Abstract
Hepatitis B virus, a widespread and serious human pathogen, replicates by reverse transcription of an RNA intermediate. The virus consists of an inner nucleocapsid or core, surrounded by a lipid envelope containing virally encoded surface proteins. Using electron cryomicroscopy, we compare the structures of the bacterially expressed RNA-containing core particle and the mature DNA-containing core particle extracted from virions. We show that the mature core contains 240 subunits in a T = 4 arrangement similar to that in expressed core (T is the triangulation number and the icosahedral shell contains 60 T subunits). During the infective cycle, the core assembles in an immature state around a complex of viral pregenomic RNA and polymerase. After reverse transcription with concomitant degradation of the RNA, the now mature core buds through a cellular membrane containing the surface proteins to become enveloped. Envelopment must not happen before reverse transcription is completed, so it has been hypothesized that a change in capsid structure may signal maturation. Our results show significant differences in structure between the RNA- and DNA-containing cores. One such difference is in a hydrophobic pocket, formed largely from residues that, on mutation, lead to abnormal secretion. We suggest that the changes we see are related to maturation and control of envelopment, and we propose a mechanism based on DNA synthesis for their triggering.
Keywords: cryomicroscopy, envelopment, nucleocapsid, viral maturation
Hepatitis B virus (HBV) is a major human pathogen and member of the Hepadnaviridae, a family of viruses that replicate by reverse transcription in the hepatoctyes of their hosts (1). During virus assembly, the core protein polymerizes around a complex consisting of pregenomic mRNA and viral polymerase to form an immature core. The RNA is reverse transcribed into single-stranded DNA and concomitantly degraded, then the partial second DNA strand is synthesized. Cores are then mature and can interact with the cytoplasmic parts of the surface proteins, already inserted into an inner cellular membrane, through which the core buds to form a virion (see Bruss, ref. 2). Budding does not occur before DNA second-strand synthesis, so it has been hypothesized that the envelopment signal must be propagated from the interior of the core where the DNA is located to the surface of the core where interaction with surface proteins and membrane takes place (3).
Core protein expressed in bacteria (4) assembles into icosahedral particles closely resembling those seen in infected liver (5, 6). Two sizes of particle contain either 180 (T = 3) or 240 (T = 4) copies of the core protein, clustered into dimers generating 90 or 120 spikes on the surface of the shell (7). The core protein is 183-aa long, with a very basic C-terminal region, believed to interact with nucleic acid. This region can be deleted back to amino acid 140, with the expressed protein still assembling but not able to package nucleic acid (8). Using empty T = 4 shells of core protein truncated at amino acid 149, the largely α-helical fold of the core protein was solved by electron cryomicroscopy (9) and an atomic structure determined by x-ray crystallography (10). The surface spikes are formed by bundles of four α-helices, two from each contributing monomer (9–11).
Relating this structure to the core in the mature virion raises several questions: is the core in the virion a T = 3 or T = 4 structure, how closely does the mature core in the virion resemble the expressed core isolated from bacteria, and, most interestingly, can differences between RNA- and DNA-containing cores be detected, which might indicate the nature of the maturation signal? We addressed these questions by electron cryomicroscopy. We computed a map at 7.6-Å resolution of the full-length T = 4 expressed core particle-containing RNA. This map shows a set of slender, approximately radial, connections linking the inside surface of the protein to an inner shell that contains RNA. These connections are probably parts of the basic tail of the core protein. We also isolated DNA-containing mature cores by stripping the envelopes from virions from infected blood samples. The DNA-containing cores have a T = 4 structure and a map computed at 8.8-Å resolution shows very similar features to the full length RNA-containing shell. However, some significant differences in structure may relate to the control of envelopment. In particular, there is a difference in a hydrophobic pocket in the spike, formed largely by residues that, on mutation, have been shown to lead to abnormal viral secretion (12–16).
Materials and Methods
Purification of Expressed Core Shells. Empty core shells made from protein truncated at residue 149 were prepared as described in ref. 9. Expression of the full-length core protein in Escherichia coli gave RNA-containing particles that were isolated essentially as before (9).
Viral Core Preparation. Authentic DNA-containing cores were prepared from virions isolated from blood from HBV carriers. Frozen serum containing HBV was obtained from the Diagnostics, Development, and Research Division, National Blood Service, Colindale Centre, London.
