Abstract
Cyclotides are plant-derived miniproteins that have the unusual features of a head-to-tail cyclized peptide backbone and a knotted arrangement of disulfide bonds. It had been postulated that they might be an especially large family of host defense agents, but this had not yet been tested by field data on cyclotide variation in wild plant populations. In this study, we sampled Australian Hybanthus (Violaceae) to gain an insight into the level of variation within populations, within species, and between species. A wealth of cyclotide diversity was discovered: at least 246 new cyclotides are present in the 11 species sampled, and 26 novel sequences were characterized. A new approach to the discovery of cyclotide sequences was developed based on the identification of a conserved sequence within a signal sequence in cyclotide precursors. The number of cyclotides in the Violaceae is now estimated to be >9000. Cyclotide physicochemical profiles were shown to be a useful taxonomic feature that reflected species and their morphological relationships. The novel sequences provided substantial insight into the tolerance of the cystine knot framework in cyclotides to amino acid substitutions and will facilitate protein engineering applications of this framework.
INTRODUCTION
The cyclotides are a distinctive class of plant defense peptides produced by various species in the Violaceae and Rubiaceae (Craik et al., 1999; Craik, 2001; Göransson et al., 2003; Gustafson et al., 2004; Trabi and Craik, 2004). They are characterized by their head-to-tail cyclization and cystine knot motif (Craik et al., 2001), which contribute to their exceptional resistance to physical, chemical, and biochemical degradation (Gran, 1973a; Colgrave and Craik, 2004). Topologically related macrocyclic peptides containing a cyclic cystine knot also occur in a plant from the Cucurbitaceae family (Hernandez et al., 2000; Felizmenio-Quimio et al., 2001), and a range of other circular proteins have been discovered in bacteria, plants, and animals over recent years (Trabi and Craik, 2002; Craik et al., 2003). The cyclotides display antiviral, uterotonic, and other biological activities (Gran, 1973b; Gustafson et al., 1994; Witherup et al., 1994; Tam et al., 1999; Craik, 2001) but are believed to function in vivo as insecticidal agents (Jennings et al., 2001). The mechanism by which they act is currently under investigation but is believed to be mediated through disruption of biological membranes (Nourse et al., 2004; Kamimori et al., 2005). Because of their great stability and well-defined molecular architecture, the cyclotides have attracted attention as potential frameworks for peptide-based drug design (Craik et al., 2002).
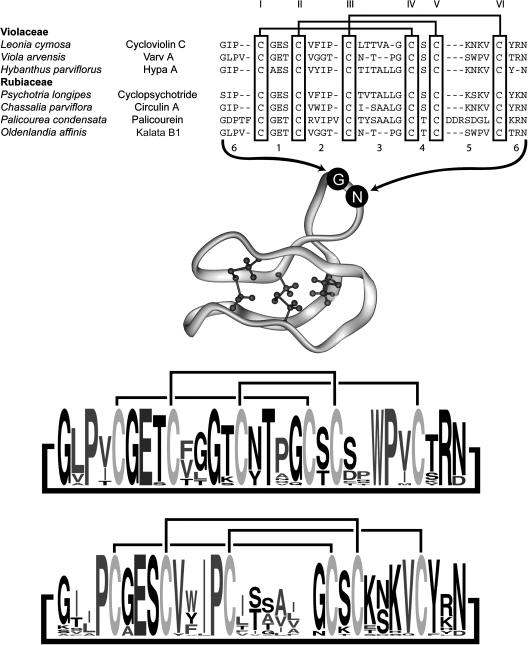
At present, ∼50 cyclotide sequences and nine three-dimensional structures have been reported (Saether et al., 1995; Daly et al., 1999; Skjeldal et al., 2002; Rosengren et al., 2003; Barry et al., 2004; Trabi and Craik, 2004; Chen et al., 2005; Jennings et al., 2005; Koltay et al., 2005; Mulvenna et al., 2005). Structures of linear analogues (Barry et al., 2003) and a disulfide-deficient mutant (Daly et al., 2003) are also available. Figure 1 shows a selection of cyclotide sequences from a range of genera together with a typical three-dimensional structure. The sequences are aligned based on the six conserved Cys residues and show that certain backbone loops between Cys residues are highly conserved. Also shown are sequence logos (Crooks et al., 2004) to graphically depict current knowledge of cyclotide sequences. Preliminary analysis of available plant material has shown a wealth of sequence diversity within the cyclotides, indicating that thousands of unique sequences may exist (Trabi and Craik, 2004). This diversity represents a significant potential resource for the development of environmentally friendly crop protection agents that might avoid the development of resistance by target insects (Craik et al., 2004). However, contrary observations, such as the presence of near-identical cyclotide sequences in widely diverged species such as Oldenlandia affinis (Rubiaceae) and Viola arvensis (Violaceae), as shown in Figure 1, call into question the potential range of variation of this class.
Figure 1.
Representative Sequences of Cyclotides from the Violaceae and Rubiaceae.
All peptides are head-to-tail cyclized, and the sequences are written starting from the presumed N-terminal cleavage point in the linear precursors (Jennings et al., 2001; Dutton et al., 2004). Sequences are aligned based on the conserved Cys residues, which are numbered in Roman numerals according to the order in which they appear in the linear precursor. Loops corresponding to the backbone segments between Cys residues are numbered for reference. Loop 6 is formed at the cyclization point of the linear precursor. References for the listed cyclotides are as follows: cycloviolin C (Hallock et al., 2000), varv A (Claeson et al., 1998), Hypa A (Broussalis et al., 2001), cyclopsychotride (Witherup et al., 1994), circulin A (Derua et al., 1996), palicourein (Bokesch et al., 2001), and kalata B1 (Saether et al., 1995). A ribbon diagram of the cyclotide framework based on the structure of kalata B1 (PDB ID 1NB1) with the cystine knot core rendered and numbering of loops is shown in the middle of the figure. At the bottom of the figure are two sequence logos (Crooks et al., 2004) to graphically illustrate sequence conservation within known cyclotides. The size of the one-letter amino acid code reflects the relative abundance of that residue in known cyclotides. Separate logos are shown for Möbius (top) and bracelet (bottom) subfamilies of cyclotides (Craik et al., 1999).
To date, most of the plant material studied for cyclotide isolation has been of cultivated origin and of limited genetic diversity. Additionally, there has been a bias toward more readily available Viola species (Schöpke et al., 1993; Göransson et al., 2003; Svangård et al., 2003, 2004; Trabi and Craik, 2004) despite the many other genera in the Violaceae. To address these deficiencies, an extensive examination of cyclotide diversity in wild populations of Australian Hybanthus (Violaceae) was conducted in order to gauge the level of cyclotide diversity within populations, within currently defined species across their geographical distributions, and between species. It was hoped that this information would shed light on the cyclotide variation of individual Hybanthus populations and the potential utility of cyclotide profiles as a chemotaxonomic feature in the Violaceae. Furthermore, knowledge of sequence diversity will assist in the utilization of the cyclotide framework as a protein engineering tool.
More generally, the number of reported antimicrobial and defense peptides has increased significantly in the last few years, with >700 sequences being sourced from organisms ranging from bacteria to plants and animals. In plants, although investigations into variations in small secondary metabolites, such as alkaloids and terpenes, in wild populations have been reported (Turner, 1967; Da Costa et al., 2005), even in Viola itself (Flamini et al., 2003), similar studies for peptide variation in wild populations are lacking. Among the few examples of studies of peptide variation, the diversity of Bowman-Birk protease inhibitors in cultivated lentils and their wild relatives (Sonnante et al., 2005) has been examined, as has the chemotaxonomic utility of antimicrobial peptides in frogs (Apponyi et al., 2004). In contrast with most peptides, the unusual stability, high levels of expression, and hydrophobicity of cyclotides permit straightforward isolation and characterization, making them ideal candidates for evaluating peptide diversity in nature. In this study, cyclotides in Hybanthus from across the Australian continent were sampled to evaluate the diversity of this peptide family and, more broadly, to give insights into the diversity of peptides in nature. A new approach to the identification of cyclotide sequences was developed, and insights into the role of different amino acids at specific locations in the structure were derived.
