Abstract
Evidence for an error-free DNA damage tolerance process in eukaryotes (also called postreplication repair) has existed for more than two decades, but its underlying mechanism, although known to be different from that in prokaryotes, has remained elusive. We have investigated this mechanism in Saccharomyces cerevisiae, in which it is the major component of the RAD6/RAD18 pathway, by transforming an isogenic set of rad1Δ excision-defective strains with plasmids that carry a single thymine-thymine pyrimidine (6-4) pyrimidinone photoadduct in each strand at staggered positions 28 base pairs apart. C-C mismatches placed opposite each of the T-T photoproducts permit unambiguous detection of the events that can lead to the completion of replication: sister-strand recombination or translesion replication on one or the other strand. Despite the severe block to replication that these lesions impose, we find that more than half of the plasmids were fully replicated in a rad1Δ strain and that >90% of them achieved this end by recombination between partially replicated sister strands within the interlesion region. Approximately 60-70% of these events depended on the error-free component of the RAD6/RAD18 pathway, with the remaining events depended on RAD52; these two processes account for almost all of the recombination, which depended neither on DNA polymerase ζ nor on mismatch repair. We conclude that the error-free component of the RAD6/RAD18 pathway completes replication by a mechanism employing recombination between partially replicated sister strands, possibly by means of transient template strand switching or copy choice.
Keywords: postreplication repair, Saccharomyces cerevisiae
DNA damage tolerance processes, also called postreplication repair, are a major part of the repertoire of mechanisms used by organisms to counter the continuous onslaught of genomic damage. Unlike mechanisms that repair the damage, however, damage tolerance processes are concerned with overcoming one of its serious consequences, namely, the ability of such damage to block the progress of the DNA replicases. At least two processes are used to achieve this end: translesion replication, in which specialized DNA polymerases replicate past lesions, often generating mutations but accounting for only a minor fraction of the damage tolerance, and an error-free process that accounts for the major fraction. The existence of the latter process has been known for >30 years from studies in excision repair-deficient strains of the molecular size of DNA synthesized at various times after UV irradiation; although DNA synthesized soon after irradiation is of smaller size than its counterpart from unirradiated cells, indicating the presence of gaps and interruptions, its size corresponds to that of control DNA when isolated from the irradiated cells after a period of incubation, showing that the gaps have been filled and interruptions sealed (1). Although initially discovered in Escherichia coli (1), similar results have also been found in other organisms, including mammalian cells (2-4) and budding yeast, Saccharomyces cerevisiae (5, 6). Even though the phenomena are similar, however, different mechanisms appear to be used in E. coli and the eukaryotes. In E. coli, the gaps in the DNA synthesized initially are filled by a RecA-dependent recombination mechanism (7) that results in the transfer of segments of template DNA into the nascent strand (8, 9), but such a transfer does not appear to occur in eukaryotes (3, 10), and the mechanism they use is unknown.
In budding yeast, DNA damage tolerance depends on the RAD6 pathway (11), and the high sensitivity of rad6 and rad18 mutants to a very broad range of DNA-damaging agents attests to the high frequency with which replication forks stall and to the substantial contribution damage tolerance processes make toward cell survival. Rad6 is an E2 ubiquitin conjugase (12) which, as a heterodimer with Rad18 (13), monoubiquitinates lysine 164 of proliferating cell nuclear antigen (14), a modification that promotes translesion replication (15). The latter process employs DNA polymerase ζ, whose two subunits are encoded by REV3 and REV7 (16, 17), DNA polymerase η (18), encoded by RAD30 (19), and Rev1, a deoxycytidyl transferase (20) that also has another function in translesion replication (21). In contrast, polyubiquitination of lysine 164, in which the Rad5/Ubc13/Mms2 complex conjugates ubiquitin to lysine 63 of ubiquitin itself (22), promotes the error-free DNA damage tolerance process (14). Conversion of nascent DNA from a small to large size during postirradiation incubation is much reduced in rad6, rad18, and rad5 mutants (5, 6, 23), indicating that it depends substantially on the error-free component of the RAD6/RAD18 pathway but is not detectably dependent on DNA polymerase ζ (6). The conversion from small to large molecular size is also partially reduced in rad52 mutants (6), suggesting a minor role for the Rad52 protein in this process.
The nature of the mechanism underlying the RAD6/RAD18-dependent error-free DNA damage tolerance process has proved difficult to investigate in vivo, in part perhaps because genes directly involved in it, as opposed to its regulation, have not been identified. Moreover, if as suggested in ref. 24, the mechanism depends on transient template strand switching of the kind proposed by Higgins and coworkers (25), which entails recombination by informational exchange between partially replicated sister strands, an additional difficulty is the inability of monitoring such an event by conventional genetic methods. We have therefore investigated the mechanism by transforming an isogenic series of excision defective yeast strains with plasmid constructs that carry a single thymine-thymine pyrimidine (6-4) pyrimidinone photoadduct in each strand at staggered positions 28 base pairs (bp) apart. C-C mismatches placed opposite each of the T-T photoproducts permit unambiguous detection of the events that can lead to the completion of replication: sister-strand recombination or translesion replication on one or the other strand. Despite the serious block to replication that these lesions appear to impose, we find that the plasmids are fully replicated with >50% efficiency, and that in >90% of cases, this outcome is achieved by a recombination-dependent mechanism, possibly of the transient template strand switching or copy-choice variety.
