Abstract
Cofilin-actin rods are inclusion-like structures that are induced by certain chemical or physical stresses in cultured cells, and the rods formed in neurons are thought to be associated with neurodegeneration. Here, we cloned an Aplysia cofilin homolog and overexpressed it in cultured neurons. Overexpressed cofilin formed rod-like structures that included actin. The overall neuronal morphology was unaffected by cofilin overexpression; however, a decrease in number of synaptic varicosities was observed. Consistent with this structural change by cofilin overexpression, the synaptic strength was reduced, and furthermore, the long-term facilitation elicited by repeated pulses of 5-hydroxytryptamine was impaired in sensory-to-motor synapses. However, cofilin overexpression did not induce programmed cell death. These findings suggest that the formation of cofilin-actin rod-like structures can lead to neurodegeneration, and this might be a mechanism of rundown of neuronal and synaptic function without cell death in neurodegenerative diseases.
Keywords: neurodegeneration, Hirano body, long-term facilitation
Cofilin and actin-depolymerizing factor (ADF) are both actin-binding proteins with similar amino acid sequences and functions. They are very diverse between different species, but their overall structures and functions are comparably conserved. ADF/cofilin can bind to either G-actin or F-actin. The binding of ADF/cofilin to G-actin leads to sequestering of actin monomers, and binding to F-actin accelerates the dissociation of actin monomers from the filament and/or leads to severing of F-actin (1, 2). The destabilization of F-actin by ADF/cofilin binding is due to the structural modifications that alter the twist of actin-actin binding in F-actin helices. Thus, ADF/cofilin-bound F-actin does not bind to phalloidin, whose binding site in F-actin is modified by ADF/cofilin binding (3, 4).
ADF/cofilin activity is regulated by the phosphorylation of a serine residue at its N terminus, which results in the inactivation of ADF/cofilin. LIM-kinase and TES-kinase are known as kinases of ADF/cofilin (5-7), and slingshot and chronophin (8, 9) are known as phosphatases that reactivate cofilin (10). The phosphorylation or dephosphorylation state of cofilin can be mimicked in recombinant proteins by replacing the Ser-3 residue with glutamate (S3E) or alanine (S3A), respectively (11).
Actins and actin-binding proteins are critically involved in regulating the cytoskeleton structures (6, 7, 11, 12). Besides these general functions in regulating the actin cytoskeleton, ADF/cofilin forms a typical structure called an ADF/cofilin-actin rod or cofilin-actin rod. Cofilin-actin rods were initially called actin rods; these were later proved to contain cofilin (13). Cofilin-actin rods can be induced to form within the nucleus or cytoplasm by incubating cultured cells under specific chemical or physical conditions. In cultured neurons, neurodegenerative stimuli induce ADF/cofilin-actin rod formation, which results in the disruption of distal neurite function (11). A number of neurodegenerative diseases are known to be correlated with the formation of protein inclusions containing cytoskeletal proteins in the central nervous system. Thus, it is interesting to note that ADF/cofilin and actin can form inclusion-like rod structures. In fact, ADF/cofilin-actin rods have been suggested to be precursors of protein inclusions such as Hirano bodies, which have been found in many neurodegenerative diseases, including Alzheimer's disease (14-16), Parkinson's disease (17), and amyotrophic lateral sclerosis (17). Although many types of inclusions or protein aggregates are found in the brains of patients with neurodegeneration or in aged people, it is still unclear whether these structures actually lead to neurodegeneration or are only the byproducts produced in the process of neurodegeneration.
To determine the effect of cofilin-actin-rod formation on synaptic function and plasticity in a single neuron or at a single synapse level, the Aplysia neuron is thought to be one of the most optimal model systems because of the ease of manipulation at a single neuron level. Here, we showed that cofilin overexpression induced rod formation and led to synapse loss and impairment of synaptic plasticity, suggesting that these inclusion-like structures are involved in neurodegeneration.
Materials and Methods
Cloning of Aplysia Cofilin and Plasmid Construction. Three of the partially sequenced Aplysia EST clones have sequences homologous to known ADF/cofilin homology domains, and further sequence analysis predicted that full-length ORF sequences are included in these partial sequence. Based on the ORF sequence, we performed PCR with the following primers: Cofilin-S with an XbaI site, 5′-TGC TCT AGA GCC ACC ACC ATG TCT TCT GGG ATT AAG, and Cofilin-A with a BamHI site and without a stop codon, 5′-CGC GGA TCC CGG GAG TGG CTT TCC ACC. The PCR product was subcloned into the neuronal expression vector pNEXδ-GFP (18).
