Abstract
To understand the gene network controlling tolerance to cold stress, we performed an Arabidopsis thaliana genome transcript expression profile using Affymetrix GeneChips that contain ∼24,000 genes. We statistically determined 939 cold-regulated genes with 655 upregulated and 284 downregulated. A large number of early cold-responsive genes encode transcription factors that likely control late-responsive genes, suggesting a multitude of transcriptional cascades. In addition, many genes involved in chromatin level and posttranscriptional regulation were also cold regulated, suggesting their involvement in cold-responsive gene regulation. A number of genes important for the biosynthesis or signaling of plant hormones, such as abscisic acid, gibberellic acid, and auxin, are regulated by cold stress, which is of potential importance in coordinating cold tolerance with growth and development. We compared the cold-responsive transcriptomes of the wild type and inducer of CBF expression 1 (ice1), a mutant defective in an upstream transcription factor required for chilling and freezing tolerance. The transcript levels of many cold-responsive genes were altered in the ice1 mutant not only during cold stress but also before cold treatments. Our study provides a global picture of the Arabidopsis cold-responsive transcriptome and its control by ICE1 and will be valuable for understanding gene regulation under cold stress and the molecular mechanisms of cold tolerance.
INTRODUCTION
Low temperature is one of the major environmental stresses that many plants have to cope with during their life cycle. Plants from temperate regions have the capacity to cold acclimate, that is, to develop increased freezing tolerance after being exposed to low nonfreezing temperatures (Guy, 1990). Many physiological and molecular changes occur during cold acclimation (Thomashow, 1999). Among them, the transcriptional activation or repression of genes by low temperature is of primary importance (Thomashow, 1999). Early studies have identified a number of genes in plants that change expression under cold stress (Thomashow, 1994, 1999). A subset of the cold-responsive genes have in their promoters the dehydration-responsive element (DRE; 5′-TACCGACAT-3′)/C-repeat (CRT; 5′-TGGCCGAC-3′) with the common core motif (5′-CCGAC-3′). Transcriptional activators (DEHYDRATION-RESPONSIVE ELEMENT BINDING FACTOR 1/C-REPEAT BINDING FACTOR [DREB1/CBF]) that are capable of binding to DRE/CRT have been isolated from Arabidopsis thaliana using the yeast one-hybrid approach (Stockinger et al., 1997). Three members of the CBF gene family are rapidly and transiently induced by cold stress (Gilmour et al., 1998; Medina et al., 1999). Ectopic expression of CBF1/3 (DREB1B/A) activated the expression of genes with the DRE/CRT promoter element at warm temperatures, which resulted in constitutive freezing tolerance (Stockinger et al., 1997; Jaglo-Ottosen et al., 1998; Shinwari et al., 1998; Kasuga et al., 1999). Interestingly, Arabidopsis mutants with loss-of-function in CBF2/DREB1C show enhanced cold induction of CBF/DREB1 target genes and increased freezing tolerance, suggesting a complex interplay among the CBF/DREB1 family members and possibly also with other transcription factors (Novillo et al., 2004).
Recently, a constitutive transcription factor, INDUCER OF CBF EXPRESSION 1 (ICE1), which acts upstream of the CBFs in the cold-response pathway, was identified (Chinnusamy et al., 2003). ICE1 binds to the CBF3 promoter and may activate CBF3 expression upon cold treatment. The dominant ice1 mutation blocks the cold induction of CBF3 but not CBF1 or CBF2 (Chinnusamy et al., 2003).
The completion of the Arabidopsis genome sequence and technical advances in microarray analysis have allowed for the study of gene expression on a large scale. Several studies have used cDNA microarrays or Affymetrix GeneChips to identify cold-responsive genes in Arabidopsis (Seki et al., 2001, 2002; Fowler and Thomashow, 2002; Kreps et al., 2002). Expression profiling studies have also been performed with mutant plants or plants that overexpress a certain regulatory gene to understand the role of the genes in cold-responsive gene expression (Fowler and Thomashow, 2002; Goda et al., 2002; Osakabe et al., 2002). However, the microarrays or GeneChips used in these studies contained no more than one-third of the Arabidopsis genome, and much of the Arabidopsis genome has not been statistically examined for transcript responses to cold stress.
In this study, we used the Affymetrix Arabidopsis 24K GeneChip representing ∼24,000 genes to profile gene expression under cold stress. We identified 655 genes that are statistically cold upregulated and 284 genes that are downregulated. Our results suggest that cold stress triggers a multitude of transcriptional cascades because many of the early cold-responsive genes encode transcription factors that likely activate the genes induced late in the cold response. A number of genes important for the biosynthesis or signaling of plant hormones, such as abscisic acid, gibberellic acid, and auxin, are regulated by cold stress. The regulation of these genes might be important in coordinating cold tolerance with growth and development. We also determined the transcript profiles and their responses to cold stress in the ice1 mutant. The ice1 mutation affects the cold induction of a large number of genes, including many transcription factors. In addition, the ice1 mutation alters the basal transcript levels of many cold-responsive genes. Our study provides a broad picture of the Arabidopsis cold-responsive transcriptome and its control by ICE1.
RESULTS AND DISCUSSION
Cold-Regulated Genes in Arabidopsis
We used the Affymetrix Arabidopsis ATH1 genome GeneChip, which contains >22,500 probe sets representing ∼24,000 genes, to identify cold-regulated genes in Arabidopsis. Total RNA was prepared from Arabidopsis seedlings after 0, 3, 6, or 24 h cold treatment at 0°C. The 3- and 6-h time points were chosen to capture early responsive genes, and the 24-h point for late-responsive genes. A total of 100 to 150 seedlings from three plates was used to create pools of RNA used at each time point. As each plate can hold >300 plants, half of each plate contained the wild type and the other ice1 seedlings. Thus, at each time point, the three plates produced one wild-type pool and one ice1 pool of RNA. This procedure was repeated at each time point to produce a total of two biologically independent pools of RNA at each time point. The wild-type Arabidopsis used had a CBF3 promoter-driven luciferase transgene, the background line of the ice1 mutant (Chinnusamy et al., 2003).
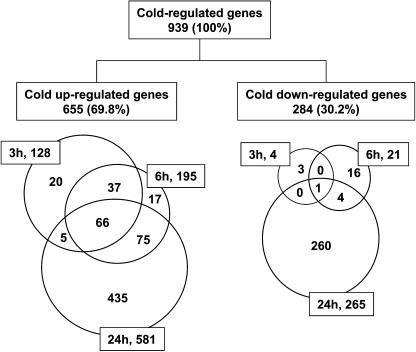
To determine cold-regulated genes in Arabidopsis, we performed statistical analyses of the GeneChip data. Signal intensity data were first obtained by the Affymetrix Microarray Suite 5.0 program, where each cell signal intensity was background subtracted, weighted using one-step Tukey's biweight algorithm, averaged, and scaled to a globally normalized intensity of 500 (the manufacturer-suggested arbitrary value) for each chip. For statistical analysis, the signal intensity data were then analyzed with use of a two-stage linear statistical model and robust test statistics with the statistical package R (R Development Core Team, 2003; http://www.R-project.org) and the Limma package (Smyth, 2005). The 10,000 bootstrap simulations (Efron and Tibshirani, 1993) were used to obtain nonparametric P values for testing a null hypothesis of no difference for gene-specific contrasts of expression levels between the different time points (see Methods for details). False discovery rates (FDRs) for various P value thresholds were later determined by the method of Benjamini and Yekutieli (2001) on the observed distribution of P values. Genes with <1% of FDR at any time point were considered significantly cold responsive. When this threshold was applied, 939 genes were determined to be cold regulated with 655 upregulated and 284 downregulated (Figure 1). Thus, ∼3.9% of all Arabidopsis genes were determined to be cold responsive.
Figure 1.
Venn Diagrams of Cold-Regulated Genes.
Percentages in parentheses were calculated with the total numbers of cold-regulated genes (939). Figures in rectangles indicate cold treatment hours (h) and total number of cold-regulated genes at each time point.
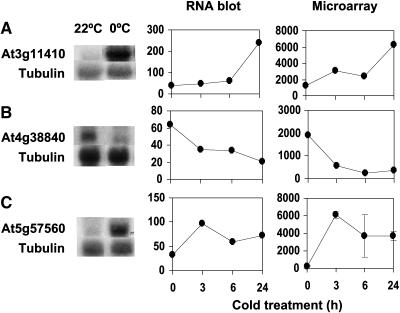
To validate the microarray data, we performed RNA gel blot analysis for three cold-regulated genes from our list. These included a protein phosphatase 2C (At3g11410), a small auxin upregulated RNA (SAUR) gene (At4g38840), and TOUCH4 (At5g57560). The RNA gel blot results showed that the three genes are all cold regulated, and their expression kinetics from the RNA gel blots is very similar to those obtained from microarray analysis (Figure 2). These results support the validity of the Arabidopsis cold-regulated transcriptome from the GeneChip analysis.
Figure 2.
RNA Gel Blot Analysis of Cold-Regulated Genes Selected from Microarray Analysis.
Cold-responsive genes were tested by RNA gel blot analysis. For expression pattern comparisons, each corresponding gene transcript level was plotted using intensities from RNA gel blots and microarrays. The x axis shows time points during cold treatment (0°C), and y axis shows signal intensities in arbitrary units. Gene names and treatments for RNA gel blot pictures: (A) At3g11410 (protein phosphatase 2C, 0°C 24 h), (B) At4g38840 (SAUR, 0°C 6 h), and (C) At5g57560 (TCH4, 0°C 3 h). Tubulin was used as a loading control.
Among the 655 cold upregulated genes, 128 were upregulated at 3 h, 195 at 6 h, and 581 at 24 h of cold treatment. Thus, most of the cold-upregulated genes are late-response genes (Figure 1). Indeed, Venn diagram analysis shows that 435 (66.4%) genes were induced exclusively at 24 h of cold treatment (Figure 1), while only 20 (3.1%) and 17 (2.6%) genes were upregulated exclusively at 3 or 6 h of cold treatment, respectively. Out of the cold upregulated genes, 66 genes (10.1%) had a high level of cold induction at all time points.
Most of the 284 downregulated genes were downregulated also only after 24 h of cold treatment. Only four genes (At1g05610, glucose-1-phosphate adenylyltransferase; At2g23900, polygalacturonase; At5g34880, hypothetical protein; At5g38035, copia-like retrotransposon) showed a substantial downregulation at 3 h, and the downregulation of one of the four (At5g38035, copia-like retrotransposon) was also observed at 6 and 24 h (Figure 1). The contrasting results among the numbers of cold downregulated genes at each time point suggest that although cold downregulated genes may play a role in cold responses in Arabidopsis, the downregulation is not a major part of the early response to cold stress. The upregulation of early genes by low temperature may be important for both upregulation and downregulation of late-response genes.
Functional Categorization of Cold-Regulated Genes
To distinguish the kinetics of regulation under cold stress, we designated genes with transcript level changes only at 3 or 6 h or at both 3 and 6 h as early and transiently upregulated or downregulated, respectively. Genes that changed exclusively at 24 h were considered late upregulated/downregulated genes. Genes with expression changes at 3 and 24 h, 6 and 24 h, or 3, 6, and 24 h were grouped as early and continually upregulated or downregulated, respectively.
Our cold-regulated genes were categorized into 19 functional groups using the Functional Catalogue at http://mips.gsf.de/projects/funcat, with manual adjustment when necessary (Table 1; Ruepp et al., 2004). Interestingly, different functional categories appeared to be activated at different time points after cold treatment. Although genes without significant similarity to known genes or functions were in fact the largest group of cold-regulated genes, our analysis here focused on genes with sequence homology to known genes. Among the early transiently upregulated genes, the largest group were genes involved in transcription and the next largest were signal transduction genes; 19 genes are involved in transcription, and 12 are in signal transduction (Table 1). On the other hand, out of the early continually upregulated genes, 26 and 22 are involved in transcription and cell defense, respectively (Table 1). Genes involved in transcription (68 genes) and metabolism (67 genes) were the major groups among late upregulated genes. The large number of genes involved in metabolism suggests that plants start to undergo substantial changes in metabolism later in the cold, after an initial inhibition of metabolism due to reduced enzymatic activities under cold. Previous physiological and metabolomic studies have shown many metabolic changes during cold acclimation, such as increased accumulation of soluble sugars and other compatible osmolytes (Wanner and Junttila, 1999; Cook et al., 2004). In summary, genes involved in transcription (113 genes) and metabolism (92 genes) are the two major groups of cold upregulated genes, which are followed by genes involved in defense (67 genes) and signal transduction (63 genes).
Table 1.