Methods
Yeast Strains and Plasmids. The isogenic series of strains used was constructed by deleting RAD18, RAD52, RAD5, REV3, or MSH2 in a rad1Δ derivative of CL1265-7C (MATα arg4-17 his3Δ-1 leu2-3,112 trp1 ura3-52). A RAD6 deletion was also constructed in this background but proved to be unsuitable for experimental use because of its extremely poor viability and the propensity to rapidly acquire srs2 suppressor mutations, which partially suppress the growth defect. Each mutant strain was checked by PCR to verify that they contained the correct genomic alteration, and their phenotype was examined to ensure it was consistent with expectation. The plasmids pYPOG1 and pYPOG2 were derived from pUC19 by modification of the polylinker sequence and insertion of the URA3-2micron ori cassette from pYDV1 (26) into the unique AatI site. Plasmids pYPOG1 and pYPOG2 are identical except with regard to the two base-pairs immediately flanking the PstI and EcoRI restriction sites, which are T:A, G:C and G:C, C:G in pYPOG1, but C:G, A:T and G:C, A:T in pYPOG2. These sequence markers were used to detect interplasmid recombination when strains were simultaneously transformed with both pYPOG1 and pYPOG2 constructs.
Assembly of the Plasmid Insert. The complementary 80- and 72-nucleotide strands that form the duplex sequence inserted into either pYPOG1 or pYPOG2 were assembled individually by ligating together oligonucleotides annealed to complementary scaffold oligomers as described in ref. 27, followed by purification of the required single-stranded species by electrophoresis through a 12% denaturing polyacrylamide gel. The 72-mer strand was assembled from the following oligonucleotides, in this order: 5′-TCCACGGTACCTTAG-3′, 5′-GCAAGTTGGAG-3′, 5′-GGTTACCAGTAGCTCGTACCCTCCCCCTTGCGAGTCCAGCAAGACC-3′. For the 80-mer, they were 5′AATTGGTCTTGCGGACTC-3′; 5′-GCAAGTTGGAG-3′; 5′-GGTACGAGCTACTGGTAACCCTCCCCCTTGCTAAGGTACCGTGGATGCA-3′. A T-T (6-4) photoadduct was placed at the unique T-T site in the 11-mer 5′-GCAAGTTGGAG-3′ as described in ref. 27 and incorporated into both the 72-mer and 80-mer. A complete set of strands were also assembled by using lesion-free 11-mer to act as controls. The duplex insert molecules resulting from the annealing of full-length complementary strands possessed NsiI and MfeI ends, compatible with PstI and EcoRI cohesive ends. All oligonucleotides used to assemble full-length strands were phosphorylated, except for those forming the MfeI terminus. These molecules also possessed a double C-C mismatch opposite either the T-T (6-4) lesion or the lesion-free T-T doublet that acted as sequence markers.
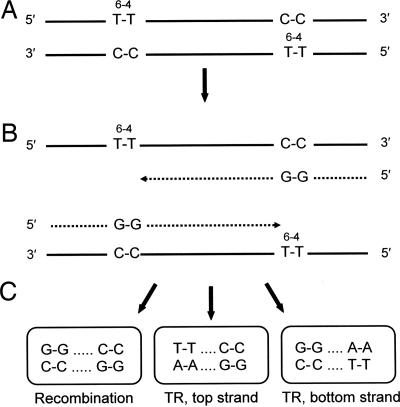
Construction of Lesion-Containing Plasmids, Transformation, and Analysis. The 80-mer/72-mer insert was ligated into the PstI and EcoRI sites of either pYPOG1 or pYPOG2 by using a two-step procedure designed to promote ligation efficiency and maximize production of the desired construct. In the first step, high DNA concentration, excess insert, and the presence of restriction enzymes were used to very efficiently ligate an insert to each end of PstI cut plasmid. In the second step, an insert was cleaved from one arm of the plasmid by EcoRI digestion, and recircularization was promoted by decreasing the DNA concentration >100-fold. For the first step, a 5-fold molar excess of the insert was ligated to a PstI-linearized plasmid DNA at a DNA concentration of ≈600 ng/μl by using 330 units of high concentration T4 ligase/μl and 1 unit/μl each of PstI and NsiI. Under these conditions, an insert was ligated to both ends of the linearized plasmid in the first step with an efficiency that was close to 100%, as shown by the detection of PvuII restriction fragments 209 and 273 bp in length but not those that were 76 bp shorter. After the first-step ligation, the linear plasmid product was digested with EcoRI to generate a plasmid end compatible with the MfeI terminus of the insert and to release a fragment containing one of the inserts. Recircularization of the plasmid was achieved by phosphorylation of the MfeI end of the construct and ligation at a DNA concentration of <5 ng/μl by using 2 units/μl T4 ligase in the presence of 0.08 units/μl of MfeI and EcoRI. The covalently closed circular species was purified by electrophoresis through a 0.7% agarose gel in Tris-acetate-EDTA buffer containing ethidium bromide, run in the dark to prevent DNA damage. To excise the desired covalently closed circular species, all but the outermost lanes of the gel (which were discarded) were entirely shielded from the brief exposure to 300 nm of UV used to visualize the bands and minimize UV damage in the plasmid. Such damage can appreciably reduce transformation frequencies, particularly in rad1Δ rad18Δ rad52Δ strains. Covalently closed circular DNA was isolated from the gel slices with a QIAQuick Gel Extraction Kit (Qiagen, Valencia, CA) by following the manufacturer's protocol but with but with additional column washes and greater