The buffer for all work was 20 mM Tris chloride, pH 7.4 (at 20°C)/140 mM NaCl (Tris-buffered saline, TBS). Individual packs were thawed and centrifuged (3,700 × g for 20 min). Supernatants were layered over 500 μl of 20% sucrose in TBS and centrifuged (266,000 × g for 30 min). The pellets were resuspended in 50 μl of TBS (after leaving wetted overnight at 4°C to soften), pooled, and subjected to equilibrium centrifugation with CsCl (266,000 × g for 72 h). Fractions (≈250 μl) from the gradient were monitored by electron microscopy and DNA analysis. Virions were found on the higher density side of the major peak of surface antigen, and the appropriate fractions were dialyzed against TBS at 20°C. Fractions were pooled, a sample was taken for assay of anti-core antibodies, and the virus suspension was added to 100 μl of equal amounts of Protein A and Protein G-Sepharose, prewashed into TBS, and then rolled (1 h) to remove any contaminating anti-core antibodies that otherwise precipitated the cores when the envelopes were stripped from the virions. The Sepharose was pelleted (12,000 × g for 10 min), and the supernatant taken for purification of cores. A further sample was taken for assay of anti-core antibodies to confirm their removal.
Virus was concentrated by pelletting in 1.5-ml tubes (266,000 × g for 30 min) and resuspended in TBS, bringing the volume to 100 μl. Cores were prepared by removal of the lipid envelope and surface antigen by the addition of 100 μl of 2 mM EDTA/40 mM DTT in TBS, incubation for 5 min, then the addition of 200 μl of 1 mM EDTA/3%(wt/vol) dodecylmaltoside in TBS and incubation for 10 min. This treatment also disrupts contaminating surface antigen particles. Cores were concentrated by pelleting in 1.5-ml tubes (266,000 × g for 1 h) and resuspended in 50 μl of TBS (leaving at 4°C overnight to soften the pellet). The isolated DNA-containing cores have not been exposed to CsCl, because it is the enveloped virions that were fractionated on the gradient, thus making the preparation of DNA cores comparable with that of RNA cores.
Electron Cryomicroscopy. RNA-containing expressed core shells were vitrified and imaged over holes of a holey carbon film by standard methods (9). For the DNA-containing cores from virions, which were at a lower concentration, particles were captured on a thin carbon film supported on a thicker holey carbon film (17) before blotting and freezing.
Image Processing. Selected micrographs were digitized and particles were selected automatically by using the findem program (18). Initial maps were calculated from class averages generated by multivariate statistical analysis and classification by using imagic (19). Orientations for the class averages were determined by self-common lines (20). Refinement of particle origins and orientations was done by cross-common lines against the current best RNA or DNA map (7). Final RNA and DNA maps were computed from 1,472 and 1,791 particles, respectively, with a sampling of 1.17 Å per voxel and 1.21 Å per voxel before magnification rescaling. Agreement between maps, values of defocus, and estimates of effective resolution were computed from Fourier shell correlations as in ref. 9.
The resolution of the final RNA core map was 7.6 Å, (Fourier shell correlation = 0.33, corresponding to a signal-to-noise ratio of 1 in the full map; ref. 21) and 8.8 Å (Fourier shell correlation = 0.33) for the final DNA core map (Fig. 6, which is published as supporting information on the PNAS web site). An improved contrast transfer function (CTF) correction procedure was used to merge maps computed on a per micrograph basis. A CTF with an envelope indicating a resolution-dependent decay in signal-to-noise ratio was derived for each micrograph by calculating the Fourier shell correlation between the 3D map from that micrograph and the best current merge of all of the other maps. Application of the procedure gives a reciprocal-space weighting of images from each film, which takes into account resolution-dependent and film-specific (defocus, imaging quality, and specimen instability) variation in the quality of the signal. The final merged maps were sharpened to restore the correct reciprocal-space amplitude profile, derived from the known crystal structure (10). The amplitude correction was determined with the nucleic acid excluded from the maps. Magnification scaling by using the dockem program (22), now based on the Fast Local Correlation Function (18), took the crystal structure as reference, with a mask defined so that only the protein shell contributed to the scaling. The various maps were aligned by using the known icosahedral symmetry axes. In a difference map, the signal-to-noise ratio is lower than in the individual maps, so the RNA and DNA maps were low-pass Fourier filtered with a cosine edge tapering to zero over the band 9.5–8.5 Å. Student's t test was used to assess the significance of differences between the RNA and DNA maps, based on variances calculated by splitting both RNA and DNA data sets into three parts (23).