Cell Culture. Cell culture was performed as described in ref. 19. Aplysia kurodai was purchased from a local supplier in Pusan, South Korea, and maintained in recirculating seawater tanks at 14°C before use. Pleural sensory neurons were isolated from the pleural ganglion and cocultured with an identified LFS (Left F cluster innervating to the Siphon) motor cell, which was isolated from the abdominal ganglia and maintained at 18°C in an incubator for 4 days.
Microinjection. The various DNA constructs (1 mg/ml DNA) were dissolved in a buffer containing 0.1% Fast Green, 10 mM Tris·Cl (pH 7.3), and 100 mM NaCl and microinjected into Aplysia cultured sensory neurons by applying a positive air pressure, as described in refs. 18 and 20. The microinjected cells were incubated at 18°C for 24 h and used for electrophysiological measurement and immunocytochemistry.
Electrophysiology. Experiments were performed on culture day 4 after plating. LFS motor neurons cocultured with sensory neurons were impaled with sharp microelectrodes (10∼20 MΩ) filled with 2 M K-acetate, 0.5 M KCl, and 10 mM K-Hepes and hyperpolarized to -80 mV to prevent the cells from firing action potentials. Intracellular signals were amplified by using the Axoclamp-2B amplifier (Axon instruments, Union City, CA). Synaptic potentials were evoked in the LFS motor cell by stimulating each sensory cell with a brief (0.1∼0.5 ms) depolarizing pulse, using an extracellular electrode placed near the cell body of a sensory neuron. Data were stored on VCR tapes by using a digital data recorder (model VR-10B, InstruTECH, Port Washington, NY). To examine basal synaptic transmission, the excitatory postsynaptic potential (EPSP) was measured before and 24 h after microinjection. To investigate the effect of cofilin overexpression on synaptic plasticity, the initial EPSP value was measured 24 h after microinjection. The cultures then received five pulses of 5-hydroxytryptamine (5-HT) for 5 min at 15-min intervals to induce long-term facilitation. The amount of synaptic facilitation was calculated as the percentage change in EPSP amplitude recorded after the 5-HT treatment versus its initial value before treatment. All data are presented as the mean ± SEM of percentage change in the EPSP amplitude.
Cell Imaging. For immunostaining of actin, a previously described method by Schaefer et al. (21) was slightly modified. The cultures were washed with low Ca2+ and low-ionic-strength ASW (100 mM NaCl/10 mM KCl/5 mM MgCl2/15 mM Hepes/60 g/liter glycine, pH 7.9) containing 5 mM EGTA for 1 min, and then permeabilized with 1% Triton X-100 in stabilizing buffer (SB) containing 80 mM Pipes, 5 mM EGTA, and 1 mM MgCl2 plus 4% polyethylene glycol (PEG) (molecular weight 3,350) for 1 min. Cells were fixed with 3.7% formaldehyde in SB and washed with PBS. This preparation method for immunostaining extracts soluble proteins and improves visualization of the cytoskeletal structures (22). After blocking nonspecific binding by preincubating cells with 3% BSA in PBS, the cells were incubated for 1 h with the anti-actin Ab (1st Ab; Sigma), diluted 1:100 in blocking solution. After washing out unbound 1st Ab, the cells were incubated for 1 h with the Cy3-conjugated anti-rabbit IgG (2nd Ab), diluted 1:100∼1:500 in blocking solution. Immunofluorescence was observed under a confocal microscope (Radiance 2000, Bio-Rad; LSM 510, Zeiss). For phalloidin staining, a method for labeling F-actin, fixed cells were treated with Texas red phalloidin (Molecular Probes) in PBS (100 nM) for 30 min and observed with a confocal microscope.
For iontophoresis, the cultured neurons were impaled with microelectrodes (20∼30 MΩ) filled with 100 mM Alexa 594 staining dye in 200 mM KCl solution (Molecular Probes) followed by hyperpolarization with a 1-Hz, -0.5-nA current pulse of 500 ms duration for 5 min. To count the varicosity number, first images were taken shortly after microinjection of DNA and second images were taken 24 h after the injection. Varicosities were identified by the method described in ref. 23. The total amount of varicosity in the contact region between a sensory and motor neuron was counted.