elution volume. The purified plasmid vector was quantitated with a PicoGreen dsDNA Quantitation Kit (Molecular Probes, Eugene, OR) and a Turner Quantech Digital Filter Fluorometer (Barnstead/Thermolyne, Dubuque, IA) according to the kit protocols. Yeast strains were made competent as described in ref. 28, and either two or three parallel transformation reactions were used per experiment. In each reaction, 50 μl of competent cells was transformed with 7 ng of purified plasmid construct. After suitable dilution, the transformation mixture was spread over either two or four plates, depending on the number of transformant colonies expected, containing synthetic dextrose medium lacking uracil. From 4 to 11 replicate experiments were carried out per strain, and in all cases, three independently assembled batches of construct were used. Even though the amount of spontaneous damage to the plasmid constructs during assembly was very small, collecting data from independently assembled batches minimized any influence of such damage on inherent transformability and, hence, on the estimates of the fraction of replicated plasmids. This fraction was estimated by normalizing the number of URA3+ colonies arising from transformation with the lesion-containing construct to the number of URA3+ colonies arising from transformation with lesion-free control plasmid. Analyses of the types of DNA damage tolerance mechanism used to complete replication were carried out by DNA sequence determinations of the insert region by using fluorescent-tagged cycle sequencing reactions. Differentiation of events entailing recombination or translesion replication on one or the other strand was made possible by tandem double-cytosine mismatches opposite the T-T (6-4) photoadducts (Fig. 1).
Fig. 1.
Expected replication products and sequence motifs for recombination and translesion replication on one or the other strand. (A) T-T (6-4) photoadducts in each template strand at staggered positions 28 bp apart and opposing C-C mismatch strand markers. (B) Proposed replication intermediates after stalling of replicases at the photoadducts. (C) Sequence motifs indicating recombination or translesion replication (TR) on one or the other template strand. A-A is most frequently inserted opposite the T-T (6-4) photoadduct during TR, but A-C and other insertions are also observed. Solid lines, original templates; dotted lines, replicated strands.
Results
Experimental Design. The goal of this work was to investigate the mechanism underlying the error-free component of the RAD6/RAD18 DNA damage tolerance pathway, which enables the completion of replication in genomes that contain unrepaired damage. To achieve this end, we transformed an isogenic series of excision-repair deficient (rad1Δ) yeast strains with plasmids that carried a specifically located T-T (6-4) photoadduct in each strand at staggered positions 28 bp apart, under conditions in which almost all transformants contained plasmids originating from a single transforming molecule. Estimates of the fraction of fully replicated plasmids were obtained by normalizing the number of URA3+ colonies resulting from transformation with the photoproduct-containing construct to the number obtained from transformation with an equal amount of lesion-free construct; transformant colonies can be established only if plasmid replication is completed. The types of events that were used to overcome the block to fork progression and allow replication to be completed were determined by sequence analysis. C-C mismatches placed opposite the T-T photoadduct enabled unambiguous identification of the DNA strand, or part of a strand, from which replicated plasmids originated and differentiated between recombination events within the 28 bp interlesion region and translesion replication events on either the leading or lagging strands (Fig. 1); these three events were the only ones detected. We chose T-T (6-4) lesions because they cannot be bypassed by DNA polymerase η and are bypassed only very inefficiently by DNA polymerase ζ and, therefore, severely inhibit replication fork progression; <4% of plasmids carrying this lesion within a single-stranded region can be replicated (28). As a consequence, duplex plasmids carrying such a lesion in each strand are particularly suitable tools for detecting tolerance mechanisms by using sister-strand recombination. The plasmid construct used is not intended to simulate the result of UV irradiation, because the close proximity of T-T (6-4) photoproducts is unlikely after such a treatment but rather is intended to model any circumstance where both strands are damaged. In view of the high sensitivity of rad6 and rad18 mutants to UV irradiation and many other DNA-damaging agents, replication forks must stall with high frequency, an event that double-strand damage is likely to cause with particular efficiency. Further, such damage is specifically known to be produced by some agents such as ionizing radiation (29).