RESULTS
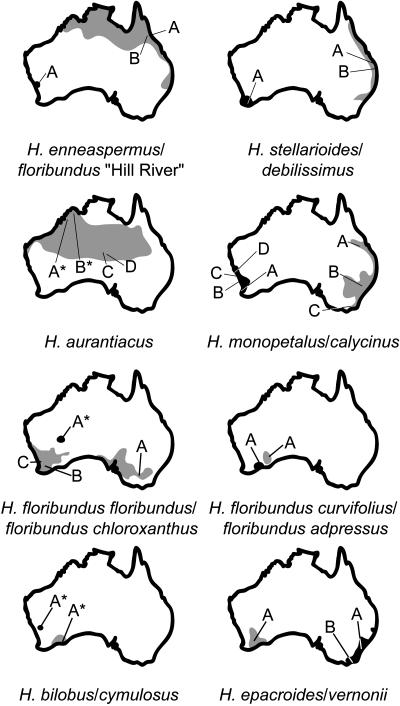
Aerial plant material was collected from wild populations of 9 of the 12 Australian Hybanthus species as defined by Bennett (1972), and another two species were sampled from dried herbarium specimens. With the exception of H. floribundus (subsp floribundus (eastern) and subsp adpressus) and H. enneaspermus, plants were typically observed to form small isolated colonies of three to five plants, limiting the breadth of sampling. To minimize ecological impact, root material was not collected, although previous studies have clearly established that cyclotides are present in roots of Violaceae species (Trabi and Craik, 2004). Collection localities are summarized in Figure 2, with the total distribution of each species (adapted from Bennett, 1972) shown in gray or black (first and second listed species per map, respectively), with letters indicating the position of each population collected within its geographical range. Specimens obtained from dried material are indicated with an asterisk.
Figure 2.
Distributions and Collection Sites of Australian Hybanthus.
Distributions of each species are shown in gray for the first species listed for each map and in black for the second. Collections are coded with letters and precise locations indicated with lines. The original location of collections of dried material from herbarium specimens is indicated with an asterisk. Hybanthus are seen to inhabit all but the most arid deserts in Australia.
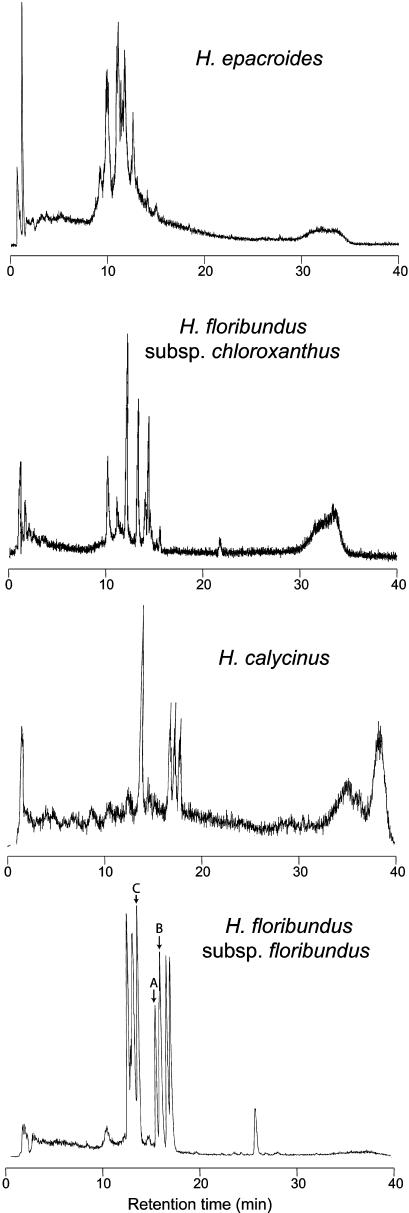
Extraction and liquid chromatography–mass spectrometry (LC-MS) of each sample gave a series of traces with a range of masses and retention times consistent with, but extending, those seen in previously studied cyclotides. Representative traces are shown in Figure 3. Interestingly, the dried herbarium sample of H. floribundus subsp chloroxanthus gave LC-MS data of similar quality to that obtained in freshly collected samples. In part, this reflects the exceptional stability of cyclotides. The masses and retention times of each trace were recorded for peaks with a signal-to-noise ratio greater than five. A total of 540 retention times and masses was recorded across all samples. Thirty-six of the identified components had masses within experimental error of those of cyclotides previously reported in other plants, including V. odorata, V. arvensis, V. cotyledon, Leonia cymosa, Hybanthus parviflorus, Psychotria longipes, Chassalia parviflora, and O. affinis. Retention times for the previously reported cyclotides were not available under comparable LC conditions, limiting assessment of the likelihood that Hybanthus contains these characterized peptides. However, it is possible, and indeed likely, that some of these 36 peptides reflect already known cyclotides found in other species. When possible common compounds with masses within 1 D and retention times within 1 min were aggregated for individual species, it was deduced that at least 246 unique cyclotides are present. The data were compiled for each species in a graph of retention time versus mass.
Figure 3.
Sample LC-MS Traces of Selected Collections.
Total ion count is graphed against retention time. Variations in peptide concentration and diversity are seen, and the dried herbarium specimen of H. floribundus subsp chloroxanthus is shown to give comparable data quality to fresh material. The peaks of Hyfl A-C sequenced in this study are indicated in the trace of H. floribundus subsp floribundus with corresponding letters.
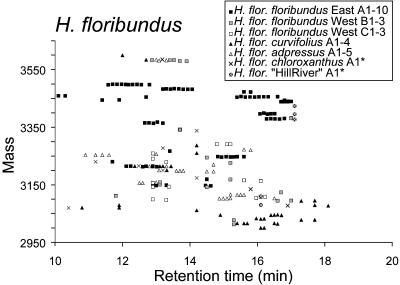
The chemical markers (mass and retention time) for individual cyclotides from H. floribundus and its subspecies are shown in Figure 4, with each population shown with a different symbol. For well-sampled species such as H. floribundus subsp floribundus (Victoria), several peaks originating from different plant samples are clustered together in groups within 1 D and 1 min retention time. As these clusters are presumably attributable to single peptide sequences, these levels of variability in mass and retention time were taken as the limits of experimental error in subsequent analyses. For clarity, overlapping points in Figure 4 were separated in the time dimension, giving horizontal clusters reflecting individual peptides. A complete list of species, masses, and retention times is available in Supplemental Table 1 online.
Figure 4.
Scatterplot of H. floribundus Peptide Masses and Retention Times.
Individual species are indicated with different symbols. Masses range from ∼3000 to 3500 D and retention times from 10 to 20 min. Horizontal clusters of peaks reflect possible common peptides. Overlapping peaks have been horizontally dispersed for clarity.
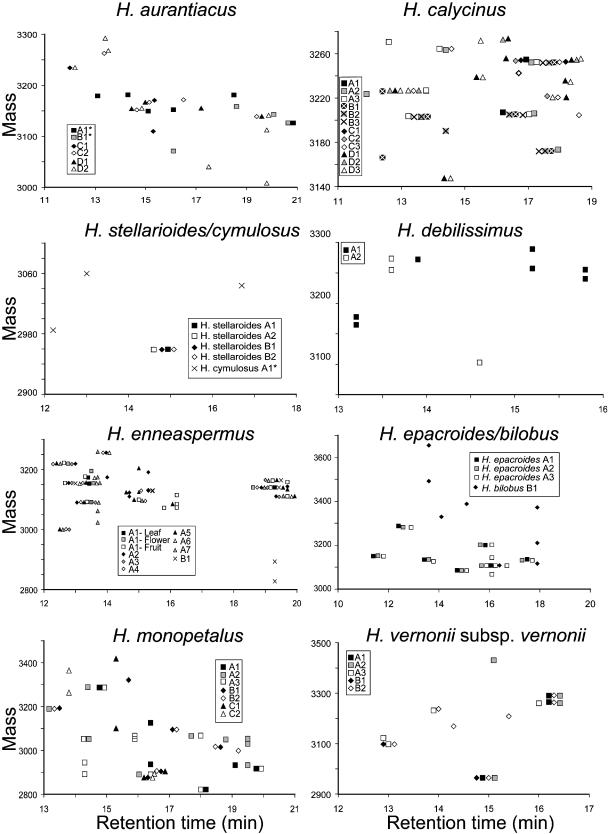
LC-MS data for the remaining species are similarly shown in Figure 5, with each individual plant of each species represented with a unique symbol. The possibility of isobaric (e.g., Leu for Ile) or near isobaric (e.g., Asp for Asn) amino acid substitutions leading to broadening of the observed clusters along the retention time axis must be considered given that such substitutions are known from previously published sequences. However, as these represent minor sequence variations, their impact on the utility of such clusters to infer relatedness between samples is not significant. The important finding that emerges from Figures 4 and 5 is that cyclotides are widespread in the Hybanthus genus and that sequence variation appears to be substantial, as assessed by variations in masses and physicochemical properties.
Figure 5.
Scatterplots of Hybanthus Peptide Masses and Retention Times.
Individual species and collection locations are indicated with different symbols in each plot. Collections of individual plants from one location are differentiated by shading. Masses range from 2800 to 3700 D and retention times from 10 to 20 min. Horizontal clusters of peaks reflect possible common peptides. Overlapping peaks have been horizontally dispersed for clarity.