Functional Categorization of Cold-Regulated Genes
| Cold Upregulated
|
Cold Downregulated
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETU
|
ECU
|
LU
|
Subtotal
|
ETD
|
ECD
|
LD
|
Subtotal
|
Total
|
||||||||||
| Functional Category | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % |
| Metabolism | 6 | 7.5 | 19 | 12.0 | 67 | 13.7 | 92 | 12.6 | 2 | 10.0 | 1 | 20.0 | 67 | 24.1 | 70 | 23.1 | 162 | 15.7 |
| Energy | 1 | 1.3 | 5 | 3.2 | 18 | 3.7 | 24 | 3.3 | 0.0 | 0.0 | 6 | 2.2 | 6 | 2.0 | 30 | 2.9 | ||
| Storage protein | 0.0 | 0.0 | 1 | 0.2 | 1 | 0.1 | 0.0 | 0.0 | 0.0 | 0 | 0.0 | 1 | 0.1 | |||||
| Cell cycle and DNA processing | 0.0 | 0.0 | 9 | 1.8 | 9 | 1.2 | 0.0 | 1 | 20.0 | 6 | 2.2 | 7 | 2.3 | 16 | 1.6 | |||
| Transcription | 19 | 23.8 | 26 | 16.5 | 68 | 13.9 | 113 | 15.5 | 1 | 5.0 | 0.0 | 19 | 6.8 | 20 | 6.6 | 133 | 12.9 | |
| Protein synthesis | 0.0 | 0.0 | 4 | 0.8 | 4 | 0.5 | 2 | 10.0 | 0.0 | 2 | 0.7 | 4 | 1.3 | 8 | 0.8 | |||
| Protein fate (folding, modification, and destination) | 1 | 1.3 | 8 | 5.1 | 22 | 4.5 | 31 | 4.3 | 3 | 15.0 | 1 | 20.0 | 13 | 4.7 | 17 | 5.6 | 48 | 4.7 |
| Protein with binding function or cofactor requirement | 0.0 | 1 | 0.6 | 4 | 0.8 | 5 | 0.7 | 0.0 | 0.0 | 5 | 1.8 | 5 | 1.7 | 10 | 1.0 | |||
| Cellular transport, transport facilitation, and transport routes | 4 | 5.0 | 8 | 5.1 | 26 | 5.3 | 38 | 5.2 | 0.0 | 0.0 | 13 | 4.7 | 13 | 4.3 | 51 | 4.9 | ||
| Cellular communication/signal transduction mechanism | 12 | 15.0 | 11 | 7.0 | 40 | 8.2 | 63 | 8.7 | 0.0 | 1 | 20.0 | 18 | 6.5 | 19 | 6.3 | 82 | 8.0 | |
| Cell rescue, defense, and virulence | 6 | 7.5 | 22 | 13.9 | 39 | 8.0 | 67 | 9.2 | 0.0 | 0.0 | 13 | 4.7 | 13 | 4.3 | 80 | 7.8 | ||
| Interaction with the cellular environment | 0.0 | 1 | 0.6 | 0.0 | 1 | 0.1 | 0.0 | 0.0 | 0.0 | 0 | 0.0 | 1 | 0.1 | |||||
| Interaction with the environment (systemic) | 0.0 | 1 | 0.6 | 4 | 0.8 | 5 | 0.7 | 0.0 | 0.0 | 5 | 1.8 | 5 | 1.7 | 10 | 1.0 | |||
| Transposable elements and viral and plasmid proteins | 0.0 | 1 | 0.6 | 1 | 0.2 | 2 | 0.3 | 1 | 5.0 | 1 | 20.0 | 1 | 0.4 | 3 | 1.0 | 5 | 0.5 | |
| Cell fate | 0.0 | 0.0 | 7 | 1.4 | 7 | 1.0 | 0.0 | 0.0 | 0.0 | 0 | 0.0 | 7 | 0.7 | |||||
| Development (systemic) | 0.0 | 1 | 0.6 | 3 | 0.6 | 4 | 0.5 | 0.0 | 0.0 | 2 | 0.7 | 2 | 0.7 | 6 | 0.6 | |||
| Biogenesis of cellular components | 2 | 2.5 | 3 | 1.9 | 7 | 1.4 | 12 | 1.6 | 1 | 5.0 | 0.0 | 6 | 2.2 | 7 | 2.3 | 19 | 1.8 | |
| Subcellular localization | 4 | 5.0 | 6 | 3.8 | 14 | 2.9 | 24 | 3.3 | 0.0 | 0.0 | 6 | 2.2 | 6 | 2.0 | 30 | 2.9 | ||
| No clear classification/unclassified | 25 | 31.3 | 45 | 28.5 | 156 | 31.8 | 226 | 31.0 | 10 | 50.0 | 0.0 | 96 | 34.5 | 106 | 35.0 | 332 | 32.2 | |
| Total | 80 | 100.0 | 158 | 100.0 | 490 | 100.0 | 728 | 100.0 | 20 | 100.0 | 5 | 100.0 | 278 | 100.0 | 303 | 100.0 | 1031 | 100.0 |
Note that among 939 cold-responsive genes, 132 belong to multiple functional categories and 10 are missing in the Functional Catalogue. The missing 10 genes were manually grouped into different functional categories following database searches. ETU/D, early transiently upregulated/downregulated; ECU/D, early continually upregulated/downregulated; LU/D, late upregulated/downregulated.
Cold downregulated genes showed slower kinetics. Only 20 genes were downregulated early and transiently, and five genes were downregulated early and continually under cold stress, while most of the genes (278 genes) were downregulated late by low temperature (Table 1). As the majority of cold downregulated genes were down after 24 h of cold treatment, late downregulated genes represent the major groups of cold downregulated genes. Among the late downregulated genes, genes involved in metabolism are the largest group (67), with fewer genes in other categories. Thus, low temperature stress mainly downregulates metabolism genes, and the downregulation happens late in the cold.
Cold Regulation of Signal Transduction Components
Consistent with a role of Ca2+ in early cold signal transduction, Ca2+ binding proteins were the main signaling components induced at the early time points during cold stress (Table 2; see Supplemental Tables 3, 4, and 10 online). In fact, two of the four early transiently upregulated protein kinases require indirect or direct Ca2+ involvement for kinase activity (At1g01140, SOS2-like protein kinase 6/CBL-interacting protein kinase 9; At5g66210, calcium-dependent protein kinase), making eight genes Ca2+ related out of 12 early transiently upregulated genes. Protein phosphorylation and dephosphorylation have been implicated in cold signal transduction. Indeed, the number of protein kinases and phosphatases with altered expression was the largest (total of 32 genes among 82 cold-regulated signaling genes) (Table 2; see Supplemental Table 10 online). The induction of protein kinases was observed to exhibit all types of kinetics, whereas protein phosphatase gene induction occurred preferably at the late time point (Table 2; see Supplemental Table 10 online). In addition, the only signaling component that was early and continually downregulated was a protein phosphatase 2C (At5g02760).
Table 2.
Cold-Regulated Signaling Genes
| Cold Upregulated
|
Cold Downregulated
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETU
|
ECU
|
LU
|
Subtotal
|
ETD
|
ECD
|
LD
|
Subtotal
|
Total
|
||||||||||
| Category | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % |
| Calcium binding protein | 6 | 50.0 | 3 | 27.3 | 5 | 12.5 | 14 | 22.2 | 0.0 | 0.0 | 1 | 5.6 | 1 | 5.3 | 15 | 18.3 | ||
| Protein phosphatase | 1 | 8.3 | 1 | 9.1 | 8 | 20.0 | 10 | 15.9 | 0.0 | 1 | 100.0 | 1 | 5.6 | 2 | 10.5 | 12 | 14.6 | |
| Protein kinase | 4 | 33.3 | 4 | 36.4 | 8 | 20.0 | 16 | 25.4 | 0.0 | 0.0 | 4 | 22.2 | 4 | 21.1 | 20 | 24.4 | ||
| Receptor-like kinase | 0.0 | 0.0 | 7 | 17.5 | 7 | 11.1 | 0.0 | 0.0 | 7 | 38.9 | 7 | 36.8 | 14 | 17.1 | ||||
| Blue light teceptor | 0.0 | 0.0 | 1 | 2.5 | 1 | 1.6 | 0.0 | 0.0 | 0.0 | 0.0 | 1 | 1.2 | ||||||
| Histidine kinase | 0.0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 | 0.0 | 1 | 5.6 | 1 | 5.3 | 1 | 1.2 | |||||
| Respone regulator | 0.0 | 1 | 9.1 | 1 | 2.5 | 2 | 3.2 | 0.0 | 0.0 | 3 | 16.7 | 3 | 15.8 | 5 | 6.1 | |||
| Lipid signaling protein | 1 | 8.3 | 1 | 9.1 | 6 | 15.0 | 8 | 12.7 | 0.0 | 0.0 | 1 | 5.6 | 1 | 5.3 | 9 | 11.0 | ||
| GTP-related protein | 0.0 | 1 | 9.1 | 4 | 10.0 | 5 | 7.9 | 0.0 | 0.0 | 0.0 | 0 | 0.0 | 5 | 6.1 | ||||
| Total | 12 | 100.0 | 11 | 100.0 | 40 | 100.0 | 63 | 100.0 | 0 | 0.0 | 1 | 100.0 | 18 | 100.0 | 19 | 100.0 | 82 | 100.0 |
Note that three pseudoresponse regulators (APRR1, At5g61380; APRR2, At4g18020; APRR5, At5g24470) are included in the response regulator category.
Fourteen genes for receptor-like kinases (RLKs) were cold regulated (Table 2; see Supplemental Table 10 online). Notably, all these cold-regulated RLKs responded late in the cold, which suggests that they might perceive secondary signals generated by early cold responses. Among the cold-regulated RLKs, two RLKs have been studied in detail. The leucine-rich repeat transmembrane protein kinase, RKL1 (At1g48480), was predominantly expressed in stomatal cells and was induced by wounding, pathogen, and salicylic acid (Tarutani et al., 2004a, 2004b). The RECEPTOR-LIKE PROTEIN KINASE 1 (RPK1) gene (At1g69270) has been reported to be induced by abscisic acid (ABA), dehydration, high salt, and cold (Hong et al., 1997). Recent work indicates that RPK1 plays an important role in ABA signal transduction (Osakabe et al., 2005). These results suggest that cold signal transduction shares at least in part some common pathways with other biotic, abiotic, and ABA signaling through these RLKs.
Intriguingly, the expression of a type-A response regulator, ARR16 (At2g40670), and a type-B response regulator, ARR10 (At4g31920), was reduced at the late time point during cold stress (Table 2; see Supplemental Table 10 online). These response regulators of the two-component system have been implicated in cytokinin signaling (Hwang and Sheen, 2001; Kiba et al., 2002). Thus, this observation indicates an interaction between cytokinin signaling and cold response. Consistent with this notion, histidine kinase2 (At5g35750), another component of two-component systems, is also cold regulated (Table 2; see Supplemental Table 10 online).
The pseudoresponse regulator APRR1 (At5g61380) was cold upregulated late, while another, APRR5 (At5g24470), was upregulated early and continually. A third pseudoresponse regulator, APRR2 (At4g18020), was cold downregulated late (Table 2; see Supplemental Table 10 online). Pseudoresponse regulators are proteins that lack the conserved Asp residue that in typical response regulators is the phosphorylation target of the upstream kinase in two-component systems (Hwang et al., 2002). APRR1 is also called TIMING OF CAB EXPRESSION1 and is an essential component of the central oscillator for circadian rhythms in Arabidopsis (Strayer et al., 2000; Mizuno, 2004). APRR5 has also been implicated in circadian rhythms (Yamamoto et al., 2003; Fujimori et al., 2005). It is worth noting that our cold treatment started at noon, that is, 6 h after dawn, and the seedlings were grown in a 16-h-light and 8-h-dark cycle for 2 weeks before the treatments. A previous report showed that the peaks of mRNA accumulation of APRR1 and APRR5 occurred at 12 and 8 h after dawn with 16-h-light/8-h-dark photoperiod conditions (Matsushika et al., 2000). Therefore, our control (i.e., 0 h treatment at noon) expression level should already have been at least half or close to the maximum expression level. Significant cold induction of APRR1 and APRR5 genes took place mainly at 24 h (i.e., at noon after one full circadian cycle) under cold treatment. Thus, the higher expression levels of APRR1 and APRR5 genes at 0°C appear to be the consequence of cold regulation rather than circadian effects.
Nine genes involved in phospholipid signaling were cold regulated, and almost all (eight genes) were upregulated by cold stress (Table 2; see Supplemental Table 10 online). These include four phospholipase Cs (ATPLC1, At5g58670; ATPLC4, At5g58700; ATPLC5, At5g58690; a putative phospholipase C [PLC], At4g34920), one phospholipase D (PLDα1, At3g15730), two diacylglycerol kinases (ATDGK1, At5g07920; ATDGK2, At5g63770), one inositol polyphosphate 5-phosphatase II (IP5PII; At4g18010), and one 3′(2′),5′-bisphosphate nucleotidase/inositol polyphosphate 1-phosphatase (IPPase; At5g63990) (see Supplemental Table 10 online). All of these genes were cold regulated at the late time point except for IP5PII (At4g18010) and ATDGK1 (At5g07920), which are early and transiently and early and continuously induced under cold stress, respectively (see Supplemental Table 10 online). Because all of these genes function in phospholipid-based signaling, their induction/repression by cold stress strongly suggests that phospholipid second messengers are an important part of cold signaling. PLCs act on phosphatidylinositol 4,5-bisphosphate, generating inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG). IP3 is known to trigger the release of Ca2+ from internal stores, and DAG has been shown to activate protein kinase C in animal cells (Irvine, 1992). There are no homologs of protein kinase C in Arabidopsis, and DAG is converted to phosphatidic acid (PA) by diacylglycerol kinases (Munnik, 2001). PA can also be generated by direct hydrolysis of membrane phospholipids, such as phosphatidylcholine and phophatidylethanolamine by phospholipase D (PLD). There are accumulating lines of evidence that PA is involved in cold signaling. During cold stress, PA content increased in Arabidopsis suspension cell cultures, and this increase was due to the activation of PLC and PLD (Ruelland et al., 2002). Studies with Arabidopsis mutant or overexpression lines of PLDs supported their involvement in plant cold tolerance (Welti et al., 2002; Li et al., 2004). Consistent with these reports, cold induction of genes involved in PA production, such as diacylglycerol kinases and a PLD, suggests that PA is an important part of plant cold responses as reported for wounding and osmotic stress (Frank et al., 2000; Wang et al., 2000; Katagiri et al., 2001).
The FRY1 bifunctional 3′(2′),5′-bisphosphate nucleotidase/inositol polyphosphate 1-phophatase was shown to play an important role as an IPPase (Xiong et al., 2001). Loss-of-function mutations in FRY1 resulted in a significant increase in the expression of cold-responsive genes, which supports a role of IPPase in attenuating the IP3 signal triggered by cold (Xiong et al., 2001). The expression of one of its paralogs (Atg563990) located next to FRY1 on chromosome 5 was significantly increased during cold (see Supplemental Table 10 online), suggesting its role in cold stress as an IPPase. Because most of these IP3-related genes were induced late, IP3 signaling is likely to be a late event during cold stress. Nevertheless, the early and transient induction of IP5PII (At4g18010) suggests that an early involvement of IP3 in cold signaling cannot be ruled out completely.
Cold stress also induced two Rab GTPase genes (ATRABC1/ATRAB18, At1g43890; ATRABG3d, At1g52280) (Table 2; see Supplemental Table 10 online). Although no in planta function of these genes has been established in Arabidopsis, Rab GTPases are known to function in intracellular membrane trafficking (Zerial and McBride, 2001). Thus, cold upregulation of the two Rab GTPase indicates that active membrane trafficking might take place under cold stress.