Replication of the Lesion-Carrying Plasmids Is Highly Efficient, Depends Almost Entirely on Sister-Strand Recombination, and Is Largely the Consequence of the Error-Free Component of the RAD6/RAD18 Pathway. Despite the apparently severe impediment to replication imposed by the placing of a T-T (6-4) photoadduct in each strand of the construct at closely opposed positions, we found that a surprisingly high fraction of plasmids could nonetheless be fully replicated, indicating that yeast has a highly efficient damage tolerance pathway for reaching this goal. In the rad1Δ strain, this fraction was 55% (Table 1) and, moreover, an astonishing ≈51% of these plasmids achieved this end by a mechanism that depended on recombination between partially replicated sister strands within the 28-bp interlesion region. The error-free component of the RAD6/RAD18 pathway was responsible for most, but not all, of this activity. As shown by the data from the rad1Δ rad18Δ strain (Table 1), this component was responsible for the recombination-dependent replication of ≈70% of the plasmids ((50.7 - 15.5)/50.7 = 69.4%). In almost all of the remaining plasmids, recombination-dependent replication required the activity of RAD52, as indicated by the results from the rad1Δ rad18Δ rad52Δ strain, in which only 2.3% of plasmids were replicated in this manner, and by the data from the rad1Δ rad52Δ strain in which the frequency of recombination-dependent replication was 24.2% rather than 50.7% as in the rad1Δ strain. The amounts of recombination attributed to the RAD18- and RAD52-dependent processes do not appear to be additive, suggesting that these two processes compete for at least some of the stalled replication forks. After subtracting the background of 2.3% from the recombination frequencies observed in the rad1Δ, rad1Δ rad18Δ, and rad1Δ rad52Δ strains (giving frequencies of 48.4%, 13.2%, and 21.9% respectively), the RAD6/RAD18-dependent fraction appears to be 73% [(48.4 - 13.2)/48.4], and the RAD52-dependent fraction 45% [(48.4 - 21.9)/48.4]. Because these fractions sum to 118%, the data imply that at least one of these two processes can cope with an additional 18% of the blocked replication forks when the other is absent. Because the product of the RAD5 gene is known to be involved in the error-free component of the RAD6/RAD18 pathway, we also investigated recombination-dependent replication in a rad1Δ rad5Δ strain. As can be seen (Table 1), the fraction of plasmids making use of this process was reduced to 21.9% from the 50.7% seen in the rad1Δ strain. Although this reduced frequency clearly supports the involvement of the RAD6/RAD18 pathway in overcoming the blocks to the progress of replication forks, the reduction is less than found in the rad1Δ rad18Δ strain. This difference, even though small, is nonetheless significant (P = 0.02), indicating either that Rad18 has a function (or functions) in the recombination process in addition to that carried out jointly with Rad5 or that the amount of Rad52-dependent recombination is influenced by the presence of proliferating cell nuclear antigen monoubiquitination in the rad5, but not rad18, mutant. After correction of the results from the rad1Δ rad5Δ strain for the 2.3% background frequency, these data indicate that the RAD5-dependent process accounts for 60% of the recombination-dependent replication compared with the estimate of 73% based on the results from the rad1Δ rad18Δ strain. As expected, DNA polymerase ζ plays no role in recombination-dependent replication; in the rad1Δ rev3Δ strain, 49.6% of the plasmids used this process (Table 1), a frequency no different from that in the rad1Δ strain. More importantly, 49.2% of plasmids replicated in the rad1Δ msh2Δ strain used the recombination-dependent process, a fraction also no different from that in the rad1Δ strain, indicating that mismatch repair plays no part in this activity. Even though the C-C mismatches placed opposite the T-T (6-4) photoadducts may well be substrates for the binding of mismatch-repair proteins, such binding is not responsible for the production of the sequence motif attributed to recombination. Although the majority of plasmids completed replication by using the recombination-dependent process, a minority did so by means of translesion replication. In the rad1Δ strain, 93% of fully replicated plasmids achieved this end by recombination and 7% by translesion replication. Because translesion replication past the T-T (6-4) photoadduct is unusually inefficient in vivo, a higher proportion of plasmids might employ such bypass when different lesions are encountered that are better substrates for translesion replication.
