Abstract
We describe the use of a series of gradually expanded thymine nucleobase analogs in probing steric effects in DNA polymerase efficiency and fidelity. In these nonpolar compounds, the base size was increased incrementally over a 1.0-Å range by use of variably sized atoms (H, F, Cl, Br, and I) to replace the oxygen molecules of thymine. Kinetics studies with DNA Pol I (Klenow fragment, exonuclease-deficient) in vitro showed that replication efficiency opposite adenine increased through the series, reaching a peak at the chlorinated compound. Efficiency then dropped markedly as a steric tightness limit was apparently reached. Importantly, fidelity also followed this trend, with the fidelity maximum at dichlorotoluene, the largest compound that fits without apparent repulsion. The fidelity at this point approached that of wild-type thymine. Surprisingly, the maximum fidelity and efficiency was found at a base pair size significantly larger than the natural size. Parallel bypass and mutagenesis experiments were then carried out in vivo with a bacterial assay for replication. The cellular results were virtually the same as those seen in solution. The results provide direct evidence for the importance of a tight steric fit on DNA replication fidelity. In addition, the results suggest that even high-fidelity replicative enzymes have more steric room than necessary, possibly to allow for an evolutionarily advantageous mutation rate.
Keywords: fidelity, mutagenesis, replication bypass, steady-state kinetics
Ahigh fidelity for DNA replication is required to maintain proper transfer of genetic information during cell division. The first and most influential step that determines this fidelity is synthesis of a new base pair by a replicative DNA polymerase (1-4). This choice, which occurs dozens of times per second, involves the selection of one nucleotide among four for insertion into the growing primer strand, opposite each DNA template base as it is addressed in turn. In eukaryotes, the replicative enzymes are DNA polymerases δ, α, and ε (5). In eubacteria, the replicative polymerases are Pol III, which synthesizes the leading strand, and Pol I, which assists Pol III with the lagging strand (6). These latter polymerases make an error (synthesis of a mismatched pair) only once in ≈104 to 105 nucleotide insertions (6).
The biophysical origin of this fidelity is a long-standing topic of research on polymerases. Early studies often focused on matching of Watson-Crick hydrogen bonds; however, it was subsequently recognized that at the terminus of DNA, base pairing selectivity in the absence of enzymes is too low to account for the observed enzymatic fidelity (7). More recently, it has been shown that a nonpolar isostere of thymine (difluorotoluene) can be replicated with nearly wild-type fidelity despite its lack of hydrogen bonding ability (8-10). Such observations, in conjunction with structural (11-15) and mutational (16-19) studies, have led to the hypothesis that geometry of DNA base pairs may be regulated by a close fit in polymerase active sites (1, 2, 20-22). Although substitutions on nucleoside sugars have been made (23), systematic tests of this steric concept are lacking at the level of the base pairs, where the genetic information is carried.
Here we describe such a systematic test, wherein the size of a DNA base analog is gradually increased over a 1.0-Å range. We examine the in vitro efficiency and fidelity of base pair synthesis by Escherichia coli DNA Pol I (Klenow fragment) with these analogs, and we then examined the same questions in vivo with a bacterial replication assay. We find that both the DNA Pol I active site and the entire bacterial replication machinery are exquisitely sensitive to very small changes in base pair size. In addition, we find that the optimum size is larger than that of natural DNA, suggesting a simple steric mechanism for influencing evolutionary adaptability.
Materials and Methods
Nucleotide Analogs and Modified Oligonucleotides. Modified nucleoside analogs were prepared as described in ref. 24, and the 5′-triphosphate derivatives were prepared and characterized by standard procedures. Phosphoramidite derivatives of the modified nucleosides were prepared as described in ref. 24. DNA sequences containing modified bases were prepared by following the procedures outlined in ref. 25. Details are given in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
In Vitro Steady-State Kinetics and Bacterial Replication Assay. We used 28-mer/23-mer template-primer duplexes having the sequence (5′-ACTGXTCTCCCTATAGTGAGTCGTATTA)·(5′-TAATACGACTCACTATAGGGAGA) as polymerase substrates. Kinetics were measured at 37°C in a buffer containing 10 mM Mg2+. Buffer details are given in the legend of Table 1. The primer was 5′ end-labeled and extended by the polymerase in the presence of a single dNTP species over varied concentration and time. Products of single-nucleotide insertions were resolved from unreacted primer by 20% denaturing gel electrophoresis and were quantitated by autoradiography. The bacterial replication assay (26) for bypass efficiency and fidelity is briefly described in the text. Details of the kinetics experiments and bacterial replication assay are given in Supporting Materials and Methods.