A putative GDP/GTP pyrophosphokinase (RELA/SPOT HOMOLOG 2 [RSH2], At3g14050) was early and continually upregulated by cold (Table 2; see Supplemental Table 10 online). This gene encodes a plant homolog of RelA and SpoT (van der Biezen et al., 2000). In bacteria, RelA (p)ppGpp synthase and SpoT (p)ppGpp hydrolase determine the level of guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp) in response to starvation and other stress conditions. Accumulation of (p)ppGpp in response to unfavorable conditions initiates the stringent response. Among three Arabidopsis RelA and SpoT homologs, RSH1 interacts with a pathogen resistance protein (RPP5) in the yeast two-hybrid system and confers phenotypes associated with (p)ppGpp synthesis in Escherichia coli and Streptomyces coelicolor (van der Biezen et al., 2000). Recently, it was reported that RSH from the halophyte Suaeda japonica, a RelA and SpoT homolog more closely related to RSH2 and RSH3 than RSH1, confers salt tolerance when expressed in E. coli and Saccharomyces cerevisiae (Yamada et al., 2003). Thus, despite the lack of functional analysis in plants, the induction of RSH2 under cold stress suggests a possible occurrence of (p)ppGpp-mediated signal transduction under cold stress.
Cold Regulation of Transcription Factors
Studies of the cold-induced DREB1/CBF family of transcription factors have demonstrated the importance of transcription factors in plant freezing tolerance (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999). To understand the regulation of transcription factors by cold, we surveyed their expression levels during cold stress (Tables 3 and 4; see Supplemental Table 11 online). Among the 655 cold upregulated genes, 113 (17.3%) genes are annotated to function in transcription. By contrast, only 20 transcription-related genes (7.0%) were found among the 284 downregulated genes (Table 3). Transcription category genes include both transcription factors that have DNA binding domain(s) and transcription activation/repression domain(s) and other factors that modulate the activity of transcription factors. There are 95 transcription factors and 18 other transcriptional regulators among cold upregulated transcription category genes (Tables 3 and 4; see Supplemental Table 11 online). Eighteen transcription factors and two other transcriptional regulators belong to the cold downregulated transcription category (Tables 3 and 4; see Supplemental Table 11 online).
Table 3.
Cold-Regulated Genes Involved in Transcription: Transcription Factors with Known DNA Binding Domains
| Cold Upregulated
|
Cold Downregulated
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETU
|
ECU
|
LU
|
Subtotal
|
ETD
|
ECD
|
LD
|
Subtotal
|
Total
|
||||||||||
| Category | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % |
| AP2/ERF | 7 | 36.8 | 11 | 42.3 | 3 | 4.4 | 21 | 18.6 | 0.0 | 0.0 | 1 | 5.3 | 1 | 5.0 | 22 | 16.5 | ||
| B3 (ARF) | 0.0 | 0.0 | 1 | 1.5 | 1 | 0.9 | 0.0 | 0.0 | 0.0 | 0 | 0.0 | 1 | 0.8 | |||||
| bZIP | 0.0 | 1 | 3.8 | 5 | 7.4 | 6 | 5.3 | 0.0 | 0.0 | 0.0 | 0 | 0.0 | 6 | 4.5 | ||||
| CCAAT | 0.0 | 0.0 | 1 | 1.5 | 1 | 0.9 | 0.0 | 0.0 | 0.0 | 0 | 0.0 | 1 | 0.8 | |||||
| CG-1 (CAMTA4) | 0.0 | 0.0 | 1 | 1.5 | 1 | 0.9 | 0.0 | 0.0 | 0.0 | 0 | 0.0 | 1 | 0.8 | |||||
| GRAS | 0.0 | 0.0 | 2 | 2.9 | 2 | 1.8 | 0.0 | 0.0 | 1 | 5.3 | 1 | 5.0 | 3 | 2.3 | ||||
| Homeodomain | 0.0 | 0.0 | 1 | 1.5 | 1 | 0.9 | 0.0 | 0.0 | 1 | 5.3 | 1 | 5.0 | 2 | 1.5 | ||||
| HSF | 1 | 5.3 | 2 | 7.7 | 1 | 1.5 | 4 | 3.5 | 0.0 | 0.0 | 0.0 | 0 | 0.0 | 4 | 3.0 | |||
| MADS | 0.0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 | 0.0 | 1 | 5.3 | 1 | 5.0 | 1 | 0.8 | |||||
| MYB | 1 | 5.3 | 0.0 | 10 | 14.7 | 11 | 9.7 | 0.0 | 0.0 | 1 | 5.3 | 1 | 5.0 | 12 | 9.0 | |||
| bHLH | 1 | 5.3 | 0.0 | 0.0 | 1 | 0.9 | 0.0 | 0.0 | 7 | 36.8 | 7 | 35.0 | 8 | 6.0 | ||||
| NAC | 1 | 5.3 | 1 | 3.8 | 7 | 10.3 | 9 | 8.0 | 0.0 | 0.0 | 1 | 5.3 | 1 | 5.0 | 10 | 7.5 | ||
| SPB | 0.0 | 1 | 3.8 | 0.0 | 1 | 0.9 | 0.0 | 0.0 | 1 | 5.3 | 1 | 5.0 | 2 | 1.5 | ||||
| Trihelix | 0.0 | 0.0 | 1 | 1.5 | 1 | 0.9 | 0.0 | 0.0 | 2 | 10.5 | 2 | 10.0 | 3 | 2.3 | ||||
| WRKY | 5 | 26.3 | 1 | 3.8 | 2 | 2.9 | 8 | 7.1 | 0.0 | 0.0 | 0.0 | 0 | 0.0 | 8 | 6.0 | |||
| Zn | 2 | 10.5 | 7 | 26.9 | 18 | 26.5 | 27 | 23.9 | 0.0 | 0.0 | 2 | 10.5 | 2 | 10.0 | 29 | 21.8 | ||
| ETC | 1 | 5.3 | 2 | 7.7 | 15 | 22.1 | 18 | 15.9 | 1 | 100.0 | 0.0 | 1 | 5.3 | 2 | 10.0 | 20 | 15.0 | |
| Total | 19 | 100.0 | 26 | 100.0 | 68 | 100.0 | 113 | 100.0 | 1 | 100.0 | 0 | 0.0 | 19 | 100.0 | 20 | 100.0 | 133 | 100.0 |
Note that ETC includes the transcription regulatory proteins, one transcription factor with unclassified DNA binding domain (At3g61260), and general transcription factor TFIIB (At2g41630). AP2, APETALA2; ERF, ethylene-responsive element binding factor; ARF, auxin response factor; HSF, heat shock transcription factor; CAMTA, calmodulin binding transcription activators; CCAAT, CCAAT binding motif; SPB, squamosa promoter binding.
Table 4.
Cold-Regulated Genes Involved in Transcription: Transcription Regulators Indirectly Involved in Transcription Processes
| Category | AGI IDa | Gene Name | Kinetics |
|---|---|---|---|
| Chromatin | At1g63020 | RNA polymerase IV (RNA polymerase D) subunit; NRPD1a | ECU |
| At5g63330 | DNA binding bromodomain-containing protein | ECU | |
| At5g50320 | GCN5-related N-acetyltransferase (GNAT) family | LU | |
| At3g12980 | Histone acetyltransferase 5 (HAC5) | LU | |
| At5g26040 | Histone deacetylase, putative | LU | |
| At5g14270 | Bromodomain-containing protein | LU | |
| At5g10550 | Bromodomain protein like | ETD | |
| Splicing | At1g28060 | snRNP; similarity to U4/U6 small nuclear ribonucleoprotein hPrp3 | LU |
| At4g03430 | Putative pre-mRNA splicing factor | LU | |
| At4g05410 | U3 snoRNP-associated–like protein | LU | |
| At4g25500 | STA1; Arg/Ser-rich splicing factor RSp40 | LU | |
| At5g46920 | Intron maturase, type II family protein | LU | |
| Turnover | At3g44260 | CCR4-associated factor 1-like protein | ETU |
| At2g22090 | UBP1 interacting protein, putative | LD | |
| Other | At1g69250 | Nuclear transport factor 2 (NTF2) family | LU |
| At5g59950 | RNA and export factor binding protein, putative | LU | |
| At2g42520 | DEAD box RNA helicase, putative | LU | |
| At4g34910 | DEAD/DEAH box helicase (RH16) | LU |
AGI, Arabidopsis Genome Initiative.
When classified by their characteristic DNA binding domains, except for one that encodes a DNA binding protein with a nonclassified binding domain (At3g61260), the 95 cold upregulated transcription factors fall into 15 families (Table 3). Transcription factors with APETALA 2/ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR (AP2/ERF) and Zn finger domains are the two major families, accounting for 21 genes and 27 genes of the 95 upregulated transcription factors, respectively. Interestingly, these two groups of transcription factors appear to act in different time frames. The AP2/ERF family was mainly induced early transiently and continually during cold, whereas the Zn finger family was mostly late induced under cold stress. Therefore, the AP2/ERF transcription factors may play a major role early in the cold response, as is also supported by the fact that DREBs/CBFs are in the AP2/ERF family and are important for the activation of many late cold-responsive genes (Fowler and Thomashow, 2002).
Our microarray data revealed that cold induction of basic leucine zipper (bZIP) transcription factors occurred mainly at a late time point (Table 3; see Supplemental Table 11 online). bZIP transcription factors play a role in plant pathogen responses, light signaling, and ABA and abiotic stress signaling (Jakoby et al., 2002). We found that two ABA-responsive element binding factors (ABF1, At1g49720; ABF4/AREB2, At3g19290) and ABA INSENSITIVE 5 (ABI5, At2g36270) genes were cold induced. These ABFs/ABREBs are highly similar to ABI5 and belong to group A bZIP transcription factors (Jakoby et al., 2002). Although they are all ABA responsive, the expression patterns of ABFs/ABREBs in response to various stress treatments are different (Choi et al., 2000; Uno et al., 2000). In addition, Arabidopsis plants overexpressing ABF2/AREB1, ABF3, or ABF4/AREB2 display distinct stress responses (Kang et al., 2002; Kim et al., 2004), suggesting specific or partially overlapping functions of ABFs/ABREBs in stress responses.
Several transcription factors involved in plant development (e.g., ARF, GRAS, Homeodomain, MADS, and NAC) were regulated by cold stress (Table 3; see Supplemental Table 11 online). For example, auxin response factor 7/NONPHOTOTROPHIC HYPOCOTYL 4 (ARF7/NPH4, At5g20730) was cold upregulated at the late time point. ARF7/NPH4 functions in auxin-dependent differential cell expansion of hypocotyls (Stowe-Evans et al., 1998; Harper et al., 2000; Tatematsu et al., 2004; Okushima et al., 2005). Other examples include NAC transcription factors that are mainly cold induced at the late time point. Plant-specific NAC family transcription factors have a conserved NAC domain at the N termini of the proteins (Olsen et al., 2005) and have been implicated in plant development (Souer et al., 1996; Aida et al., 1997; Sablowski and Meyerowitz, 1998; Xie et al., 2000; Takada et al., 2001; Vroemen et al., 2003). It was recently reported that several NAC transcription factors are also involved in biotic and abiotic stress responses (Collinge and Boller, 2001; Hegedus et al., 2003; Fujita et al., 2004; Tran et al., 2004). Thus, these transcription factors might be effectors of cold signaling and function in reprogramming plant development to cope with cold stress.
Interestingly, four heat shock transcription factors (HSFs) and eight WRKY transcription factors were also cold responsive and were all upregulated (Table 3; see Supplemental Table 11 online). HSF and WRKY plant transcription factors have been implicated in heat stress and pathogen responses, respectively (Eulgem et al., 2000; Nover et al., 2001). These results suggest that several HSF and WRKY factors can be involved in plant responses to multiple stresses. Recently, it was shown that WRKY factors are important in senescence, morphogenesis, and ABA and gibberellin signaling in aleurone cells (Robatzek and Somssich, 2001; Johnson et al., 2002; Zhang et al., 2004; Xie et al., 2005). Also, WRKY expression was induced by various abiotic stresses (Pnueli et al., 2002; Rizhsky et al., 2002; Seki et al., 2002).
Most of the MYB transcription factors (10 out of 12 genes) were cold induced late, while most of the basic helix-loop-helix (bHLH) transcription factors (seven out of eight genes) were cold downregulated late (Table 3; see Supplemental Table 11 online). As MYB and bHLH proteins often interact with each other to control transcription (Stracke et al., 2001; Heim et al., 2003; Ramsay and Glover, 2005), this differential expression of MYB and bHLH transcription factors suggests that the regulation of some cold-responsive genes may be achieved by modulating the ratio of these partner factors.
Cold Regulation of Other Transcription Regulators
Our microarray data revealed that the expression levels of two mitochondrial genes (cox1 and nad6) increased during cold stress (see Supplemental Table 2 online). cox1 (AtMg01360) encodes cytochrome c oxidase subunit 1 and nad6 (AtMg00270) encodes NADH dehydrogenase subunit 6. Related to this, we noticed that seven genes encoding a protein with a pentatricopeptide repeat (At1g74750, At3g57430, At3g60980, At4g36680, At5g02860, At5g25630, and At5g61370) are cold regulated (see Supplemental Tables 1, 5, and 9 online). All but At5g25630, which shows late downregulated kinetics, were late induced by cold. Proteins with a pentatricopeptide repeat are known to be involved in organellar RNA metabolism (Lurin et al., 2004). We reported previously that a mutation in nuclear-encoded mitochondrial complex I subunit resulted in impaired cold acclimation and reduced cold-responsive gene expression (B.-h. Lee et al., 2002), suggesting the importance of coordination of nuclear cold-responsive gene expression and mitochondrial functions. Our microarray data further support the notion that proper organellar functions, especially mitochondrial electron transport, are important for plant cold responses, including cold-regulated gene expression.