Cytochalsin D was purchased from Sigma (catalog no. C8273), and the stock solution was made up to 1 mM in DMSO. Treatment was performed by dilution of stock solution in L15/ASW media (24).
TUNEL staining was performed as described by Lim et al. (25). As a positive control, cultured neurons were treated with 1 mM hydrogen peroxide for 3 h.
Results
Cloning of Aplysia Cofilin. cDNAs encoding the Aplysia cofilin homolog were obtained from the Aplysia kurodai EST database (24). Three of the partially sequenced EST clones contained sequences homologous to ADF/cofilin, and these had been proved to have the same 444-bp-long ORF (a protein with 147 aa of ≈16 kDa). The gene expression in the Aplysia neuron was confirmed by RT-PCR with RNA purified from cultured Aplysia pleural mechanosensory neuron as a template (data not shown).
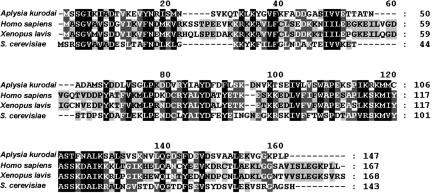
ADF/cofilins are composed of a single ADF homology domain (26) and have a phosphorylation site at their N terminus. Aplysia cofilin is also composed of a single ADF homology domain and has putative phosphorylation sites at Ser-2/Ser-3. The deduced amino acid sequence of Aplysia cofilin has 30% or more identity and ≈60% similarity with mammalian cofilin1 (nonmuscle isoform) and other eukaryotic homologs (Fig. 1).
Fig. 1.
Alignment of Aplysia cofilin amino acid sequences with ADF/cofilin proteins in other species. Proteins of the ADF/cofilin family are composed of a single ADF/cofilin homology domain. Aplysia cofilin has 32% identity (degree of exact match) and 54% similarity (degree of match including partially positive match between amino acids with similar property, from NCBI blastp results) with Homo sapiens cofilin1, 31% identity and 55% similarity Xenopus laevis Xac1, and 33% identity and 57% similarity with Saccharomyces cerevisiae cofilin1.
Previously, an Aplysia ADF/cofilin homologous protein was detected by staining with antisera to XAC1 and ADF/cofilin phosphoprotein antibody (27). This protein is slightly heavier than 15 kDa in molecular mass, and it is possible that the cloned protein of ≈16 kDa is the same as the Aplysia ADF/cofilin homologous protein.
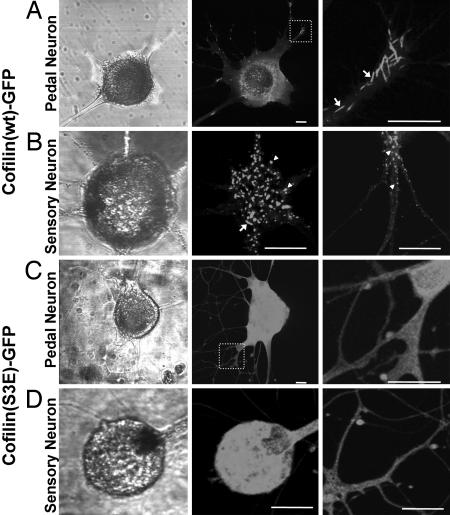
Expression Pattern of GFP-Fused Aplysia Cofilin Shows a Rod-Like Structure. To examine the expression pattern of Aplysia cofilin, we fused GFP to the C terminus of cofilin (pNEXδ-cofilin-GFP) and microinjected into primary cultured neurons dissociated from the central ganglia or pleural sensory cluster of Aplysia. At the initial stage of expression, when GFP signal was beginning to appear ≈6 to 12 h after microinjection, diffuse fluorescence of GFP was observed throughout the cell body and neurites (data not shown). However, fluorescent thick filaments or rod-like structures appeared mostly around the soma after ≈12 h, and these appeared extensively in both soma and neurites after 24 h (Fig. 2A). The rod-shaped patterns varied in length (1∼20 μm), and some of them had cylindrical shape that was similar to cofilin-actin rod found in mammalian systems (11, 13, 28, 29). The expression pattern in cultured pleural mechanosensory neuron was similar to that in other neurons cultured from the central ganglia, although the rod-like structures were relatively shorter (<10 μm), and some of them rather looked punctuated in sensory neurons (Fig. 2B). These results suggest that cloned Aplysia cofilin can form the rod-like structures similar to those previously reported for ADF/cofilin-actin rods.
