Abstract
Cloning the whole 3.5-megabase (Mb) genome of the photosynthetic bacterium Synechocystis PCC6803 into the 4.2-Mb genome of the mesophilic bacterium Bacillus subtilis 168 resulted in a 7.7-Mb composite genome. We succeeded in such unprecedented large-size cloning by progressively assembling and editing contiguous DNA regions that cover the entire Synechocystis genome. The strain containing the two sets of genome grew only in the B. subtilis culture medium where all of the cloning procedures were carried out. The high structural stability of the cloned Synechocystis genome was closely associated with the symmetry of the bacterial genome structure of the DNA replication origin (oriC) and its termination (terC) and the exclusivity of Synechocystis ribosomal RNA operon genes (rrnA and rrnB). Given the significant diversity in genome structure observed upon horizontal DNA transfer in nature, our stable laboratory-generated composite genome raised fundamental questions concerning two complete genomes in one cell. Our megasize DNA cloning method, designated megacloning, may be generally applicable to other genomes or genome loci of free-living organisms.
Keywords: bacterial genomes, DNA assembly, genome symmetry
The advent of whole-genome sequencing made possible the comparison of sequenced bacterial genomes and provided evidence for horizontal gene transfer (HGT) (1-3) which, because it is not restricted to genus, is reflected in the diversity of bacterial genome sizes and structure. Examples of HGT are found in certain bacteria that carry multiple chromosomes. The smaller of the two chromosomes of Vibrio cholerae may derive from a giant plasmid and participate in the gene capture system (4). In the Gram-negative strain Deinococcus radiodulans, (5) one large [2.43-megabase (Mb)] chromosome may be derived from the Gram-negative obligatory Thermus thermophilus (6) and the second 0.45-Mb chromosome may be derived from an archae. An increase by 1.34-Mb in the genome of Escherichia coli O157:H7 (7, 8) compared with the laboratory domesticated strain K-12 (9) was unveiled by the integration of a number of phage DNA. A highly probable candidate for a symbiotic island in the Mesorhizobium loti strain MAFF303099 is a DNA segment as large as 611 kb that includes most of the nitrogen-fixing genes as a cluster (10). These examples indicate that DNA of significantly large size can transfer naturally beyond the species barrier and play a key role in the creation of genome variations.
DNA transfer performed in the laboratory is collectively referred to as DNA cloning. Although DNA cloning is one of the most basic tools used in biology to understand genes and gene function(s), cloning is not considered representative of HGT because the process occurs under well regulated laboratory conditions. The handling of large numbers of genes and, consequently, giant DNA requires innovative technologies with applicability to a broad range of basic research areas. Our technology allows the cloning of giant DNAs as are suggested in HGT. In 1995, our group suggested the use of the Bacillus subtilis 168 genome as a stable cloning vehicle for giant DNA (11). We termed the cloning vehicle BGM (for B. subtilis genome) vector and demonstrated the successful reconstruction of long contiguous DNAs (12-14). Our cloning principle took advantage of features inherent to this bacterium, i.e., the development of natural competence and the subsequent homologous recombination activity in the cytoplasm. Both features are induced because of their association with growth-phase transition (15, 16). The target DNA is guided in the BGM vector by simultaneous homologous recombination at two small flanking DNAs called landing pad sequences (LPS), integrated at the BGM cloning locus before cloning. The two LPS, ordered and oriented correctly, are termed the LPS array (LPA) (13, 14). Sliding the LPA results in elongation of the adjacent target DNA (Fig. 1). We offer such elongation-coupled cloning in the BGM vector, hereafter called inchworm elongation (IWe), as an elegant alternative to current cloning methods. We applied this method in the complete cloning of the whole 3.5-Mb genome of the photosynthetic bacterium Synechocystis PCC6803 (17) into the 4.2-Mb genome of the mesophilic bacterium B. subtilis 168 (Fig. 2).