Table 1. Percent plasmids replicated.
| Strain | Relevant genotype | Plasmids replicated, % | Percent by recombination | Percent by translesion replication | No. of plasmids sequenced | No. of replicate experiments |
|---|---|---|---|---|---|---|
| POGY9 | rad1Δ | 54.7 ± 3.2 | 50.7 | 4.0 | 182 | 5 |
| POGY12 | rad1Δ rad18Δ | 15.5 ± 1.6 | 15.5 | 0.0 | 84 | 6 |
| HSZY9 | rad1Δ rad52Δ | 28.6 ± 3.6 | 24.2 | 4.4 | 169 | 5 |
| HSZY7 | rad1Δ rad18Δ rad52Δ | 2.3 ± 0.8 | 2.3 | 0.0 | 22 | 11 |
| POGY13 | rad1Δ rad5Δ | 23.5 ± 1.9 | 21.9 | 1.6 | 165 | 5 |
| POGY3 | rad1Δ rev3Δ | 50.1 ± 3.0 | 49.6 | 0.5 | 174 | 4 |
| POGY17 | rad1Δ msh2Δ | 53.0 ± 2.3 | 49.2 | 3.8 | 125 | 4 |
Shown are the percent of total plasmids replicated, the percent replicated by a recombination-dependent process, and the percent replicated by using translesion replication in an excision repair-defective (rad1Δ) strain and its isogenic rad18Δ, rad52Δ, rad18Δ rad52Δ, rad5Δ, rev3Δ, and msh2Δ deletion derivatives.
Replicated Plasmids Are Derived from a Single Transforming Molecule and Result in Homogeneous Sequences. We investigated the possibility that transformant arose from the uptake of more than one plasmid by looking for sequence heterogeneity among replicated plasmids within a transformant colony and also by transforming strains with a mixture of T-T (6-4) carrying constructs generated in pYPOG1 and pYPOG2. These two vectors are identical except for the sequence of a base pair doublet flanking each side of the 80-mer/72-mer insertion site (see Materials and Methods), which constitute markers that can be used to detect interplasmid recombination within the insert sequences. We found no evidence indicating that transformants originated from more than one plasmid or that interplasmid recombination occurred. No sequence heterogeneity was found among replicated plasmids present in 20 subclones from each of 15 colonies of the rad1Δ msh2Δ strain transformed with T-T (6-4)-containing construct, all resulting from recombination-dependent replication, or in 25 subclones from four colonies of the rad1Δ strain transformed with the same construct. Moreover, no sequences indicating interplasmid recombination were detected among replicated plasmids present in 10 subclones from each of nine colonies resulting from the transformation of the rad1Δ strain with a mixture of 3.5 ng pYPOG1 construct DNA and 3.5 ng pYPOG1 construct DNA, rather than the 7.0 ng construct DNA used in other experiments, or in 6 subclones from an additional colony transformed by this mixture. We conclude that transformants rarely originate from more than one construct molecule, and, hence, the occurrence of any possible interplasmid recombination must also be rare.
Discussion
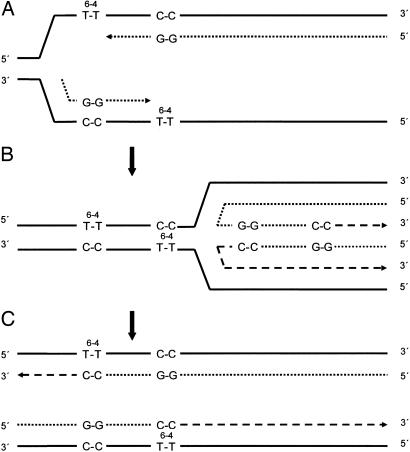
We find that excision-defective strains of budding yeast can very efficiently complete the replication of duplex plasmids that carry a T-T (6-4) photoadduct in each strand at staggered positions 28 bp apart, despite the potential of these lesions to severely inhibit elongation by any of the polymerases present in this organism. Astonishingly, ≈55% of the plasmids can be fully replicated, and in >90% of these transformants, replication depended on a mechanism employing recombination within the 28-bp interlesion region with the remainder resulting from translesion replication on one or the other strand. Most of this recombination must have taken place between partially replicated sister strands, because we found no evidence for the uptake and replication of more than one plasmid molecule per transformant. Results from isogenic strains carrying deletions of RAD18, RAD52, RAD5, or RAD18 and RAD52, in addition to the necessary RAD1 deletion, indicated that 60-70% of the recombination-dependent replication resulted from the action of the error-free component of the RAD6/RAD18 DNA damage tolerance pathway, with the remaining fraction dependent on RAD52 (Table 1). The molecular mechanism responsible for recombination is unknown in either of these cases. Because the RAD6/RAD18 pathway does not repair double-strand breaks (30), the recombination mechanism responsible for its error-free damage tolerance component presumably operates on intact stalled forks rather than those that have generated these breaks. The model proposed by Higgins and coworkers (25), modified to include lesions on both DNA strands as in the present case, appears to be a plausible candidate for such a process (Fig. 2). After the stalling of nascent strand elongation when the lesions are encountered (Fig. 2 A), regression of the replication fork leads to annealing of the displaced nascent strands and the formation of a chicken-foot structure (Fig. 2B). Because each nascent strand is lesion-free, both 3′ OH termini can now be extended, and replication can be completed on both templates after the resumption of fork progression (Fig. 2C). Complete replication generates duplex molecules that each carry a single lesion in the conserved strand. In plasmids, almost all duplicated genomes in the succeeding cycle of replication are likely to be derived from the undamaged strand. If this recombination mechanism occurs in yeast chromosomes, and there is no obvious reason why it should not, discarding of strands is presumably not possible, but a further round of recombination could occur in the succeeding cycle of replication to generate a lesion-free duplex. However, such an additional round of recombination might not be required if, unlike the model shown in Fig. 2, a lesion occurred in only one strand. Moreover, in excision-proficient strains, replicated molecules containing lesions, such as, for example, those depicted in Fig. 2C, could also be substrates for nucleotide excision repair. A transient template strand switching or copy-choice mechanism of the kind illustrated in Fig. 2 presumably requires such gene products as helicases to drive replication fork regression (Fig. 2B) to restore the forward progression of the fork and, perhaps, to facilitate nascent strand annealing. However, no genes encoding activities of these kinds that are associated with the RAD6-dependent error-free tolerance process have yet been identified. The annealing of nascent strands may well be possible without replication fork regression, obviating the need for the helicases.
