Abstract
The selective movement of ions between intracellular compartments is fundamental for eukaryotes. Arabidopsis thaliana Na+/H+ exchanger 1 (AtNHX1), the most abundant vacuolar Na+/H+ antiporter in A. thaliana, has important roles affecting the maintenance of cellular pH, ion homeostasis, and the regulation of protein trafficking. Previously, we have shown that the AtNHX1 C-terminal hydrophilic region localized in the vacuolar lumen plays an important role in regulating the antiporter's activity. Here, we have identified A. thaliana calmodulin-like protein 15 (AtCaM15), which interacts with the AtNHX1 C terminus. When expressed in yeast, AtCaM15 is localized in the vacuolar lumen. The transient expression of AtCaM15 in Arabidopsis leaf protoplasts showed that AtCaM15 is present in the central vacuole. The binding of AtCaM15 to AtNHX1 was Ca2+- and pH-dependent and decreased with increasing pH values. Our results also show that the binding of AtCaM15 to AtNHX1 modified the Na+/K+ selectivity of the antiporter, decreasing its Na+/H+ exchange activity. Taken together, the presence of a vacuolar calmodulin-like protein acting on the vacuolar-localized AtNHX1 C terminus in a Ca2+- pH-dependent manner suggests the presence of signaling entities acting within the vacuole.
Keywords: ion homeostasis, pH regulation
Na+/H+ antiporters play a major role in pH and Na+ homeostasis of cells throughout the biological kingdom, including bacteria, algae, fungi, worms, higher plants, and mammals, including humans (1-3). These ion exchangers are integral membrane proteins residing in the plasma membranes and endomembranes of many different cell types. In plants, vacuolar Na+/H+ antiporters use the proton electrochemical gradient generated by the vacuolar H+-translocating enzymes H+-ATPase and H+-PPiase (4) to couple the movement of H+ down its electrochemical potential with the movement of Na+ against its electrochemical potential (5). In Arabidopsis thaliana, the cation proton antiporter 1 family of vacuolar cation/H+ transporters comprises six members, A. thaliana Na+/H+ exchanger 1 (AtNHX1)-AtNHX6 (6, 7), which have significant similarity to the yeast endosomal Nhx1 (8, 9) and the Caenorhabditis elegans endosomal NHXs (10). Plant vacuolar Na+/H+ antiporters have been shown to play important roles in cellular ion homeostasis, including the sequestration of Na+ ions into the vacuole (11, 12), and vacuolar pH regulation (13). Analyses of the transcriptional profile of AtNHX1 insertional knockout mutant plants growing in the absence and presence of salt (14) revealed changes in gene expression supporting the notion that, as in the yeast ortholog Nhx1p (15), AtNHX1 plays a significant role in protein trafficking and protein targeting, probably via the regulation of intra-vesicular acidic pH (16). Topological analyses revealed that, whereas the N terminus of AtNHX1 is facing the cytosol, almost the entire C-terminal hydrophilic region of the protein resides in the vacuolar lumen (17). Moreover, the deletion of the C terminus of AtNHX1 doubled the Na+/K+ selectivity ratio of the antiporter, suggesting a regulatory role of the C terminus of the antiporter (17). The vacuolar localization of the AtNHX1 C terminus, together with the regulation of the antiporter selectivity by its C terminus, implied that regulatory determinants of the antiporter activity could exert their effects from within the vacuole (17).
In the current study, we characterized the interaction of the C terminus of AtNHX1 with A. thaliana calmodulin (CaM)-like protein 15 (AtCaM15) and established that it binds to the antiporter's C terminus from within the vacuole. Our results also show that the binding of AtCaM15 to AtNHX1 is Ca2+- and pH-dependent and that AtCaM15 modifies the cation selectivity of the antiporter.
Materials and Methods
Materials. All restriction enzymes and DNA polymerases were purchased from New England Biolabs (Beverly, MA). Most chemicals were purchased from Sigma-Aldrich, unless otherwise specified. Cellulase and pectolyase were purchased from Kyowa Chemical Products (Osaka, Japan).
Two-Hybrid Method. Interaction trap screening in yeast was performed essentially as described by Finley and Brent (18). The DNA fragment corresponding to the AtNHX1 C-terminal hydrophilic region (120 aa) was amplified by PCR and cloned into the EcoRI and XhoI sites of the pEG202 vector (18) (designated pEG202-X1CT) and used as the bait construct. pEG202-X1CT was transformed into the Saccharomyces cerevisiae EGY48 strain (MATα trp1 his3 ura3 lexAops::LEU2) along with the pSH18-34 and pJG4-5 Arabidopsis cDNA libraries (kindly provided by Hong Zhang, Texas Tech University, Lubbock). Plasmids from positive clones for the LEU2 reporter and LacZ reporter assays were confirmed by PCR analysis followed by HaeIII digestion. Putative interactors were then recovered and sequenced as described (18). BCO1 and BCO2 primers (18), which correspond to the coding sequence of the B42 activation domain and the ADH1 terminator sequence of pJG4-5, respectively, were used for PCR analysis. One of the positive clones contained a cDNA corresponding to a CaM-like protein AtCaM15 (MIPS code At3g03000). Two hybrid assays were also performed with CaM-binding protein (CBP; MIPS code At2g18750) and petunia CaM81 (identical to AtCaM7; MIPS code At3g43810) (both were kindly provided by W. A. Snedden, Queens University, Kingston, ON, Canada). CBP and CaM81 were cloned into pEG202 and pJG4-5 vectors, respectively.