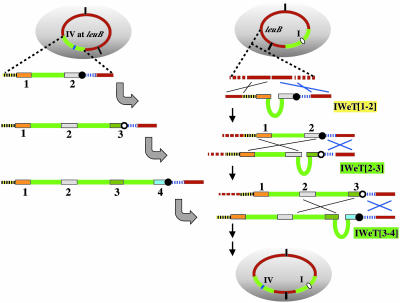
Fig. 1.
IWe steps to megacloning. A set of LPS was prepared by PCR and arranged in tandem in pBR322-derived vector in E. coli to yield a set of LPA plasmids. LPA plasmids, constructed from pCISP310B and pCISP311B (12) or pCISP334 and pCISP335 (21), have either a cat (filled circle) or an erm (open circle) marker used alternatively. The LPA[1/2] plasmid was delivered to the GpBR sequence by means of double-homologous recombination as indicated by X. IWe[1/2] indicates that Synechocystis DNA taken up by competent B. subtilis cells recombines at LPA and the internal continuous green-colored sequence integrates. An LPA[2/3] plasmid is delivered to conduct the next IWe[2/3]. Repeats of LPA[n/n + 1] and IWe[n/n + 1] elongate the continuous Synechocystis genome DNA in GpBR. Delivery of LPA was selected by cat or erm alternatively. IWe selection is by an internal positive selection system (13) briefly described in Methods. The heavy gold arrow in LPA indicates that the λ cI857 repressor gene linked with the spectinomycin resistance gene (spc) was repeatedly used. Strains for all IWe are described in Fig. 6. I-PpoI sites at both ends of GpBR, indicated by vertical lines inside BGM at the bottom, always remain unchanged and were used to produce cloned DNA in GpBR as shown in Fig. 4.
Fig. 2.
Structure of the megacloned Synechocystis genome in the B. subtilis genome. Schematic drawings of the genomes of Synechocystis PCC6803 (green circle) (17), B. subtilis 168 (brown circle), and strain BEST7613 (mosaic with green and brown). Seven plasmids reported for Synechocystis are omitted. B. subtiis 168 has no indigenous plasmid. Regions A, B, and C are from the B. subtilis genome and regions [I]-[IV] are from the Synechocystis genome. The BEST7613 genome (7,700 kb) shows approximate size because the B. subtilis strain used in this study, BEST7003 (13), had certain deletions and was shorter than the reported strain (26). The locations of oriC and terC are known for B. subtilis only. Antibiotic resistance markers inserted in the BEST7613 genome are shown with relevant genes and locations: resistance to blasticidin S (bsr), tetracycline (tet), spectinomycin (spc), neomycin (neo), and phleomycin (phl). The GpBR (indicated by the striped boxes) is the megacloning locus prepared at the proB, leuB and ytqB loci of the B. subtilis genome. I-PpoI sites indicated by bars inside the BEST7003 and BEST7163 circle are created at the ends of GpBRs and of three megacloned Synechocystis genomes. The GpBR sequences that remained in BEST7613 after removal manipulations are shown. GpBR between regions A and [I] was removed after BEST7155 (Fig. 3). The GpBR sequence between regions [II] and B was replaced by tet before BEST7566 (Fig. 3) to reduce the overlap of regions [II] and [III]. GpBR between regions B and [IV] was replaced by spc before BEST7374 (Fig. 3). The I-PpoI site is left behind upon all of these removals. Overlaps of 5.984 kb for regions [II] and [III], 4.479 kb for regions [III] and [IV], and 0.334 kb for regions [IV] and [I] remained in BEST7613. Six fragments yielded from BEST7613 by I-PpoI digestion are shown in Fig. 4.
Methods
Bacterial Strains and Plasmids. The preparation and transformation of competent E. coli and B. subtilis were as described in ref. 18. LB broth was used to grow B. subtilis and E. coli. The bacteria were grown at 37°C; cI gene selection by antibiotics was at 30°C because cI857 encodes proteins that are labile at the higher temperature.