To compare cyclotide profiles of geographically separated populations of the same species, H. calycinus is a good reference example because this species is morphologically well defined and occupies a consistent ecological niche along the coastal plains of southwestern Australia. Four populations were sampled across 500 km of coastline, revealing subsets of common peptides between the groups (Figure 5). The southern and central collections (samples A and B, respectively, in Figure 2) appeared to have more overlap with each other than with the two northern collections (samples C and D). An increase in the incidence of peptides unique to a population was observed with decreasing latitude (Figure 5, where there are two, three, three, and seven unique compounds for collections A to D, respectively).
A similar pattern is found in the more diverse H. monopetalus in Figure 5. In comparison with H. calycinus, this species has fewer common peptides between sites, consistent with its greater morphological and ecological variation and the greater separation of populations. Once again, the number of unique peptides in each population was observed to increase with decreasing latitude (4, 3, and 18 peptides from populations C, B, and A, respectively).
The cyclotides of H. epacroides and H. bilobus, two species with an unusual spiny growth habit, were investigated, as the separation of these species is contentious (Bennett, 1972). The dried herbaria specimens of H. bilobus were found to give reliable LC-MS traces. The clustering of a mass from a dried herbarium sample of H. floribundus subsp chloroxanthus with other H. floribundus subspecies collected from live material indicated that the cyclotides were unchanged by preservation and storage. The analysis of H. epacroides and H. bilobus suggested only limited affinity between the two forms through a single clustered peak.
The less thoroughly sampled species studied showed comparable patterns. For H. aurantiacus, there was little overlap between northern and central populations, with only a single set of coincident peptides in Figure 5. H. stellarioides contained a single cyclotide in the limited sample studied, but as fruiting material was not available, further diversity may exist. The profiles for H. vernonii subsp vernonii were found to be very consistent between northern and southern populations despite marked morphological and ecological differences (from montane sandstone to coastal scrub, respectively).
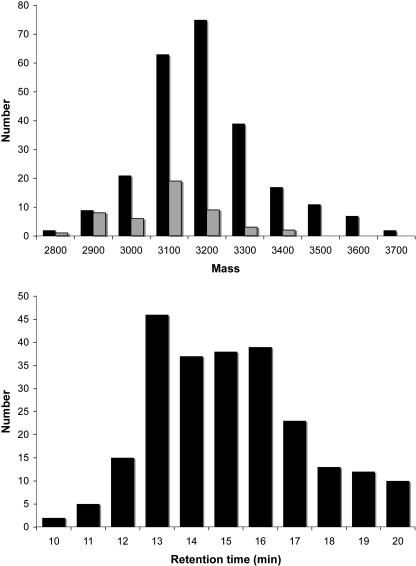
Across all species, the distribution of masses and retention times both followed approximately bell curve distributions as shown in Figure 6. Compounds were observed to vary in mass from ∼2800 to 3800 D and in retention time from 10 to 20 min. The average mass was slightly skewed lower than the midpoint of the range of masses, and the average retention times appear roughly grouped into two sets. Overall, the new data significantly expanded the range of masses seen previously for cyclotides.
Figure 6.
Mass and Retention Time Distributions for Australian Hybanthus Cyclotides.
Peptides vary in mass from 2800 to 3700 D and in retention time from 10 to 20 min. Also shown is the mass distribution for published cyclotides (gray bars).
The small size of individual plants and populations of most Australian Hybanthus prevented the collection of sufficient material for isolation and chemical sequencing of all individual peptides. One exception was H. floribundus subsp floribundus, which is a large plant that was collected from an extensive colony in southeastern Australia. Bulk plant material of H. floribundus subsp floribundus was extracted and purified to give several chromatographically pure cyclotides. From these, three new sequences were determined by reduction, digestion, and tandem mass spectrometry (MS-MS) analysis as described in Methods. The sequences of the new peptides, named Hyfl A-C, are summarized in Table 1. The peptide naming convention established for H. parviflorus is adopted in this work (Broussalis et al., 2001). Hyfl A is similar to the previously reported cyclotides circulin A and Hypa A (68 and 74% identity, respectively). By contrast, Hyfl B and C both contain amino acid substitutions previously unseen in cyclotides, namely a Gln and Met in loop 6, Lys in loop 2, and tandem Glu residues in loop 3 (see Figure 1 for the definitions and locations of the backbone loops in cyclotides). Interestingly, the Met residue in Hyfl B was found in the oxidized state only, while both oxidized and unmodified forms were found in Hyfl C. The presence of the tripeptide sequence AET in loop 1 is also novel. Previously reported cyclotides have the sequences GES, GET, or AES in this highly conserved loop.
Table 1.
Novel Cyclotide Sequences Derived from Hybanthus Species
| Name | Hybanthus sp | 6 | 1 | 2 | 3 | 4 | 5 | 6 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | |||||||||
| Hyfl A | floribundus E | -SIS- | C | GES | C | VYIP- | C | TVT-ALVG | C | T | C | --KDKV | C | YLN |
| Hyfl B | floribundus E | GSPIQ | C | AET | C | FIGK- | C | Y-TEEL-G | C | T | C | --TAFL | C | MKN |
| Hyfl C | floribundus E | GSPRQ | C | AET | C | FIGK- | C | Y-TEEL-G | C | T | C | --TAFL | C | MKN |
| Hyfl D | floribundus E | GSVP- | C | GES | C | VYIP- | C | F-TGIA-G | C | S | C | --KSKV | C | YYN |
| Hyfl E | floribundus E | GEIP- | C | GES | C | VYLP- | C | F----LPN | C | Y | C | --RNHV | C | YLN |
| Hyfl F | floribundus E | -SIS- | C | GET | C | TTFN- | C | WI----PN | C | K | C | NHHDKV | C | YWN |
| Hyfl G | floribundus E | XXXXX | C | AET | C | VVLP- | C | FI---VPG | C | S | C | --KSSV | C | YFN |
| Hyfl H | floribundus E | XXXXX | C | AET | C | IYIP- | C | F-TEAV-G | C | K | C | --KDKV | C | YKN |
| Hyfl I | floribundus W | G-IP- | C | GES | C | VFIP- | C | I-SGVI-G | C | S | C | --KSKV | C | YRN |
| Hyfl J | floribundus W | G-IA- | C | GES | C | AYFG- | C | WI----PG | C | S | C | --RNKV | C | YFN |
| Hyfl K | floribundus W | GT-P- | C | GES | C | VYIP- | C | F-T-AVVG | C | T | C | --KDKV | C | YLN |
| Hyfl L | floribundus W | GT-P- | C | AES | C | VYLP- | C | F-TGVI-G | C | T | C | --KDKV | C | YLN |
| Hyfl M | floribundus W | GNIP- | C | GES | C | IFFP- | C | F----NPG | C | S | C | --KDNL | C | YYN |
| Hyfl N | floribundus W | XXXXX | C | GET | C | VILP- | C | I-SAAL-G | C | S | C | --KDTV | C | YKN |
| Hyfl O | floribundus W | XXXXX | C | GET | C | VIFP- | C | I-SAAF-G | C | S | C | --KDTV | C | YKN |
| Hyfl P | floribundus W | XXXXX | C | XXX | C | VWIP- | C | I-SGIA-G | C | S | C | --KNKV | C | YLN |
| Hymo A | monopetalus | XXXXX | C | GET | C | LFIP- | C | IFS--VVG | C | S | C | --SSKV | C | YRN |
| Hymo B | monopetalus | XXXXX | C | GET | C | VTGT- | C | Y-T---PG | C | A | C | --DWPV | C | KRD |
| Hyst A | stellarioides | XXXXX | C | GET | C | IWGR- | C | Y-SENI-G | C | H | C | --GFGI | C | TLN |
| Hyve A | vernonii | XXXXX | C | GET | C | LFIP- | C | LTS--VFG | C | S | C | --KNRG | C | YKI |
| Hyca A | calycinus | XXXXX | C | GET | C | VVDTR | C | Y-T---KK | C | S | C | --AWPV | C | MRN |
| Hyde A | debilissimus | XXXXX | X | XXX | C | VWIP- | C | I-S-AAIG | C | S | C | --KSKV | C | YRN |
| Hyen A | enneaspermus | XXXXX | C | GES | C | VYIP- | C | TVT-ALLG | C | S | C | --KDKV | C | YKN |
| Hyen B | enneaspermus | XXXXX | C | GET | C | KVTKR | C | --SGQ--G | C | S | C | -LKGRS | C | Y-D |
| Hyep A | epacroides | XXXXX | C | GET | C | VVLP- | C | FI---VPG | C | S | C | --KSSV | C | YFN |
| Hyep B | epacroides | XXXXX | C | GET | C | IYIP- | C | F-TEAV-G | C | K | C | --KDKV | C | YKN |
Sequences are aligned based on the conserved Cys residues (I to VI), and loops are numbered as in Figure 1. Hyfl A-C were identified by peptide sequencing. An oxidized Met in Hyfl B is shown underlined to indicate that only the oxidized form was detected. By contrast, both oxidized and reduced forms were detected for Hyfl C. The remaining sequences were identified by PCR as full or partial mRNA transcripts. XXXXX represents an undetermined section of sequence, outside that detected by the primer.