Expression of many genes involved in RNA metabolism and chromatin remodeling was altered by cold stress (Table 4; see Supplemental Table 11 online). Expression of four pre-mRNA splicing factors (At1g28060, At4g03430, At4g05410, and At4g25500), two DEAD box RNA helicases (At2g42520 and At4g34910), and two RNA transport factors (At1g69250 and At5g59950) are increased by cold stress. These genes are known to be involved in RNA metabolism, including splicing, ribosome biogenesis, mRNA export, translational regulation, and RNA degradation (Farina and Singer, 2002; Jurica and Moore, 2003; Rocak and Linder, 2004). In addition, CCR4-associated factor 1 (CAF1)-like protein (At3g44260) is early and transiently cold induced. CAF1 is a critical component of the major cytoplasmic deadenylase in yeast, which initiates mRNA turnover by shortening of the poly(A) tail (Tucker et al., 2001). Also, UBP1-interacting proteins are thought to be involved in a complex that recognizes U-rich sequences in plant 3′ untranslated regions for the stabilization of mRNAs in the nucleus (Lambermon et al., 2002). Thus, induction of the CAF1-like protein and repression of the UPB1-interacting protein under cold stress might enhance the turnover of specific mRNA transcripts.
We have shown that mutations in an Arabidopsis DEAD box RNA helicase (LOS4, AT3G53110) affect mRNA export, cold sensitivity, and cold-responsive gene expression (Gong et al., 2002, 2005). Moreover, an Arabidopsis sta1 mutant defective in one of our cold upregulated pre-mRNA splicing factors (At4g03430) displayed hypersensitivity to cold stress and mis-splicing of at least one cold-responsive gene, while it enhanced the transcript stability of some genes (B.-h. Lee and J.-K. Zhu, unpublished data). Taken together, the cold induction of several genes for RNA metabolism supports a strong connection between RNA metabolism and cold responses.
Interestingly, many genes that alter the chromatin status showed cold-regulated expression (Table 4; see Supplemental Table 11 online). One histone deacetylase (At5g26040), two histone acetylases (At3g12980 and At5g50320), and three bromodomain-containing proteins (At5g10550, At5g14270, and At5g63330) were among the cold-regulated genes involved in chromatin remodeling. Histone acetylases add acetyl groups to histone tails, whereas histone deacetylases remove acetyl groups from the histone tail. The bromodomain recognizes the acetylated Lys residues in histone tails (Eberharter and Becker, 2002). The kinetics of cold regulation of these genes differs, suggesting a complex and dynamic nature of cold-responsive gene regulation at the chromatin level.
We found that NRPD1a (At1g63020) was early and continually cold upregulated (Table 4; see Supplemental Table 11 online). NRPD1a encodes a plant-specific RNA polymerase IV subunit and is important in small interfering RNA-directed de novo DNA methylation important for silencing of transposon and other repetitive DNA in plants (Herr et al., 2005; Onodera et al., 2005). Recently, many small RNAs that are potentially associated with gene silencing were isolated from a small RNA library constructed from abiotic stress–treated Arabidopsis (Sunkar and Zhu, 2004). Taken together, our finding suggests that some endogenous small RNAs might be increased by cold treatment, and NRPD1a may play an important role in this process.
In conclusion, cold-regulated gene expression is not only controlled by promoter binding transcription factors but may also be regulated at the posttranscriptional and chromatin levels.
Cold Regulation of Genes Related to Plant Hormone Biosynthesis and Signaling
Plant hormones are crucial regulators of growth and development. Plants grown under cold stress display growth and development patterns different from those under normal growth conditions, which might have to do with an altered hormone homeostasis and/or signal transduction in cold-stressed plants. To investigate this possibility, we inspected the expression pattern of hormone-related genes under cold treatment.
ABA is an important stress hormone that mediates abiotic stress signal transduction and tolerance. ABA accumulates in response to abiotic stress, such as drought and salt (Xiong and Zhu, 2003). Cold stress also increases endogenous ABA levels in plants but to a much lesser extent (Lang et al., 1994). Because ABA biosynthesis is mainly regulated at the transcriptional level (Xiong and Zhu, 2003), it is of interest to see whether ABA biosynthesis genes are regulated by cold. According to our microarray data, none of the known ABA synthesis genes were cold regulated, suggesting that ABA biosynthesis is not a major event in cold stress, consistent with previous studies (Lang et al., 1994).
Although ABA levels may not change greatly under cold stress, ABA signal transduction could be involved in cold responses. Cold stress induces many genes that are also induced by ABA and osmotic stress (Thomashow, 1999). Furthermore, many ABA-induced genes were induced by cold according to our microarray data (Table 5). As described above, a bZIP transcription factor, ABF1 (At1g49720), which can bind to the ABA-responsive element in ABA-responsive promoters (Choi et al., 2000), is early and continually induced by cold (Table 5). A related gene, ABF4/AREB2 (At3g19290), was also induced by cold. Thus, it is possible that the cold induction of some ABA-inducible genes might be mediated by ABF1 and ABF4.
Table 5.
Cold Upregulated Genes with Hormone-Related Roles
| Hormone | AGI ID | Gene Name | Kinetics |
|---|---|---|---|
| ABA | At4g17615 | Calcineurin B-like protein 1 (CBL1) | ETU |
| At5g52310 | Low-temperature-induced protein 78 | ECU | |
| At1g20440 | Dehydrin (COR47) | ECU | |
| At1g20450 | Dehydrin (ERD10) | ECU | |
| At4g24960 | ABA-induced–like protein | ECU | |
| At5g15960 | Stress-induced protein KIN1 | ECU | |
| At1g49720 | ABA-responsive elements binding factor (ABF1) | ECU | |
| At2g36270 | ABA-responsive element binding protein (ABI5) | LU | |
| At5g58670 | Phosphoinositide-specific phospholipase C | LU | |
| At4g15910 | Drought-induced protein (Di21) | LU | |
| At1g29395 | Similar to the cold acclimation protein WCOR413 in wheat | LU | |
| At1g28200 | GRAM domain-containing protein | LU | |
| At3g19290 | ABA-responsive elements binding factor (ABF4/ABREB2) | LU | |
| At3g11410 | Protein phosphatase 2C (PP2C), putative | LU | |
| At4g26080 | Protein phosphatase ABI1 | LU | |
| At5g50720 | ABA-responsive protein (HVA22e) | LU | |
| At2g19450 | Acyl CoA:diacylglycerol acyltransferase (DGAT) | LU | |
| At1g69270 | Receptor protein kinase-related (RPK1) | LU | |
| At5g13200 | GRAM domain-containing protein | LU | |
| Auxin | At5g54490 | PINOID binding protein containing putative calcium binding motifs | ETU |
| At1g13260 | AP2 domain transcription factor, putative (RAV1) | ETU | |
| At5g35735 | Auxin-induced protein family | LU | |
| At5g20730 | Auxin response factor (ARF7) | LU | |
| At5g53590 | Auxin-induced (indole-3-acetic acid–induced) protein family | LU | |
| Ethylene | At1g28370 | Ethylene-responsive element binding factor 11, putative | ETU |
| At4g17490 | Ethylene-responsive element binding factor (ERF6) | ETU | |
| At5g51190 | AP2 domain transcription factor, putative | ETU | |
| At4g11280 | 1-Aminocyclopropane-1-carboxylate synthase 6 (ACS6) | ECU | |
| At3g15210 | Ethylene-responsive element binding factor 4 (ERF4) | ECU | |
| At5g47230 | Ethylene-responsive element binding factor 5 (AtERF5) | ECU | |
| At1g67310 | Calmodulin binding protein | LU | |
| At3g21420 | Oxidoreductase, 2OG-Fe(II) oxygenase family | LU | |
| At4g20880 | Ethylene-regulated transcript 2 (ERT2) | LU | |
| GA | At1g02400 | GA 2-oxidase, putative | LU |
| At5g17490 | GA response modulator, putative (RGL3) | LU |
Gibberellins (GAs) are involved in many plant developmental processes, including seed development, stem elongation, flowering, and fruit development (Richards et al., 2001). Many genes involved in GA biosynthesis have been isolated and characterized (Hedden and Kamiya, 1997). In several plants, such as rice (Oryza sativa), pea (Pisum sativum), and Arabidopsis, the transcript level of GA 20-oxidase, which catalyzes the conversion of C20-GA into C19-GA, is downregulated by GA. By contrast, treatment with GA biosynthesis inhibitor resulted in an increased level of GA 20-oxidase transcripts. In addition to this GA-induced negative feedback regulation of GA biosynthesis, positive regulation of a GA catabolic enzyme, GA 2-oxidase, was also reported (Thomas et al., 1999). Therefore, changes in the expression of these genes can indicate GA homeostasis under cold stress.
In this study, we found that two GA 20-oxidases (At3g60290 and At4g03060) were downregulated, and one, GA 2-oxidase (At1g02400), was cold induced (Tables 5 and 6). These expression changes suggest the possibility of increased GA accumulation under cold stress. Although this potential GA accumulation under low temperature conditions might be part of the vernalization process that promotes flowering, it is also possible that GA might affect cold stress signaling and tolerance. Recently, it was demonstrated that in imbibed seeds, cold treatment enhances the biosynthesis of GA via the activation of GA3Ox1 (or GA 3β hydroxylase, GA4, At1g15550) to promote seed germination (Yamauchi et al., 2004).
Table 6.
Cold Downregulated Genes with Hormone-Related Roles
| Hormone | AGI ID | Gene Name | Kinetics |
|---|---|---|---|
| Auxin | At2g45210 | Putative auxin-regulated protein; SAUR gene family | ETD |
| At4g38840 | Auxin-induced protein-like; SAUR gene family | ETD | |
| At4g39950 | CYP79B2 | LD | |
| At4g31500 | CYP83B1 | LD | |
| At5g54510 | Auxin-responsive–related protein | LD | |
| At3g25290 | Auxin-induced protein family | LD | |
| At2g01420 | Auxin transporter splice variant B (PIN4) | LD | |
| At1g29430 | Auxin-induced protein, putative; SAUR gene family | LD | |
| At1g29500 | Auxin-induced protein, putative; SAUR gene family | LD | |
| At4g15550 | UDP-glucose:indole-3-acetate β-d-glucosyltransferase (iaglu) | LD | |
| At1g24100 | Putative indole-3-acetate β-glucosyltransferase | LD | |
| At4g34770 | Auxin-induced protein family; SAUR gene family | LD | |
| At1g72430 | Auxin-induced protein-related; SAUR gene family | LD | |
| At1g28130 | Auxin-regulated GH3 protein-related | LD | |
| At2g47750 | Auxin-responsive protein-related | LD | |
| At2g36910 | Multidrug resistance P-glycoprotein (pgp1) | LD | |
| Brassinosteroid | At3g61460 | BRH1 RING finger protein | ETD |
| At5g05690 | CPD, DWF3 | LD | |
| At3g50750 | Expressed protein | LD | |
| Cytokinin | At5g35750 | Histidine kinase-related protein; AHK2 | LD |
| At3g63440 | Cytokinin oxidase family protein | LD | |
| At4g31920 | ARR10; type B two-component response regulator protein | LD | |
| At2g40670 | ARR16; type A two-component response regulator protein | LD | |
| Ethylene | At5g25190 | Ethylene-responsive element binding protein, putative | LD |
| GA | At3g60290 | SRG1-like protein; GA 20-oxidase | LD |
| At1g14920 | GA response modulator (GAI/RGA2) | LD | |
| At4g03060 | Putative oxidoreductase; GA 20-oxidase | LD | |
| Jasmonate | At1g17990 | 12-Oxophytodienoate reductase, putative | LD |
| Salicylic acid | At1g21270 | Wall-associated kinase 2 (WAK2) | LD |
GA signaling is mediated through DELLA proteins that act as negative regulators (Olszewski et al., 2002). Upon GA perception, some DELLA protein is removed from the nucleus by degradation, releasing the repression of GA signaling. For example, in rice, the level of nuclear-localized SLR1-green fluorescent protein fusion decreased with GA treatment (Itoh et al., 2002). The Arabidopsis DELLA protein REPRESSOR OF GA (RGA) was shown to be regulated in a similar way (Silverstone et al., 2001). Rice has only one DELLA protein (Ikeda et al., 2001), but Arabidopsis has five: GA INSENSITIVE (GAI, At1g14920), RGA (At2g01570), RGA-LIKE PROTEIN 1 (RGL1, At1g66350), RGL2 (At3g03450), and RGL3 (At5g17490). Not all of these proteins are regulated by degradation. GA treatment did not lead to GAI and RGL1 degradation (Fleck and Harberd, 2002; Wen and Chang, 2002). RGL2 appears to be transcriptionally regulated by GA during seed germination (S. Lee et al., 2002). We found that GAI (At1g14920) was downregulated and RGL3 (At5g17490) upregulated under cold, possibly by cold-induced GA (Tables 5 and 6). Thus, GAI and RGL3 might be regulated at the mRNA level. Since the physiological function of RGL3 is unknown, our observation that it is upregulated by cold provides a clue to its function.
CONSTITUTIVE PHOTOMORPHOGENIC DWARF/DWARF3 (CPD/DWF3, At5g05690) was cold downregulated 24 h after cold treatment (Table 6). CPD/DWF3 encodes a member of the cytochrome P450 90A family required for C23 hydroxylation of cathasterone to teasterone in brassinolide biosynthesis (Szekeres et al., 1996). CPD/DWF3 expression is specifically downregulated by brassinolide treatment but not by other plant hormones (Mathur et al., 1998). Our observation of CPD/DWF3 downregulation by cold suggests that brassinosteroid level might be lowered in the cold, or cold signaling might interfere with brassinosteroid regulation of the CPD/DWF3 gene. Consistent with this, expression of BRASSINOSTEROID-RESPONSIVE RING-H2 1 (BRH1, At3g61460), whose transcript is downregulated by brassinolide (Molnar et al., 2002), was also downregulated at 6 h after cold stress.
We noticed that the expression of many genes with auxin-related roles was affected by cold stress. Twenty-one auxin-related genes showed cold upregulation or downregulation, and the majority of them (16 genes) were cold downregulated, while only five genes were cold upregulated (Tables 5 and 6). Among the 16 cold downregulated auxin genes, six belong to the SAUR gene family (Table 6). SAUR gene transcripts are very unstable (Hagen and Guilfoyle, 2002). Thus, cold stress may either prevent the stabilization of the SAUR gene transcripts (e.g., by reducing auxin levels) or decrease their transcription.