Cloning of Long Continuous DNA by IWe. The BGM vector has at least two inserts derived from B. subtilis RM125, a restriction-modification-deficient strain (12, 13). One insert is a 4.3-kb pBR322 sequence, the genomic pBR (GpBR) sequence (19), at the indicated genome locus proB, leuB, or ytqB (Figs. 2 and 3). During the cloning, the GpBR sequence is separated into 2.3-kb (the amp half of the sequence) (Fig. 2, yellow and brown striped boxes) and 2.1-kb (the tet half of the sequence) (Fig. 2, blue and white striped boxes) in the genome that always remain at both ends of the insert. The rare-cutting enzyme I-PpoI recognition sites in the two halves provide the internal cloned segments upon I-PpoI cleavage (Figs. 2 and 4). The other insert is a neomycin resistance gene (neo) between yvfC and yveP (20). Because the Pr promoter of the neo gene is regulated by binding of a cI-repressor gene product, the presence or absence of the cI gene in GpBR renders BGM derivatives sensitive or resistant to neomycin (13, 20). Screening of the selected neomycin-resistant colonies by the concomitant loss of the linked marker (spc) was followed by colony PCR with a primer set designed to amplify the newly elongated region (data not shown). The DNA structure of candidate recombinants was analyzed by digestion with the I-PpoI, which always cut at the junction of the Synechocystis and B. subtilis genome. The entire length of the insert was isolated and resolved by contour-clamped homogeneous electric field (CHEF) gel electrophoresis. In certain IWe cases, the use of pCISP334 and pCISP335 as LPA construction vectors (Fig. 1) instead of the previously used pCISP310B and pCISP311B (12, 13) greatly facilitated our work because it made unnecessary the preparation of two LPS containing the elongation plasmid (21). Primers for LPS are listed in Table 1, which is published as supporting information on the PNAS web site. Type II restriction enzymes and T4 DNA ligase were obtained from Toyobo (Tokyo, Japan) except for I-PpoI, which was from Promega. The PCR primers were made by DATE Concept (Sapporo, Japan).
Fig. 3.
All of the steps to complete megacloning of the Synecohsystis genome. (Upper Left and Upper Center) Four Synechocystis genome regions (Upper Left) with sll1652 (filled oval; open oval if deleted) and rrn genes (filled box, open box if deleted) were megacloned by IWe (Upper Center). (Right) The four megacloned recombinants were used to complete BEST7613 by IWeT; oval and box symbols correspond to those for Upper Left and Upper Center. For Upper Center and Right, the two short bars connected to the outside of the genome circle represent oriC (top) and terC (bottom), schematically indicating genome symmetry. (Lower Center) BGM vectors are schematically drawn with the GpBR at three loci. (Lower Left) Colonies formed on LB plates during 17 h at 37°C.
Fig. 4.
IWe and IWeT steps in megacloning. I-PpoI fragments from indicated BGM strains run on CHEF gels with size markers indicated to the right. (Upper Left) I-PpoI fragments of cloned region [I] indicated by green arrowheads. (Upper Right) Strains of IWeT-mediated integration and elongation of region [IV] in BEST7324 (BEST7155 plus sll1652 disrupted by bsr) to BEST7374. (Lower Left) Strains of IWeT-mediated integration and elongation of region [II] in BEST7527 to BEST7566. (Lower Right) IWeT-mediated integration and elongation of region [III]. Indicated strains of IWeT of region [III] are separated into preintegration (lanes 1-4) and postintegration (lanes 5-9) of region [II]. Synechocystis genome DNA and the BGM vector part are indicated by differently colored arrowheads. Six I-PpoI fragments produced from BEST7613 are mapped to those in Fig. 2. Running conditions are shown. Yeast chromosomal DNA was run as a size marker.