More details on core preparation and imaging are given in Supporting Text, which is published as supporting information on the PNAS web site.
Results
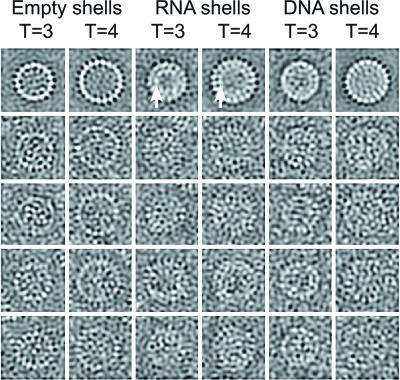
Comparison of Various Core Particles. Images from cryomicrographs of various kinds of core (Fig. 1) show their general appearance. The top row contains averaged images where the enhanced contrast makes the overall features clear, whereas the lower rows show individual images. The T = 3 and T = 4 particles made from recombinant truncated core protein (left two columns) look empty, whereas the particles made from full-length protein (middle two columns) have a clear additional inner shell of material (arrows). This material consists of heterogeneous RNAs packaged from the E. coli cells in which the core protein was expressed (24). These particles contain ≈15% RNA, i.e., an average of ≈2,650 RNA bases per shell. Hybridization of the gel with a probe for the core protein gene shows that about one-third of the packaged RNA is overexpressed, but partially degraded, core protein mRNA (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 1.
Images from cryomicrographs of various core structures. The top row of averaged aligned images shows the general features with high contrast and reduced noise. The remaining four rows show individual particles. From the left, the columns represent empty particles of core protein truncated at residue 149 (T = 3 and T = 4), particles made from full-length core protein containing RNA (T = 3 and T = 4) (arrows indicate the additional shell of density) and mature DNA-containing core particles extracted from virions (T = 3 and T = 4). Box size = 480 Å.
DNA-containing cores from virions (Fig. 1, right two columns) appear fuller than the truncated particles, but the inner material is not organized into as well defined a shell as in the RNA-containing expressed core particles. Importantly, cores from virions were almost exclusively (≈98%) of the T = 4 size and the T = 4 symmetry was confirmed from the computed map. A targeted search through all of the micrographs of virally derived cores by using an automatic particle finding program (18) with templates for T = 3 and T = 4 particles found only 2% of T = 3 sized particles, confirming a visual inspection of the micrographs. A map computed from the few T = 3 sized particles, although noisy, confirmed that they are T = 3 (data not shown).
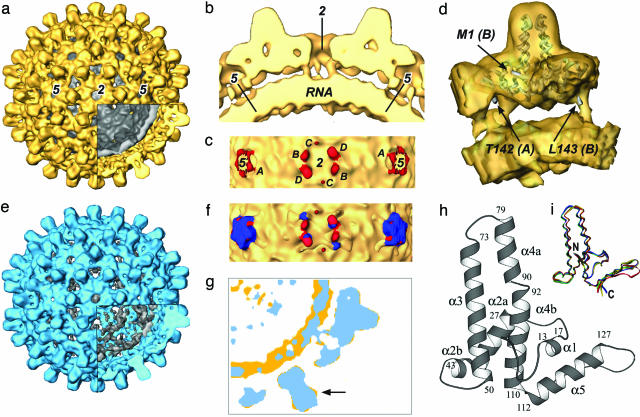
Structure of the T = 4 RNA-Containing Shell. The map of the T = 4 RNA-containing particle made from full-length core protein (Fig. 2a) is very similar to that computed from truncated core protein (9). The atomic model from x-ray crystallography of the truncated core protein (10) fits very well into this map (Fig. 8, which is published as supporting information on the PNAS web site). The close agreement between these structures indicates that no gross alteration has occurred in the outer protein shell as a result of the basic C-terminal tails and the consequent RNA packaging. There are, however, considerable differences on the inside, where a strong shell of density between radii 90–125 Å is now present (Fig. 2 a and b). This inner shell corresponds to the additional ring of density seen in the images of full-length core particles (Fig. 1). There are also slender links between the inside surface of the core protein and the inner shell of density. Five such links occur around each fivefold axis and six around each strict twofold (local sixfold) axis (Fig. 2c). They emerge from positions at or very close to those corresponding to the last ordered residues (amino acid 142/143) seen in the x-ray map of the truncated protein (Fig. 2d). We think that they correspond to parts of the basic tails of the core protein arranged close to the T = 4 quasi-equivalent positions. The links for subunits A, B, and D (as defined in refs. 10 and 25) are fairly strong, whereas that for subunit C appears rather weak, perhaps because it is less ordered. The inner shell of density represents the packaged RNA plus those parts of the C-terminal basic tails that interact closely with it.