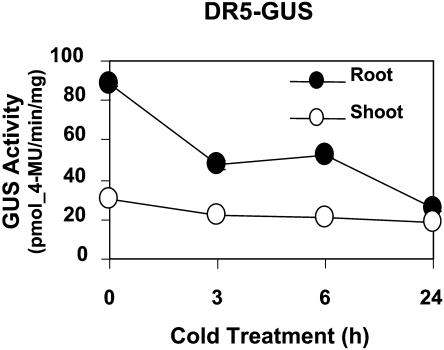
To examine the possibility of cold regulation of auxin levels, we monitored β-glucuronidase (GUS) activity in Arabidopsis seedlings expressing the DR5-GUS transgene. The DR5-GUS Arabidopsis plant has the GUS reporter gene driven by the synthetic auxin-responsive DR5 promoter, which has been widely used as a reporter for endogenous auxin levels (Ulmasov et al., 1997). The GUS activity in DR5-GUS seedlings decreased after cold treatment (Figure 3), particularly in roots. This suggests that cold may decrease endogenous auxin levels, especially in roots, or interfere with auxin sensitivity. Other cold-affected, auxin-related genes include those in signaling and transcription (At5g20730 and At5g54490), auxin conjugation/inactivation (At1g24100, At4g15550, At1g28130, At2g47750, and At5g54510), auxin biosynthesis (At4g31500 and At4g39950), and polar transport (At2g01420 and At2g36910).
Figure 3.
Quantification of DR5-GUS Activity during Cold Stress.
GUS quantification is shown in pmol·4-MU/min/mg on the y axis during cold treatment (h, x axis) of DR5-GUS–expressing Arabidopsis.
Obviously, cold is not the only factor that downregulates auxin-inducible genes. Wounding has a similar effect (Cheong et al., 2002). One proposed mechanism for downregulation by wounding is suppression of auxin signal transduction by a NPK1-like gene, a putative homolog of ANP1 that negatively regulates auxin-responsive genes (Kovtun et al., 1998). In our microarray study, the NPK1-like gene (At2g30040) was early and transiently upregulated by cold, which suggests that cold repression of auxin-inducible genes might be mediated in a similar way as wounding (see Supplemental Table 2 online). Another proposed possibility of auxin-related gene downregulation by wounding was potentially lower levels of active auxin triggered by wounding (Cheong et al., 2002). This was based on the observation that the gene expression of a nitrilase, an auxin biosynthetic enzyme that catalyzes the conversion of indole-3-acetonitrile to indole-3-acetic acid (IAA), was downregulated and the transcript levels of two IAA glucosyltransferases, auxin conjugating enzymes, were upregulated by wounding (Cheong et al., 2002). In our microarray data, nitrilase genes were not significantly changed by cold, but IAA glucosyltransferase genes (At1g24100 and At4g15550) were downregulated. Therefore, although the disturbance of auxin homeostasis, transport, and signaling might all contribute to the downregulation of auxin-inducible genes by cold, the mechanisms appear to differ between the stresses.
Downregulation of auxin transport and auxin-responsive genes may eventually contribute to reduced plant growth rate under cold stress. Related to the growth regulation, we found that two members of the α-expansin gene family (Lee et al., 2001) were repressed at the late time point by cold (see Supplemental Table 9 online): expansin A1 (At1g69530) and A6 (At2g28950). Interestingly, three expansin-like genes (EXLA1, At3g45970; EXLA2, At4g38400; EXLA3, At3g45960) were cold upregulated (see Supplemental Table 2 online), which suggests that these expansin-like genes may have a distinct function in plant growth.
Cold-Regulated Gene Expression in the ice1 Mutant
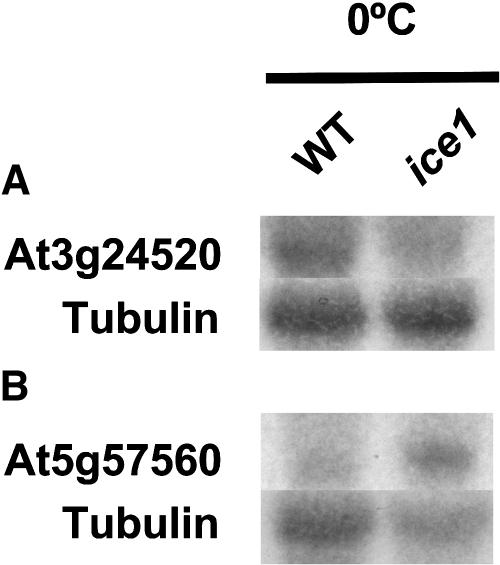
We have previously reported that the bHLH transcription factor ICE1 functions as an upstream regulator of CBF genes (Chinnusamy et al., 2003). The dominant ice1 mutant shows reduced cold induction of CBF3 and several COR (for cold responsive) genes that are under control of CBFs. To understand the role of ICE1 in global gene expression in the cold, the expression of the 939 cold-regulated genes in the wild type was compared with that in the ice1 mutant at each time point during cold stress. As the ice1 mutant was originally isolated due to its lower expression of CBF3 (At4g25480) during cold stress, the CBF3 expression level in both the wild type and ice1 mutants was examined first. With the FDR of 1%, CBF3 expression in ice1 was statistically lower than the wild type only at 3 h after cold treatment. Using RNA gel blot analysis, we have previously observed that CBF3 transcript levels were clearly lower in ice1 than in the wild type at least until 24 h after cold treatment (Chinnusamy et al., 2003). Thus, the FDR of 1% appears to be too strict to be used as a threshold for the comparison of cold-responsive gene expression between the wild type and ice1. Another conventional FDR of 5% appeared to be an appropriate standard, as it rendered CBF3 transcript levels to be significantly lower in ice1 at 3, 6, and 24 h under cold stress. Therefore, 5% FDR was used here for the comparison of 939 cold-responsive genes between the wild type and ice1. The details of the statistical methods are described in Methods, and results from this analysis were confirmed by RNA hybridization with two randomly selected genes showing significantly different transcript levels between the wild type and ice1 (Figure 4).
Figure 4.
RNA Gel Blot Analysis of Cold-Regulated Genes in the Wild Type and ice1.
(A) At3g24520 (HSF1, 0°C 24 h).
(B) At5g57560 (TCH4, 0°C 0 h). Tubulin was used as a loading control.
Among the 939 cold-regulated genes, the expression of 369 cold-regulated genes was affected in the ice1 mutant (Table 7; see Supplemental Tables 12 and 13 online). Among them were 240 cold upregulated and 129 downregulated genes (Table 7; see Supplemental Tables 12 and 13 online). Thus, the expression of ∼40% of cold-regulated genes was altered in the ice1 mutant in comparison to the wild type. To see if the altered expression affected primarily cold-regulated genes, 939 non-cold-regulated genes were randomly selected, and the gene expression of the random 939 genes was analyzed. The analysis revealed that only the expression of 120 of these genes was significantly different between the wild type and ice1, with 64 being higher and 56 significantly lower in ice1 (Table 7). In summary, the expression of many of cold-regulated genes was affected by the ice1 mutation, and the ice1 mutation has a differential effect on the expression of cold-regulated genes than on that of genes in general.
Table 7.
Comparison of Cold-Regulated Gene Expression between the Wild Type and ice1
| Comparison Time PointsTranscript Level in ice1
|
0 to 24 h
|
0 h
|
3 to 24 h (during Cold)
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Higher | Lower | Subtotal | Higher | Lower | Subtotal | Higher | Lower | Subtotal | |
| Cold upregulated (655) | 142 | 101 | 240a | 132 | 10 | 142 | 56 | 101 | 157 |
| Cold downregulated (284) | 72 | 58 | 129b | 13 | 49 | 62 | 72 | 41 | 112b |
| Cold regulated (939) | 214 | 159 | 369 | 145 | 59 | 204 | 128 | 142 | 269 |
| Random (939) | 64 | 56 | 120 | 41 | 30 | 71 | 50 | 44 | 94 |
Three genes (At1g09070, At1g76590, and At4g15910) belong to both higher and lower categories at a certain time point.
One gene (At4g26950) belongs to both the higher and lower categories at a certain time point.
Numbers in parentheses are total numbers of genes in each category.
We then examined the basal level (i.e., at 0 h) of expression of cold-regulated genes in the wild type and ice1. Basal transcript levels of 204 of the 939 cold-regulated genes were affected in ice1 (Table 7). Interestingly, among ice1-affected cold upregulated genes, a substantially larger number of genes had higher basal transcript levels in ice1 than lower basal levels compared with the wild type (132 genes versus 10 genes). By contrast, ice1-affected cold downregulated genes had more genes with lower basal levels in ice1 than higher basal levels relative to the wild type (49 versus 13) (Table 7). The results suggest that the wild-type ICE1 protein plays a critical role in maintaining the basal expression levels for both cold upregulated and downregulated genes.
A comparison of the expression levels of cold-regulated genes after cold treatment revealed that there are approximately twofold more cold upregulated genes in ice1 with lower expression levels than those with higher expression levels (101 versus 56) (Table 7). By contrast, the number of cold downregulated genes with higher transcript levels in ice1 was higher than that with lower expression levels (72 versus 41) (Table 7). These are opposite tendencies to those observed in the basal transcript levels in ice1. It suggests that ICE1 plays a large role in the cold activation of gene expression as well as in the downregulation of genes.
Because ICE1 is the most upstream transcription factor known in the cascades of gene expression under cold stress (Chinnusamy et al., 2003), we examined in more detail the effect of the ice1 mutation on cold regulation of transcription factors (Tables 8 and 9; see Supplemental Table 11 online). Including CBF3 (At4g25480), the expression of 52 (39.1%) out of the 133 cold-regulated genes involved in transcription was affected by the ice1 mutation (Tables 8 and 9; see Supplemental Table 11 online). Before cold treatments (i.e., 0 h), expression levels of most cold-responsive transcription-related genes were higher in ice1, and all of them belong to cold upregulated genes (Table 8). Interestingly, the expression level of these genes, except for one Zn finger domain gene (At3g28210), became similar to the wild-type level as the duration of cold stress became longer. In other words, at 24 h after cold stress, these transcription-related genes with higher basal transcript levels in ice1 did not show significant difference from the wild-type levels. This suggests that the responsiveness of cold induction (i.e., the rate of gene induction) and the basal levels of these genes are affected in ice1. Our results thus indicate that ICE1 regulates cold tolerance by controlling the basal expression and induction of many cold-responsive transcription factors.
Table 8.
Cold-Regulated Transcription Genes Altered in ice1
| Transcript Level in ice1 at 0°C
|
|||||||
|---|---|---|---|---|---|---|---|
| Category | AGI ID | Kinetics | 0 h | 3 h | 6 h | 24 h | Gene Name |
| AP2 | At1g28370 | ETU | H | Ethylene-responsive element binding factor 11 | |||
| At1g77640 | ETU | H | H | AP2 domain transcription factor, putative | |||
| At5g51990 | ETU | L | L | AP2 domain transcription factor-like protein | |||
| At5g53290 | ETU | H | AP2 domain transcription factor, putative | ||||
| At2g23340 | ECU | L | AP2 domain transcription factor, putative | ||||
| At2g40350 | ECU | H | H | AP2 domain transcription factor, putative (DREB2) | |||
| At3g50260 | ECU | H | AP2 domain transcription factor, putative | ||||
| At4g25480 | ECU | L | L | L | DRE binding protein (DREB1A/CBF3) | ||
| At5g07310 | ECU | H | AP2 domain putative transcription factor | ||||
| At2g28550 | LU | L | AP2 domain transcription factor RAP2.7 | ||||
| At4g36900 | LU | L | AP2 domain protein RAP2.10 | ||||
| At4g36920 | LU | H | H | Floral homeotic protein APETALA2 | |||
| Myb | At5g67300 | ETU | H | Myb-related protein, 33.3K | |||
| At5g01200 | LU | L | L | Myb family transcription factor | |||
| At5g16560 | LD | H | Myb family transcription factor | ||||
| bHLH | At1g32640 | ETU | H | bHLH protein (RAP-1); ATMYC2 | |||
| At2g23760 | LU | L | L | BEL1-like homeobox 4 protein (BLH4) | |||
| At1g09250 | LD | L | bHLH protein; tRNA processing; chloroplast | ||||
| At1g18400 | LD | L | L | L | L | Helix-loop-helix protein homolog–related | |
| At1g73830 | LD | L | L | bHLH protein family | |||
| At3g06120 | LD | H | H | bHLH protein family | |||
| bZIP | At2g36270 | LU | H | H | bZIP transcription factor AtbZip39 | ||
| At4g01120 | LU | L | L | G-box binding bZip transcription factor GBF2/AtbZip54 | |||
| At4g34590 | LU | H | H | H | bZIP transcription factor ATB2/Atbzip11 | ||
| GRAS | At1g14920 | LD | L | L | L | Signal response protein (GAI) | |
| HSF | At3g24520 | ECU | L | L | L | HSF1 | |
| MADS | At3g66656 | LD | L | MADS box protein | |||
| NAC | At5g24590 | ETU | L | No apical meristem (NAM) protein family | |||
| At3g10500 | LU | H | H | NAM family protein | |||
| At2g33480 | LD | H | NAM protein family | ||||
| SPB | At1g76580 | ECU | L | L | SPL1-Related3 protein | ||
| Trihelix | At5g01380 | LU | H | Transcription factor GT-3a | |||
| WRKY | At2g38470 | ETU | H | Putative WRKY-type DNA binding protein | |||
| At2g46400 | ETU | H | WRKY family transcription factor | ||||
| At4g01250 | ETU | H | H | WRKY family transcription factor | |||
| At4g31800 | ETU | H | H | WRKY family transcription factor | |||
| At2g30250 | ECU | H | WRKY family transcription factor | ||||
| At1g62300 | LU | H | H | H | Transcription factor WRKY6 | ||
| Zn | At3g55980 | ETU | H | Zn finger (CCCH-type) family protein | |||
| At1g27730 | ECU | H | Salt tolerance Zn finger protein; ZAT10 | ||||
| At5g59820 | ECU | H | H | Zn finger protein Zat12 | |||
| At1g10170 | LU | H | H | NF-X1–type Zn finger family protein | |||
| At1g25250 | LU | L | Zn finger (C2H2 type) family protein | ||||
| At3g28210 | LU | H | H | H | H | Zn finger protein (PMZ)–related | |
| At5g18550 | LU | H | Zn finger–like protein | ||||
| At2g44380 | LD | H | CHP-rich Zn finger protein, putative | ||||
| Not classified | At3g61260 | LU | L | L | L | Putative DNA binding protein | |
| Chromatin | At1g63020 | ECU | H | RNA polymerase IV (RNA polymerase D) subunit; NRPD1a | |||
| At5g10550 | ETD | H | Bromodomain protein-like | ||||
| RNA Metab. | At4g03430 | LU | H | Putative pre-mRNA splicing factor (STA1) | |||
| At4g05410 | LU | H | H | U3 snoRNP-associated–like protein | |||
| At5g46920 | LU | H | Intron maturase, type II family | ||||
H or L indicates that the transcript level is higher or lower, respectively, in ice1 compared with the wild type. SPB, squamosa promoter binding; RNA Metab., RNA metabolism.