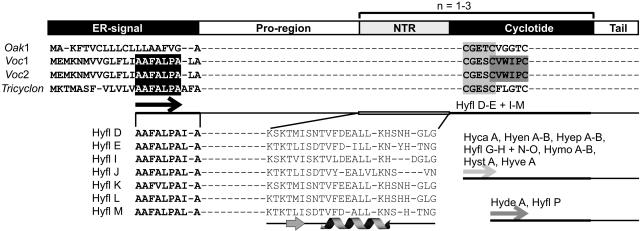
Although it is clear that the approach described above, involving the direct isolation and sequencing of cyclotide peptides, provides a route to new sequence information, one of the goals of this study was to develop a faster method of exploring cyclotide diversity. The Hybanthus samples were therefore probed for cyclotide sequences by reverse transcription of cyclotide precursor mRNA. Using forward primers designed to anneal to sequences of loop 1 of previously characterized cyclotides (Jennings et al., 2001; Dutton et al., 2004), 16 additional partial cyclotide sequences were determined, as shown in Table 1. A further seven complete cyclotide domain sequences (Hyfl D-E and Hyfl I-M) were determined using a forward primer annealing to a newly identified conserved element within the endoplasmic reticulum (ER) signal of previously characterized cyclotide precursors from the Violaceae family (Dutton et al., 2004; Mulvenna et al., 2005). The strategy used for obtaining these sequences is outlined in Figure 7 and is based on knowledge of the generic cyclotide precursor structure, which comprises regions coding for an ER signal sequence, a propeptide domain, the mature peptide domain, and a small hydrophobic tail C-terminal to the mature domain. A region within the propeptide sequence just upstream of the mature domain is referred to as the N-terminal repeat (NTR) region, as some cyclotide precursors have up to three tandem copies of it and an adjacent mature cyclotide domain.
Figure 7.
Schematic Representation of the General Organization of Cyclotide Precursors.
Each precursor protein consists of an ER signal, a pro-region, one to three copies of a mature cyclotide domain preceded by an NTR sequence within the pro-region, and a short tail region. The latter typically comprises four to seven hydrophobic amino acids. The various elements are drawn roughly to scale, with the ER signal typically 22 amino acids, the propeptide (including NTR) typically 40 to 60 amino acids, and the cyclotide domain typically 30 amino acids. A selection of precursor sequences is shown to illustrate the strategy used for designing primers to detect cyclotides by RT-PCR. For clarity, only the ER signal sequences and part of the cyclotide sequences are shown. Oak1 is the precursor for kalata B1 (Jennings et al., 2001) and represents cyclotide precursors from the Rubiaceae family. Voc1, Voc2 (Dutton et al., 2004), and Tricyclon (Mulvenna et al., 2005) from the Violaceae family produce the mature cyclotides cycloviolacin O8, cycloviolacin O11, and tricyclon A, respectively. In this study, a conserved sequence element, AAFALPA (black box), was identified within the ER signal of the Violaceae precursors and used as a new primer binding site. Other conserved elements used as primer targets were the CGETC and CVWIPC sequences within the cyclotide domain (light and dark gray boxes, respectively). The new Hybanthus cyclotides that were identified using these primers are indicated after each primer binding site (arrows). Hyfl F was identified using a primer based on the SISC sequence in loop 1 of Hyfl A (data not shown in this figure). The partial ER signal and the most conserved part of the NTR region are shown for the seven cyclotides that were identified using the AAFALPA primer. Secondary structure prediction of the pro-region by the program PROFsec identified a short β-sheet and an α-helix within the NTR (illustrated underneath the sequences) and a further two helical segments in the pro- region upstream of the NTR (data not shown).
The predicted masses for six of the seven peptide sequences were found to have corresponding LC-MS points for H. floribundus in the complete list of peptide masses and retention times (see Supplemental Table 1 online) within 1 D. The identification of additional sequences without corresponding masses in the LC-MS traces of the same plant may reflect the amplification of rare transcripts. Alignment of the NTR sequences of the seven partial precursors of H. floribundus with those of V. odorata (Dutton et al., 2004) and V. tricolor (Mulvenna et al., 2005) showed a high degree of sequence similarity (see Supplemental Table 2 online), pointing to a vital role of this domain in the processing of precursors to produce mature cyclotides. Secondary structure prediction identified a helical motif in the NTR region that is conserved in other known cyclotide precursors and is believed to play a role in folding, processing, or detoxification of cyclotide domains from the precursor (Dutton et al., 2004).
DISCUSSION
In this study, we have successfully investigated cyclotide diversity in wild populations of Australian Hybanthus, leading to a description of the degree of diversity within populations, species, and the genus. A comparison with known cyclotide masses suggests that the plants sampled contain at least 246 new cyclotides. A new approach to the discovery of cyclotide sequences was developed, and 26 novel cyclotide sequences were characterized, substantially increasing the sequence knowledge base for this family and clearly establishing cyclotides as a large family of plant proteins. The utility of cyclotide physicochemical profiles as a chemotaxonomic feature was also demonstrated with common sets of peptides consistent with morphologically defined species.
Isolation and analysis of the peptides from one Hybanthus species by MS-MS sequencing characterized three novel cyclotides, namely Hyfl A-C. Comparison of Hyfl A with the previously determined sequence of Hypa A from the Argentine H. parviflorus (Broussalis et al., 2001) indicates high sequence similarity. The other two new peptides displayed residues previously unseen in their respective loops. These peptides demonstrate that nontrivial variations in cyclotide sequence remain to be discovered and highlight the importance of continuing investigations into other genera in the Violaceae. The expanding diversity of sequences also increases the likelihood that different cyclotides will possess different activities in vivo. The observed diversity demonstrates the versatility of the cyclic cystine knot as a combinatorial template, reinforcing the suggestion that it has exciting potential applications in drug design (Craik et al., 2002). Of the 54 published cyclotide sequences, only two contain Met residues. Two more Met-containing cyclotides are presented in this study (Hyfl B and C), and in both cases oxidized forms of Met were detected. This result is consistent with a previous study (Chen et al., 2005), where an oxidized Met residue was reported to be very solvent exposed, accounting for 7.5% of the total surface area of the cyclotide.
We have previously isolated partial cDNA clones from O. affinis (Jennings et al., 2001), V. odorata (Dutton et al., 2004), and V. tricolor (Mulvenna et al., 2005) by RT-PCR using forward primers based on known cyclotide sequences. The partial cDNA clones were sequenced and used either directly to screen a cDNA library (Jennings et al., 2001; Dutton et al., 2004) or for design of gene-specific primers to screen a rapid amplification of cDNA ends library (Mulvenna et al., 2005) for full-length clones. In this study, we have used a similar approach to amplify partial cDNA clones from Hybanthus but without subsequent cDNA or rapid amplification of cDNA ends library construction to obtain the full-length clones. Although this strategy allows for rapid assessment of the diversity within mature cyclotide domains, the first three to four amino acids of this domain fall outside the primer region and are therefore not detected. This prevents comparison of predicted mass for the cDNA isolated peptide sequences with peptide masses found by LC-MS analysis of plant extracts.
In a new approach, we therefore designed a forward primer that targets a newly identified conserved element (AAFALPA) in the precursor sequence upstream of the mature cyclotide domain, identified by comparison of previously isolated full-length cyclotide precursors in the Violaceae family (Dutton et al., 2004; Mulvenna et al., 2005). Figure 7 shows the architecture of typical cyclotide precursors and highlights the previously unreported conserved sequence element in the ER signal domain. Using primers targeted to this region and to oligo(dT) in RT-PCR proved successful for obtaining the full sequence of the mature domain as well as most of the precursor without the need for cDNA library construction and screening. Also, using a primer that binds upstream of the mature domain sequence rather than to known sequences within the cyclotide itself increases the likelihood of isolating cyclotides with hitherto unseen sequences. Cyclotides with rare sequences that are less abundant in the plant may be an indication of increased biological activity, as the plant would need smaller amounts of a more active peptide.