Table 1. Steady-state kinetic efficiencies for single-nucleotide insertions by DNA Pol I (Klenow fragment, exonuclease-deficient) with variably sized template bases.
| dNTP | Template base | Vmax, %·min–1 | KM, μM | Efficiency, Vmax/KM | Relative efficiency |
|---|---|---|---|---|---|
| dATP | H | 0.90 ± 0.18 | 20 ± 2 | 4.5 × 104 | 3.5 × 10–3 |
| F | 24 ± 3 | 9.4 ± 2.9 | 2.6 × 105 | 2.0 × 10–1 | |
| L | 27 ± 5 | 3.3 ± 2.8 | 8.2 × 106 | 6.5 × 10–1 | |
| B | 12 ± 1 | 21 ± 5 | 5.7 × 105 | 4.4 × 10–2 | |
| I | 1.3 ± 0.2 | 26 ± 11 | 5.0 × 104 | 3.8 × 10–3 | |
| T | 24 ± 1 | 1.9 ± 0.5 | 1.3 × 107 | 1 | |
| dGTP | H | 0.025 ± 0.007 | 600 ± 210 | 4.2 × 101 | 3.2 × 10–6 |
| F | 0.0071 ± 0.0006 | 36 ± 12 | 2.0 × 102 | 1.5 × 10–5 | |
| L | 0.029 ± 0.007 | 190 ± 90 | 1.5 × 102 | 1.2 × 10–5 | |
| B | 0.023 ± 0.005 | 220 ± 100 | 1.0 × 102 | 7.7 × 10–6 | |
| I | 0.017 ± 0.001 | 58 ± 18 | 2.9 × 102 | 2.2 × 10–5 | |
| T | 0.41 ± 0.15 | 56 ± 34 | 7.3 × 103 | 5.6 × 10–4 | |
| dCTP | H | 0.031 ± 0.006 | 100 ± 60 | 3.1 × 102 | 2.4 × 10–5 |
| F | 0.071 ± 0.011 | 33 ± 11 | 2.2 × 103 | 1.7 × 10–4 | |
| L | 0.021 ± 0.003 | 25 ± 13 | 8.4 × 102 | 6.5 × 10–5 | |
| B | 0.038 ± 0.005 | 79 ± 33 | 4.8 × 102 | 3.7 × 10–5 | |
| I | 0.036 ± 0.001 | 20 ± 8 | 1.8 × 103 | 1.4 × 10–4 | |
| T | 0.022 ± 0.006 | 46 ± 28 | 4.8 × 102 | 3.7 × 10–5 | |
| dTTP | H | 0.062 ± 0.022 | 61 ± 37 | 1.0 × 103 | 7.7 × 10–5 |
| F | 0.12 ± 0.03 | 22 ± 13 | 5.5 × 103 | 4.2 × 10–4 | |
| L | 0.13 ± 0.04 | 18 ± 18 | 7.2 × 103 | 5.5 × 10–4 | |
| B | 0.20 ± 0.03 | 37 ± 12 | 5.4 × 103 | 4.2 × 10–4 | |
| I | 0.085 ± 0.037 | 48 ± 39 | 1.8 × 103 | 1.4 × 10–4 | |
| T | 0.012 ± 0.004 | 38 ± 38 | 3.2 × 102 | 2.5 × 10–5 |
The template contained variably sized thymidine analogs. The kinetics were measured in a buffer containing 5 mM template–primer DNA, 0.005 or 0.1 unit/ml enzyme, 50 mM Tris-HCl buffer (pH 7.5), 10 mM MgCl2, 1 mM dithiothreitol, and 50 mg/ml bovine serum albumin at 37°C. Vmax was normalized to the lowest enzyme concentration used. Relative efficiency is relative to insertion of dATP opposite T.