Mapping of the AtCaM15 binding site of C-terminal AtNHX1 was also performed by two-hybrid assays with a series of deletion constructs of the AtNHX1 C terminus. Deletions of 30 bp of DNA each, constructed in a 3′-to-5′ direction, were generated by PCR amplification and cloned into pEG202 as described above.
Protease Protection Assays. N terminus and C terminus 3xFLAG-tagged AtCaM15 constructs were generated by a fusion PCR method (17) and cloned into the SpeI and PstI site of p425GPD and p425CYC1 plasmids (American Type Culture Collection). The resulting constructs were transformed into the yeast strain W303-1B (MATα ura3-1 leu2-3,112 his3-11,15 trp1-1 ade2-1 can1-100) independently. Yeast vacuoles were isolated as reported earlier (17). Vacuoles (100 μg of protein) were incubated with 10 μg/ml proteinase K in 10 mM Tris/Mes, pH 6.9, in the presence or absence of 1% Triton X-100 for 30 min at room temperature. Proteolysis was terminated by the addition of PMSF (20 mM final concentration). Proteins were resolved with 12% SDS/PAGE and analyzed by Western blot analyses using a FLAG M2 antibody (Sigma-Aldrich) and anti-mouse IgG antibody horseradish peroxidase conjugate (Molecular Probes). The blots were visualized by enhanced chemioluminiscence (ECL) plus Western blotting detection reagents (Amersham Biosciences).
Arabidopsis Leaf Protoplast Transient Transformation. Approximately 10-20 leaves from ≈3-week-old Arabidopsis plants were placed in cold (4°C) wash buffer (0.5 M mannitol/10 mM Mes-KOH/0.2 mM CaCl2/2 mM DTT, pH 5.5) and cut into small (≈1-5 mm2) pieces using a clean, single-edged razor blade. The leaf pieces were gently pressed against the bottom of the Petri dish to remove excess liquid and transferred to digestion buffer (0.5 M mannitol/25 mM Mes-KOH/1 mM CaCl2/2 mM DTT/0.5% BSA/1% cellulase/0.05% pectolyase, pH 5.5) for ≈2 h with gentle shaking at 60 rpm in an Orbit shaker model 3520 (Labline, Melrose Park, IL). The protoplasts were diluted in approximately three volumes of cold wash buffer before filtering through Miracloth (CN Biosciences, La Jolla, CA).
Protoplast transformation was performed according to Sheen and colleagues (19). After centrifugation for 2 min at 100 × g, protoplasts were resuspended in cold W5 solution (154 mM NaCl/125 mM CaCl2/5 mM KCl/2 mM Mes, pH 5.7) and spun again at 100 × g before resuspension in 3 ml of W5. After incubation for 30 min on ice, the protoplasts were collected by centrifugation at 100 × g, and resuspended in 2 ml MMg solution (0.4 M mannitol/15 mM MgCl2/4 mM Mes, pH 5.7). Protoplasts (≈40,000) were used for single transformation with 20 μg of plasmid DNA. An equal volume of polyethylene glycol/Ca solution (40% PEG 4000/200 mM mannitol/100 mM CaCl2) was mixed with the protoplast/DNA solution and incubated at room temperature for 30 min. The transformed protoplasts were diluted with two volumes of W5 solution and collected by centrifugation at 100 × g for 2 min. The supernatant was carefully removed, and the protoplast were resuspended in 800 μl of W5 solution and incubated for ≈24 h in the dark at room temperature. Thirty minutes before visualization, protoplasts were treated with 20 μM FM4-64 to visualize membranes (20). A Leica DMR series fluorescent microscope equipped with a Chroma 86013 filter set (Chroma Technology, Rockingham, VT) and CoolSNAP-HQ (Roper Scientific, Tucson, AZ) was used to visualize the transformed protoplasts. EGFP was visualized by using filters S484/15x and S517/30m, FM4-64 was visualized with S555/25x and S605/40m. All images were taken at ×200 magnification. Images were pseudocolored and merged with metamorph software (Universal Imaging, Downingtown, PA).
Coimmunoprecipitation. AtNHX1 was tagged with a 3xHA tag at the C terminus in the p426TEF vector (pHpl-13) (17), and AtCaM15 and CaM81 (AtCaM7) were tagged with a 3xFLAG tag at the C terminus in p425GPD. The 3xHA tagged C-terminally truncated AtNHX1 (440 aa from N terminus) was generated by fusion PCR (17) and cloned into EcoRI/SalI site of p426TEF. The resulting plasmids were introduced into the yeast strain TY002 (MATα pep4-3 leu2 trp1 ura3-52 prb1-1122 nhx1Δ::TRP1 ena1-5Δ::KanMX6). Exponentially grown cells were harvested, washed twice with distilled and deionized H2O, and homogenized by vortex with 200 μg of glass beads (Biospec Products, Bartlesville, OK) in solubilization buffer (10 mM Tris/5 mM Mes/5 mM acetic acid/2 mM DTT/1% C12E9, pH 5.5, 6.5, or 7.5) with 5 μg/ml leupeptine, 5 μg/ml pepstasin A, and 1 mM PMSF. Lysates (corresponding to 500 μg of protein) were incubated with rabbit anti-FLAG antibody (Sigma) for 1 h, followed by incubation with 35 μl of 40% preequilibrated protein A Sepharose (Sigma) in solubilization buffer for 1 h at 4°C with gentle rocking. Immunocomplexes were precipitated by centrifugation at 5,000 × g and washed five times with solubilization buffer. The proteins were resolved by SDS/PAGE followed by Western blot analyses using a mouse anti-HA monoclonal antibody (Covance, Richmond, CA) and FLAG M2 antibody (primary) with anti-mouse IgG antibody horseradish peroxidase conjugate. CaCl2 (10 mM) and Na-EGTA (5 mM), pH 7.5, were included throughout the protein extraction and wash procedures where indicated.