Table 9.
Numbers of Cold-Regulated Transcription Genes Altered in ice1
| Category | Gene Number Altered in ice1 |
|---|---|
| AP2/ERF (22) | 12 |
| B3 (1) | 0 |
| bZIP (6) | 3 |
| CCAAT (1) | 0 |
| CG-1/CAMTA4 (1) | 0 |
| GRAS (3) | 1 |
| Homeodomain (2) | 1 |
| HSF (4) | 1 |
| MADS (1) | 1 |
| MYB (12) | 3 |
| bHLH (8) | 5 |
| NAC (10) | 3 |
| SPB (2) | 1 |
| Trihelix (3) | 1 |
| Unclassified DNA domain (1) | 1 |
| WRKY (8) | 6 |
| Zn (29) | 8 |
| Chromatin remodeling (4) | 2 |
| RNA metabolism (6) | 3 |
| Total | 52 |
Numbers in parentheses indicate the total number of cold-regulated genes in each category.
Among the transcription-related genes with altered expression in ice1, AP2 domain transcription factors were the major group, followed by bZIPs and WRKYs (Table 9). In particular, the expression of most of the cold-inducible WRKY factors (six out of eight) was enhanced in ice1, and the alteration mostly occurred at either 0 h or early time points (Tables 8 and 9). In fact, WRKY transcription factors and AP2 domain transcription factors were the major early cold-regulated transcription factors (Table 3). Thus, ICE1 seems to preferentially target early or upstream transcription factors in cold-regulated gene expression.
The ice1 mutation also affected the expression of 31 (37.8%) of the 82 cold-regulated genes involved in signal transduction (Tables 10 and 11; see Supplemental Table 10 online). Genes encoding Ca2+-signaling proteins, RLKs, and lipid-signaling proteins were the major genes whose expression was altered in ice1 (Table 11). Expression of other cold-regulated protein kinases and protein phosphatase was largely not changed in ice1. The results suggest that the ICE1 pathway may involve lipid and RLK-mediated processes.
Table 10.
Cold-Regulated Signaling Genes Altered in ice1
| Transcript Level in ice1 at 0°C
|
|||||||
|---|---|---|---|---|---|---|---|
| Category | AGI ID | Kinetics | 0 h | 3 h | 6 h | 24 h | Gene Name |
| Calcium binding | At2g43290 | ETU | H | Putative calcium binding protein | |||
| At5g37770 | ETU | H | Calmodulin-related protein 2, Touch-induced (TCH2) | ||||
| At3g10300 | ECU | H | Calcium binding EF-hand family protein | ||||
| At4g27280 | ECU | H | Calcium binding EF-hand family protein | ||||
| At5g49480 | ECU | H | Sodium-inducible calcium binding protein | ||||
| At3g51920 | LU | H | Putative calmodulin | ||||
| At5g55990 | LU | L | Calcineurin B-like protein 2 (gb|AAC26009.1) | ||||
| At4g16350 | LD | H | Calcineurin B-like protein 6 (CBL6) | ||||
| Protein phosphatase | At3g16800 | LU | L | Protein phosphatase 2C (PP2C), putative | |||
| At5g02760 | ECD | L | L | Protein phosphatase-like protein | |||
| Protein kinase | At1g01140 | ETU | L | SOS2-like protein kinase PKS6/CBL-interacting protein kinase 9 (CIPK9) | |||
| At3g24550 | ETU | L | Protein kinase-related | ||||
| At1g07150 | ECU | H | Protein kinase family; MAPKKK13 | ||||
| At3g57760 | ECU | H | Protein kinase family protein | ||||
| At2g28930 | LU | H | Protein kinase (APK1b) | ||||
| RLK | At1g61380 | LU | H | S-like receptor protein kinase | |||
| At3g53810 | LU | H | H | H | H | Receptor lectin kinase, putative | |
| At1g12460 | LD | L | L | Leucine-rich repeat transmembrane protein kinase, putative | |||
| At1g34210 | LD | H | Somatic embryogenesis receptor-related kinase | ||||
| At1g68780 | LD | L | L | L | Leucine-rich repeat protein family | ||
| At3g23110 | LD | H | Disease resistance family protein, contains leucine-rich repeat | ||||
| At4g30520 | LD | L | RLK homolog | ||||
| Histine kinase | At5g35750 | LD | L | L | L | Histidine kinase (AHK2) | |
| Response regulator | At4g18020 | LD | H | Pseudoresponse regulator 2 (APRR2) | |||
| Lipid signaling | At4g18010 | ETU | H | IP5PII | |||
| At5g07920 | ECU | L | Diacylglycerol kinase (ATDGK1) | ||||
| At5g58670 | LU | H | H | H | Phosphoinositide-specific phospholipase C (ATPLC1) | ||
| At5g58690 | LU | L | Phosphoinositide-specific phospholipase C-line (MZN1.13) | ||||
| At5g58700 | LU | H | Phosphoinositide-specific phospholipase C4 (PLC4) | ||||
| At5g63770 | LU | H | Diacylglycerol kinase | ||||
| GTP-related | At1g30960 | LU | H | GTP binding protein, ERG-related | |||
H or L indicates that the transcript level is higher or lower, respectively, in ice1 compared with the wild type. Note that pseudoresponse regulators are included in the response regulator category.
Table 11.
Numbers of Cold-Regulated Signaling Genes Altered in ice1
| Category | Gene Number Altered in ice1 |
|---|---|
| Calcium binding protein (15) | 8 |
| Protein phosphatase (12) | 2 |
| Protein kinase (20) | 5 |
| RLK (14) | 7 |
| Blue light receptor (1) | 0 |
| Histidine kinase (1) | 1 |
| Response regulator (5) | 1 |
| Lipid-signaling protein (9) | 6 |
| GTP-related protein (5) | 1 |
| Total (82) | 31 |
Numbers in parentheses indicate the total number of cold-regulated genes in each category.
An examination of genes with plant hormone-related roles in ice1 revealed that ABA- and auxin-related genes were predominantly affected in ice1 (Tables 12 and 13). This indicates that the activation of ICE1 may be upstream of the hormone-related changes during cold regulation.
Table 12.
Cold Upregulated Plant Hormone-Related Genes Altered in ice1
| Transcript Level Altered in ice1
|
|||||||
|---|---|---|---|---|---|---|---|
| Hormone | AGI ID | Kinetics | 0 h | 3 h | 6 h | 24 h | Gene Name |
| ABA | At5g52310 | ECU | L | Low-temperature-induced protein 78 | |||
| At1g20440 | ECU | L | Dehydrin (COR47) | ||||
| At1g20450 | ECU | L | L | Dehydrin (ERD10) | |||
| At4g24960 | ECU | L | ABA-induced–like protein | ||||
| At5g15960 | ECU | L | L | Stress-induced protein KIN1 | |||
| At2g36270 | LU | H | H | ABA-responsive element binding protein, putative | |||
| At5g58670 | LU | H | H | H | Phosphoinositide-specific phospholipase C | ||
| At4g15910 | LU | H | L | Drought-induced protein (Di21) | |||
| At1g29395 | LU | L | Similar to the cold acclimation protein WCOR413 in wheat | ||||
| Auxin | At5g35735 | LU | H | Auxin-induced protein family | |||
| Ethylene | At1g28370 | ETU | H | Ethylene-responsive element binding factor 11, putative | |||
H or L indicates that the transcript level is higher or lower, respectively, in ice1 compared with the wild type.
Table 13.
Cold Downregulated Plant Hormone-Related Genes Altered in ice1
| Transcript Level Altered in ice1
|
|||||||
|---|---|---|---|---|---|---|---|
| Hormone | AGI ID | Kinetics | 0 h | 3 h | 6 h | 24 h | Gene Name |
| Auxin | At2g45210 | ETD | H | H | H | Putative auxin-regulated protein; SAUR gene family | |
| At4g38840 | ETD | L | Auxin-induced protein like; SAUR gene family | ||||
| At4g39950 | LD | H | H | H | CYP79B2 | ||
| At4g31500 | LD | H | H | CYP83B1 | |||
| At5g54510 | LD | H | Auxin-responsive-related protein | ||||
| At3g25290 | LD | H | H | H | H | Auxin-induced protein family | |
| At2g01420 | LD | L | L | L | L | Auxin transporter splice variant B (PIN4) | |
| At1g29430 | LD | L | L | L | L | Auxin-induced protein, putative; SAUR gene family | |
| At1g29500 | LD | L | L | L | Auxin-induced protein, putative; SAUR gene family | ||
| Cytokinin | At5g35750 | LD | L | L | L | Histidine kinase-related protein; AHK2 | |
| GA | At3g60290 | LD | L | L | SRG1-like protein; GA 20-oxidase | ||
| At1g14920 | LD | L | L | L | GA response modulator (GAI/RGA2) | ||
H or L indicates that the transcript level is higher or lower, respectively, in ice1 compared with the wild type.
Recently, putative CBF3 and CBF2 target genes were identified using Arabidopsis transgenic lines overexpressing CBF3 or CBF2 (Maruyama et al., 2004; Vogel et al., 2005). We compared the reported CBF3- and CBF2-targeted cold-responsive genes with ice1-affected cold-responsive genes. Out of 102 genes identified as cold upregulated CBF3 and CBF2 target genes, 74 are in our cold upregulated gene list. These 74 genes include six CBF3-specific target genes, 18 genes targeted by both CBF2 and CBF3, and 50 CBF2-specific target genes. Four of the six CBF3-specific target genes (67%) were altered in ice1, whereas 19 of the 50 CBF2-specific target genes (38%) were affected by the ice1 mutation (see Supplemental Tables 14 to 16 online). Fifteen of the eighteen genes (83%) targeted by both CBF2 and CBF3 showed altered expression in the ice1 mutant (see Supplemental Tables 14 to 16 online). In addition, we found that only one (At4g02330) out of the 24 cold-responsive ZAT12 target genes (Vogel et al., 2005) was affected by ice1. The results show that the ice1 mutation has a more pronounced effect on the target genes of CBFs, particularly of CBF3, which is consistent with the observation that the dominant ice1 mutation has a very strong effect on the cold induction of CBF3.
Conclusions
In this study, we identified 655 cold upregulated and 284 downregulated genes in Arabidopsis using the Affymetrix Arabidopsis 24,000-gene GeneChip. Many transcription factors were induced during cold stress, particularly during early cold stress. By contrast, only one transcription factor was downregulated early during cold stress. These results suggest that cold responses in plants are initiated mainly by transcriptional activation rather than repression of genes. The downregulation of other transcription factors later in the cold may be the result of early activation of transcription factors. In addition, a number of RNA metabolism genes and chromatin remodeling proteins were also cold regulated, suggesting their involvement in cold-responsive gene regulation.
Many metabolism-related genes were cold induced and some were cold repressed, consistent with dynamic changes in metabolism observed under cold stress (Cook et al., 2004). Another large number of cold-regulated genes are in the cell rescue, defense, and virulence category. Many of these are induced not only by cold but also by other abiotic and biotic stresses and by ABA. Different signal transduction pathways may crosstalk and converge in the activation of these stress genes.
One notable observation was the downregulation of many auxin-inducible SAUR genes by cold stress. These SAUR genes were also reported to be downregulated by wounding (Cheong et al., 2002). A database search at www.genevestigator.ethz.ch revealed that many SAUR genes are downregulated by many other stresses, including osmotic stress and heat stress (Zimmermann et al., 2004). Therefore, it seems that the downregulation of many SAUR genes is one of the general stress responses that may be responsible for altered plant growth and development in response to stress. How are SAUR genes repressed by cold? According to our cold-regulated gene profiles, both auxin homeostasis and signaling appear to be disturbed by cold stress, as the expression of auxin polar transporter genes and one NPK1-like gene (At2g30040, see above) was changed in response to cold stress. Consistent with this, we found that DR5-GUS reporter activity was decreased in cold-treated Arabidopsis.
Under cold conditions, plants grow more slowly, and some even show growth defects or damage. Some of these cold-induced growth changes might be attributed to the slowing of photosynthesis and generally low metabolic activities in the cold (Kubien et al., 2003). Our microarray data revealed other potential causes of altered plant growth and development at low temperatures. We observed the downregulation of two expansin genes. Therefore, one possibility is that the downregulation causes reduced cell expansion, which in turn affects plant growth in the cold. In addition, the altered homeostasis of auxin and possibly other plant hormones such as ethylene, GA, and brassinosteroid might also perturb plant development at low temperatures. Related to this, some development-relevant transcription factors (e.g., ARF, GRAS, Homeodomain, MADS, and NAC) were cold responsive, suggesting their potential involvement in reprogramming plant development under cold stress.
Our comparison of cold-responsive gene expression profiles between the wild type and ice1 mutant supports the important role of ICE1 in cold response gene regulation and cold tolerance in plants. ICE1 regulates the expression of many transcription factors, which in turn may activate or repress other downstream cold-responsive genes. ICE1 is a MYC-like transcription factor in the bHLH family (Chinnusamy et al., 2003). Our survey of MYC recognition sites in cold-responsive genes with altered expression in the ice1 mutant did not reveal any significant differences in the occurrence of MYC binding sites between promoters from randomly chosen genes and the ice1-affected cold-responsive genes. This is probably because the consensus MYC recognition site (CANNTG) is not specific enough for computational detection, and the specific ICE1 binding site needs better definition.