Results and Discussion
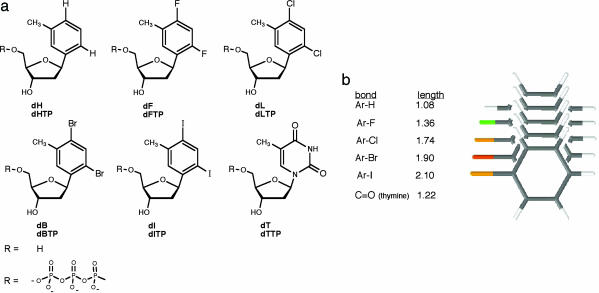
We sought to test the active site tightness hypothesis directly and systematically by preparing a series of nucleobase analogs having gradually increasing size. The nucleosides 3-toluene-1-β-d-deoxyriboside (dH), 2,4-difluoro-5-toluene-1-β-d-deoxyriboside (dF), 2,4-dichloro-5-toluene-1-β-d-deoxyriboside (dL), 2,4-dibromo-5-toluene-1-β-d-deoxyriboside (dB), and 2,4-diiodo-5-toluene-1-β-d-deoxyriboside (dI) are all shape analogs of thymidine (Fig. 1) (24). Because they have varied substituents at the 2,4 positions (namely, H, F, Cl, Br, and I), they vary in size in 0.2- to 0.4-Å increments, giving a total size difference of 1.0 Å from the smallest to largest (Fig. 1b). To evaluate the biophysical effects of the steric series in a polymerase active site, we prepared nucleoside triphosphate derivatives of the five compounds (Fig. 1a), thus allowing them to act as incoming nucleotide analogs of dTTP; conversely, we prepared template DNAs containing the five nucleoside analogs downstream from a primer binding site.
Fig. 1.
Structures of thymidine mimics having gradually increasing size. (a) Nucleoside and nucleoside triphosphate structures and abbreviations, with the natural nucleoside (dT) for comparison. (b) Bond lengths for aryl-halogen and aryl-hydrogen bonds. The thymine C=O bond length is shown for comparison. The nucleoside analogs were prepared as described in ref. 24, and the 5′-triphosphate derivatives were prepared and characterized by standard procedures. Details are given in Supporting Materials and Methods. Phosphoramidite derivatives of the modified nucleosides were prepared as described in ref. 31, and DNA sequences containing modified bases were prepared by following the literature procedure. Details are available in Supporting Materials and Methods.
We chose DNA polymerase I (Klenow fragment, exonuclease-deficient) as an ideal candidate for initial study because it is enzymatically well characterized among replicative enzymes and yet is relatively small and acts as a single subunit (27). We carried out kinetics studies of single-nucleotide insertions in the steady state by using a gel electrophoresis-based assay (28). Kinetic data are given in Tables 1 and 2. Table 1 shows results for insertion of natural dNTPs in separate experiments opposite each of the five analogs in a template strand. Table 2 shows the opposite case: insertion of compounds dH, dF, dL, dB, and dI as dNTP analogs opposite the natural template bases A, C, T, or G. As a reference, we compared the results with parallel data for natural thymidine as dTTP (the incoming nucleotide) and dT, the natural nucleoside template (Fig. 1). Fig. 2 shows all of the efficiency data in a histogram comparison.
Table 2. Steady-state kinetic efficiencies for single nucleotide insertions by DNA Pol I (Klenow fragment, exonuclease-deficient) with variably sized nucleoside triphosphates.
| dNTP | Template base | Vmax, %·min–1 | KM, μM | Efficiency, Vmax/KM | Relative efficiency |
|---|---|---|---|---|---|
| dHTP | A | 0.11 ± 0.20 | 110 ± 20 | 1.0 × 103 | 6.3 × 10–5 |
| dFTP | 2.9 ± 0.1 | 29 ± 10 | 1.0 × 105 | 6.3 × 10–3 | |
| dLTP | 39 ± 4 | 18 ± 5 | 2.2 × 106 | 1.4 × 10–1 | |
| dBTP | 13 ± 2 | 23 ± 6 | 5.7 × 105 | 3.6 × 10–2 | |
| dITP | 1.8 ± 0.1 | 18 ± 2 | 1.0 × 105 | 6.3 × 10–3 | |
| dTTP | 30 ± 3 | 1.9 ± 0.4 | 1.6 × 107 | 1 | |
| dHTP | G | 0.0065 ± 0.0016 | 200 ± 90 | 3.3 × 101 | 2.0 × 10–6 |
| dFTP | 0.0067 ± 0.0017 | 300 ± 120 | 2.2 × 101 | 1.4 × 10–6 | |
| dLTP | 0.035 ± 0.004 | 110 ± 30 | 3.2 × 102 | 2.0 × 10–5 | |
| dBTP | 0.0094 ± 0.0004 | 50 ± 10 | 1.9 × 102 | 1.2 × 10–5 | |
| dITP | 0.0020 ± 0.0001 | 43 ± 7 | 4.7 × 101 | 2.9 × 10–6 | |
| dTTP | 0.42 ± 0.18 | 140 ± 90 | 3.0 × 103 | 1.9 × 10–4 | |