Transport Assays. The AtNHX1 full-length ORF cDNA was cloned into pYPGE15 as described in ref. 17. The AtCaM15 ORF was amplified by PCR and cloned into the BamHI and SalI sites of p425GPD vector. pYPGE15, p425GPD, pYPGE15-AtNHX1, and p425GPD-AtCaM15 plasmids were introduced into yeast strain TY001 (17) to generate strains harboring pYPGE15/p426GPD, pYPGE15/p425GPD-AtCaM15, pYPGE15-AtNHX1/p426GPD, and pYPGE-AtNHX1/p425GPD-AtCaM15.
Vacuole isolation and transport assays were performed as described in ref. 17. Rates of transport of vacuoles isolated from the yeast strains harboring AtNHX1 and p426GPD or AtNHX1 and AtCaM15 were subtracted by the rates of transport of vacuoles originated from the yeast strains harboring pYPGE15 and p425GPD or pYPGE15 and p425GPD-AtCaM15, respectively. There were no significant differences between the transport kinetics from vacuoles isolated from the TY001 strain and vacuoles from the TY001 strain expressing AtCaM15. Curves were fitted to the mean values of rates at each concentration and measured with kaleidegraph software (Synergy, Reading, PA).
Results
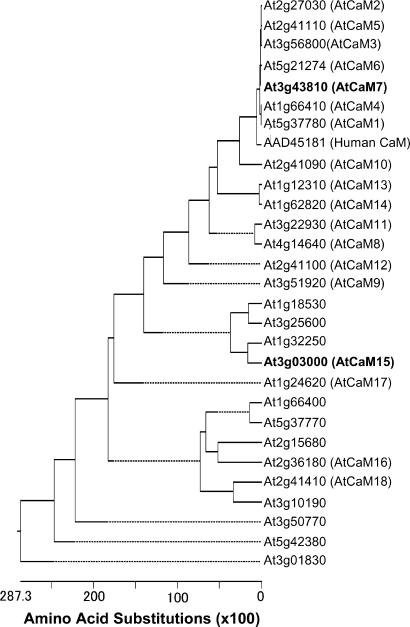
The AtNHX1 C Terminus Interacts with AtCaM15 in Yeast. To identify regulatory proteins affecting AtNHX1 function, we performed a yeast two-hybrid screen, with the C terminus of AtNHX1 as bait. The C-terminal 120 amino acids of AtNHX1 were cloned into the plasmid pEG202 (18). An Arabidopsis seedlings cDNA library was cloned into pJG4-5 to encode the “prey” proteins. A total of 4 × 105 colonies were screened on Gal/Raffinose His-Ura-Trp-Leu- selection media. One thousand colonies that induced transcription of the LexAops::LacZ promoter were selected for further classification by PCR and restriction enzyme digestion analysis. Based on restriction enzyme profiles, cDNAs were chosen for DNA sequencing. Among the positive clones identified, CaM-like protein, matching the predicted sequence of AtCaM15 (At3g03000), was chosen for further analysis. CaM are small Ca2+-binding proteins that can transduce secondary messenger signals into a variety of cellular responses in a wide range of organisms, including mammals, fungi, and plants (21, 22). Plants possess a relatively large family of CaM-like proteins compared with other organisms, and 28 CaM-like isoforms have been annotated in the Arabidopsis genome (Fig. 1). AtCaM15, with belongs to a subgroup of CaM somewhat divergent from the more conserved CaM (e.g., AtCaM1-AtCaM7) (Fig. 1), was the only CaM identified from the initial two-hybrid screening.
Fig. 1.
Phylogenetic analysis of AtCaM. Protein sequences were aligned and a phylogenetic tree was generated by dnastar (DNASTAR, Madison, WI) with clustalw. AtCaM were annotated by following the methods of R. E. Zielinski (personal communication). AtCaM15 and AtCaM7 are indicated by bold letters.
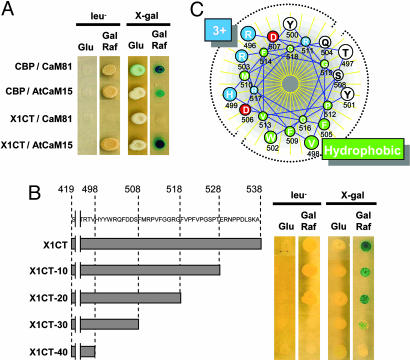
To test the apparent specificity of the interaction between the AtNHX1 C terminus and AtCaM15, a yeast two-hybrid assay was used. For this assay, AtCaM15 or petunia CaM81, a protein with conserved CaM motifs and identical with Arabidopsis AtCaM7 (At3g43810), was used as the prey along with the AtNHX1 C terminus or a known Arabidopsis CBP (At2g18750) (22, 23) as the bait. As expected, CaM81 and CBP interacted with each other, allowing the yeast cells expressing both proteins to grow in the absence of leucine and to develop a blue color in the presence of X-Gal (Fig. 2A). AtCaM15 and CBP also displayed interaction, with their combined expression activating the Leu2 and LacZ reporter genes (Fig. 2 A). These results suggested that AtCaM15 could interact with known CaM-binding targets. However, although AtCaM15 interacted with the AtNHX1 C terminus, CaM81 did not (Fig. 2 A), suggesting a unique structural specificity in the AtCaM15-AtNHX1 interaction or recognition. CaM have the ability to activate a large number of target proteins that share very little amino acid sequence similarity in their binding sites, and the CaM-binding domains of a target protein must be empirically determined (22, 24). Known CaM targets are composed of a stretch of 17-25 aa having a positively charged amphiphilic region and a tendency to form an α-helical structure upon binding to CaM (22, 24). To characterize the AtCaM15-binding region within the AtNHX1 C terminus, serial deletions of 10 aa were constructed from the C terminus of AtNHX1, and the interaction of the different truncated proteins with AtCaM15 was analyzed by a two-hybrid assay (Fig. 2B). Although the deletion of 20 aa (residues 538-519) did not affect the interaction between AtNHX1 and AtCaM15, the deletion of 30 aa abolished the Leu2 reporter gene and reduced the LacZ reporter activity. The deletion of 40 aa abolished the activation of Leu2 and LacZ, indicating that the AtCaM15 binding site corresponds to the amino acids at positions Val-498 to Gly-518 within the AtNHX1 C terminus (Fig. 2B). Furthermore, the stretch of AtNHX1 amino acids from Arg-496 to Gly-518 has the potential to form a positively charged amphiphilic α-helix characteristic of a CaM-binding domain that could correspond to the AtCaM15 target site (Fig. 2C).
