Blake, M.E., Arntzen, C.J., and Gutterman, J.U. (2001). Avicins: Triterpenoid saponins from Acacia victoriae (Bentham) induce apoptosis by mitochondrial perturbation. Proc. Natl. Acad. Sci. USA 98, 5821–5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y., Chen, L., Ha, S., Gross, B., Falcone, B., Walker, D., Mokhtarzadeh, M., and Walker, S. (2003). Crystal structure of the MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases. Proc. Natl. Acad. Sci. USA 100, 845–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y., and Walker, S. (2002). Remarkable structural similarities between diverse glycosyltransferases. Chem. Biol. 9, 1287–1296. [DOI] [PubMed] [Google Scholar]
- Hughes, J., and Hughes, M.A. (1994). Multiple secondary plant product UDP-glucose glucosyltransferase genes expressed in cassava (Manihot esculenta Crantz) cotyledons. DNA Seq. 5, 41–49. [DOI] [PubMed] [Google Scholar]
- Huhman, D.V., and Sumner, L.W. (2002). Metabolic profiling of saponins in Medicago sativa and Medicago truncatula using HPLC coupled to an electrospray ion-trap mass spectrometer. Phytochemistry 59, 347–360. [DOI] [PubMed] [Google Scholar]
- Jones, G., Willett, P., Glen, R.C., Leach, A.R., and Taylor, R. (1997). Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267, 727–748. [DOI] [PubMed] [Google Scholar]
- Jones, P., and Vogt, T. (2001). Glycosyltransferases in secondary plant metabolism: Tranquilizers and stimulant controllers. Planta 213, 164–174. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.-Y., Cowan, S.W., and Kjeldgaard, M. (1991). Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A47, 110–119. [DOI] [PubMed] [Google Scholar]
- Jorgensen, K., Rasmussed, A.V., Morant, M., Nielsen, A.H., Bjarnholt, N., Zagrobelny, M., Bak, S., and Moller, B.L. (2005). Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr. Opin. Plant Biol. 8, 280–291. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. (1991). MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- Kubo, A., Arai, Y., Nagashima, S., and Yoshikawa, T. (2004). Alteration of sugar donor specificities of plant glycosyltransferases by a single point mutation. Arch. Biochem. Biophys. 429, 198–203. [DOI] [PubMed] [Google Scholar]
- Lamzin, V.S., Perrakis, A., and Wilson, K.S. (2001). The ARP/WARP suite for automated construction and refinement of protein models. In International Tables for Crystallography, M.G. Rossmann and E. Arnold, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 720–722.
- Lariviere, L., Gueguen-Chaignon, V., and Morera, S. (2003). Crystal structures of the T4 phage beta-glucosyltransferase and the D100A mutant in complex with UDP-glucose: Glucose binding and identification of the catalytic base for a direct displacement mechanism. J. Mol. Biol. 330, 1077–1086. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. (1993). PROCHECK—A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- Li, Y., Baldauf, S., Lim, E.K., and Bowles, D.J. (2001). Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J. Biol. Chem. 276, 4338–4343. [DOI] [PubMed] [Google Scholar]
- Lim, E., Baldauf, S., Li, Y., Elias, L., Worrall, D., Spencer, S.P., Jackson, R.G., Taguchi, G., Ross, J., and Bowles, D.J. (2003). Evolution of substrate recognition across a multigene family of glycosyltransferases in Arabidopsis. Glycobiology 13, 139–145. [DOI] [PubMed] [Google Scholar]
- Lobsanov, Y.D., Romero, P.A., Sleno, B., Yu, B., Yip, P., Herscovics, A., and Howell, P.L. (2004). Structure of Kre2p/Mnt1p: A yeast alpha1,2-mannosyltransferase involved in mannoprotein biosynthesis. J. Biol. Chem. 279, 17921–17931. [DOI] [PubMed] [Google Scholar]
- Merritt, E.A., and Bacon, D.J. (1997). Raster3D: Photorealistic molecular graphics. In Methods in Enzymology, C.W.J. Carter and R.M. Sweet, eds (New York: Academic Press), pp. 505–524. [DOI] [PubMed]
- Mulichak, A.M., Losey, H.C., Lu, W., Wawrzak, Z., Walsh, C.T., and Garavito, R.M. (2003). Structure of the TDP-epi-vancosaminyltransferase GtfA from the chloroeremomycin biosynthetic pathway. Proc. Natl. Acad. Sci. USA 100, 9238–9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulichak, A.M., Losey, H.C., Walsh, C.T., and Garavito, R.M. (2001). Structure of the UDP-glucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics. Structure 9, 547–557. [DOI] [PubMed] [Google Scholar]