| dHTP | C | 0.0067 ± 0.0023 | 310 ± 170 | 2.2 × 101 | 1.4 × 10–6 |
| dFTP | 0.012 ± 0.004 | 200 ± 110 | 6.0 × 101 | 3.8 × 10–6 | |
| dLTP | 0.050 ± 0.010 | 160 ± 66 | 3.1 × 102 | 1.9 × 10–5 | |
| dBTP | 0.016 ± 0.001 | 74 ± 22 | 2.2 × 102 | 1.4 × 10–5 | |
| dITP | 0.0023 ± 0.0004 | 23 ± 1 | 1.0 × 102 | 6.3 × 10–6 | |
| dTTP | 0.067 ± 0.040 | 510 ± 400 | 1.3 × 102 | 8.3 × 10–6 | |
| dHTP | T | 0.0046 ± 0.0011 | 59 ± 47 | 7.8 × 101 | 4.9 × 10–6 |
| dFTP | 0.0066 ± 0.0010 | 37 ± 15 | 1.8 × 102 | 1.1 × 10–5 | |
| dLTP | 0.54 ± 0.10 | 62 ± 21 | 8.7 × 103 | 5.4 × 10–4 | |
| dBTP | 0.8 ± 0.16 | 38 ± 21 | 2.1 × 104 | 1.3 × 10–3 | |
| dITP | 0.23 ± 0.02 | 37 ± 7 | 6.2 × 103 | 3.9 × 10–4 | |
| dTTP | 0.010 ± 0.002 | 63 ± 18 | 1.6 × 102 | 1.0 × 10–5 |
Incoming nucleotide analogs of variable size were used. Kinetic measurements and conditions were as described for Table 1.
Fig. 2.
Histogram of nucleotide insertion efficiencies vs. varied base pair size. Steady-state efficiencies (as Vmax/KM) using DNA Pol I (exonuclease-deficient) are shown on a log scale. (a) Insertion of natural nucleotides opposite template base analogs of increasing size (with a template T for comparison). (b) Insertion of nucleoside triphosphate analogs of increasing size, with data for the natural dTTP shown for comparison. Template-primer duplexes had the sequence (5′-ACTGXTCTCCCTATAGTGAGTCGTATTA)·(5′-TAATACGACTCACTATAGGGAGA). Kinetics were measured at 37°C in a buffer containing 10 mM Mg2+. Buffer details are given in the legend of Table 1. The primer was 5′ end-labeled and was extended by the polymerase in the presence of a single dNTP species over various concentrations and times. Products of single-nucleotide insertions were resolved from unreacted primer by 20% denaturing gel electrophoresis and were quantitated by autoradiography. Details of the kinetics experiments are given in Supporting Materials and Methods.
Kinetic efficiency was measured as Vmax/KM for nucleotide insertions at 37°C. A comparison of efficiencies for the steric series in the template with insertion of natural dATP (the presumably preferred partner) showed that, as size increased from the H to F to Cl analogs, a total increase of 0.66 Å, efficiency increased markedly by a factor of 180. The data are compared graphically with respect to relative size in Fig. 3a. Surprisingly, the Cl-substituted analog, whose “base” is larger than thymine by 0.5 Å, is the most efficient of the entire series. The efficiency with this chlorinated thymidine analog is the same within experimental error as that with the natural template, dT, despite the nonpolar nature of dL (24). With increasing size beyond the dichloro-substituted compound (dB and dI, respectively), the efficiency dropped markedly by a factor of 164-fold for the largest in response to a subtle size increase of only 0.35 Å over the optimum.
Fig. 3.
The effects of expanding base pair size in a polymerase active site, shown by plotting thymidine analog bond lengths (the variable bond) versus efficiency and fidelity. (a) Efficiency for insertion of natural dATP as a function of template bond lengths; various thymidine mimics (H, F, L, B, and I) are in the template. (b) Efficiency for insertion of dNTPs of various sizes (as a function of bond lengths) against adenine in the template. (c) Fidelity for insertion of natural dATP (rather than dCTP, dGTP, or dTTP) opposite template thymine analogs of increasing size. Fidelity was defined as efficiency for the correct T analog:A pair divided by the efficiency of the most efficient mismatch. (d) Selectivity for insertion of thymidine nucleotide analogs opposite a template A (rather than C, G, or T) as a function of incoming nucleotide size. The dashed line in each plot shows the value for the corresponding natural T-A/A-T base pair.