Fig. 2.
Interaction between AtNHX1 and AtCaM15. (A) Two-hybrid screen. CBP or AtNHX1 C terminus were introduced into the yeast strain EGY48 strain harboring pSH18-34 with either CaM81 or AtCaM15. (B) Search for AtCaM15-binding domain. A series of deletion constructs of AtNHX1 C terminus were introduced into EGY48 harboring AtCaM15. (C) Helical-wheel model of 496- to 518-aa region of AtNHX1. Charged residues are colored red (acidic) and blue (basic), respectively, and hydrophobic residues are colored green.
The Interaction Between AtCaM15 and AtNHX1 C Terminus Is Ca2+- and pH-Dependent. The interaction between AtCaM15 and AtNHX1 was also confirmed by coimmunoprecipitation (pull-down) assays. The 3xHA-tagged AtNHX1 (AtNHX1::3xHA) and 3xFLAG-tagged AtCaM15 (AtCaM15::3xFLAG) and 3xFLAG-tagged CaM81 (CaM81::3xFLAG) were coexpressed in yeast cells, and whole-cell lysates were incubated with anti-FLAG antibody, followed by incubation with protein A-Sepharose to pull down the IgG-AtCaM15::3xFLAG protein complexes. The expression levels of tagged proteins were similar among all strains used in this experiment (Fig. 3 A-C). As shown in Fig. 3, AtNHX1::3xHA-tagged was successfully pulled down only in the presence of AtCaM15 and Ca2+, whereas CaM81::3xFLAG could not pull down AtNHX1::3xHA even in the presence of Ca2+, and the C-terminally truncated AtNHX1 did not show any interaction with AtCaM15 (Fig. 3D), confirming the results obtained in the two-hybrid assays. The interaction appeared to be Ca2+-dependent. The interaction between AtNHX1::3xHA and AtCaM15::3xFLAG was also observed with Ca2+ concentrations ranging from 1 to 10 mM (data not shown). No interaction was detected in the absence of Ca2+ and the presence of EGTA. Interestingly, the recognition between AtNHX1 and AtCaM15 was also pH-dependent because the amount of AtNHX1 pulled down increased when the pH of the reaction decreased (Fig. 3 B and C). Whereas very little interaction was seen when the coimmunoprecipitation was performed at pH 7.5, the amount of AtNHX1 pulled down by AtCaM15 increased when the pH of the reaction was lowered to 6.5 and increased even further at pH 5.5. Similar results were obtained when N-terminal AtCaM15::3xFLAG or CaM81::3xFLAG were used in the pull-down reactions (data not shown). The recognition of the FLAG epitope by the FLAG antibody was not affected by the pH of the incubation buffer, because the amount of immunoprecipitated CaM did not change with changes in the pH of the reaction (Fig. 3). Moreover, the direct interaction between AtCaM15 and the AtNHX1 C terminus, and the pH dependency and Ca2+ dependency of the interaction were further confirmed by an in vitro pull-down assay using affinity-purified GST-AtCaM15 and His6-S-tagged AtNHX1 C terminus (Supporting Materials and Methods and Fig. 6, which are published as supporting information on the PNAS web site).
Fig. 3.
Coimmunoprecipitation of AtNHX1 and AtCaM15. (A-C) AtNHX1::3xHA and AtCaM15::3xFLAG or CaM81::3xFLAG were coexpressed in the yeast strain TY002. Total lysates were incubated with rabbit anti-FLAG antibody followed by protein A-Sepharose beads, then washed and subjected to Western blot analysis. Antibody incubations were carried out at pH 7.5 (A), 6.5 (B), and 5.5 (C), respectively. (D) Coimmunoprecipitation of C-terminally truncated AtNHX1 (ΔC) and AtCaM15 at pH 6.5. Arrowheads shown in anti-HA Western blots indicate background signals due to protein A and rabbit IgG. Two or 0.5 μg of protein were loaded to each blot to serve as untreated control for anti-HA and anti-FLAG Western blot, respectively. Data are representative of five independent experiments.