Preparation of Genomic DNA. Analytical grade DNA for CHEF gel electrophoresis was prepared in agarose plugs as described in ref. 18, with the minor modification that proteinase-K digestion was at 50°C. The running conditions for CHEF gel electrophoresis are listed in Fig. 4. Genomic DNA for analysis by conventional gel electrophoresis was prepared by a liquid isolation method (22). Preparation of DNA probes and Southern hybridization was carried out by the DNA labeling and detection kit (Boehringer-Mannheim, which is now Roche Molecular Biochemicals).
Results and Discussion
Why Is the Synechocystis Genome the Target DNA? Synechocystis PCC6803 (17) was chosen as the target for megacloning for several reasons. The preparation of LPS by PCR-mediated amplification requires sequence information on the whole genome for the design of primers. The preparation of high-molecular-weight genomic DNA is necessary for technical reasons described elsewhere (13). More importantly, the possible expression of genes not detrimental to the B. subtilis host but otherwise hazardous had to be avoided (23). Therefore, the sequenced 3,573-kb genome of the unicellular photosynthetic bacterium Synechocystis, thought to be nonpathogenic, was the only available choice when this work started in 1997, 1 year after its sequence was reported (17).
Megacloning of the Synechocystis Genome. The procedure, hereafter termed as the megacloning method, is summarized in Fig. 3. After many trials and errors, we were able to establish a standard protocol for IWe. For isolation of the correct recombinant shown in Fig. 1, an ≈5-kb LPS is required for the 30-kb cloning target region. We divided IWe-mediated cloning of the Synechocystis genome into four regions, [I]-[IV], because of the reason described below. IWe-mediated megacloning of region [I] proceeded clockwise in BGM (Fig. 6, which is published as supporting information on the PNAS web site), and it yielded BEST7155, which carried the 908-kb segment (Fig. 3). The success of stepwise elongation was confirmed by the increase in fragment sizes upon digestion with I-PpoI of selected intermediate strains resolved by CHEF gel electrophoresis (Fig. 4). The structure of the 908-kb DNA in BEST7155 was identical to that of the Synechocystis genome examined with six restriction enzymes (Fig. 6).
Three Megacloning Procedures for the Rest of the Synechocystis Genome. After two more IWe on BEST7155, all further IWe attempts involving the resultant 980 kb proved unsuccessful (data not shown). Although the reason(s) for this failure was not obvious, it suggests that the symmetry of the B. subtilis genome architecture may be involved (24-26). All B. subtilis 168 derivatives (24, 27) and related species (28) have circular genomes that manifest excellent symmetry with respect to the two distances between the origin of DNA replication (oriC) and its termination (terC). Therefore, we changed our cloning strategy by dividing the entire Synechocystis genome into four putative regions, regions [I] (908 kb), [II] (887 kb), [III] (890 kb), and [IV] (931 kb) (Fig. 3). To minimize the imbalance introduced by megacloning, we prepared two more GpBR loci at leuB (19) and ytqB (29) on the left side of the B. subtilis genome. Megacloning of regions [II], [III], and [IV] in GpBR at proB, leuB, and ytqB yielded BEST7383, BEST8648, and BEST8817, respectively (Fig. 3). Details of these IWe procedures are shown in Fig. 6. Southern analysis shown in Fig. 6 with six restriction enzymes using the Synechocystis genome as a probe identified no apparent aberrant structures in these regions. Our results showed that the entire 3.5-Mb Synechocystis genome was covered by a BGM library comprised of the four independently megacloned recombinants shown in Fig. 3. The colony morphology of the four megacloned recombinants exhibited no distinguishing features (Fig. 3).