Fig. 2.
Expression pattern of cofilin(wt)-GFP in cultured neurons. (A) Cultured Aplysia pedal neuron expressing cofilin(wt)-GFP shows rod-like pattern in the soma and neurites. Arrows indicate the cylindrical rod structures. (B)In cultured pleural mechanosensory neuron, rod-like structures tended to be shorter compared with pedal neuron. Punctuated structures are indicated by arrowheads. (C) Cofilin(S3E)-GFP expression in Aplysia pedal neuron showing diffused pattern. (D) In cultured sensory neurons, cofilin(S3E)-GFP also shows diffused expression pattern. In A and C, Right shows the magnified images of dotted square in Middle. (Scale bars: 25 μm).
Mutation in a Putative Phosphorylation Site Results in Change in Expression Pattern. Ser-3 of Aplysia cofilin was replaced with Glu to mimic the phosphorylated state of this protein (11). This mutant protein was also fused to GFP to visualize the subcellular localization. Cofilin(S3E)-GFP showed diffuse distribution both in the cytoplasm and in the neurites when expressed in cultured Aplysia neurons, and it did not show specific localization observed in cofilin(wt)-GFP expressing neurons (Fig. 2 C and D). This result suggests that Aplysia cofilin might be regulated by phosphorylation and dephosphorylation. Therefore, the rod-shaped structures observed in cofilin(wt)-GFP expressing neurons are thought to be induced by dephosphorylated active forms.
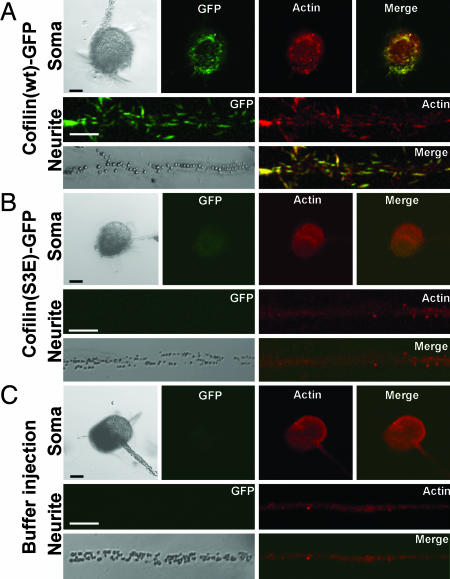
The Rod-Like Structure Induced by Cofilin Overexpression Contains Actin. To examine whether the rod-like structure contains actin, cofilin(wt)-GFP-expressing neurons were immunostained with anti-actin antibody. Anti-actin immunostaining showed the rod-like structures, which were mainly colocalized with cofilin(wt)-GFP (Fig. 3A). However, neurons expressing cofilin(S3E)-GFP (Fig. 3B) or buffer-injected control neurons (Fig. 3C) did not show thickly stained rod-like structures as did the cofilin(wt)-GFP expressing neurons. This finding suggests that the rod-shaped structures are induced by cofilin overexpression, not simply because of labeling of already existing structures by the binding of cofilin(wt)-GFPs.
Fig. 3.
Immunostaining of cultured Aplysia mechanosensory neurons with anti-actin antibody. (A) Cofilin(wt)-GFP (green) colocalized with actin staining pattern (red) in both soma and neurite. The colocalized pattern of cofilin and actin shows the rod-like structure. (B) Cofilin(S3E)-GFP shows diffused expression pattern, and actin staining did not exhibit the rod-shaped structure. The fluorescent intensity of GFP was weakened by our staining method (see Materials and Methods). (C) Actin staining pattern of buffer-injected neuron was similar to that of cofilin(S3E)-GFP-expressing neuron. (Scale bar: 50 μm.)