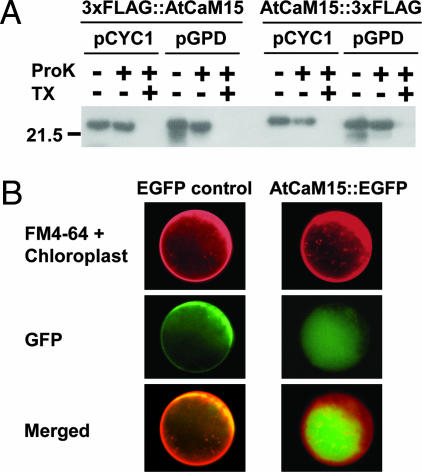
AtCaM15 Is Localized in the Vacuole in Yeast and in Planta. Because we have shown previously that the C terminus of AtNHX1 is localized in the vacuolar lumen (17), a protease protection assay (17) (Fig. 4A) was used to test whether AtCaM15 is also localized there. These experiments were based on the following rationale: If AtCaM15 is facing the outside of the vacuole (i.e., the cytosol), then AtCaM15 will not be detected when the vacuoles are treated with proteinase K in the presence or absence of Triton X-100 (added to permeabilize the vacuolar membrane). In contrast, if AtCaM15 is in the vacuolar lumen, AtCaM15 will be only detected when the vacuoles are treated with proteinase K in the absence of Triton X-100. Two types of promoters, GPD (strong promoter) and CYC1 (weak promoter) were used to test whether the expression level of the recombinant AtCaM15 could affect its cellular localization. In addition, because there is no apparent vacuolar sorting signal sequence in AtCaM15, both N- and C-terminal ends of AtCaM15 were tagged by 3xFLAG.
Fig. 4.
Vacuolar localization of AtCaM15. (A) AtCaM15 fused with 3xFLAG tag at the N terminus (3xFLAG::AtCaM15) or C terminus AtCaM15::3xFLAG) was introduced into yeast W303 strain and the isolated vacuoles were incubated with proteinase K (ProK) in the presence or absence of Triton X-100 (TX) as indicated. The data are representative of three independent experiments. (B) Arabidopsis leaf protoplasts were transiently transformed with EGFP or AtCaM15::EGFP plasmids. Transformed protoplasts were stained with FM4-64 to visualize membranes, and images were acquired as described in Materials and Methods.
Both C terminus and N terminus FLAG-tagged AtCaM15 constructs were expressed in yeast, and purified intact vacuoles were isolated from each strain expressing the different constructs. The vacuoles were incubated with proteinase K to digest proteins exposed to the vacuolar surface (17). Vacuolar proteins were then resolved by SDS/PAGE, and the 3xFLAG tags were detected by Western blotting using anti-FLAG tag antibody (Fig. 4A). Both C terminus and N terminus FLAG-tagged AtCaM15 were detected after protease treatment in the absence of Triton X-100 but not in the presence of Triton X-100, indicating that AtCaM15 was present in the vacuolar lumen. The lower molecular mass bands in the control lanes of the GPD promoter-driven AtCaM15 are likely partial degradation products due to leakage during the vacuolar preparation as they disappeared upon incubation with proteinase K. It should be noted that the lack of detection of AtCaM15 in the presence of Triton X-100 and proteinase K was not due to an increased susceptibility of AtCaM15 to proteinase K in the presence of detergent because recombinant AtCaM15 was cleaved by proteinase K in the absence of Triton X-100 (data not shown).
To confirm the localization of AtCaM15 in planta, Arabidopsis leaf protoplasts were transformed with constructs bearing EGFP-tagged AtCaM15 (AtCaM15::EGFP) or EGFP alone. The fluorescence of FM4-64 was used to visualize membranes (Fig. 4B). In cells expressing GFP alone, the GFP fluorescence was seen mainly in the cytosol, whereas, in cells expressing AtCaM15::EGFP, the fluorescence was seen in the vacuole. Some fluorescence was also seen in cytosol and nucleus (data not shown), but no fluorescence was detected in other intracellular structures, such as the Golgi or endoplasmic reticulum, because the EGFP fluorescence did not colocalize with FM4-64 staining (Fig. 4B). Vacuolar localization of AtCaM15::GFP was also confirmed by confocal microscopy (Supporting Materials and Methods; see also Fig. 7, which is published as supporting information on the PNAS web site).
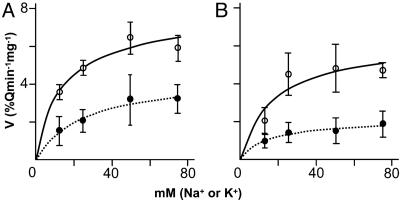
AtCaM15 Modifies the Ion Selectivity of AtNHX1. Recently, it has been shown that the C terminus of AtNHX1 modulates the antiporter's cation selectivity and that the deletion of the C terminus resulted in an increase in the rate of Na+/H+ exchange relative to the rate of K+/H+ exchange (17). The interaction of AtNHX1 C terminus and AtCaM15 led us to investigate whether the binding of AtCaM15 to AtNHX1 C terminus could affect the antiporter's activity. H+-coupled cation transport activity was measured by using purified vacuoles from yeast (17) expressing AtNHX1 alone or coexpressing AtNHX1 and AtCaM15 by monitoring vacuolar pH-dependent quenching of acridine orange fluorescence. The vacuolar H+-ATPase was activated by the addition of Mg2+ and ATP, and the ATPase activity was stopped by the addition of bafilomycin (17). Once a steady-state acidic pH inside was attained, Na+ or K+ was added and the cation/H+ exchange activity was measured (17). Both type of vacuoles displayed similar rates of H+-ATPase activity and a similar steady-state acidic-inside pH gradient (results not shown) (17). In all cases, the antiporter activity displayed Michaelis-Menten saturation kinetics (Fig. 5). The coexpression of AtCaM15 and AtNHX1 significantly lowered the Vmax of the Na+/H+ exchange activity, whereas the Vmax of the K+/H+ exchange activity was not significantly altered (Fig. 5 and Table 1), leading to a 46% decrease in the Na+/K+ selectivity ratio. The Na+/K+ selectivity ratio of vacuoles isolated from yeast coexpressing AtCaM15 and the C-terminally truncated AtNHX1 [Vmax (Na+)/Vmax (K+) = 1.59] (data not shown) was similar to the Na+/K+ selectivity ratio of vacuoles isolated from vacuoles expressing the C-terminally truncated AtNHX1 alone [Vmax (Na+)/Vmax (K+ = 1.48] (17), indicating that the changes in Na+/K+ selectivity-induced AtCaM15 were due to its binding to the AtNHX1 C terminus, further supporting the pull-down results (Fig. 3).