Fig. 2.
Maps of the RNA-containing and DNA-containing core particles. (a) The T = 4 RNA map with the outer protein parts colored yellow and the inner shell of RNA and parts of the C-terminal tails colored gray (cutaway). (b) A central slab of the map, containing twofold and fivefold axes, shows slender links between the inside surface of the protein shell and the outer surface of the inner RNA-containing shell. (c) Radial view of the links (red) and the inner shell (yellow) with the outer part of the core protein removed. There are five links around each fivefold axis and six links around each twofold (local sixfold) axis. The symmetry of the links corresponds to the T = 4 quasi-equivalence of the core protein subunits. The subunit identity of each link is indicated by the letters A, B, C, and D. (d) View of one protein dimer (the A-B dimer) cut from the map with a ribbon diagram of the x-ray structure embedded. The last ordered C-terminal residues [T142(A) and L143(B)] seen in the x-ray structure of the truncated protein and the N terminus [M1(B)] of one subunit are indicated. The links joining outer and inner shells lie very close to the last ordered residues in each subunit and probably represent the next part of each protein chain. (e) Map of the DNA-containing mature core extracted from virions, with the outer protein parts colored blue and the inner shell comprising DNA and parts of C-terminal tails colored gray (cutaway). The map represents a T = 4 structure very similar to the T = 4 RNA-containing expressed core particle. (f) Superposition of RNA and DNA maps in the link region showing the nucleic acid containing shells (yellow) and the links in the RNA (red) and DNA maps (blue). (g) Superimposed central sections from the RNA map (yellow) and the DNA map (blue). The inner RNA shell is stronger and more clearly defined than the inner DNA shell. Parts of the dimer spikes in the RNA core protein protrude sideways beyond the corresponding features in the DNA map, indicating a small change in structure of the spike (arrow). (h) Fold of the core protein (10). (i) Variation in the fold of the core protein. The four independent subunits are aligned on the central hydrophobic region. There are two classes of conformation of the helical hairpins, and each dimer is composed of one monomer from each class.
Structure of the DNA-Containing Viral Core. The map computed from the virally derived DNA-containing cores shows a T = 4 structure (Fig. 2e), closely resembling the RNA-containing expressed shell but with some small, although significant, differences. Corresponding to the images (Fig. 1), the inner material in the DNA-containing particle is more diffuse than the strongly defined shell seen in the RNA-containing particle. Nevertheless, clear radial links are seen between the inner surface of the outer protein shell and the material in the interior (Fig. 2f). These links approximate to those in the RNA map but the links closest to the fivefold axis (subunit A) in the DNA map are stronger and are rotated by ≈20° compared with those in the RNA map. The B and D links are also shifted slightly and deviate more as they go to lower radii. This behavior is expected, because the tails are likely to be flexible and, therefore, more constrained close to the main protein shell. The link to subunit C is not seen in the DNA map. The diffuse appearance of the inner material in the DNA map compared with the RNA map probably reflects differences in the properties of double-stranded DNA (dsDNA) versus ssRNA and their interactions with the different parts of the core protein.
Differences Between RNA- and DNA-Containing Cores. The fold of the monomer core protein consists of a set of closely linked α-helices (10) (Fig. 2h). The spike consists of an outward-going helix-3 and a returning helix-4 forming a hairpin. Helix-4 is linked to the most C-terminal helix-5 directly by glycine 111. Helix-5 is followed by a tight turn containing multiple proline residues and then an extended chain visible to residue 142/143. The four independent subunits solved in the T = 4 icosahedral asymmetric unit show small differences in the hairpin conformation (Fig. 2i).
Although the overall structure of the mature DNA core is very similar to that of the expressed RNA core, there are movements of a few angstroms in places. The most striking difference is in the shape of the spikes protruding outward from the spherical shells: those of the RNA capsid are bulbous or knob shaped, being wider toward the outer end, whereas in the DNA structure, the profile of the spike is more uniform and regular. These differences can be seen in the isosurface rendering of the maps (Fig. 2 a and e), and in profile in the cutaway regions. The narrower profile of the DNA spike is clear when a central section of the DNA map is overlaid on the corresponding RNA section (Fig. 2g). The change is seen most easily in the movie (Movie 1, which is published as supporting information on the PNAS web site).