Fig. 2.
Model for the completion of replication by transient template strand switching (24). (A) Replication intermediates after stalling of replicases at sites of T-T (6-4) photoadducts. (B) Replication fork regression, annealing together of nascent strands, and formation of a chicken foot structure. 3′ OH termini can now be extended by the replicase. (C) Resumption of replication fork progression, annealing of nascent strands to their original templates, and the completion of replication is now possible. Solid lines, original template strands; dotted lines, strand segments replicated before stalling at the photoadduct sites; dashed lines, strand segments replicated in the chicken foot structure and thereafter.
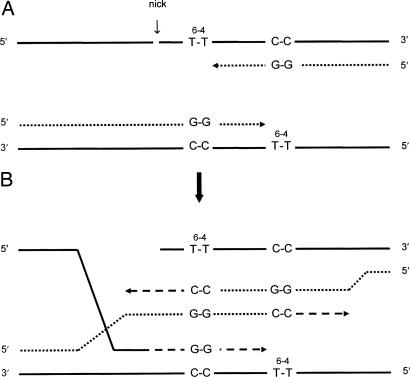
Although a majority of the stalled replication forks were restarted by a RAD6/RAD18-dependent recombination mechanism, a significant fraction were restarted by a recombination mechanism dependent on RAD52. Although Rad52 is required for the repair of double-strand breaks (30) by homologous recombination (31), such a process does not appear to be possible in the present circumstance because a damage-free homolog, as needed for such a repair pathway, is likely, at best, to be rare. However, although the nature of the Rad52-dependent recombination process is therefore unknown, it possibly entails the production of a 3′ single-stranded DNA end to which the heptomeric Rad52 ring structure can bind. A 3′ single-stranded DNA end of this kind might be generated by breakage of a template strand 5′ to one of the lesions, providing an opportunity for Rad52-facilitated strand invasion (Fig. 3A). The distorted DNA structure at the lesion site might trigger such breakage, although the identity of the endonuclease that might achieve this cut is not known. This invasion could promote displacement and annealing of nascent strands without replication fork regression (Fig. 3B). After extension by only a few nucleotides, reannealing of nascent strands to their original templates would allow the completion of replication. Strand invasion could also be initiated by breakage in the interlesion region if it occurred after the limited replication depicted in Fig. 3A had previously taken place. Although this model is obviously speculative, a link between the RAD6/RAD18-dependent and RAD52-dependent processes is nevertheless clearly indicated by the phenotype of srs2 mutations, which partially suppresses the UV sensitivity of rad6 and rad18 mutants but not their deficiency with respect to induced mutagenesis (32). This suppression is abolished in a rad52 mutant (33), indicating that some of the stalled replication forks normally processed by the RAD6/RAD18 pathway can be diverted, when this pathway is disabled, into a RAD52-dependent process by loss of SRS2 gene function. SRS2 encodes a helicase (34) that disrupts the formation of Rad51 nucleoprotein filaments, perhaps preventing blocked replication forks from generating substrates for recombination repair (35, 36). Srs2 therefore appears to act as a gatekeeper that retains some of the stalled forks in a state that permits recombination between partially replicated sister strands by transient template strand switching. Even so, some stalled forks presumably do become substrates for recombination by a RAD52-dependent mechanism even when the RAD6/RAD18 pathway is functional, as shown by the results in Table 1.
Fig. 3.
Model for recombination initiated by breakage and strand invasion. (A) Template strand breakage occurs 5′ to one or the other of the T-T (6-4) photoadducts when replication forks have stalled. (B) Strand invasion promotes annealing of nascent strands, which allows each of them to be extended and, after the reannealing of the nascent strands to their original templates, the completion of plasmid replication. Some limited extension of the invading template 3′ end can also occur, requiring resection of one or the other broken ends to reconstruct the broken strand. Full plasmid replication results in strands as depicted in Fig. 2C. Solid lines, original template strands; dotted lines, strand segments replicated before stalling at the photoadduct sites; dashed lines, strand segments replicated after strand invasion and the annealing of nascent strands.