There are two main subfamilies of cyclotides, termed Möbius and bracelet. Members of the Möbius subfamily are characterized by a conserved cis-Pro in loop 5 that creates a twist in the cyclic backbone, thereby making it conceptually similar to a Möbius strip. Bracelet cyclotides do not have the cis-Pro and, consequently, have no twist in the backbone. Moreover, in Möbius cyclotides discovered to date, there is practically no variation in loop sizes, and the net charge is generally neutral or negative, whereas known bracelet cyclotides have variable loop sizes and commonly carry a net charge of +2. Approximately two-thirds of the cyclotides reported to date belong to the bracelet subfamily. Most of the cyclotide sequences isolated from the Hybanthus spp in this study lack the cis-Pro in loop 5 and are classified as bracelet peptides, except for Hyca A and Hymo B, which are assigned to the Möbius subfamily.
Many of the new cyclotides contain sequence elements not previously observed. For example, Hyca A has an unusual loop 2 of five residues (VVDTR) as opposed to the standard four in Möbius cyclotides. Also, the amino acid content of this loop stands out, and the hydrophobic patch partially formed by this loop is most likely expanded due to the presence of two successive hydrophobic residues. With two consecutive Lys residues in loop 4, not found in any other cyclotides to date, Hyca A has a net charge of +2 and is thus more cationic than other Möbius cyclotides. Together with the increased hydrophobic region, this makes Hyca A more amphipathic. The unusual presence of a Met residue in loop 6 as seen in Hyfl B and C is also observed in Hyca A.
Solution structures of the bracelet cyclotides circulin A (Daly et al., 1999) and cycloviolacin O1 (Craik et al., 1999; Rosengren et al., 2003), and the Möbius cyclotides kalata B1 (Rosengren et al., 2003) and kalata B2 (Jennings et al., 2005), show that the typical bracelet structure, with six to seven residues in loop 3, features a short helical segment in this region, whereas in Möbius cyclotides, the four residues in loop 3 form a type II β-turn. Interestingly, the new bracelet cyclotides Hyen B, Hyep A, and Hyfl E, F, J, G, and M have a short loop 3 of only four residues, suggesting that this region forms a turn, as in Möbius cyclotides, rather than a helix. Furthermore, in Hyen B, Hyfl F/J, and Hyst A, the conserved Pro preceding Cys III in bracelets is substituted with other residues, and all but one (Hyen B) of these bracelet cyclotides with a shortened loop 3 have a Pro in position 3 of this loop, as do most Möbius cyclotides. Both Pro residues are important for the turn geometry in the respective subfamilies (Rosengren et al., 2003), further strengthening the suggestion that the bracelet cyclotides mentioned above feature a loop 3 typical of Möbius cyclotides. Hyfl F was also interesting due to an expanded loop 5 of six residues (NHHDKV) as opposed to the usual four in bracelets, showing that the cyclotide framework is capable of tolerating a range of sequences of varying length and amino acid content. This finding is very significant, as it suggests that loops 3 and 5 are potentially amenable to the grafting of foreign bioactive epitopes into the sequence in protein engineering or drug design applications (Craik et al., 2002).
In other so far known cyclotides, the one-residue loop 4 consists of a Ser, a Lys, or a Thr, but in Hyfl E, Hymo B, and Hyst A, a Tyr, an Ala, and a His, respectively, form this loop. Other unusual amino acid combinations of the newly discovered cyclotide sequences include KVTKR, IWGR, and TTFN in loop 2 of Hyen B, Hyst A, and Hyfl F and GFGI/ KNRG in loop 5 of Hyst A and Hyve A, respectively. Hyve A further lacks the conserved Asn or Asp in loop 6 thought to be involved in cyclization, raising the question whether this peptide might be an acyclic homologue.
Finally, it is interesting to note the substitution of the conserved Gly residue preceding Cys IV with a Lys in Hyca A and Asn in Hyfl E and Hyfl F. Only in three cyclotides reported to date is this residue not a Gly, namely, an Asn in circulin D and circulin E (Gustafson et al., 2000) and a Glu in Vodo M (Svangård et al., 2003). It is thought that a positive φ angle in the peptide backbone of the last residue in this loop is important for defining the type II β-turn that is needed to link loop 3 to the cystine knot (Rosengren et al., 2003). Gly residues readily adopt conformations with positive φ angles due to the absence of a side chain. Asn also has the ability to adopt positive φ angles by forming a stabilizing hydrogen bond between the side chain and main chain, thereby introducing a turn in the backbone. Most other amino acids cannot adopt this conformation due to steric hindrance by the side chain. However, Lys residues are also known to be capable of adopting positive φ angles. The substitution of the Gly with a Lys in Hyca A will therefore probably allow for turn geometry consistent with other cyclotides.
The experimental methods developed to measure cyclotide diversity in this study proved effective on minimal amounts of wild collected material (<200 mg) and gave reproducible resolution to within 1 D and 1 min retention time by LC-MS. Only three masses comparable to those of cyclotides occurred outside of the 10- to 20-min retention time band, and very few peptide masses inconsistent with cyclotides (<2000 or >4000 D) occurred in this band. This suggests that cyclotides are present at relatively high concentration in plant tissue and possess distinct mass and chromatographic properties, rendering their isolation and analysis straightforward and reliable. Although it is possible that some cyclotides may have been missed by the extraction and LC-MS procedure, the gradient used covered a wide range of hydrophilic/hydrophobic properties, and we are confident that the vast majority of cyclotides present was detected. Cyclotide diversity within an individual species varied from a single peptide (H. stellarioides) to 32 peptides (H. enneaspermus). The extremes of variation seen in these two species are noteworthy, considering they were once classified as a single taxon (Bennett, 1972). On average, 10 to 20 cyclotides were seen per species. Considering there are ∼900 species (Ballard et al., 1999) in the Violaceae (for which every species investigated to date has contained cyclotides), it is conservatively estimated that there are >9000 unique cyclotides in this plant family.
The variation seen between individual plants in a population is possibly attributable to variations in cyclotide expression rather than differences in genetics given previous observations showing the dependence of cyclotide profile on plant tissue and season (Trabi and Craik, 2004; Trabi et al., 2004). In the well-sampled population of H. enneaspermus (Figure 5), tissues were separated for one plant and indicated increasing cyclotide diversity from leaves to flowers to fruits (3, 4, and 13 different cyclotides, respectively). This is consistent with cyclotides serving as defense molecules and is comparable to the localization of specific plant defensins to the stigma of Nicotiana (Atkinson et al., 1993; Nielsen et al., 1995) in that the most critical tissues for plant reproduction are the most heavily protected. The pattern of increasing incidence of unique cyclotides with decreasing latitude in H. calycinus and H. monopetalus may be a reflection of greater numbers or diversity of pathogens and herbivores in warmer climates.
The influence of geographical isolation of Hybanthus populations is seen in the cyclotide profiles of H. floribundus and its subspecies in Figure 4. This species has been described as consisting of a type form (subsp floribundus) present in southeastern and southwestern Australia and several additional morphologically distinct forms in the southwest (subsp adpressus, chloroxanthus, curvifolius, and “Hill River”). The Nullarbor desert represents a genetic barrier that separates the Hybanthus populations of these two regions. Analysis of the cyclotide profiles of this group reveals a tendency for western collections of all subspecies to cluster more highly together than to the eastern population. This suggests that the eastern and western populations of the type subspecies are at least as distinct as the described morphological variants in the southwest and that H. floribundus subsp floribundus could be further split to account for this difference.
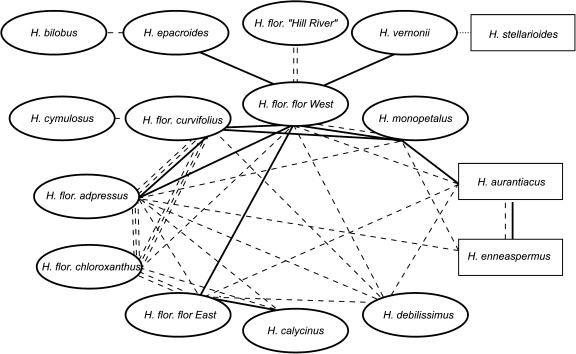
The data generated in this study allow some conclusions about taxonomic relationships between Hybanthus species to be derived. A comparison of the cyclotide physicochemical data across all species (see Supplemental Figure 1 online) using similar criteria for collating peptides within species revealed 42 peptides that could be shared between species. In Figure 8, each species is represented as an oval or a rectangle (Hybanthus section Variabiles, E.M. Bennett, or Suffruticosi, G.K. Schulze, respectively; Bennett, 1972). This analysis demonstrated a network of relationships through possible common peptides. The western Australian subspecies of H. floribundus clustered closely together and displayed well-defined links to H. epacroides, H. cymulosus, H. monopetalus, and H. vernonii. Connections between H. monopetalus and H. aurantiacus, and H. aurantiacus and H. enneaspermus were also seen. The latter relatively isolated grouping is noteworthy, as these species are the section Suffruticosi of Hybanthus (along with H. stellarioides that displayed only a single peptide with uncertain relationships), although all other Australian species are in section Variabiles (Bennett, 1972). Continuing work on cDNA isolation from the collected samples is expected to provide a clearer picture of the evolution of the Australian Hybanthus and their cyclotides.