The converse experiments, measuring efficiency for dNTP analog insertion opposite natural adenine in the template, showed very similar results (Table 2 and Fig. 2b). Once again, efficiency increased with size until a maximum was reached, and then it dropped with further size increases, showing a large range of 2,200-fold in activity (see plot in Fig. 3b). The chlorinated dNTP was inserted opposite adenine with greatest efficiency, and the efficiency was only 7-fold lower than for insertion of natural dTTP, again despite the fact that the chlorinated compound is larger than dTTP by 0.5 Å. The change from Cl to Br to I in the template base (a size difference of 0.35 Å) caused a 22-fold drop in efficiency, apparently reflecting the steep potential energy function of a steric effect in the active site.
We also evaluated fidelity systematically by comparing correctly matched and mismatched pairs across this series. The efficiency of each correctly matched A-T analog pair was divided by the efficiency for enzymatic synthesis of the corresponding mismatched T-T analog pair, which is the most efficient mismatch, thus defining fidelity. The most efficient mismatches with the nonpolar thymine mimics were those opposite T, consistent with previous studies (8, 9), whereas natural thymidine is most frequently mispaired with guanine. The fidelity data are plotted graphically with respect to size in Fig. 3 c and d.
The results for template nucleoside mimics of variable size showed that fidelity increased with increasing size from the smallest compound and reached a maximum at the dichlorotoluene compound. The magnitude of increase in fidelity was large: the fidelity with the smallest compound (dH) was only 45, whereas, with dL, it was 1,100. This apparently optimally sized compound displayed fidelity that was within experimental error of that for natural thymidine. As size increased further, the fidelity then dropped markedly (by a factor of 40 for a size increase of 0.35 Å from dL to dI). Thus, the size effects on fidelity closely mirrored those seen for efficiency, suggesting a direct relationship between the physical origins of both effects.
The converse experiments evaluating fidelity effects with increasing dNTP size were similar, although not identical (see Table 2 and Fig. 3d). In this case, the fidelity maximum was reached at a slightly smaller size, dFTP, instead of dLTP, which are different by 0.38 Å in bond lengths. Particularly striking in this case was a large increase in fidelity on increasing size from dHTP to dFTP, reflecting a difference of 0.28 Å.
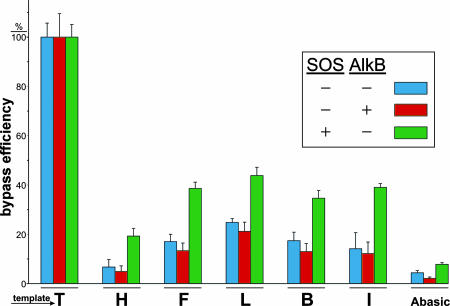
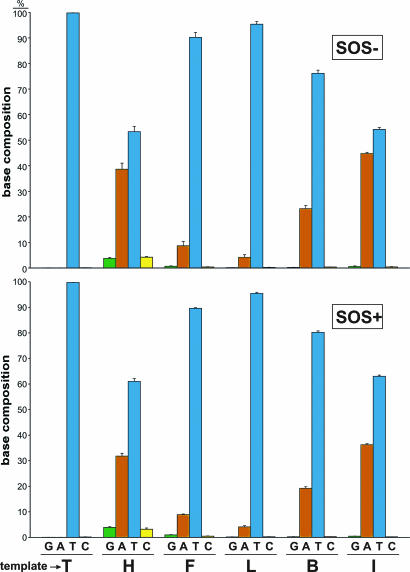
To ascertain the influence of subangstrom size increases on the template coding properties of the T analogs in vivo, we ligated oligonucleotides containing each analog into a single-stranded M13 vector, which were then passaged through E. coli and scored for bypass efficiency and mutagenesis (fidelity) as described in ref. 26. We chose E. coli deficient in AlkB to eliminate the possibility of metabolization of the H analog, which was subsequently found to have no influence on the results when compared with AlkB-proficient cells. Bypass experiments shown in Fig. 4 were performed in a competitive manner, and efficiencies were evaluated by PCR-based quantitation of successfully replicated progeny DNA (see Fig. 6, which is published as supporting information on the PNAS web site). SOS induction of bypass polymerases by prior irradiation of E. coli with UV light increased transanalog synthesis ≈2-fold for all thymine analogs. Although all five analogs hindered the progression of the normal and bypass polymerases, the bell-shaped histograms for the different cell types were similar to the above in vitro Pol I (Klenow fragment) studies, with maximum bypass for the L (dichloro) analog. Likewise, the replication fidelity experiments in E. coli shown in Fig. 5 depict bell curves for the normal and bypass polymerases with maximal in vivo fidelity for the analog L. Single-base deletions were also evident at the size extremes for the analogs H and I, which diminished when bypass polymerases were induced (Figs. 7 and 8, which are published as supporting information on the PNAS web site). It is noteworthy that the difference between the match and mismatch efficiency increased in cells expressing induced bypass polymerases [L (0.2), B (8.0), and I (17.4)], implying higher fidelity for the larger isosteres for bypass polymerases, which may contain larger or looser active sites.