Given possible constraints related to genome symmetry and considering that region [I] (908 kb) was located in the right half of BEST7155, we thought that megacloning of region [III] (887 kb) or [IV] (909 kb) in the left half of BEST7155 would correct the hypothetical imbalance and restore symmetry. In these megaclonings, we did not repeat the process used for IWe but used progressive transfer of the existing intermediate BGMs already obtained by IWe. This IWe-clone transfer (IWeT) method is shown in Fig. 5. Because of longer DNA cloning by IWeT (70 kb on average compared with 30 kb by IWe), transfer of region [IV] was completed by using 13 BGM strains indicated by the orange and green horizontal arrows in Fig. 6 to yield BEST7374. The step-by-step size increase in the I-PpoI fragments is shown in Fig. 4. The genome of BEST7374 is ≈6,000 kb, 1,800 kb of which derive from the Synechocystis genome. Genome symmetry was restored, but there were no obvious phenotypic changes in the colony (Fig. 3).
Fig. 5.
IWeT steps for assembly of the megacloned Synechocystis genome segment in BGM. IWeT of region [IV] to the leuB locus of the strain BEST7324 designated in the legend to Fig. 4 Upper Right; region [I] at proB is shown as an example. Selected IWe strains indicated by orange and green arrows from IWe region [IV] shown in Fig. 6 were used. The integration started by IWeT[1/2] is followed by alternate use of end markers indicated by cat (filled circle) or erm (open circle). The supply of longer regions for homologous recombination (indicated by a blue X) resulted in more effective elongation than was possible with IWe.
Complete Megacloning: Retention of Structural Symmetry and Lack of Ribosomal Operon Gene. The assembly of region [III] by IWeT in BEST7374 proceeded to 456 kb and resulted in strain BEST7497 (Fig. 3). Elongation of the next segment by IWeT stalled unexpectedly. Although all BGM screened with the appropriate antibiotic resistance markers elongated the predicted segment correctly, there was concomitant deletion of ≈300 kb in region [IV] (data not shown). We contemplated two scenarios to explain this phenomenon. One scenario considers incompatibility of the expressed genes, the interaction of gene products expressed from the new IWeT region, and the deleted region of [IV] exerted detrimental effects on the growth of B. subtilis. The other scenario postulates disturbance in genome symmetry that was corrected by a compensatory deletion. With respect to the first scenario, the presence of a ribosomal RNA operon gene (rrnB) in the deleted region of region [IV] drew our attention. Pinpoint removal of the 5.4-kb rrnB of BEST7497 was performed by replacing it with a phleomycin resistance gene (phl); this is a method we normally use with B. subtilis (19). Strain BEST7527 (Fig. 3) accepted the previously stalled IWeT region without inducing any deletion in region [IV]. To avoid an increase in asymmetry, we chose to move region [II] (887 kb) to the right side of BEST7527 in an effort to restore symmetry. We performed IWeT of region [II] in BEST7527 by continuous elongation of the existing region [I] and completed without encountering any obvious problems. The result was BEST7566 (Fig. 3), whose genome manifested reversed asymmetry by ≈400 kb: 1,400 kb on the left and 1,800 kb on the right. We speculate that our earlier unsuccessful attempts to elongate region [I] beyond 980 kb was ascribable to the existence of a significant imbalance.
Further elongation by IWeT of the remaining region of region [III] in BEST7566 stalled again at the segment that included the rrnA operon, the other ribosomal operon of the Synechocystis strain. As before, pinpoint deletion of the 5.4-kb rrnA and its replacement with a 1.3-kb neomycin resistance gene (neo) allowed completion of the remaining IWeT. Consequently, we were able to isolate BEST7613, a 7.7-Mb composite genome that consisted of the 3.5-Mb Synechocystis genome and the 4.2-Mb BGM vector. Our IWeT-mediated assembly process is shown in Fig. 4 and consists of a step-by-step increase in the size and number of I-PpoI fragments. We had to insert an antibiotic resistance marker gene, the blasticidin S resistance gene (bsr), to inactivate the sll1652 gene of BEST7155 (Figs. 2 and 3). This pinpoint manipulation was necessary because the expressed Sll1652 product inhibited spore formation of B. subtilis in a yet unidentified manner. Because the Synechocystis genome segments in all recombinants replicated as part of the B. subtilis genome, selection by antibiotics was unnecessary. The strains we isolated in the course of megacloning grew well on LB plates (Fig. 3).