Figure 8.
Diagram of Potential Common Peptides Shared between Different Species of Australian Hybanthus.
Individual species are denoted by ovals or rectangles (subsections Variabiles and Suffruticosi, respectively). Peptides are seen as potentially common to two collections if they fall within 1 D and 1 min. Cases of two or more individual plants of two species sharing a potential common peptide are shown with a solid line. Instances of one or more individuals of two species sharing a potential common peptide are shown with a dashed line. The single peptide of H. stellarioides only had potential overlap with a peptide from H. vernonii with a looser mass difference restraint of 6 D.
It is important to note that the relationship deductions in Figure 8 have some limitations, including the fact that it is possible that not all cyclotides in a given species will have been detected, and there may be seasonal variations in cyclotide expression. Effects of the latter were minimized by collecting all plant material while in active growth at the commencement or in the middle of flowering. Furthermore, if seasonal or developmental variation, or detection limitations, had led to the absence of any peptides from individual species, this would only remove potential relationships from Figure 8, which thus represents a conservative interpretation of the LC-MS data.
This study adds to the emerging picture of the diversity of defense peptides throughout all major phyla in nature. Initiatives in collating this emerging information have been made for knottins (Gelly et al., 2004) and antimicrobial peptides (Tossi and Sandri, 2002; Brahmachary et al., 2004; Wang and Wang, 2004) in the form of online databases of the expanding list of cDNA and peptide sequences and their structures and activities. A database of cyclotide sequences (www.cyclotide.com) will soon be substantially updated to accommodate this rapidly growing family.
In conclusion, this study indicates that an extensive range of cyclotides remains to be identified in natural populations of plants in the Violaceae. The estimated >9000 unique cyclotides in this plant family represents an enormous wealth of peptide diversity for potential use in crop protection, based on the insecticidal activity of the prototypic cyclotides kalata B1 (Jennings et al., 2001) and B2 (Jennings et al., 2005). The novel sequences indicate that nontrivial variations on cyclotide sequences await discovery. The pattern of cyclotides potentially common to multiple species of Australian Hybanthus corresponded with previously described relationships derived from morphological features. The low impact of field collection and the simplicity of measuring cyclotide LC-MS profiles indicate that they provide a convenient chemotaxonomic feature to assist the classification of the Violaceae.
METHODS
Field Collection
Aerial plant material of nine species of Hybanthus (H. aurantiacus, calycinus, debilissimus, enneaspermus, epacroides, floribundus [subsp adpressus, curvifolius, and floribundus], monopetalus, stellarioides, and vernonii subsp vernonii) was collected during active growth from various locations throughout Australia directed by herbarium records. Samples of 50 to 200 mg were stored on ice in 3 mL of RNAlater solution (Ambion). Additional dried material from H. aurantiacus, H. cymulosus, H. bilobus, H. floribundus subsp chloroxanthus, and H. floribundus subsp “Hill River” was obtained from previous herbarium collections due to the inaccessibility of these populations. A larger sample (150 g) of H. floribundus subsp floribundus was collected in Victoria from an extensive colony of this robust species.
Extraction
Samples, including flowering or fruiting material (∼20 to 50 mg) where available, were frozen in liquid nitrogen and ground to a fine powder and then extracted overnight into 2 mL of methanol:dichloromethane (50:50). Next, 1 mL of water was added and the aqueous layer collected and then another 1 mL of methanol was added for overnight extraction. The extract was similarly diluted with 1 mL of water and separated the following day. The resulting methanolic aqueous extract was freeze-dried and subjected to reverse-phase chromatography on a Strata-X 33-μM polymeric sorbent column (30 mg/mL; Phenomenex). Samples were loaded in 1 mL of 20% acetonitrile/water with 0.1% formic acid to a preequilibrated column, washed with 1 mL of the same solution, then eluted with 1 mL of 90% acetonitrile with 0.1% formic acid and freeze-dried to give a cyclotide-enriched fraction.
LC-MS Analysis
Peptide samples were dissolved in 50 μL of 25% acetonitrile/water with 0.1% formic acid. Analysis was performed on an HP Series 1100 LC/auto-injector coupled to a Micromass LCT mass spectrometer (Waters) equipped with an electrospray ionization source. Samples of 25 μL were subjected to a gradient of 90% acetonitrile with 0.1% formic acid over 0.1% aqueous formic acid (25 to 75% over 30 min at 0.3 mL/min) on a Grom column (150 × 2 mm, 3 μm, equipped with a security guard column, flow rate 300 μL/min). Mass spectra were acquired over a mass range of 800 to 1800 D with a capillary voltage of 3.5 kV and a cone voltage of 30 V. Data were analyzed and processed using MassLynx software.
MS-MS Sequencing
The 150 g sample of H. floribundus subsp floribundus was homogenized and extracted into dichloromethane:methanol following published procedures (Trabi et al., 2004). Individual peptides were isolated through repeated reverse-phase HPLC using gradients of 90% acetonitrile:10% water:0.5% trifluoroacetic acid against 100% water:0.5% trifluoroacetic acid. Purified cyclotides were reduced at a ratio of ∼6 nmol of peptide in 20 μL of 0.1 M NH4HCO3, pH 8.0, to 1 μL of 0.1 M Tris-carboxyethylphosphine at 65°C for 10 min. The reduction was confirmed by matrix-assisted laser desorption ionization time of flight MS after desalting using ZipTip purification (Millipore), which involved several washing steps followed by elution in 10 μL of 80% acetonitrile (0.5% formic acid). The desalted samples were mixed in a 1:1 ratio with matrix consisting of a saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile (0.5% formic acid). Two hundred shots per spectrum were acquired in positive ion reflector mode on a Voyager DE-STR mass spectrometer (Applied Biosystems). The laser intensity was 1800, the accelerating voltage was 20,000 V, the grid voltage was 64% of the accelerating voltage, and the delay time was 165 ns. The low mass gate was 500 D. Data were collected between 500 and 5000 D. Calibration was undertaken using a peptide mixture obtained from Sigma-Aldrich (MSCal1). The mixture contained bradykinin, angiotensin, and insulin B chain.
Enzymatic Digestion and Nanospray MS-MS Sequencing
To the reduced peptide, trypsin, endoGlu-C, or both was added to give a final peptide-to-enzyme ratio of 50:1. Trypsin and endoGlu-C digestions were performed for 1 and 3 h, respectively, while in the combined digestion, initial trypsin incubation for 1 h was followed by addition of endoGlu-C for a further 3 h. The digestions were quenched by the addition of an equal volume of 0.5% formic acid and desalted using ZipTip technology (Millipore). Samples were stored at 4°C prior to analysis. The digestion products were examined by matrix-assisted laser desorption ionization time of flight MS followed by sequencing by nanospray MS-MS on a QStar mass spectrometer (Chen et al., 2005). A capillary voltage of 900 V was applied, and spectra were acquired between mass-to-charge ratios of 60 to 2000 for both time of flight spectra and product ion spectra. The collision energy for peptide fragmentation was varied between 10 and 50 V, depending on the size and charge of the ion. The Analyst QS software program was used for data acquisition and processing. The MS-MS spectra were examined and sequenced based on the presence of both b- and y-series of ions present (N- and C-terminal fragments). Chymotrypsin digests using the same conditions as for trypsin were also conducted to confirm the results obtained for each of the peptide sequences. Amino acid composition was confirmed by amino acid analysis. The disulfide connectivity of CysI-IV, CysII-V, and CysIII-VI was assumed based on homology with previously reported cyclotides (Göransson and Craik, 2003).
RNA Extraction, RT-PCR, and Secondary Structure Prediction
Plant material stored in RNAlater solution was subjected to mRNA isolation using the RNAqueous kit (Ambion). Single-stranded cDNA was prepared from RNA using the OneStep RT-PCR kit (Qiagen), which also facilitated amplification of partial clones by oligo(dT) and degenerate forward primers (Proligo) designed from known cyclotide or cyclotide precursor sequences. The primers used were as follows: 5′-GGGCHGCHTTYGCHCTTCCHGC-3′, coding for the peptide sequence AAFALPA; 5′-CTNGGNAGYATHTCNTGYGG-3′ (peptide sequence GSISC); 5′-GGGGATCCTGYGCNGARACNTG-3′ (peptide sequence CAETC); 5′-CGATCGATTGYGGIGARAGTTGYGT-3′ (peptide sequence CGESCV); 5′-GGGGATCCTGYGGIGARACITG-3′ (peptide sequence CGETC); and 5′-CGATCGATTGYGTITGGATHCCITG-3′ (peptide sequence CVWIPC). The resulting DNA fragments were excised from agarose gel and cloned into the pCR 2.1-TOPO vector using the TOPO Cloning technology (Invitrogen) for sequencing. The amino acid sequences corresponding to the NTR of identified precursors were analyzed for secondary structure using the PROFsec program at the PredictProtein server (Rost et al., 2004) at http://www.predictprotein.org.