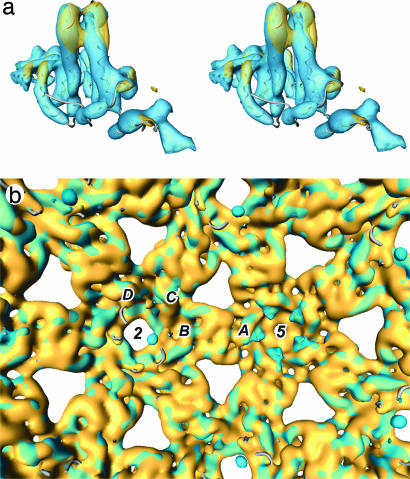
A difference map summarizes the differences between the two core structures. The level of significance of the differences was determined by using Student's t test, and only the strongest, most significant differences are shown (Fig. 3; see also Fig. 9, which is published as supporting information on the PNAS web site). The conformational changes at the tips of the spikes are highlighted in red, indicating significant density present in the RNA map but not in the DNA map. Also highlighted in red are contacts between the dimers, indicating that these interdimer interactions in RNA cores are weakened on transition to DNA cores.
Fig. 3.
Strongest significant differences in the protein shells between the DNA and RNA maps superimposed on part of the RNA map with the x-ray model embedded. The most significant differences (t test: P < 0.0001) are shown in red where the RNA map is stronger and in blue where the DNA map is stronger. (a and b) a is viewed from outside and b is from inside the particle. The inner shells of density in each map have been removed for clarity. a shows mainly differences in the outer part of the dimer spikes (red), whereas b shows differences inside the lower part of the spike around the fivefold axes and in the interface region between dimers (blue). There are small distributed changes within the subunits and in the packing between dimers.
Closer comparison of the A-B dimer in the RNA and DNA maps reveals that helix-4 of subunit A is straighter in the DNA structure (Fig. 4a). Because helix-4 of subunit A is in contact with helix-3 from subunit B, a complementary movement from helix-3 is required. Similarly, because there is close contact of all four of the helices in the spike, changes in orientation of helix-3 (subunit A) and helix-4 (subunit B) must occur. The changes occurring in subunits A and B are different. The straightening and movement of helix-4 of subunit B is more apparent toward the inside of the shell rather than at the tip as for subunit A. In forming the dimer, the hairpin motifs from each subunit cross at an angle, with helix-3 creating the knob shape at the tip of the RNA spike and protruding beyond the equivalent density in the DNA spike (arrow in Fig. 2g). By analogy to a pair of scissors, each hairpin forms one of the blades. A small rotation of each of the units forming the blades can close them, creating a more parallel alignment and generating the observed narrower profile of the DNA spike. Thus, the change in shape of the dimer spikes is explained by a local change in conformation of the protein chains (including unkinking of helix-4) and their reorientation relative to one other.
Fig. 4.
Superposition of the RNA (yellow) and DNA (blue) maps. (a) Stereoview of the A-B dimer spike with concerted changes in the helix bundle shown by the way patches of blue or yellow protrude on the surface of the outer half of the spike. The crystallographic chain trace is embedded in the density. (b) View of the inner surfaces of the protein shell with the nucleic acid shell removed. Note the additional (blue) density around the fivefold axes in the DNA map, corresponding with the C-terminal region of subunit A.
It was evident from the crystal structure of the core that the dimers A-B and C-D do not have exact twofold symmetry. Two distinct conformations of the monomer (Fig. 2i) are present in each dimer, related by a 178° rotation. At the level of resolution in the electron microscope maps the RNA core spikes appear twofold symmetric, but the DNA core spike density appears to deviate from twofold (Fig. 2g and Movie 1). Asymmetry in the size of the pocket on either side of the DNA core spike indicates a further departure from quasi-symmetry in the DNA structure. Although the C-D dimer makes a similar change in shape (Movie 1), the small conformational changes within each subunit appear to be different in detail.