Although >90% of the replicated plasmids circumvented the lesions by means of recombination, a minority achieved this outcome by translesion replication. Averaging results from the rad1Δ, rad1Δ rad52Δ, rad1Δ rad5Δ, and rad1Δ msh2Δ strains (Table 1), in which translesion replication frequencies are expected to be normal, the actual bypass frequency per transforming plasmid was 3.5%. Because the number of translesion replication events on the two strands is approximately equal (29:31), the frequency per strand is ≈1.8%. This value is ≈2-fold lower than the 3.3% bypass frequency on a single strand observed in experiments with the identical strains (28). In these experiments, only translesion replication, but not recombination, can occur, because the T-T (6-4) lesion is placed within a 28-nt single-stranded region within an otherwise duplex plasmid. This discrepancy might suggest that translesion replication and the error-free recombination mechanism compete for stalled replication forks and that the frequency of translesion replication increases when recombination is impossible. Although at least some of the discrepancy between the estimates from the present and previous work depends on the low frequency of translesion replication (1.6%) observed in the rad1Δ rad5Δ strain, it does not appear to eliminate it. If this result is excluded, the estimate for the frequency of translesion replication per strand is ≈2%, which is a little more than 60% of the previously observed value. It is unclear why the frequency of translesion replication in the rad1Δ rad5Δ strain is lower than in the other translesion replication-competent strains. The role of Rad5 in the error-free component of the RAD6/RAD18 pathway is well established by the epistatic interaction between rad5 and rad6 mutations (37) together with the absence of any marked effect of rad5 mutations on induced mutagenesis (38). It is also supported by the discovery that the Rad5/Ubc13/Mms2 complex polyubiquitinates lysine 164 in proliferating cell nuclear antigen by conjugating ubiquitin to lysine 63 of ubiquitin itself (14), whereas monoubiquitination of lysine 164 promotes translesion replication (15) and polyubiquitination promotes the RAD6/RAD18-dependent error-free process of DNA damage tolerance (14). Nevertheless, some RAD5 alleles, designated rev2, were isolated in a screen for mutants deficient in UV-induced mutagenesis (39), although, for reasons that remain unknown, only the reversion of certain ochre alleles are reduced in rad5/rev2 mutants; the reversion of amber, initiation, missense, and frameshift alleles are unaffected (38). It is therefore unlikely that the reduced frequency of translesion replication in the rad1Δ rad5Δ strain can be explained on this basis, and, overall, the frequency of translesion replication may therefore be as much as 2-fold lower in the two circumstances, possibly because such bypass is less frequent when competing with the recombination-dependent process for stalled replication forks.
Interestingly, the high efficiency with which the plasmid constructs are replicated in yeast is in marked contrast to our results from E. coli by using an essentially identical experimental method (27). Although a very similar recombination mechanism, which is RecA and RecF independent, appears to be used in the bacterium, it can promote the replication of only ≈8-10% of the plasmids, rather than ≈50% as in yeast. E. coli therefore has two recombination-dependent damage tolerance mechanisms, the major one is RecA dependent and entails the physical exchange of DNA, whereas the minor is RecA independent and entails the exchange of genetic information. Viewed in conjunction with earlier information (3, 10), our results suggest that yeast, and perhaps all eukaryotes, predominantly make use of the latter mechanism.
Acknowledgments
We thank Dr. Paul O'Grady for construction of deletion mutants and Dr. Peter Gibbs for advice and technical help. This work was supported by National Institutes of Health Public Health Service Grant GM60652.