Accession Numbers
Sequence data from this article can be found in the UniProt Knowledgebase under accession numbers P84647 to P84649 (Hyfl A to Hyfl C, respectively) and in the GenBank/EMBL data libraries under accession numbers DQ192575 to DQ192577 (Hyfl D to Hyfl F, respectively) and DQ187926 to DQ187945 (Hyfl G to Hyep B, as listed in Table 1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Peptide Masses and Retention Times.
Supplemental Table 2. Proregion in Cyclotide Precursors from Violaceae.
Supplemental Figure 1. Aggregate Scatterplot Data for All Sampled Australian Hybanthus.
Supplementary Material
Acknowledgments
This work was supported by grants from the Australian Research Council, by an Australian Postgraduate Award, and a University of Queensland Graduate School Research Travel Award. D.J.C. is an Australian Research Council Professorial Fellow. We owe great debts to the following individuals for making plant collecting expeditions not just possible but immensely enjoyable. Wayne Woods, Svetlana Micic, and Rodrigo Esparza-Salas all played essential roles. The helpful and enthusiastic staffs of the herbaria of Western Australia, Queensland, New South Wales, and Victoria were also crucial for the success of this project, and we owe them heartfelt thanks.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: David J. Craik (d.craik@imb.uq.edu.au).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.034678.
References
- Apponyi, M.A., Pukala, T.L., Brinkworth, C.S., Maselli, V.M., Bowie, J.H., Tyler, M.J., Booker, G.W., Wallace, J.C., Carver, J.A., Separovic, F., Doyle, J., and Llewellyn, L.E. (2004). Host-defence peptides of Australian anurans: Structure, mechanism of action and evolutionary significance. Peptides 25, 1035–1054. [DOI] [PubMed] [Google Scholar]
- Atkinson, A.H., Heath, R.L., Simpson, R.L., Clarke, A.E., and Anderson, M.A. (1993). Proteinase inhibitors in Nicotiana alata are derived from a precursor protein which is processed into five homologous inhibitors. Plant Cell 5, 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard, H.E., Sytsma, K.J., and Kowal, R.R. (1999). Shrinking the violets: Phylogenetic relationships of infrageneric groups in Viola (Violaceae) based on internal transcribed spacer DNA sequences. Syst. Bot. 23, 439–458. [Google Scholar]
- Barry, D.G., Daly, N.L., Bokesch, H.R., Gustafson, K.R., and Craik, D.J. (2004). Solution structure of the cyclotide palicourein: Implications for the development of a pharmaceutical framework. Structure 12, 85–94. [DOI] [PubMed] [Google Scholar]
- Barry, D.G., Daly, N.L., Clark, R.J., Sando, L., and Craik, D.J. (2003). Linearization of a naturally occurring circular protein maintains structure but eliminates hemolytic activity. Biochemistry 42, 6688–6695. [DOI] [PubMed] [Google Scholar]
- Bennett, E.M. (1972). A revision of the Australian species of Hybanthus Jacquin (Violaceae). Nuytsia 1, 218–241. [Google Scholar]
- Bokesch, H.R., Pannell, L.K., Cochran, P.K., Sowder II, R.C., McKee, T.C., and Boyd, M.R. (2001). A novel anti-HIV macrocyclic peptide from Palicourea condensata. J. Nat. Prod. 64, 249–250. [DOI] [PubMed] [Google Scholar]
- Brahmachary, M., Krishnan, S.P.T., Koh, J.L., Khan, A.M., Seah, S.H., Tan, T.W., Brusic, V., and Bajic, V.B. (2004). ANTIMIC: A database of antimicrobial sequences. Nucleic Acids Res. 32, 586–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broussalis, A.M., Göransson, U., Coussio, J.D., Ferraro, G., Martino, V., and Claeson, P. (2001). First cyclotide from Hybanthus (Violaceae). Phytochemistry 58, 47–51. [DOI] [PubMed] [Google Scholar]
- Chen, B., Colgrave, M.L., Daly, N.L., Rosengren, K.J., Gustafson, K.R., and Craik, D.J. (2005). Isolation and characterization of novel cyclotides from Viola hederaceae: Solution structure and anti-HIV activity of vhl-1, a leaf-specific expressed cyclotide. J. Biol. Chem. 280, 22395–22405. [DOI] [PubMed] [Google Scholar]
- Claeson, P., Göransson, U., Johansson, S., Luijendijk, T., and Bohlin, L. (1998). Fractionation protocol for the isolation of polypeptides from plant biomass. J. Nat. Prod. 61, 77–81. [DOI] [PubMed] [Google Scholar]
- Colgrave, M.L., and Craik, D.J. (2004). Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: The importance of the cyclic cystine knot. Biochemistry 43, 5965–5975. [DOI] [PubMed] [Google Scholar]
- Craik, D.J. (2001). Plant cyclotides: Circular, knotted peptide toxins. Toxicon 39, 1809–1813. [DOI] [PubMed] [Google Scholar]
- Craik, D.J., Daly, N.L., Bond, T., and Waine, C. (1999). Plant cyclotides: A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J. Mol. Biol. 294, 1327–1336. [DOI] [PubMed] [Google Scholar]
- Craik, D.J., Daly, N.L., Mulvenna, J., Plan, M.R., and Trabi, M. (2004). Discovery, structure and biological activities of the cyclotides. Curr. Protein Pept. Sci. 5, 297–315. [DOI] [PubMed] [Google Scholar]
- Craik, D.J., Daly, N.L., Saska, I., Trabi, M., and Rosengren, K.J. (2003). Structures of naturally occurring circular proteins from bacteria. J. Bacteriol. 185, 4011–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craik, D.J., Daly, N.L., and Waine, C. (2001). The cystine knot motif in toxins and implications for drug design. Toxicon 39, 43–60. [DOI] [PubMed] [Google Scholar]
- Craik, D.J., Simonsen, S., and Daly, N.L. (2002). The cyclotides: Novel macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discov. Dev. 5, 251–260. [PubMed] [Google Scholar]
- Crooks, G.E., Hon, G., Chandonia, J.-M., and Brenner, S.E. (2004). WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa, F.B., Terfloth, L., and Gasteiger, J. (2005). Sesquiterpene lactone-based classification of three Asteraceae tribes: A study based on self-organizing neural networks applied to chemosystematics. Phytochemistry 66, 345–353. [DOI] [PubMed] [Google Scholar]
- Daly, N.L., Clark, R.J., and Craik, D.J. (2003). Disulfide folding pathways of cystine knot proteins. Tying the knot within the circular backbone of the cyclotides. J. Biol. Chem. 278, 6314–6322. [DOI] [PubMed] [Google Scholar]
- Daly, N.L., Koltay, A., Gustafson, K.R., Boyd, M.R., Casas-Finet, J.R., and Craik, D.J. (1999). Solution structure by NMR of circulin A: A macrocyclic knotted peptide having anti-HIV activity. J. Mol. Biol. 285, 333–345. [DOI] [PubMed] [Google Scholar]
- Derua, R., Gustafson, K.R., and Pannell, L.K. (1996). Analysis of the disulfide linkage pattern in circulin A and B, HIV-inhibitory macrocyclic peptides. Biochem. Biophys. Res. Commun. 228, 632–638. [DOI] [PubMed] [Google Scholar]
- Dutton, J.L., Renda, R.F., Waine, C., Clark, R.J., Daly, N.L., Jennings, C.V., Anderson, M.A., and Craik, D.J. (2004). Conserved structural and sequence elements implicated in the processing of gene-encoded circular proteins. J. Biol. Chem. 279, 46858–46867. [DOI] [PubMed] [Google Scholar]
- Felizmenio-Quimio, M.E., Daly, N.L., and Craik, D.J. (2001). Circular proteins in plants: Solution structure of a novel macrocyclic trypsin inhibitor from Momordica cochinchinensis. J. Biol. Chem. 276, 22875–22882. [DOI] [PubMed] [Google Scholar]