Fig. 5.
Effect of AtCaM15 on Na+/H+ (•) or K+/H+ antiporter (○) activity. Shown are vacuoles isolated from yeast harboring pYPGE15-AtNHX1 and p425GPD empty vector (A) and vacuoles isolated from yeast strains harboring pYPGE15-AtNHX1 and p425GPD-AtCaM15 (B). Each point is the mean ± SD (n > 5).
Table 1. Kinetic property of the vacuoles harboring AtNHX1 with or without AtCaM15.
| Parameters | AtNHX1 | AtNHX1 + AtCaM15 |
|---|---|---|
| Na+, Vmax | 4.50 ± 0.38 | 2.05 ± 0.17 |
| K+, Vmax | 7.42 ± 0.6 | 6.13 ± 1.2 |
| Na+/K+, Vmax | 0.61 | 0.33 |
| Na+, Km | 24.3 ± 5.49 | 15.4 ± 4.19 |
| K+, Km | 12.2 ± 3.78 | 16.8 ± 10.3 |
Apparent Vmax (%Q per min per mg of protein) and Km (mM) values were calculated from data in Fig. 5. Values are the mean ± SD (n > 5).
Discussion
Plant vacuolar Na+/H+ antiporters have been shown to play important roles in intracellular trafficking (14), vacuolar pH regulation (13), and the sequestration of Na+ ions into the vacuole (11, 12). Recently, it has been shown that the deletion of the C terminus of the A. thaliana vacuolar Na+/H+ antiporter AtNHX1 increased the relative Na+/H+ exchange activity of the antiporter, suggesting that its C terminus plays a role in modulating the ion selectivity of the antiporter (17). To identify proteins interacting with the AtNHX1 C terminus, we conducted a two-hybrid assay with the C terminus as bait. The assay identified a number of putative interactors (E.B., unpublished results), among them was the CaM-like protein AtCaM15. Because of the significant role of CaM in the transduction of secondary messenger signals into a variety of cellular responses (21, 22) and the role of CaM in regulating the activity of the mammalian Na+/H+ exchanger NHE1 (25, 26), we proceeded to characterize the AtNHX1/AtCaM15 interaction. Plants possess a relatively large family of CaM-like proteins (28 members in Arabidopsis), as compared with other organisms, and AtCaM15 was the only CaM-like protein identified in our screen. The AtNHX1 C terminus did not interact with CaM81, a protein identical with AtCaM7, which is a representative of the more conserved plant CaM, suggesting that the AtCaM15/AtNHX1 interaction is specific. By using amino acid serial deletions of the AtNHX1 C terminus, we identified a stretch of amino acids (Arg-496 to Gly-518) that has the ability to form an amphiphilic α-helical structure having positively charged and hydrophobic halves, a common structural feature of CaM target peptides, suggesting that this region of AtNHX1 could correspond to the binding site of the AtCaM15 binding site to the AtNHX1 C terminus. The AtNHX1/AtCaM15 interaction was also confirmed by coimmunoprecipitation. By using a nhx1-null mutant yeast strain coexpressing AtNHX1 and AtCaM15 or AtNHX1 and CaM81, we showed that AtNHX1 was successfully immunoprecipitated by the IgG-AtCaM15 protein complex but not by the IgG-CaM81 protein complex, indicating a specific interaction between AtCaM15 and the AtNHX1 C terminus. Topological studies have demonstrated that the entire hydrophilic C terminus of AtNHX1 resides in the vacuolar lumen (17). The protease protection assays indicated that AtCaM15 was also present in the vacuoles of yeast strains expressing AtNHX1 and AtCaM15 (Fig. 3). Moreover, Arabidopsis leaf protoplasts expressing AtCaM15::EGFP displayed vacuolar-localized GFP fluorescence (Fig. 3). Although CaM have been shown to be present in the nucleus, the cytosol (21, 22), and peroxisomes (27), we report the presence of a CaM in a vacuolar compartment. AtCaM15::EGFP fluorescence was detected in the central vacuole. Our assays did not detect fluorescence in smaller vesicles, and the presence of AtCaM15 in smaller lytic or storage vacuoles (28) cannot be ruled out. Nevertheless, although speculative, the absence of a known sorting signal sequence in AtCaM15, together with its localization in the vacuole but not in the endoplasmic reticulum or Golgi, might suggest that AtCaM15 could reach the vacuole by means of a cytoplasm-to-vacuole targeting (Cvt) pathway similar to S. cerevisiae aminopeptidase I (29) or α-mannosidase (30).
The plant vacuole serves as a primary pool of Ca2+, and total vacuolar Ca2+ concentrations have been estimated to be in the range of 1-10 mM (31). Most of the vacuolar Ca2+ ions are bound to organic acids and several types of low-affinity Ca2+-binding proteins (32, 33). Although the concentration of free Ca2+ in higher plant vacuoles is still unknown, the concentration of vacuolar free Ca2+ has been estimated to be in the micromolar range (34, 35) in algae cells and between 1-2 mM in higher plant cells (36). The ability of AtCaM15 to bind Ca2+ in the micromolar range would suggest that AtCaM15 could play a role as a vacuolar high-affinity Ca2+-binding protein and that, at the basal intravacuolar Ca2+ concentrations, Ca2+ would be constitutively bound to AtCaM15 in the cells.