Genetic Constraints on the Species Barrier by Ribosomes. Phylogenic analyses are based on the concept of one ribosomal RNA species per genome (30). The isolation of stable Synechocystis rrn carriers, BEST8817, BEST8648, BEST7374, and BEST7497 (Fig. 3), led us to postulate that the exclusion of the two Synechocystis rrn operons with identical sequences, rrnA and rrnB, was due to the coexistence in the cells of currently unidentified genes. Our attempts to deliver the Synechocystis rrnA operon to BEST7163 were unsuccessful because of the occurrence of deletions that are currently under investigation (our unpublished observations). The formation of a Synechocystis ribosome complex in the cytoplasm of B. subtilis may stimulate the translation of potentially transcribed Synechocystis genes. Consequently, some translated products may directly or indirectly exert detrimental effects on the viability of the B. subtilis bacterium. A number of transcripts, but very few proteins, originating from Synechocystis genes present in BEST7613 support this hypothesis (our unpublished observations). An inducible Synechocystis rrn operon is required to investigate this phenomenon.
Growth of BGM Recombinants. BEST7613 grew well on LB plates (Fig. 3) and in LB medium but not in pure BG11 medium used for the cultivation of Synechocystis (31). Several factors and components, including the cell membrane and cell wall, have to be processed for a strain to convert to a different stable metabolic state in response to changes in the culture medium. This observation raises the question as to what the regulatory factors besides ribosomal RNA are. BEST7613 lacks seven plasmids indigenous to Synechocystis encoding 397 genes (17). Because we were unable to identify essential genes among the plasmid-borne genes, we did not pursue IWe cloning of these plasmids. Differences in the copy number of genomes in each cell, 12 for PCC6803 (31) vs. one for B. subtilis (18, 24, 26), may reflect a basic requirement for this Synechocystis derivative strain to grow as a photosynthetic bacterium in BG11 medium. We have not yet located the origin of DNA replication (ori) in this Synechocystis genome (17), and it may be disordered in the genome structure of BEST7613. In addition, mutations not reflected in the altered length of restriction fragments may have accumulated by not being used during cultivation in LB media (32).
Comparison of Our Cloning Vehicle and Conventional Cloning Vehicles. Two systems aimed at the cloning of giant DNAs have been developed: bacterial artificial chromosomes in E. coli (33) and yeast artificial chromosomes in Saccharomyces cerevisiae (34). Despite the nomenclature, these vectors are not chromosomes but independent replicons in the respective hosts. Selection is necessary to sustain recombinants, and the clones are inherently unstable genetically because of the nature of the cloning vectors. Our megacloning method not only extends the size limit of stable DNA cloning but provides for inherent structural stability and allows the performance of highly sophisticated pinpoint manipulations as demonstrated by sequence editing of the selective disruption of sll1652 and of the deletions of rrnA or rrnB. Thus, our method is readily available for the cloning of smaller genomes or possibly genome (segments) of >3.5 Mb.
Impact of Megacloning on Genomics. The demonstration of Synechocystis genome-cloning in the BGM vector raises questions crucial for an understanding of genome dynamics. Megacloning unveils basic principles that underlie HGT and raises issues that pertain to the plasticity of genome structures. In addition, advances in megacloning techniques open opportunities not only for basic genome research but also for the construction of beneficial microbes by the bioindustry. Because megacloning requires only a certain amount of DNA and sequence information, its targets need not be restricted to bacterial genomes but may include organisms as large as the 1.2-Mb Mimivirus (35). Elsewhere (14), our group documented the flexible reconstruction of large-sized mouse genome DNA. Those findings indicated that appropriately edited DNA cloned in the BGM vector yields DNA suitable for use in different research undertakings. The retrieval of megacloned DNA by the recovery system (14, 36) or the genome surgery system (37) renders the BGM vector a tool with which technical breakthroughs may be achieved.