- Flamini, G., Cioni, P.L., and Morelli, I. (2003). Variability of the essential oil of Viola etrusca. Ann. Bot. (Lond.) 91, 493–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelly, J.C., Gracy, J., Kaas, Q., Le-Nguyen, D., Heitz, A., and Chiche, L. (2004). The KNOTTIN website and database: A new information system dedicated to the knottin scaffold. Nucleic Acids Res. 32, 156–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göransson, U., Broussalis, A.M., and Claeson, P. (2003). Expression of Viola cyclotides by liquid chromatography-mass spectrometry and tandem mass spectrometry sequencing of intercysteine loops after introduction of charges and cleavage sites by aminoethylation. Anal. Biochem. 318, 107–117. [DOI] [PubMed] [Google Scholar]
- Göransson, U., and Craik, D.J. (2003). Disulfide mapping of the cyclotide kalata B1. Chemical proof of the cystic cystine knot motif. J. Biol. Chem. 278, 48188–48196. [DOI] [PubMed] [Google Scholar]
- Gran, L. (1973. a). Oxytocic principles of Oldenlandia affinis. Lloydia 36, 174–178. [PubMed] [Google Scholar]
- Gran, L. (1973. b). On the effect of a polypeptide isolated from “Kalata-Kalata” (Oldenlandia affinis DC) on the oestrogen dominated uterus. Acta Pharmacol. Toxicol. (Copenh.) 33, 400–408. [DOI] [PubMed] [Google Scholar]
- Gustafson, K.R., McKee, T.C., and Bokesch, H.R. (2004). Anti-HIV cyclotides. Curr. Protein Pept. Sci. 5, 331–340. [DOI] [PubMed] [Google Scholar]
- Gustafson, K.R., Sowder II, R.C., Henderson, L.E., Parsons, I.C., Kashman, Y., Cardellina II, J.H., McMahon, J.B., Buckheit, R.W., Jr., Pannell, L.K., and Boyd, M.R. (1994). Circulins A and B: Novel HIV-inhibitory macrocyclic peptides from the tropical tree Chassalia parvifolia. J. Am. Chem. Soc. 116, 9337–9338. [Google Scholar]
- Gustafson, K.R., Walton, L.K., Sowder, R.C.I., Johnson, D.G., Pannell, L.K., Cardellina, J.H.I., and Boyd, M.R. (2000). New circulin macrocyclic polypeptides from Chassalia parvifolia. J. Nat. Prod. 63, 176–178. [DOI] [PubMed] [Google Scholar]
- Hallock, Y.F., Sowder II, R.C., Pannell, L.K., Hughes, C.B., Johnson, D.G., Gulakowski, R., Cardellina II, J.H., and Boyd, M.R. (2000). Cycloviolins A-D, anti-HIV macrocyclic peptides from Leonia cymosa. J. Org. Chem. 65, 124–128. [DOI] [PubMed] [Google Scholar]
- Hernandez, J.F., Gagnon, J., Chiche, L., Nguyen, T.M., Andrieu, J.P., Heitz, A., Trinh Hong, T., Pham, T.T., and Le Nguyen, D. (2000). Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry 39, 5722–5730. [DOI] [PubMed] [Google Scholar]
- Jennings, C., West, J., Waine, C., Craik, D., and Anderson, M. (2001). Biosynthesis and insecticidal properties of plant cyclotides: The cyclic knotted proteins from Oldenlandia affinis. Proc. Natl. Acad. Sci. USA 98, 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings, C.V., Rosengren, K.J., Daly, N.L., Plan, M., Stevens, J., Scanlon, M.J., Waine, C., Norman, D.G., Anderson, M.A., and Craik, D.J. (2005). Isolation, solution structure, and insecticidal activity of kalata B2, a circular protein with a twist: Do Möbius strips exist in nature? Biochemistry 44, 851–860. [DOI] [PubMed] [Google Scholar]
- Kamimori, H., Hall, K., Craik, D.J., and Aguilar, M.I. (2005). Studies on the membrane interactions of the cyclotides kalata B1 and kalata B6 on model membrane systems by surface plasmon resonance. Anal. Biochem. 337, 149–153. [DOI] [PubMed] [Google Scholar]
- Koltay, A., Daly, N.L., Gustafson, K.R., and Craik, D. (2005). Structure of circulin B and implications for antimicrobial activity of cyclotides. Int. J. Pept. Res. Ther. 11, 51–58. [Google Scholar]
- Mulvenna, J., Sando, L., and Craik, D. (2005). Processing of a 22 kDa precursor protein to produce the novel circular protein tricyclon A. Structure 13, 691–701. [DOI] [PubMed] [Google Scholar]
- Nielsen, K.J., Heath, R.L., Anderson, M.A., and Craik, D.J. (1995). Structures of a series of 6-kDa trypsin inhibitors isolated from the stigma of Nicotiana alata. Biochemistry 34, 14304–14311. [DOI] [PubMed] [Google Scholar]
- Nourse, A., Trabi, M., Daly, N.L., and Craik, D.J. (2004). A comparison of the self-association behavior of the plant cyclotides kalata B1 and kalata B2 via analytical ultracentrifugation. J. Biol. Chem. 279, 562–570. [DOI] [PubMed] [Google Scholar]
- Rosengren, K.J., Daly, N.L., Plan, M.R., Waine, C., and Craik, D.J. (2003). Twists, knots, and rings in proteins. Structural definition of the cyclotide framework. J. Biol. Chem. 278, 8606–8616. [DOI] [PubMed] [Google Scholar]
- Rost, B., Yachdav, G., and Liu, J. (2004). The ProteinPredict server. Nucleic Acids Res. 32, W321–W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saether, O., Craik, D.J., Campbell, I.D., Sletten, K., Juul, J., and Norman, D.G. (1995). Elucidation of the primary and three-dimensional structure of the uterotonic polypeptide kalata B1. Biochemistry 34, 4147–4158. [DOI] [PubMed] [Google Scholar]
- Schöpke, T., Hasan, A.M.I., Kraft, R., Otto, A., and Hiller, K. (1993). Hamolytisch aktive komponenten aus Viola tricolor L. and Viola arvensis Murray. Sci. Pharm. 61, 145–153. [Google Scholar]
- Skjeldal, L., Gran, L., Sletten, K., and Volkman, B.F. (2002). Refined structure and metal binding site of the kalata B1 peptide. Arch. Biochem. Biophys. 399, 142–148. [DOI] [PubMed] [Google Scholar]
- Sonnante, G., De Paolis, A., and Pignone, D. (2005). Bowman-Birk inhibitors in Lens: Identification and characterization of two paralogous gene classes in cultivated lentil and wild relatives. Theor. Appl. Genet. 110, 596–604. [DOI] [PubMed] [Google Scholar]
- Svangård, E., Göransson, U., Hocaoglu, Z., Gullbo, J., Larsson, R., Claeson, P., and Bohlin, L. (2004). Cytotoxic cyclotides from Viola tricolor. J. Nat. Prod. 67, 144–147. [DOI] [PubMed] [Google Scholar]
- Svangård, E., Göransson, U., Smith, D., Verma, C., Backlund, A., Bohlin, L., and Claeson, P. (2003). Primary and 3-D modelled structures of two cyclotides from Viola odorata. Phytochemistry 64, 135–142. [DOI] [PubMed] [Google Scholar]
- Tam, J.P., Lu, Y.A., Yang, J.L., and Chiu, K.W. (1999). An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc. Natl. Acad. Sci. USA 96, 8913–8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tossi, A., and Sandri, L. (2002). Molecular diversity in gene-encoded, cationic antimicrobial polypeptides. Curr. Pharm. Des. 8, 743–761. [DOI] [PubMed] [Google Scholar]
- Trabi, M., and Craik, D.J. (2002). Circular proteins—No end in sight. Trends Biochem. Sci. 27, 132–138. [DOI] [PubMed] [Google Scholar]
- Trabi, M., and Craik, D.J. (2004). Tissue-specific expression of head-to-tail cyclized miniproteins in Violaceae and structure determination of the root cyclotide Viola hederacea root cyclotide1. Plant Cell 16, 2204–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabi, M., Svangård, E., Herrmann, A., Göransson, U., Claeson, P., Craik, D.J., and Bohlin, L. (2004). Variations in cyclotide expression in Viola species. J. Nat. Prod. 67, 806–810. [DOI] [PubMed] [Google Scholar]
- Turner, B.L. (1967). Plant chemosystematics and phylogeny. Pure Appl. Chem. 14, 189–213. [PubMed] [Google Scholar]
- Wang, Z., and Wang, G. (2004). APD: The antimicrobial peptide database. Nucleic Acids Res. 32, 590–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherup, K.M., Bogusky, M.J., Anderson, P.S., Ramjit, H., Ransom, R.W., Wood, T., and Sardana, M. (1994). Cyclopsychotride A, a biologically active, 31-residue cyclic peptide isolated from Psychotria longipes. J. Nat. Prod. 57, 1619–1625. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.