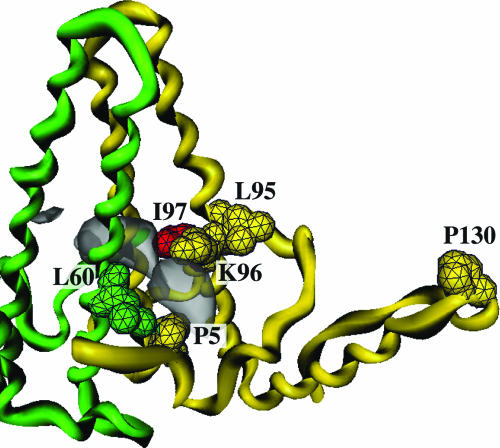
Two main regions show additional density in the DNA map, shown in blue on the difference map (Fig. 3). First, the region of amino acids 140–145 of the A subunit, arranged around the fivefold axis, appears more ordered than in the RNA map, and the interaction with helix-5 of the same subunit is stabilized (Fig. 3b, view from inside the shell, and Fig. 4b). Second, the DNA structure has strong significant additional density in externally accessible pockets on either side of the spikes, at the interface between the two subunits forming each dimer (Fig. 5). Each pocket is bounded by residues P5, K96, and I97 of one subunit and L60 from the other subunit of the dimer (10).
Fig. 5.
The hydrophobic pocket in the spike. The two chains forming the dimer are colored yellow and green and various functionally important residues (P5, L60, L95, K96, and P130; see Discussion) are shown, including I97 in red. Density present in the pocket in the DNA map but not in the RNA map is indicated by the gray surface.
Discussion
These results give a detailed picture of authentic DNA-containing cores of HBV from mature virions. The core shells are of T = 4 type containing 240 subunits arranged in dimers with a tiny minority of T = 3 shells. Nonenveloped cores extracted from infected liver also showed a majority of T = 4 size (6). The overall protein structure in the mature core is similar to that of the empty or of the RNA-containing bacterially expressed cores, made respectively from truncated protein or full-length protein, but with some significant differences. The mature core may have a slightly different protein sequence from the expressed core, because it was from an anonymous donor with an unknown viral genotype, but core protein is highly conserved (>95% identity over genotypes A–F) so the genotype is unlikely to affect the structure. Also, cores from virions may have packaged cellular factors not present in expressed cores, but it is unlikely that such factors would cause changes in the core protein itself, and in any case, would be present from the time of core assembly and so are unlikely to be the trigger for envelopment. The major distinction is that expressed cores contain ssRNA and the mature cores dsDNA, and because envelopment depends on DNA synthesis, we believe the differences we see are related to core maturation during virion assembly.
We see two differences between full-length expressed core particles containing RNA and the previously solved empty particles of the truncated protein. There is a strong inner shell of density corresponding to the encapsidated RNA and those parts of the C-terminal tails that interact closely with the RNA. More interestingly, we see four links between the inside of the protein shell and the RNA layer, corresponding to the four independent subunits in the T = 4 shell. Moreover the links start very close to the last ordered residues (amino acid 142/143) seen in the crystal structure. These links almost certainly correspond to the next segment of the polypeptide chain, which must be icosahedrally ordered to be visible after the averaging applied during computation of the map. A difference map between empty core particles consisting of protein truncated at either amino acid 149 or at amino acid 140 showed similar features, and a similar interpretation was made (26). There are no interpretable features in the inner shell itself, suggesting that this material is not icosahedrally symmetric. The C-terminal tails are probably flexible and, therefore, increasingly disordered away from the folded part of the core protein.
Comparing the structures of the RNA- and DNA-containing cores, we find numerous small differences throughout the protein. The whole molecule acts as a system of α-helical rods and flexible links by which a change on the inside is transmitted to various parts of the surface. These rather small changes must be sufficient to initiate envelopment at an appropriate stage of second-strand synthesis, probably triggered by the extended C-terminal tails, because these are in closest contact with the nucleic acid. Because the C-terminal links to the A subunits around the fivefold axes appear much stronger in the DNA than the RNA map, the change may start there. The residues in the ordered C-terminal of the protein (amino acids 132–142) both stabilize the monomer fold by interacting with helix-5 and provide part of the interface between dimers in forming the shell. Changes in this region can propagate to the outside of the shell, because helix-5 is linked to the base of helix-4 by a single glycine residue and helix-4 runs out to the tip of the spike. In addition, in the A subunit of the DNA core, the closer packing of amino acids 132–142 against helix-5 brings residues near 142 into closer contact with the base of helix-4, providing a more direct route for DNA synthesis to affect helix-4. Changes in the position of helix-4 must, in turn, move helix-3. The combined effect of the movement of helices-3 and -4 alters the shape of the spike and changes the nature of a hydrophobic pocket, marked by extra density present in the DNA map, on the side of the spike (Fig. 5).