Fig. 4.
Replication bypass efficiency of thymidine analogs of various sizes by normal and bypass (SOS-induced) polymerases in E. coli. The transanalog DNA synthesis experiment was performed in triplicate as described in Supporting Materials and Methods, with one standard deviation shown.
Fig. 5.
Fidelity of replication of thymidine analogs of various sizes by normal (Upper) and SOS-induced bypass (Lower) polymerases in E. coli. The distribution of G (green), A (red), T (blue), and C (yellow) at the analog site from the phage progeny is shown. Because the analogs function like T in vivo, T replaces them in the progeny. Base compositions were obtained in triplicate as described in Supporting Materials and Methods, with one standard deviation shown.
The above experiments give evidence for a number of significant mechanistic aspects of DNA replication. First, as base pairs increase in bulk, they can clearly reach a size at which they are rejected as efficient substrates. This result confirms the importance of steric rejection of many DNA lesions that increase nucleobase size, such as methylated bases and exocyclic adducts. Our data show that this rejection begins to be seen at size increases that are smaller than known DNA lesions. For example, with a small, ≈0.4-Å size increase over the optimum (only about a quarter of a single bond length), we observed a steric rejection amounting to over two orders of magnitude on efficiency, suggesting that DNA base lesions (which are larger) could easily achieve biologically relevant rejection levels with steric effects alone. The results are also consistent with studies involving deoxyribose variants with substituents added at the 4′ position, where overly large substitutions showed lowered polymerase activity (23).
Second, our observation that efficiency increases markedly with pair size up to the maximum suggests that there is a positive energetic influence of size (and/or a negative influence of insufficient size) at the transition state for phosphodiester bond formation. This result may be due to two factors: first, unfilled voids are energetically unfavorable in aqueous solution (29) and, second, too small of a size may lead to an incorrect positioning of the triphosphate moiety for phosphodiester bond formation. Natural base pairings that are smaller than the canonical Watson-Crick size, such as pyrimidine-pyrimidine mispairs, might easily be rejected by such mechanisms. Importantly, we observe that the natural base pair size is less than optimum in efficiency for Pol I (and indeed, for the replication machinery in intact E. coli), which demonstrated an efficiency maximum at a size 0.5-0.7 Å larger than a Watson-Crick pair taking into account hydrogen bonding contraction (see below). This finding is consistent with recent observations that low fidelity and low efficiency tend to correlate in the known DNA polymerases (30). If low fidelity is caused by a large active site, then the unfilled volume associated with such an active site should lower efficiency, as observed here with the smallest substrate sizes. Note that the current experiments would not distinguish between the physical resting space in the polymerase active site and induced space, whereby larger substrates might be accommodated by low-thermodynamic-cost movements of side chains or helices in the enzyme. Thus, we define the current steric space as the “functional space” rather than necessarily as a defined, static void; future structural studies of polymerases at high resolution may help make this distinction.
It is worthwhile to consider whether other chemical and physical factors in this nucleotide series might explain these results. For example, hydrophobicity and stacking propensity also change with increasing size (25). However, the changes in these two properties are relatively small across the series and increase from smallest to largest, a trend that clearly does not correlate with the observed effects. One may also consider possible electrostatic effects, because the electronegativity of the substituents also varies. The results show a relatively poor correlation; for example, H is electropositive relative to C, and I is neutral, yet the analogs containing them have similar activities. In addition, the difluoro analog has a considerably higher net dipole than the dichloro case, but the dichlorinated species is considerably more active. Previous studies with compounds having stronger net dipoles have also failed to note an effect with Pol I (31). Thus, the data are not well explained by these effects and are most consistent with the sized-based hypothesis.