- Mulichak, A.M., Lu, W., Losey, H.C., Walsh, C.T., and Garavito, R.M. (2004). Crystal structure of vancosaminyltransferase GtfD from the vancomycin biosynthetic pathway: Interactions with acceptor and nucleotide ligands. Biochemistry 43, 5170–5180. [DOI] [PubMed] [Google Scholar]
- Oh, S.R., Kinjo, J., Shii, T., Ikeda, T., Nohara, T., Ahn, K.S., Kim, J.H., and Lee, H.K. (2000). Effects of triterpenoids from Pueraria lobata on immunohemolysis: Beta-d-glucuronic acid plays an active role in anticomplementary activity in vitro. Planta Med. 66, 506–510. [DOI] [PubMed] [Google Scholar]
- Oleszek, W., Junkuszew, M., and Stochmal, A. (1999). Determination and toxicity of saponins from Amaranthus cruentus seeds. J. Agric. Food Chem. 47, 3685–3687. [DOI] [PubMed] [Google Scholar]
- Osbourn, A.E. (2003). Molecules of interest. Saponins in cereals. Phytochemistry 62, 1–4. [DOI] [PubMed] [Google Scholar]
- Osbourn, A.E., Bowyer, P., and Daniels, M.J. (1996). Saponin detoxification by plant pathogenic fungi. Adv. Exp. Med. Biol. 404, 547–555. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z., and Minor, W. (1997). Processing of X-ray diffraction data collected in oscillation mode. In Methods in Enzymology, C.W.L. Carter and R.M. Sweet, eds (New York: Academic Press), pp. 307–326. [DOI] [PubMed]
- Papadopoulou, K., Melton, R.E., Leggett, M., Daniels, M.J., and Osbourn, A.E. (1999). Compromised disease resistance in saponin-deficient plants. Proc. Natl. Acad. Sci. USA 96, 12923–12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette, S., Moller, B.L., and Bak, S. (2003). On the origin of family 1 plant glycosyltransferases. Phytochemistry 62, 399–413. [DOI] [PubMed] [Google Scholar]
- Pedersen, L.C., Dong, J., Taniguchi, F., Kitagawa, H., Krahn, J.M., Pedersen, L.G., Sugahara, K., and Negishi, M. (2003). Crystal structure of an alpha 1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis. J. Biol. Chem. 278, 14420–14428. [DOI] [PubMed] [Google Scholar]
- Pedersen, L.C., Tsuchida, K., Kitagawa, H., Sugahara, K., Darden, T.A., and Negishi, M. (2000). Heparan/chondroitin sulfate biosynthesis: Structure and mechanism of human glucuronyltransferase I. J. Biol. Chem. 275, 34580–34585. [DOI] [PubMed] [Google Scholar]
- Persson, K., Ly, H.D., Dieckelmann, M., Wakarchuk, W.W., Withers, S.G., and Strynadka, N.C. (2001). Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs. Nat. Struct. Biol. 8, 166–175. [DOI] [PubMed] [Google Scholar]
- Ross, J., Li, Y., Lim, E., and Bowles, D.J. (2001). Higher plant glycosyltransferases. Genome Biol. 2, 3004.1–3004.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandermann, H., Jr. (1992). Plant metabolism of xenobiotics. Trends Biochem. Sci. 17, 82–84. [DOI] [PubMed] [Google Scholar]
- Small, E. (1996). Adaptations to herbivory in alfalfa (Medicago sativa). Can. J. Bot. 74, 807–822. [Google Scholar]
- Sprang, S., Standing, T., Fletterick, R.J., Stroud, R.M., Finer-Moore, J., Xuong, N.H., Hamlin, R., Rutter, W.J., and Craik, C.S. (1987). The three-dimensional structure of Asn102 mutant of trypsin: role of Asp102 in serine protease catalysis. Science 237, 905–909. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C. (2000). Maximum likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T.C., and Berendzen, J. (1999). Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unligil, U.M., and Rini, J.M. (2000). Glylcosyltransferase structure and mechanism. Curr. Opin. Struct. Biol. 10, 510–517. [DOI] [PubMed] [Google Scholar]
- Unligil, U.M., Zhou, S., Yuwaraj, S., Sarkar, M., Schachter, H., and Rini, J.M. (2000). X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: Catalytic mechanism and a new protein superfamily. EMBO J. 19, 5269–5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt, T., and Jones, P. (2000). Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 5, 380–386. [DOI] [PubMed] [Google Scholar]
- Vrielink, A., Ruger, W., Driessen, H.P., and Freemont, P.S. (1994). Crystal structure of the DNA modifying enzyme β-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose. EMBO J. 13, 3413–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]