The likely functional importance of the hydrophobic pocket (27) is highlighted by the clustering there of mutations with various secretion phenotypes (Fig. 5). A frequent natural mutation I97L in the adr subtype leads to immature virions containing ssDNA (12). Two other natural mutations, P5T and L60V, give very low secretion of virions (13), and P5T corrects the immature secretion phenotype of I97L (14). Two other residues close by (L95 and K96) and also L60, when changed to alanine, give intracytoplasmic cores that do not become enveloped or secreted (15). Another natural mutation (P130T), although distant from the pocket, has a slow secretion phenotype alone and compensates for the immature secretion of I97L (16). Finally, a point mutation (A119F, adr subtype) in the preS1 region of the envelope protein can also compensate for the I97L mutant (28).
These observations suggest a model in which changes in capsid structure associated with dsDNA synthesis alter the hydrophobic pocket, allowing a productive interaction with the preS1 region of surface protein, including residue 119 (and other parts of the surface protein; ref. 2). Mutations in the core protein may both modulate the affinity for surface protein and affect the way changes propagate through the core. Thus, mutants P5T and L60V (and, similarly, mutants L60A, L95A, and K96A) may either create a pocket that does not interact well with the surface protein or form structures in which the pocket does not open properly, resulting in low secretion. In mutant I97L, the pocket may be constitutively open, or more easily opened by changes from inside the capsid, leading to immature secretion. I97L and P5T could mutually compensate by creating a pocket with appropriate affinity or time of opening. The P130T mutation cannot affect the pocket directly but may act by failing to transmit effectively the changes occurring inside the core. The diverse nature and position of these functional mutations suggests that the core is a finely tuned mechanism for controlling appropriate envelopment. Disturbing this balance, possibly by targeting the pocket, could provide an antiviral strategy.
A clue to what might trigger the observed changes in capsid structure comes from the packaging of DNA into phage heads. In phage φ29, as the head is filled with dsDNA, the portal motor has been shown to work increasingly hard against the bending, electrostatic, and entropic forces generated by close packing the DNA within the preformed head (29). A similar situation is created within the HBV core as the RNA pregenome is reverse transcribed by the viral polymerase, first into ssDNA with concomitant degradation of the RNA template and, thus, with no change in charge density or nucleic acid flexibility, but then into partially dsDNA. At some point during second-strand synthesis, the force on the inside surface of the capsid could build to a point where it triggers the change in core structure that allows envelopment. Analysis of individual DNA molecules from virions shows a range of lengths for the partial strand from one-half to three-quarters of the genome, with a minimum length for the single stranded region of 650–700 bases (30), indicating that triggering has occurred by this stage of second-strand synthesis. The force versus percentage genome packaged curve for φ29 (29) suggests that the estimated packing density of DNA in the HBV core (≈70% of that in φ29), coupled with the smaller internal radius and, thus, greater bending of dsDNA in cores, is sufficient to generate considerable internal force. This idea is supported by the observation that, in a constructed mutant lacking the RNase H activity of polymerase, the resulting RNA-DNA double-stranded hybrid becomes enveloped and secreted, whereas in a mutant lacking polymerase activity, no secreted ssRNA-containing virion-like particles could be detected (31). Triggering based on the amount of dsDNA synthesized accounts for the fact that wild-type virus gets enveloped after sufficient partial second-strand synthesis and allows a role for mutations in the core protein to modulate the structural transitions associated with maturation and envelopment.
Supplementary Material
Acknowledgments
We thank Drs. R. Eglin, D. Howell, and D. Lee (National Blood Service, London), for providing the blood samples used in this work and Prof. K. Murray (University of Edinburgh, Edinburgh) for providing plasmid pHBV130.
Author contributions: A.M.R., J.A.B., S.A.W., P.J.G.B., and R.A.C. designed research, performed research, contributed new reagents/analytic tools, analyzed data, and wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: dsDNA, double-stranded DNA; HBV, hepatitis B virus.
References
- 1.Ganem, D. & Schneider, R. J. (2001) in Fields Virology, eds. Knipe, D. & Howley, P. (Lippincott, Philadelphia), 4th Ed., pp. 2923-2969.