Supplementary Material
Acknowledgments
We thank Drs. M. Ikeuchi (University of Tokyo, Tokyo) and H. Yoshikawa (Tokyo University of Agriculture, Tokyo) for the Synechocystis strains and useful comments. We also thank Drs. T. Tanaka, S. Kaneko, T. Kaneko, S. Tabata, H. Yanagawa, T. Tsuji, and the late H. Miyake for continuous encouragement and many discussions.
Author contributions: M.I. designed research; M.I., K.T., M.K., and K.F. performed research; M.I. contributed new reagents/analytic tools; M.I., K.T., M.K., and K.F. analyzed data; and M.I. and K.T. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HGT, horizontal gene transfer; Mb, megabase(s); IWe, inchworm elongation; IWeT, IWe transfer; GpBR, genomic pBR; LPS, landing pad sequence; LPA, LPS array; CHEF, contour-clamped homogeneous electric field.
References
- 1.Doolittle, R. F. (1998) Nature 392, 339-342. [DOI] [PubMed] [Google Scholar]
- 2.Jain, R., Rivera, M. C. & Lake, J. A. (1999) Proc. Natl. Acad. Sci. USA 96, 3801-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gophna, U., Charlebois, R. L. & Doolittle, W. F. (2004) Trends Microbiol. 12, 213-219. [DOI] [PubMed] [Google Scholar]
- 4.Heidelberg, J. F., Eisen, J. A., Nelson, W. C., Clayton, R. A., Gwinn, M. L., Dodson, R. J., Haft, D. H., Hickey, E. K., Peterson, J. D., Umayam, L, et al. (2000) Nature 406, 477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White, O., Eisen, J. A., Heidelberg, J. F., Hickey, E. K., Peterson, J. D., Dodson, R. J., Haft, D. H., Gwinn, M. L., Nelson, W. C., Richardson, D. L., et al. (1999) Science 286, 1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henne, A., Brüggemann, H., Raasch, C., Wiezer, A., Hartsch, T., Liesegang, H., Johann, A., Lienard, T., Gohl, O., Martinez-Arias, R., et al. (2004) Nat. Biotechnol. 22, 547-553. [DOI] [PubMed] [Google Scholar]
- 7.Perna, N. T., Plunkett, G., III, Burland, V., Mau, B., Glasner, J. D., Rose, D. J., Mayhew, G. F., Evans, P. S., Gregor, J., Kirkpatrick, H. A., et al. (2001) Nature 409, 529-533. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi, T., Makino, K., Ohnishi, M., Kurokawa, K., Ishii, K., Yokoyama, K., Han, C. G., Ohtsubo, E., Nakayama, K., Murata, T., et al. (2001) DNA Res. 8, 11-22. [DOI] [PubMed] [Google Scholar]
- 9.Blattner, F. R., Plunkett, G., III, Bloch, C. A., Perna, N. T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J. D., Rode, C. K., Mayhew, G. F., et al. (1997) Science 277, 1453-1474. [DOI] [PubMed] [Google Scholar]
- 10.Kaneko, T., Nakamura, Y., Sato, S., Asamizu, E., Kato, T., Sasamoto, S., Watanabe, A., Idesawa, K., Ishikawa, A., Kawashima, K., et al. (2000) DNA Res. 7, 331-338. [DOI] [PubMed] [Google Scholar]
- 11.Itaya, M. (1995) Mol. Gen. Genet. 248, 9-16. [DOI] [PubMed] [Google Scholar]
- 12.Itaya, M., Nagata, T., Shiroishi, T., Fujita, K. & Tsuge, K. (2000) J. Biochem. 128, 869-875. [DOI] [PubMed] [Google Scholar]
- 13.Itaya, M., Fujita, K., Ikeuchi, M., Koizumi, M. & Tsuge, K. (2003) J. Biochem. 134, 513-519. [DOI] [PubMed] [Google Scholar]