It has been previously shown that the AtNHX1 C terminus plays a role in regulating the antiporter's cation selectivity and that the deletion of the C terminus resulted in a 2-fold increase in the Na+/K+ selectivity ratio (17). In contrast, CaM have been shown to play roles in the regulation of ion transporters. The binding of Ca2+/CaM releases the autoinhibitory domain of plant and animal type IIB Ca2+-ATPases (37, 38). There is also increasing evidence demonstrating the modulation of several animal and plant ion channels by CaM binding (21, 22). The mammalian NHE1 Na+/H+ exchanger is also regulated by interaction with regulatory proteins (25), among them CaM. The cytosolic C-terminal tail of NHE1 contains two domains that are capable of binding CaM with high affinity (CaM-A, Kd ≈ 20 nM, and CaM-B, Kd ≈ 350 nM) (26), with the unoccupied CaM-binding site exerting an autoinhibitory effect that is relieved upon Ca2+/CaM binding (36). The coexpression of AtCaM15 and AtNHX1 in yeast resulted in a significant decrease in the Vmax of the Na+/H+ exchange, resulting in a decrease of the Na+/K+ selectivity ratio of the antiporter. These differences cannot be attributed to differences in the leakiness of the vacuoles, because the steady-state pH gradient attained (after the inhibition of the vacuolar H+-ATPase by bafilomycin) was similar in vacuoles containing AtNHX1 alone or AtNHX1 and AtCaM15. Thus, the difference in cation/H+ exchange activity was due to the interaction between AtCaM15 and the C terminus of AtNHX1. These results, together with previous findings showing changes in AtNHX1 cation selectivity by the C terminus truncation (17), supports the notion of AtCaM15 playing a regulatory role modulating the antiporter's cation selectivity ratio within the vacuole by interacting with the AtNHX1 C terminus.
It is noteworthy that the interaction between the AtNHX1 C terminus and AtCaM15 was Ca2+- and pH-dependent, and the binding of Ca2+-AtCaM15 to the AtNHX1 C terminus decreased with increased vacuolar pH. Our results would suggest that, at physiological conditions (i.e., high, vacuolar, free Ca2+ concentrations and acidic vacuolar pH), AtCaM15 would be bound to the AtNHX1 C terminus and AtNHX1 would display a relatively low Na+/H+ exchange with respect to K+/H+ exchange.
It has been shown previously that vacuoles isolated from transgenic A. thaliana leaves overexpressing AtNHX1 displayed increased AtNHX1 Na+/H+ exchange activity (11) and that the increase in activity was higher than the relative increase in AtNHX1 protein abundance. Apse et al. (11) suggested that, in wild-type plants under normal growth conditions, AtNHX1 function was repressed and that the overexpression of AtNHX1 helped to overcome the endogenous repression mechanism. It is possible to speculate that AtCaM15, bound to AtNHX1, repressed the Na+/H+ exchange activity in the wild-type plants and that the increase in AtNHX1 protein provided the increased Na+/H+ exchange activity seen in the transgenic plants. Our results might also suggest a relationship between transient changes of vacuolar pH and sequestration of Na+ ions into the vacuole. It has been shown that, during the exposure of salt-sensitive plants to salt stress, the pH of the vacuole rises (39, 40). An increased vacuolar pH would release AtCaM15 from the C terminus of AtNHX1, thereby increasing Na+/H+ exchange activity and, thus, favoring an increased vacuolar Na+ accumulation.
In summary, the findings reported here are evidence of a CaM isoform (AtCaM15) localized in the plant vacuolar compartment with a function of binding and modifying the activity of a tonoplast transporter (AtNHX1) from within the vacuole. The pH-dependence of the interaction between AtCaM15 and AtNHX1 suggests the presence of pH-dependent signaling components in the vacuole.
Supplementary Material
Acknowledgments
We thank Dr. Maris Apse for helpful comments. This work was supported by National Science Foundation Grants IBN-0110622 and MCB-0343279 and by Arcadia Biosciences.
Author contributions: T.Y., G.S.A., J.B.S., and E.B. designed research, performed research, contributed new reagants/analytic tools, analyzed data, and wrote the paper.
Abbreviations: AtNHX, Arabidopsis thaliana Na+/H+ exchanger; CaM, calmodulin; AtCaM15, A. thaliana CaM-like protein 15; CBP, CaM-binding protein.