- 2.Bruss, V. (2004) Virus Res. 106, 199-209. [DOI] [PubMed] [Google Scholar]
- 3.Summers, J. & Mason, W. S. (1982) Cell 29, 403-415. [DOI] [PubMed] [Google Scholar]
- 4.Pasek, M., Goto, T., Gilbert, W., Zink, B., Schaller, H., MacKay, P., Leadbetter, G. & Murray, K. (1979) Nature 282, 575-579. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, B. J. & Richmond, J. E. (1982) Nature 296, 677-678. [DOI] [PubMed] [Google Scholar]
- 6.Kenney, J. M., von Bonsdorff, C.-H., Nassal, M. & Fuller, S. D. (1995) Structure (London) 3, 1009-1019. [DOI] [PubMed] [Google Scholar]
- 7.Crowther, R. A., Kiselev, N. A., Böttcher, B., Berriman, J. A., Borisova, G. P., Ose, V. & Pumpens, P. (1994) Cell 77, 943-950. [DOI] [PubMed] [Google Scholar]
- 8.Gallina, A., Bonelli, F., Zentilin, L., Rindi, G., Muttini, M. & Milanesi, G. (1989) J. Virol. 63, 4645-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Böttcher, B., Wynne, S. A. & Crowther, R. A. (1997) Nature 386, 88-91. [DOI] [PubMed] [Google Scholar]
- 10.Wynne, S. A., Crowther, R. A. & Leslie, A. G. W. (1999) Mol. Cell 3, 771-780. [DOI] [PubMed] [Google Scholar]
- 11.Conway, J. F., Cheng, N., Zlotnick, A., Wingfield, P. T., Stahl, S. J. & Steven, A. C. (1997) Nature 386, 91-94. [DOI] [PubMed] [Google Scholar]
- 12.Yuan, T. T.-T., Tai, P.-C. & Shih, C. (1999) J. Virol. 73, 10122-10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Pogam, S., Yuan, T. T.-T., Kumar Sahu, G., Chatterjee, S. & Shih, C. (2000) J. Virol. 74, 9099-9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chua, P. K., Wen, Y.-M. & Shih, C. (2003) J. Virol. 77, 7673-7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ponsel, D. & Bruss, V. (2003) J. Virol. 77, 416-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan, T. T.-T. & Shih, C. (2000) J. Virol. 74, 4929-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crowther, R. A., Berriman, J. A., Curran, W. L., Allan, G. M. & Todd, D. (2003) J. Virol. 77, 13036-13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roseman, A. M. (2003) Ultramicroscopy 94, 225-236. [DOI] [PubMed] [Google Scholar]
- 19.van Heel, M., Harauz, G., Orlova, E. V., Schmidt, R. & Schatz, M. (1996) J. Struct. Biol. 116, 17-24. [DOI] [PubMed] [Google Scholar]
- 20.Crowther, R. A. (1971) Philos. Trans. R. Soc. London B 261, 221-230. [DOI] [PubMed] [Google Scholar]
- 21.Rosenthal, P. B. & Henderson, R. (2003) J. Mol. Biol. 333, 721-745. [DOI] [PubMed] [Google Scholar]
- 22.Roseman, A. M. (2000) Acta Crystallogr. D 56, 1332-1340. [DOI] [PubMed] [Google Scholar]
- 23.Milligan, R. A. & Flicker, P. F. (1987) J. Cell Biol. 105, 29-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birnbaum, F. & Nassal, M. (1990) J. Virol. 64, 3319-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zlotnick, A., Cheng, N., Conway, J. F., Booy, F. P., Steven, A. C., Stahl, S. J. & Wingfield, P. T. (1996) Biochemistry 35, 7412-7421. [DOI] [PubMed] [Google Scholar]
- 26.Watts, N. R., Conway, J. F., Cheng, N., Stahl, S. J., Belnap, D. M., Steven, A. C. & Wingfield, P. T. (2002) EMBO J. 21, 876-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ning, B. & Shih, C. (2004) J. Virol. 78, 13653-13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Pogam, S. & Shih, C. (2002) J. Virol. 76, 6510-6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, D. E., Tans, S. J., Smith, S. B., Grimes, S., Anderson, D. L. & Bustamante, C. (2001) Nature 413, 748-752. [DOI] [PubMed] [Google Scholar]
- 30.Delius, H., Gough, N. M., Cameron, C. H. & Murray, K. (1983) J. Virol. 47, 337-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerelsaikhan, T., Tavis, J. E. & Bruss, V. (1996) J. Virol. 70, 4269-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.