- 14.Kaneko, S., Tsuge, K., Takeuchi, T. & Itaya, M. (2003) Nucleic Acids Res. 31, e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spizizen, J. (1958) Proc. Natl. Acad. Sci. USA. 44, 1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubnau, D. A. (1999) Annu. Rev. Microbiol. 53, 217-244. [DOI] [PubMed] [Google Scholar]
- 17.Kaneko, T., Sato, S., Kotani, H., Tanaka, A., Asamizu, E., Nakamura, Y., Miyajima, N., Hirosawa, M., Sugiura, M., Sasamoto, S., et al. (1996) DNA Res. 3, 109-136. [DOI] [PubMed] [Google Scholar]
- 18.Itaya, M. & Tanaka, T. (1991) J. Mol. Biol. 220, 631-648. [DOI] [PubMed] [Google Scholar]
- 19.Itaya, M. (1993) Mol. Gen. Genet. 241, 287-297. [DOI] [PubMed] [Google Scholar]
- 20.Itaya, M. (1999) Biosci. Biotech. Biochem. 63, 602-604. [DOI] [PubMed] [Google Scholar]
- 21.Tsuge, K. & Itaya, M. (2002) Jpn. Patent 2002-055858.
- 22.Saito, H. & Miura, K. (1963) Biochim. Biophys. Acta 72, 619-629. [PubMed] [Google Scholar]
- 23.Nelson, K. E., Paulsen, I. T., Heidelberg, J. F. & Fraser, C. M. (2000) Nat. Biotechnol. 18, 1049-1054. [DOI] [PubMed] [Google Scholar]
- 24.Itaya, M. (1993) J. Bacteriol. 175, 741-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rocha, E. P. (2004) Microbiology 150, 1609-1627. [DOI] [PubMed] [Google Scholar]
- 26.Kunst, F., Ogasawara, N., Moszer, I., Albertini, A. M., Alloni, G., Azevedo, V., Bertero, M. G., Bessieres, P., Bolotin, A., Borchert, S., et al. (1997) Nature 390, 249-256. [DOI] [PubMed] [Google Scholar]
- 27.Toda, T. & Itaya, M. (1995) Microbiology 141, 1937-1945. [DOI] [PubMed] [Google Scholar]
- 28.Qiu, D., Fujita, K., Sakuma, Y., Tanaka, T., Ohashi, Y., Ohshima, H., Tomita, M. & Itaya, M. (2004) Appl. Environ. Microbiol. 70, 6247-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uotsu-Tomita, R., Kaneko, S., Tsuge, K. & Itaya, M. (2005) Biosci. Biotech. Biochem. 69, 1036-1039. [DOI] [PubMed] [Google Scholar]
- 30.Daubin, V., Moran, N. A. & Ochman, H. (2003) Science 301, 829-832. [DOI] [PubMed] [Google Scholar]
- 31.Labarre, J., Chauvat, F. & Thuriaux, P. (1989) J. Bacteriol. 171, 3449-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bergthorsson, U. & Ochman, H. (1999) J. Bacteriol. 181, 1360-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shizuya, H., Birren, B., Kim, U. J., Mancino, V., Slepak, T., Tachiiri, Y. & Simon, M. (1992) Proc. Natl. Acad. Sci. USA. 89, 8794-8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burke, D. T., Carle, G. F. & Olson, M. V. (1987) Science 236, 806-812. [DOI] [PubMed] [Google Scholar]
- 35.Raoult, D., Audic, S., Robert, C., Abergel, C., Renesto, P., Ogata, H., La Scola, B., Suzan, M. & Claverie, J. M. (2004) Science 306, 1344-1350. [DOI] [PubMed] [Google Scholar]
- 36.Tsuge, K. & Itaya, M. (2001) J. Bacteriol. 183, 5453-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Itaya, M. & Tanaka, T. (1997) Proc. Natl. Acad. Sci. USA 94, 5378-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.