References
- 1.Orlowski, J. & Grinstein, S. (1997) J. Biol. Chem. 272, 22373-22376. [DOI] [PubMed] [Google Scholar]
- 2.Padan, E., Venturi, M., Gerchman, Y. & Dover, N. (2001) Biochem. Biophys. Acta 1505, 144-157. [DOI] [PubMed] [Google Scholar]
- 3.Blumwald, E., Aharon, G. S. & Apse, P. (2000) Biochim. Biophys. Acta 1465, 140-151. [DOI] [PubMed] [Google Scholar]
- 4.Blumwald, E. & Poole, R. J. (1985) Plant Physiol. 78, 163-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumwald, E. (1987) Physiol. Plant. 69, 731-734. [Google Scholar]
- 6.Mäser, P., Thomine, S., Schroeder, J. I., Ward, J. M., Hirschi, K., Sze, H., Talke, I. N., Amtmann, A., Maathuis, F. J. M., Sanders, D., et al. (2001) Plant Physiol. 126, 1646-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ward, J. M. (2001) Bioinformatics 17, 560-563. [DOI] [PubMed] [Google Scholar]
- 8.Aharon, G. S., Apse, M. P., Duan, S. L., Hua, X. J. & Blumwald, E. (2003) Plant Soil 253, 245-256. [Google Scholar]
- 9.Yokoi, S., Quintero, F. J., Cubero, B., Ruiz, M. T., Bressan, R. A., Hasegawa, P. M. & Pardo, J. M. (2002) Plant J. 30, 529-539. [DOI] [PubMed] [Google Scholar]
- 10.Nehrke, K. & Melvin, J. E. (2002) J. Biol. Chem., 277, 29036-29044. [DOI] [PubMed] [Google Scholar]
- 11.Apse, M. P., Aharon, G. S., Snedden, W. A. & Blumwald, E. (1999) Science 285, 1256-1258. [DOI] [PubMed] [Google Scholar]
- 12.Zhang, H.-X. & Blumwald, E. (2001) Nat. Biotechnol. 19, 765-768. [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi, T., Fukada-Tanaka, S., Inagaki, Y., Saito, N., Yonekura-Sakakibara, K., Tanaka, Y., Kusumi, T. & Iida, S. (2001) Plant Cell Physiol. 42, 451-461. [DOI] [PubMed] [Google Scholar]
- 14.Sottosanto, J. B., Gelli, A. & Blumwald, E. (2004) Plant J. 40, 752-771. [DOI] [PubMed] [Google Scholar]
- 15.Ali, R., Brett, C. L., Mukherjee, S. & Rao, R. (2004) J. Biol. Chem. 279, 4498-4506. [DOI] [PubMed] [Google Scholar]
- 16.Bowers, K., Levi, B. P., Patel, F. I. & Stevens, T. H. (2000) Mol. Biol. Cell 11, 4277-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaguchi, T., Apse, M. P., Shi, H. & Blumwald, E. (2003) Proc. Natl. Acad. Sci. USA 100, 12510-12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finley, R. L. & Brent, R. (1995) in DNA Cloning: A Practical Approach, Expression Systems, eds. Glover, D. & Hames, B. D. (Oxford Univ. Press, Oxford), 2nd Ed., pp. 170-203.
- 19.Kovtun, Y., Chiu, W. L., Tena, G. & Sheen, J. (2000) Proc. Natl. Acad. Sci. USA 97, 2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolte, S., Talbot, C., Boutte, Y., Catrice, O., Read, N. D. & Satiat-Jeunemaitre, B. S. (2004) J. Microsc. (Oxford) 214, 159-173. [DOI] [PubMed] [Google Scholar]
- 21.Zielinsky, R. E. (1998) Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 697-725. [DOI] [PubMed] [Google Scholar]
- 22.Snedden, W. A. & Fromm, H. (2001) New Phytol. 151, 35-66. [DOI] [PubMed] [Google Scholar]
- 23.Reddy, V. S., Ali, G. S. & Reddy, A. S. N. (2002) J. Biol. Chem. 277, 9840-9852. [DOI] [PubMed] [Google Scholar]
- 24.O'Neil, K. T. & DeGrado, W. F. (1990) Trends Biochem. Sci. 15, 59-64. [DOI] [PubMed] [Google Scholar]
- 25.Slepkov, E. & Fliegel, L. (2002) Biochem. Cell Biol. 80, 499-508. [DOI] [PubMed] [Google Scholar]
- 26.Bertrand, B., Wakabayashi, S., Ikeda, T., Pouyssegur, J. & Shigekawa, M. (1994) J. Biol. Chem. 269, 13703-13709. [PubMed] [Google Scholar]
- 27.Yang, T. & Poovaiah, B. W. (2002) Proc. Natl. Acad. Sci. USA 99, 4097-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Surpin, M. & Raikhel, N. (2004) Nature Rev. Mol. Cell. Biol. 5, 100-110. [DOI] [PubMed] [Google Scholar]
- 29.Klionsky, D. J., Cueva, R. & Yaver, D. S. (1992) J. Cell Biol. 119, 287-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshihisa, T. & Anraku, Y. (1990) J. Biol. Chem. 265, 22418-22425. [PubMed] [Google Scholar]
- 31.Ueoka-Nakanishi, H., Nakanishi, Y., Tanaka, Y. & Maeshima, M. (1999) Eur. J. Biochem. 262, 417-425. [DOI] [PubMed] [Google Scholar]
- 32.Yuasa, K. & Maeshima, M. (2000) Plant Physiol. 124, 1069-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heyen, B. J., Alsheikh, M. K., Smith, E. A., Torvik, C. F., Seals, D. F. & Randall, S. K. (2002) Plant Physiol. 130, 675-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, A. J. & Sanders, D. (1987) Nature 326, 397-400. [Google Scholar]
- 35.Bethmann, B., Thaler, M., Simonis, W. & Schonknecht, G. (1995) Plant Physiol. 109, 1317-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Felle, H. (1988) Planta 176, 248-255. [DOI] [PubMed] [Google Scholar]
- 37.Harper, J. F., Hong, B. M., Hwang, I. D., Guo, H. Q., Stoddard, R., Huang, J. F., Palmgren, M. G. & Sze, H. (1998) J. Biol. Chem. 273, 1099-1106. [DOI] [PubMed] [Google Scholar]
- 38.Carafoli, E. & Brini, M. (2000) Curr. Opin. Chem. Biol. 4, 152-161. [DOI] [PubMed] [Google Scholar]
- 39.Gruwel, M. L. H., Rauw, V. L., Loewen, M. & Abrams, S. R. (2001) Plant Sci. 160, 785-794. [DOI] [PubMed] [Google Scholar]
- 40.Okazaki, Y., Kikuyama, M., Hiramoto, Y. & Iwasaki, N. (1996) Plant Cell Environ. 19, 569-576. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.