Notwithstanding, the expression of many cold-responsive plant genes known to be critical in cold tolerance were affected in the ice1 mutant. For example, AP2 domain transcription factors, as illustrated by the numerous studies on the CBF genes, were the major early cold-inducible transcription factors, and these were affected by ice1. In addition, two major cold-regulated genes related to ABA or auxin were altered significantly by the ice1 mutation. These results show that ICE1 plays a major role in plant cold responses.
A previous study with an 8K Affymetrix GeneChip described 306 genes, consisting of 218 cold upregulated and 88 cold downregulated genes (Fowler and Thomashow, 2002). Comparison of cold-responsive genes between Fowler and Thomashow's data set and ours revealed that only 108 genes were in common (data not shown). This might be due in part to the different experimental conditions. Fowler and Thomashow (2002) grew plants in Gamborg's B-5 medium on phytoagar, whereas our plants were grown in Murashige and Skoog (MS) medium on regular agar with 3% sucrose, and we used 0°C for cold treatment, whereas Fowler and Thomashow (2002) used 4°C for cold stress. Another cause of this discrepancy is that the analyses in the other studies (Fowler and Thomashow, 2002; Kreps et al., 2002) had no control over the Type I error rate of their reported findings. Nevertheless, these results indicate that experimental and developmental conditions significantly affect gene expression profiles, even with similar stress treatments. Certainly, the genes commonly identified by microarray studies from different groups are the most robust cold-responsive genes.
In summary, we identified many new cold-regulated genes and determined the impact of the ice1 mutation on global gene expression under cold stress. This information provides a foundation for further experiments to explore the network of gene regulation in the cold and to determine the function of cold-responsive genes in cold tolerance through mutant analysis, transgenic overexpression, and other molecular or cell biological approaches.
METHODS
Plant Materials
Bioluminescent Arabidopsis thaliana (Columbia gl1) with a CBF3 promoter-driven luciferase transgene was used as the wild type for the microarray analysis because the ice1 mutant was derived from the CBF3-LUC transgenic Arabidopsis (Chinnusamy et al., 2003). The wild-type and ice1 mutant seeds were plated on MS agar plates supplemented with 3% sucrose. Seedlings were grown at 22°C with 16-h-light and 8-h-dark cycles for 2 weeks before being harvested. To avoid variations due to circadian rhythm, all cold treatments were started at 12 pm at 0°C under light and continued for 0 (untreated control), 3, 6, and 24 h.
Microarray Analysis
For Affymetrix GeneChip analysis, 20 μg of total RNA from the wild-type and ice1 seedlings with (3, 6, and 24 h under light) or without cold treatment were extracted using the RNeasy plant mini kit (Qiagen), and the product was used to make biotin-labeled cRNA targets. The Affymetrix Arabidopsis ATH1 genome array GeneChip, which contains >22,500 probe sets representing ∼24,000 genes, was used. Hybridization, washing, and staining were performed according to the manufacturer's instructions. Microarray data were extracted from scanned GeneChip images and analyzed by use of Microarray Suite version 5.0.1 (Affymetrix). When necessary, the data produced by Mircoarray Suite were exported into Microsoft Excel or Microsoft Access and further analyzed. For cis-element analysis, the GCG/SeqLab program in the WISCONSIN package version 10.3 was used.
GUS Assay
For GUS quantification, shoots and roots of 10-d-old DR5-GUS Arabidopsis after cold treatment (0, 3, 6, and 24 h) were extracted in 800 μL of assay solution (50 mM sodium phosphate, pH 7, 10 mM EDTA, 0.1% SDS, 10 mM β-mercaptoethanol, and 0.1% Triton X-100). After protein quantification by use of the Bradford method, 20 μg of total protein extract from each sample was added to 200 μL of 2 mM 4 methyl umbelliferyl-β-d-glucuronide (4-MU) in assay solution and incubated for 10, 20, and 30 min at 37°C. The reaction was stopped by adding 800 μL of 0.2 M Na2CO3. The enzyme activity was measured by use of Titertek Fluoroskan II (Flow Laboratories) and calculated in units of pmol·4-MU/min/mg protein. The assay was performed three times. Cold treatment starting time was 12 pm as it was for the microarray.
Statistical Analysis
Data were analyzed with use of a two-stage linear statistical model and robust test statistics as described in the Limma package (Smyth, 2005) in the Bioconductor Suite of tools for the statistical package R (R Development Core Team, 2003; http://www.R-project.org). Global effects were first estimated by use of a standard linear model on the observed intensity values, fitting array, genotype, time, and genotype by time interaction as fixed effects in the analysis:
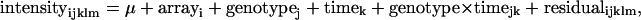 |
where i indexes arrays, j indexes genotypes, k indexes time, l indexes oligos, and m indexes individual intensity values. Residuals from this model were then used in subsequent individual gene analyses. This procedure is automated in the Limma package (Smyth, 2005) found in the Bioconductor suite of tools (http://www.bioconductor.org). The Limma package creates robust individual gene t-statistics for each contrast of interest (i.e., oligo by genotype by time) constructed by use of a shrinkage estimator for individual gene variances. The Limma model fits individual gene means, array, genotype, time, and genotype by time interaction effects regressed on the global model-adjusted intensity values (i.e., the residuals from the initial linear model).
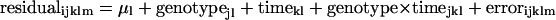 |
Parameters for the shrinkage estimator are obtained with use of an empirical Bayes method (for details, see Smyth, 2005).
Significance of the robust t-statistics was determined from P values obtained with 10,000 bootstrap simulations (Efron and Tibshirani, 1993), with a corresponding Limma analysis assuming complete exchangeability of the Limma model residuals. The Limma model residuals should contain only experimental error and thus represent values from the null hypothesis of no genotype, time, or genotype by time effects. The t-statistics from this procedure then empirically determine the distribution of the t-statistic under the null hypothesis. FDRs for various P value thresholds were later determined by use of the method of Benjamini and Yekutieli (2001) on the observed distribution of P values.
RNA Gel Blot Analysis
To validate the microarray data, biologically different RNA samples were prepared from the tissue treated as described. Twenty micrograms of total RNA was resolved in a standard RNA gel (1% agarose). RNA was then transferred onto nylon membranes (Stratagene). The RNA membranes were hybridized with appropriate probes after UV fixation. DNA fragments for probes were PCR amplified from either cDNA clones from ABRC or genomic DNA with the following primers: At3g24520, HSTF-24520F (5′-CGAGAAAGAACGGACAAAGC-3′) and HSTF-24520R (5′-AGCCACCTCGAAACAGAGAA-3′); At3g11410, PP2C-11410F (5′-GATTTGTTGCGGTGTTGTTG-3′) and PP2C-11410R (5′-CACTTCTCCGCAACATGAGA-3′); At5g57560, TCH4-57560F (5′-GAAACTCCGCAGGAACAGTC-3′) and TCH4-57560R (5′-GCACCCATCTCATCCTTTGT-3′); At4g38840, IAAind-38840F (5′-AGATTCTCCGACAAGCCAAA-3′) and IAAind-38840R (5′-TCAGTTGAAGCGAGAAGCAA-3′). We used the LabWorks program version 4.0.0.8 (UVP BioImaging Systems) for quantification of the RNA hybridization signal. Acquired band intensities were then normalized with use of respective tubulin band intensity.
Accession Number Data Deposition
Affymetrix data have been deposited in the GEO public repository under the accession number GSE3326.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Cold-Regulated Gene List.
Supplemental Table 2. Cold Upregulated Gene List.
Supplemental Table 3. Cold Early Transiently Upregulated (ETU) Gene List.
Supplemental Table 4. Cold Early Continually Upregulated (ECU) Gene List.
Supplemental Table 5. Cold Late Upregulated (LU) Gene List.
Supplemental Table 6. Cold Downregulated Gene List.
Supplemental Table 7. Cold Early Transiently Downregulated (ETD) Gene List.
Supplemental Table 8. Cold Early Continually Downregulated (ECD) Gene List.
Supplemental Table 9. Cold Late Downregulated (LD) Gene List.
Supplemental Table 10. Cold-Regulated Signaling Gene List.
Supplemental Table 11. Cold-Regulated Transcription Gene List.
Supplemental Table 12. Cold Upregulated Genes Whose Expression Is Affected by ice1.
Supplemental Table 13. Cold Downregulated Genes Whose Expression Is Affected by ice1.
Supplemental Table 14. Cold Upregulated CBF3-Specific Target Genes Whose Expression Is Affected by ice1.
Supplemental Table 15. Cold Upregulated CBF3 and CBF2 Target Genes Whose Expression Is Affected by ice1.
Supplemental Table 16. Cold Upregulated CBF2-Specific Target Genes Whose Expression Is Affected by ice1.
Supplementary Material
Acknowledgments
This work was supported by USDA National Research Initiative Grant 2003-00751 and National Science Foundation grants IBN-0212346 and MCB-0241450 to J.-K.Z. We thank David Galbraith, Vicki Chandler, Frans Tax, and Brian Larkins for helpful suggestions and critical reading of the manuscript. We also thank Kelvin Kiesler, Susan Miller, and Al Agellon for technical assistance on the Affymetrix GeneChip analysis.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jian-Kang Zhu (jian-kang.zhu@ucr.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.035568.
References
- Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Yekutieli, D. (2001). The control of the false discovery rate in multiple testing under dependency. Ann. Statist. 29, 1165–1188. [Google Scholar]
- Cheong, Y.H., Chang, H.S., Gupta, R., Wang, X., Zhu, T., and Luan, S. (2002). Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 129, 661–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B.H., Hong, X.H., Agarwal, M., and Zhu, J.K. (2003). ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, H.I., Hong, J.H., Ha, J.O., Kang, J.Y., and Kim, S.Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730. [DOI] [PubMed] [Google Scholar]
- Collinge, M., and Boller, T. (2001). Differential induction of two potato genes, Stprx2 and StNAC, in response to infection by Phytophthora infestans and to wounding. Plant Mol. Biol. 46, 521–529. [DOI] [PubMed] [Google Scholar]
- Cook, D., Fowler, S., Fiehn, O., and Thomashow, M.F. (2004). A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc. Natl. Acad. Sci. USA 101, 15243–15248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter, A., and Becker, P.B. (2002). Histone acetylation: A switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 3, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron, B., and Tibshirani, R.J. (1993). An Introduction to the Bootstrap. (New York: Chapman and Hall).