The results have important implications in the mechanisms of fidelity in replicative DNA polymerases. Our observations of increasing fidelity with increasing size suggest that active site steric tightness is an important factor contributing to this selectivity. Increases in size from the smallest substrates to the optimum yielded substantial increases in fidelity, underscoring the need to fill the available active site space before steric differences between matched and mismatched partners can be discriminated. Second, the observation that fidelity is lost when pairs are too large, even by a small amount, is also consistent with a “tight active site” model (20); if the pair cannot be accommodated in appropriate side-by-side fashion in the active site, then the enzyme cannot exert the influence of a steric wall to constrain the pair's structure. Thus, both correct pairs (involving overly large thymidine analogs) and mispairs are similarly rejected. We observed that size increases much smaller than would be the case for most lesions or mispairs led to substantial losses in fidelity, which suggests that steric effects may be sufficient to explain most or all of this replicative enzyme's observed fidelity with natural base pairs.
A final and initially somewhat surprising conclusion from these experiments is that natural Watson-Crick pairs appear to be smaller than optimum for this enzyme, both in terms of efficiency and fidelity. Taking into account the hydrogen bonding contraction (32), models suggest that a T-A pair is 0.5-0.7 Å smaller than the optimum measured here (the L-A pair without contraction). This finding suggests that Pol I has an active site that is 0.5-0.7 Å larger than is ideal and that such a replicative polymerase might be rendered more efficient and yield higher fidelity than it does if it could evolve an active site that was sterically more constricted by this amount. We presume that such a structure is possible in a folded protein and we therefore suggest that the lack of optimum tightness likely arises for evolutionary reasons. We hypothesize that selection pressures in E. coli have yielded a lower fidelity than the maximum to confer adaptability that arises from a small but influential mutation rate.
Recent structural studies of low-fidelity DNA polymerases, which function to bypass mispairs or damaged pairs, have suggested greater steric openness at their active sites (13, 14). Low-fidelity polymerases are known to have low efficiency as well (29), consistent with the current observations in the Pol I active site. The current results suggest that a general mechanism for varying fidelity may be simply related to regulation of the sterically allowable sizes of base pairings. This idea leads to a testable prediction, namely, that the base pair size optimum may be larger or broader than that with a higher fidelity enzyme, such as Pol I, and that increasing base pair size in low-fidelity enzymes may increase their efficiency and fidelity. This nucleotide series of expanding size might be generally useful in probing functional steric tightness in a systematic way for enzymes of various fidelities.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants GM072705, CA80024, and ES11399. T.W.K. was supported by a Korea Science and Engineering Foundation Postdoctoral Fellowship.
Author contributions: J.M.E. and E.T.K. designed research; T.W.K. and J.C.D. performed research; T.W.K., J.C.D., J.M.E., and E.T.K. analyzed data; and J.C.D., J.M.E., and E.T.K. wrote the paper.
Conflict of interest statement: No conflicts declared
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: dF, 2,4-difluoro-5-toluene-1-β-d-deoxyriboside; dH, 3-toluene-1-β-d-deoxyriboside; dB, 2,4-dibromo-5-toluene-1-β-d-deoxyriboside; dI, 2,4-diiodo-5-toluene-1-β-d-deoxyriboside; dL, 2,4-dichloro-5-toluene-1-β-d-deoxyriboside.