Author contributions: H.Z. performed research; C.L. designed research; C.L. analyzed data; and C.L. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rupp, W. D. & Howard-Flanders, P. (1968) J. Mol. Biol. 31, 291-304. [DOI] [PubMed] [Google Scholar]
- 2.Cleaver, J. E. & Thomas, G. H. (1969) Biochem. Biophys. Res. Comm. 36, 203-208. [DOI] [PubMed] [Google Scholar]
- 3.Lehmann, A. R. (1972) J. Mol. Biol. 66, 319-337. [DOI] [PubMed] [Google Scholar]
- 4.Buhl, S. N., Stillman, R. M., Setlow, R. B. & Regan, J. D. (1972) Biophys. J. 12, 1183-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.di Caprio, L. & Cox, B. S. (1981) Mutat. Res. 82, 69-85. [DOI] [PubMed] [Google Scholar]
- 6.Prakash, L. (1981) Mol. Gen. Genet. 184, 471-478. [DOI] [PubMed] [Google Scholar]
- 7.Smith, K. C. & Meun, D. H. C. (1970) J. Mol. Biol. 51, 459-472. [DOI] [PubMed] [Google Scholar]
- 8.Rupp, W. D., Wilde, C. E. Reno, D. L. & Howard-Flanders, P. (1971) J. Mol. Biol. 61, 25-44. [DOI] [PubMed] [Google Scholar]
- 9.Ganesan, A. K. (1974) J. Mol. Biol. 87, 103-119. [DOI] [PubMed] [Google Scholar]
- 10.Resnick, M. A., Boyce, J. & Cox, B. (1981) J. Bacteriol. 146, 285-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrence, C. W. & Hinkle, D. C. (1996) Cancer Surv. 28, 21-31. [PubMed] [Google Scholar]
- 12.Jentsch, S., McGrath, J. P. & Varshavsky, A. (1987) Nature 329, 131-134. [DOI] [PubMed] [Google Scholar]
- 13.Bailly, V., Lamb, J., Sung, P., Prakash, S. & Prakash, L. (1994) Genes Dev. 8, 811-820. [DOI] [PubMed] [Google Scholar]
- 14.Hoege, C., Pfander, B., Moldovan, G.-L., Pyrowolakis, G. & Jentch, S. (2002) Nature 419, 135-141. [DOI] [PubMed] [Google Scholar]
- 15.Stelter, P. & Ulrich, H. D. (2003) Nature 425, 188-191. [DOI] [PubMed] [Google Scholar]
- 16.Nelson, J. R., Lawrence, C. W. & Hinkle, D. C. (1996) Science 272, 1646-1649. [DOI] [PubMed] [Google Scholar]
- 17.Lawrence, C. W. (2004) Adv. Prot. Chem. 69, 167-203. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, R. E., Prakash, S. & Prakash, L. (1999) Science 283, 1001-1004. [DOI] [PubMed] [Google Scholar]
- 19.McDonald, J. P., Levine, A. S. & Woodgate, R. (1997) Genetics 147, 1557-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson, J. R., Lawrence, C. W. & Hinkle, D. C. (1996) Nature 382, 729-731. [DOI] [PubMed] [Google Scholar]
- 21.Nelson, J. R., Gibbs, P. E. M., Nowicka, A. M., Hinkle, D. C. & Lawrence, C. W. (2000) Mol. Microbiol. 37, 549-554. [DOI] [PubMed] [Google Scholar]
- 22.Ulrich, H. D. & Jentsch, S. (2000) EMBO J. 19, 3388-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torres-Ramos, C. A., Prakash, S. & Prakash, L. (2002) Mol. Cell. Biol. 22, 2419-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence, C. W. (1994) BioEssays 16, 253-258. [DOI] [PubMed] [Google Scholar]
- 25.Higgins, N. P., Kato, K. & Strauss, B. (1976) J. Mol. Biol. 101, 417-425. [DOI] [PubMed] [Google Scholar]
- 26.Gibbs, P. E. M. & Lawrence, C. W. (1995) J. Mol. Biol. 251, 229-236. [DOI] [PubMed] [Google Scholar]
- 27.Ozgenc, A. I., Szekeres, E. S. & Lawrence, C. W. (2005) J. Bacteriol. 187, 1974-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibbs, P. E. M., McDonald, J., Woodgate, R. & Lawrence, C. W. (2005) Genetics 169, 575-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yokoya, A., Cunniffe, S. M. T. & O'Neill, P. (2002) J. Am. Chem. Soc. 124, 8859-8866. [DOI] [PubMed] [Google Scholar]
- 30.Resnick, M. A. & Martin, P. (1976) Mol. Gen. Genet. 143, 119-129. [DOI] [PubMed] [Google Scholar]
- 31.Symington, L. S. (2002) Microbiol. Mol. Biol. Rev. 66, 630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawrence, C. W. & Christensen, R. B. (1979) J. Bacteriol. 139, 866-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schiestl, R. H., Prakash, S. & Prakash, L. (1990) Genetics 124, 817-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rong, L. & Klein, H. L. (1992) J. Biol. Chem. 268, 1252-1259. [PubMed] [Google Scholar]
- 35.Veaute, X., Jeusset, J., Soustelle, C., Kowalczkowski, S. C., Le Cam, E. & Fabre, F. (2003) Nature 423, 309-312. [DOI] [PubMed] [Google Scholar]
- 36.Krejci, L., Van Komen, S., Li, Y., Villemain. J., Reddy, M. S., Klein. H., Ellenburger, T. & Sung, P. (2003) Nature 423, 305-309. [DOI] [PubMed] [Google Scholar]
- 37.Johnson, R. E., Henderson, S. T., Petes, T. D., Prakash, S., Bankmann, M. & Prakash, L. (1992) Mol. Cell. Biol. 12, 3807-3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawrence, C. W & Christensen, R. B. (1978) Genetics 90, 213-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemontt, J. F. (1971) Genetics 68, 21-33. [DOI] [PMC free article] [PubMed] [Google Scholar]