- Eulgem, T., Rushton, P.J., Robatzek, S., and Somssich, I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Farina, K.L., and Singer, R.H. (2002). The nuclear connection in RNA transport and localization. Trends Cell Biol. 12, 466–472. [DOI] [PubMed] [Google Scholar]
- Fleck, B., and Harberd, N.P. (2002). Evidence that the Arabidopsis nuclear gibberellin signalling protein GAI is not destabilised by gibberellin. Plant J. 32, 935–947. [DOI] [PubMed] [Google Scholar]
- Fowler, S., and Thomashow, M.F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, W., Munnik, T., Kerkmann, K., Salamini, F., and Bartels, D. (2000). Water deficit triggers phospholipase D activity in the resurrection plant Craterostigma plantagineum. Plant Cell 12, 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori, T., Sato, E., Yamashino, T., and Mizuno, T. (2005). PRR5 (PSEUDO-RESPONSE REGULATOR 5) plays antagonistic roles to CCA1 (CIRCADIAN CLOCK-ASSOCIATED-1) in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 69, 426–430. [DOI] [PubMed] [Google Scholar]
- Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., Tran, L.S.P., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876. [DOI] [PubMed] [Google Scholar]
- Gilmour, S.J., Zarka, D.G., Stockinger, E.J., Salazar, M.P., Houghton, J.M., and Thomashow, M.F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16, 433–442. [DOI] [PubMed] [Google Scholar]
- Goda, H., Shimada, Y., Asami, T., Fujioka, S., and Yoshida, S. (2002). Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 130, 1319–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, Z.Z., Dong, C.H., Lee, H., Zhu, J.H., Xiong, L.M., Gong, D.M., Stevenson, B., and Zhu, J.K. (2005). A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17, 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, Z.Z., Lee, H., Xiong, L.M., Jagendorf, A., Stevenson, B., and Zhu, J.K. (2002). RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc. Natl. Acad. Sci. USA 99, 11507–11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy, C.L. (1990). Cold acclimation and freezing stress tolerance: Role of protein metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41, 187–223. [Google Scholar]
- Hagen, G., and Guilfoyle, T. (2002). Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385. [PubMed] [Google Scholar]
- Harper, R.M., Stowe-Evans, E.L., Luesse, D.R., Muto, H., Tatematsu, K., Watahiki, M.K., Yamamoto, K., and Liscum, E. (2000). The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12, 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden, P., and Kamiya, Y. (1997). Gibberellin biosynthesis: Enzymes, genes and their regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 431–460. [DOI] [PubMed] [Google Scholar]
- Hegedus, D., Yu, M., Baldwin, D., Gruber, M., Sharpe, A., Parkin, I., Whitwill, S., and Lydiate, D. (2003). Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol. Biol. 53, 383–397. [DOI] [PubMed] [Google Scholar]
- Heim, M.A., Jakoby, M., Werber, M., Martin, C., Weisshaar, B., and Bailey, P.C. (2003). The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747. [DOI] [PubMed] [Google Scholar]
- Herr, A.J., Jensen, M.B., Dalmay, T., and Baulcombe, D.C. (2005). RNA polymerase IV directs silencing of endogenous DNA. Science 308, 118–120. [DOI] [PubMed] [Google Scholar]
- Hong, S.W., Jon, J.H., Kwak, J.M., and Nam, H.G. (1997). Identification of a receptor-like protein kinase gene rapidly induced by abscisic acid, dehydration, high salt, and cold treatments in Arabidopsis thaliana. Plant Physiol. 113, 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, D., Chen, H.C., and Sheen, J. (2002). Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 129, 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, I., and Sheen, J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389. [DOI] [PubMed] [Google Scholar]
- Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M., Futsuhara, Y., Matsuoka, M., and Yamaguchi, J. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine, R.F. (1992). Inositol lipids in cell signalling. Curr. Opin. Cell Biol. 4, 212–219. [DOI] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14, 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen, K.R., Gilmour, S.J., Zarka, D.G., Schabenberger, O., and Thomashow, M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106. [DOI] [PubMed] [Google Scholar]
- Jakoby, M., Weisshaar, B., Droge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., and Parcy, F. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. [DOI] [PubMed] [Google Scholar]
- Johnson, C.S., Kolevski, B., and Smyth, D.R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14, 1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica, M.S., and Moore, M.J. (2003). Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell 12, 5–14. [DOI] [PubMed] [Google Scholar]
- Kang, J.Y., Choi, H.I., Im, M.Y., and Kim, S.Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14, 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. [DOI] [PubMed] [Google Scholar]
- Katagiri, T., Takahashi, S., and Shinozaki, K. (2001). Involvement of a novel Arabidopsis phospholipase D, AtPLD delta, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J. 26, 595–605. [DOI] [PubMed] [Google Scholar]
- Kiba, T., Yamada, H., and Mizuno, T. (2002). Characterization of the ARR15 and ARR16 response regulators with special reference to the cytokinin signaling pathway mediated by the AHK4 histidine kinase in roots of Arabidopsis thaliana. Plant Cell Physiol. 43, 1059–1066. [DOI] [PubMed] [Google Scholar]
- Kim, S., Kang, J.Y., Cho, D.I., Park, J.H., and Kim, S.Y. (2004). ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 40, 75–87. [DOI] [PubMed] [Google Scholar]
- Kovtun, Y., Chiu, W.L., Zeng, W.K., and Sheen, J. (1998). Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature 395, 716–720. [DOI] [PubMed] [Google Scholar]
- Kreps, J.A., Wu, Y.J., Chang, H.S., Zhu, T., Wang, X., and Harper, J.F. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130, 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubien, D.S., von Caemmerer, S., Furbank, R.T., and Sage, R.F. (2003). C4 photosynthesis at low temperature. A study using transgenic plants with reduced amounts of Rubisco. Plant Physiol. 132, 1577–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambermon, M.H.L., Fu, Y., Kirk, D.A.W., Dupasquier, M., Filipowicz, W., and Lorkovic, Z.J. (2002). UBA1 and UBA2, two proteins that interact with UBP1, a multifunctional effector of pre-mRNA maturation in plants. Mol. Cell. Biol. 22, 4346–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, V., Mantyla, E., Welin, B., Sundberg, B., and Palva, E.T. (1994). Alterations in water status, endogenous abscisic acid content, and expression of rab18 gene during the development of freezing tolerance in Arabidopsis thaliana. Plant Physiol. 104, 1341–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B.-h., Lee, H., Xiong, L., and Zhu, J.-K. (2002). A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 14, 1235–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Cheng, H., King, K.E., Wang, W., He, Y., Hussain, A., Lo, J., Harberd, N.P., and Peng, J. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16, 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Choi, D., and Kende, H. (2001). Expansins: Ever-expanding numbers and functions. Curr. Opin. Plant Biol. 4, 527–532. [DOI] [PubMed] [Google Scholar]
- Li, W.Q., Li, M.Y., Zhang, W.H., Welti, R., and Wang, X.M. (2004). The plasma membrane-bound phospholipase D delta enhances freezing tolerance in Arabidopsis thaliana. Nat. Biotechnol. 22, 427–433. [DOI] [PubMed] [Google Scholar]
- Lurin, C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama, K., Sakuma, Y., Kasuga, M., Ito, Y., Seki, M., Goda, H., Shimada, Y., Yoshida, S., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2004). Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 38, 982–993. [DOI] [PubMed] [Google Scholar]
- Mathur, J., et al. (1998). Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 14, 593–602. [DOI] [PubMed] [Google Scholar]
- Matsushika, A., Makino, S., Kojima, M., and Mizuno, T. (2000). Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 41, 1002–1012. [DOI] [PubMed] [Google Scholar]
- Medina, J., Bargues, M., Terol, J., Perez-Alonso, M., and Salinas, J. (1999). The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 119, 463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, T. (2004). Plant response regulators implicated in signal transduction and circadian rhythm. Curr. Opin. Plant Biol. 7, 499–505. [DOI] [PubMed] [Google Scholar]
- Molnar, G., Bancos, S., Nagy, F., and Szekeres, M. (2002). Characterisation of BRH1, a brassinosteroid-responsive RING-H2 gene from Arabidopsis thaliana. Planta 215, 127–133. [DOI] [PubMed] [Google Scholar]
- Munnik, T. (2001). Phosphatidic acid: An emerging plant lipid second messenger. Trends Plant Sci. 6, 227–233. [DOI] [PubMed] [Google Scholar]
- Nover, L., Bharti, K., Doring, P., Mishra, S.K., Ganguli, A., and Scharf, K.D. (2001). Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 6, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novillo, F., Alonso, J.M., Ecker, J.R., and Salinas, J. (2004). CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 101, 3985–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima, Y., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, A.N., Ernst, H.A., Lo Leggio, L., and Skriver, K. (2005). NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87. [DOI] [PubMed] [Google Scholar]
- Olszewski, N., Sun, T.-p., and Gubler, F. (2002). Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 14, S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera, Y., Haag, J.R., Ream, T., Nunes, P.C., Pontes, O., and Pikaard, C.S. (2005). Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120, 613–622. [DOI] [PubMed] [Google Scholar]
- Osakabe, Y., Maruyama, K., Seki, M., Satou, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2005). Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell 17, 1105–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe, Y., Miyata, S., Urao, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). Overexpression of Arabidopsis response regulators, ARR4/ATRR1/IBC7 and ARR8/ATRR3, alters cytokinin responses differentially in the shoot and in callus formation. Biochem. Biophys. Res. Commun. 293, 806–815. [DOI] [PubMed] [Google Scholar]
- Pnueli, L., Hallak-Herr, E., Rozenberg, M., Cohen, M., Goloubinoff, P., Kaplan, A., and Mittler, R. (2002). Molecular and biochemical mechanisms associated with dormancy and drought tolerance in the desert legume Retama raetam. Plant J. 31, 319–330. [DOI] [PubMed] [Google Scholar]
- Ramsay, N.A., and Glover, B.J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10, 63–70. [DOI] [PubMed] [Google Scholar]
- Richards, D.E., King, K.E., Ait-ali, T., and Harberd, N.P. (2001). How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 67–88. [DOI] [PubMed] [Google Scholar]
- Rizhsky, L., Liang, H.J., and Mittler, R. (2002). The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 130, 1143–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek, S., and Somssich, I.E. (2001). A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 28, 123–133. [DOI] [PubMed] [Google Scholar]
- Rocak, S., and Linder, P. (2004). Dead-box proteins: The driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 5, 232–241. [DOI] [PubMed] [Google Scholar]
- Ruelland, E., Cantrel, C., Gawer, M., Kader, J.C., and Zachowski, A. (2002). Activation of phospholipases C and D is an early response to a cold exposure in Arabidopsis suspension cells. Plant Physiol. 130, 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp, A., Zollner, A., Maier, D., Albermann, K., Hani, J., Mokrejs, M., Tetko, I., Guldener, U., Mannhaupt, G., Munsterkotter, M., and Mewes, H.W. (2004). The funCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32, 5539–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski, R.W.M., and Meyerowitz, E.M. (1998). A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92, 93–103. [DOI] [PubMed] [Google Scholar]
- Seki, M., Narusaka, M., Abe, H., Kasuga, M., Yamaguchi-Shinozaki, K., Carninci, P., Hayashizaki, Y., and Shinozaki, K. (2001). Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31, 279–292. [DOI] [PubMed] [Google Scholar]
- Shinwari, Z.K., Nakashima, K., Miura, S., Kasuga, M., Seki, M., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem. Biophys. Res. Commun. 250, 161–170. [DOI] [PubMed] [Google Scholar]
- Silverstone, A.L., Jung, H.S., Dill, A., Kawaide, H., Kamiya, Y., and Sun, T.P. (2001). Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13, 1555–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, G.K. (2005). Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor, R. Gentlemen, V. Carey, S. Dudoit, R. Irizarry, and W. Huber, eds (New York: Springer), pp. 397–420.
- Souer, E., vanHouwelingen, A., Kloos, D., Mol, J., and Koes, R. (1996). The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85, 159–170. [DOI] [PubMed] [Google Scholar]
- Stockinger, E.J., Gilmour, S.J., and Thomashow, M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94, 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe-Evans, E.L., Harper, R.M., Motchoulski, A.V., and Liscum, E. (1998). NPH4, a conditional modulator of auxin-dependent differential growth responses in Arabidopsis. Plant Physiol. 118, 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. [DOI] [PubMed] [Google Scholar]
- Strayer, C., Oyama, T., Schultz, T.F., Raman, R., Somers, D.E., Mas, P., Panda, S., Kreps, J.A., and Kay, S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771. [DOI] [PubMed] [Google Scholar]
- Sunkar, R., and Zhu, J.K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16, 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres, M., Nemeth, K., Koncz-Kalman, Z., Mathur, J., Kauschmann, A., Altmann, T., Redei, G.P., Nagy, F., Schell, J., and Koncz, C. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182. [DOI] [PubMed] [Google Scholar]
- Takada, S., Hibara, K., Ishida, T., and Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128, 1127–1135. [DOI] [PubMed] [Google Scholar]
- Tarutani, Y., Morimoto, T., Sasaki, A., Yasuda, M., Nakashita, H., Yoshida, S., Yamaguchi, I., and Suzuki, Y. (2004. a). Molecular characterization of two highly homologous receptor-like kinase genes, RLK902 and RKL1, in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 68, 1935–1941. [DOI] [PubMed] [Google Scholar]
- Tarutani, Y., Sasaki, A., Yasuda, M., Nakashita, H., Yoshida, S., Yamaguchi, I., and Suzuki, Y. (2004. b). Identification of three clones which commonly interact with the kinase domains of highly homologous two receptor-like kinases, RLK902 and RKL1. Biosci. Biotechnol. Biochem. 68, 2581–2587. [DOI] [PubMed] [Google Scholar]
- Tatematsu, K., Kumagai, S., Muto, H., Sato, A., Watahiki, M.K., Harper, R.M., Liscum, E., and Yamamoto, K.T. (2004). MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16, 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, S.G., Phillips, A.L., and Hedden, P. (1999). Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 96, 4698–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow, M.F. (1994). Arabidopsis thaliana as a model for studying mechanisms of plant cold tolerance. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 807–834.
- Thomashow, M.F. (1999). Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. [DOI] [PubMed] [Google Scholar]
- Tran, L.S.P., Nakashima, K., Sakuma, Y., Simpson, S.D., Fujita, Y., Maruyama, K., Fujita, M., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, M., Valencia-Sanchez, M.A., Staples, R.R., Chen, J.J., Denis, C.L., and Parker, R. (2001). The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104, 377–386. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 97, 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen, E.A., Sun, J.H., Coleman, M.J., Bibb, M.J., and Jones, J.D.G. (2000). Arabidopsis RelA/SpoT homologs implicate (p)ppGpp in plant signaling. Proc. Natl. Acad. Sci. USA 97, 3747–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J.T., Zarka, D.G., Van Buskirk, H.A., Fowler, S.G., and Thomashow, M.F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 41, 195–211. [DOI] [PubMed] [Google Scholar]
- Vroemen, C.W., Mordhorst, A.P., Albrecht, C., Kwaaitaal, M., and de Vries, S.C. (2003). The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15, 1563–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C.X., Zien, C.A., Afitlhile, M., Welti, R., Hildebrand, D.F., and Wang, X.M. (2000). Involvement of phospholipase D in wound-induced accumulation of jasmonic acid in Arabidopsis. Plant Cell 12, 2237–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner, L.A., and Junttila, O. (1999). Cold-induced freezing tolerance in Arabidopsis. Plant Physiol. 120, 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welti, R., Li, W.Q., Li, M.Y., Sang, Y.M., Biesiada, H., Zhou, H.E., Rajashekar, C.B., Williams, T.D., and Wang, X.M. (2002). Profiling membrane lipids in plant stress responses: Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 277, 31994–32002. [DOI] [PubMed] [Google Scholar]
- Wen, C.-K., and Chang, C. (2002). Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14, 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q., Frugis, G., Colgan, D., and Chua, N.H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14, 3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Zhang, Z.L., Zou, X.L., Huang, J., Ruas, P., Thompson, D., and Shen, Q.J. (2005). Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 137, 176–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., Lee, B.-h., Ishitani, M., Lee, H., Zhang, C.Q., and Zhu, J.-K. (2001). FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 15, 1971–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., and Zhu, J.-K. (2003). Regulation of abscisic acid biosynthesis. Plant Physiol. 133, 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, A., Tsutsumi, K., Tanimoto, S., and Ozeki, Y. (2003). Plant RelA/SpoT homolog confers salt tolerance in Escherichia coli and Saccharomyces cerevisiae. Plant Cell Physiol. 44, 3–9. [DOI] [PubMed] [Google Scholar]
- Yamamoto, Y., Sato, E., Shimizu, T., Nakamich, N., Sato, S., Kato, T., Tabata, S., Nagatani, A., Yamashino, T., and Mizuno, T. (2003). Comparative genetic studies on the APRR5 and APRR7 genes belonging to the APRR1/TOC1 quintet implicated in circadian rhythm, control of flowering time, and early photomorphogenesis. Plant Cell Physiol. 44, 1119–1130. [DOI] [PubMed] [Google Scholar]
- Yamauchi, Y., Ogawa, M., Kuwahara, A., Hanada, A., Kamiya, Y., and Yamaguchi, S. (2004). Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16, 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial, M., and McBride, H. (2001). Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2, 107–117. [DOI] [PubMed] [Google Scholar]
- Zhang, Z.L., Xie, Z., Zou, X.L., Casaretto, J., Ho, T.H.D., and Shen, Q.X.J. (2004). A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 134, 1500–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsh-Hoffmann, M., Henning, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136, 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.