References
- 1.Kunkel, T. A. & Bebenek, K. (2000) Annu. Rev. Biochem. 69, 497-529. [DOI] [PubMed] [Google Scholar]
- 2.Echols, H. & Goodman, M. F. (1991) Annu. Rev. Biochem. 60, 477-511. [DOI] [PubMed] [Google Scholar]
- 3.Kunkel, T. A. (2004) J. Biol. Chem. 279, 16895-16898. [DOI] [PubMed] [Google Scholar]
- 4.Beard, W. A. & Wilson, S. H. (2003) Structure (London) 11, 489-496. [DOI] [PubMed] [Google Scholar]
- 5.Hubscher, U., Maga, G. & Spadari, S. (2002) Annu. Rev. Biochem. 71, 133-163. [DOI] [PubMed] [Google Scholar]
- 6.Loeb, L. A. & Kunkel, T. A. (1982) Annu. Rev. Biochem. 52, 429-457. [DOI] [PubMed] [Google Scholar]
- 7.Petruska, J., Goodman, M. F., Boosalis, M. S., Sowers, L. C., Cheong, C. & Tinoco, I., Jr. (1988) Proc. Natl. Acad. Sci. USA 85, 6252-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moran, S., Ren, R. X.-F., Rumney, S. & Kool, E. T. (1997) J. Am. Chem. Soc. 119, 2056-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moran, S., Ren, R. X.-F. & Kool, E. T. (1997) Proc. Natl. Acad. Sci. USA 94, 10506-10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delaney, J. C., Henderson, P. T., Helquist, S. A., Morales, J. C., Essigmann, J. M. & Kool, E. T. (2003) Proc. Natl. Acad. Sci. USA 100, 4469-4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joyce, C. M. & Steitz, T. A. (1994) Annu. Rev. Biochem. 63, 777-822. [DOI] [PubMed] [Google Scholar]
- 12.Doublie, S., Sawaya, M. R. & Ellenberger, T. (1999) Structure (London) 7, R31-R35. [DOI] [PubMed] [Google Scholar]
- 13.Nair, D. T., Johnson, R. E., Prakash, S., Prakash, L. & Aggarwal, A. K. (2004) Nature 430, 377-380. [DOI] [PubMed] [Google Scholar]
- 14.Ling, H., Boudsocq, F., Woodgate, R. & Yang, W. (2001) Cell 107, 91-102. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, S. J. & Beese, L. S. (2004) Cell 116, 803-816. [DOI] [PubMed] [Google Scholar]
- 16.Beard, W. A. & Wilson, S. H., (2000) Mutat. Res. 460, 231-244. [DOI] [PubMed] [Google Scholar]
- 17.Kim, B., Ayran, J. C., Sagar, S. G., Adman, E. T., Fuller S. M., Tran, N. H. & Horrigan, J. (1999) J. Biol. Chem. 274, 27666-27672. [DOI] [PubMed] [Google Scholar]
- 18.Patel, P. H., Suzuki, M., Adman, E., Shinkai, A. & Loeb, L. A. (2001) J. Mol. Biol. 308, 823-837. [DOI] [PubMed] [Google Scholar]
- 19.Kraynov, V. S., Showalter, A. K., Liu, J., Zhong, X. & Tsai, M.-D. (2000) Biochemistry 39, 16008-16015. [DOI] [PubMed] [Google Scholar]
- 20.Kool, E. T. (2002) Annu. Rev. Biochem. 71, 191-219. [DOI] [PubMed] [Google Scholar]
- 21.Kool, E. T. (2001) Annu. Rev. Biophys. Biomol. Struct. 30, 1-33. [DOI] [PubMed] [Google Scholar]
- 22.Goodman, M. F. (1997) Proc. Natl. Acad. Sci. USA 94, 4469-4473.9114013 [Google Scholar]
- 23.Strerath, M., Cramer, J., Restle, T. & Marx, A. (2002) J. Am. Chem. Soc. 124, 2056-2057. [DOI] [PubMed] [Google Scholar]
- 24.Kim, T. W. & Kool, E. T. (2004) Org. Lett. 6, 3949-3952. [DOI] [PubMed] [Google Scholar]
- 25.Kim, T. W. & Kool, E. T. (2005) J. Org. Chem. 70, 2048-2053. [DOI] [PubMed] [Google Scholar]
- 26.Delaney, J. C. & Essigmann, J. M. (2004) Proc. Natl. Acad. Sci. USA 101, 14051-14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kornberg, A. (1992) in DNA Replication (Freeman, New York), 2nd Ed., pp 118-121.
- 28.Goodman, M. F., Creighton, S., Bloom, L. B. & Petruska, J. (1993) Crit. Rev. Biochem. Mol. Biol. 28, 83-126. [DOI] [PubMed] [Google Scholar]
- 29.Eriksson, A. E., Baase, W. A., Zhang, X. J., Heinz, D. W., Blaber, M., Baldwin, E. P. & Matthews, B. W. (1992) Science 255, 178-183. [DOI] [PubMed] [Google Scholar]
- 30.Beard, W. A., Shock, D. D., Vande Berg, B. J. & Wilson, S. H. (2002) J. Biol. Chem. 277, 47393-47398. [DOI] [PubMed] [Google Scholar]
- 31.Lai, J. S. & Kool, E. T. (2005) Chem. Eur. J. 11, 2966-2971. [DOI] [PubMed] [Google Scholar]
- 32.Pimentel, G. C. & McClellan, A. L. (1960) in The Hydrogen Bond (Freeman, London), pp. 282-293.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.