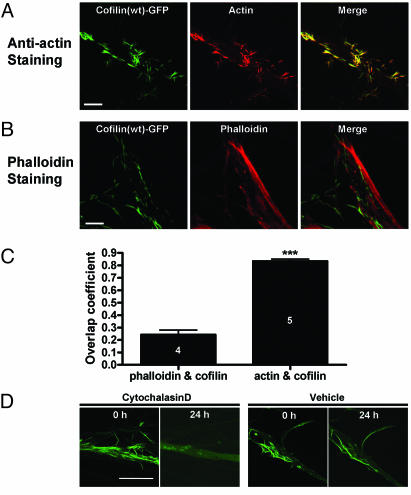
It was previously reported that cofilin-actin rods were not labeled with phalloidin, an agent which specifically labels F-actin (11, 13). Consistently, fluorescence from Texas red phalloidin did not colocalize with cofilin-GFP (Fig. 4A), whereas actin staining showed the prominent colocalization with cofilin-GFP (Fig. 4B) in Aplysia sensory neurons overexpressing cofilin(wt)-GFP. Overlap coefficient between actin and cofilin (0.834 ± 0.037, n = 5) was significantly higher than the overlap coefficient between phalloidin and cofilin (0.243 ± 0.037, n = 4; P < 0.0001, unpaired, two-tailed t test) (Fig. 4C). To further investigate the interaction between cofilin and F-actin, we incubated the cofilin-expressing neuron with cytochalasin D, an agent that disrupts F-actin (30). We found that the rod-like pattern of cofilin(wt)-GFP was disrupted 24 h after cytochalasin D incubation, whereas it was not in the vehicle control (Fig. 4D). Taken together, the rod-like structures are composed of cofilin and actin, probably F-actin, and are induced by cofilin overexpression.
Fig. 4.
Rod-like structures stain with anti-actin antibody, but not with fluorescent phalloidin. (A) Cofilin(wt)-GFP expression patterns colocalize with actin as a rod-like structure in Aplysia neurons. (B) Texas red phalloidin staining shows that the majority of cofilin(wt)-GFP do not colocalize with phalloidin. (C) Histogram shows the quantification data of colocalization between cofilin(wt)-GFP and actin or phalloidin. Overlap coefficient between actin and cofilin is 0.834 ± 0.016 (n = 5), and this is significantly higher than the overlap coefficient of cofilin and phalloidin (0.243 ± 0.037, n = 4; ***, P < 0.0001). The analysis was performed by using imagej software (National Institutes of Health, Bethesda; http://rsb.info.nih.gov/ij). (D) Cytochalasin D treatment abolished the pattern found in cofilin(wt)-GFP-expressing neuron, whereas vehicle treatment did not. (Scale bar: 20 μm.)
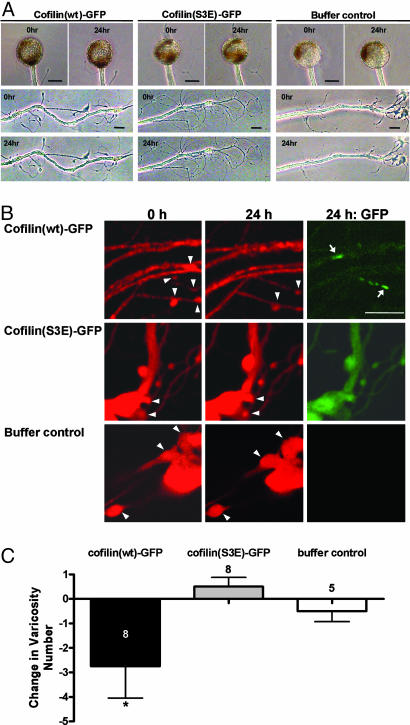
The Synaptic Structure Was Impaired by Cofilin-GFP Overexpression. Cofilin-GFP did not seem to affect the gross morphology of neurons. We could not find any change in cell body and major neurites after cofilin-GFP expression (Fig. 5A). In Aplysia, a presynaptic structure called varicosity is comparably large in size, and individual varicosities are identifiable when observed again after several days. Hence, using a sensory-to-motor neuron coculture system, we examined whether cofilin overexpression produces a change in varicosity number in sensory neurons forming synapses with motor neurons. To see the fine neuronal morphology, Alexa 594 dye was iontophoresed into the sensory neurons as a whole-cell marker. We found that the varicosity number was decreased 24 h after microinjection in cofilin(wt)-GFP-expressing neurons (-2.8 ± 1.2, n = 8) but not in cofilin(S3E)-GFP-expressing neurons (0.5 ± 0.4, n = 8) and buffer-injected control neurons (-0.5 ± 0.4, n = 5; one-way ANOVA: F = 3.90, df = 2, P < 0.05) (Fig. 5 B and C), indicating that cofilin overexpression disrupts the synaptic structure. To see whether the loss of varicosity was directly related with the rod-like structure, we compared the expression pattern of cofilin(wt)-GFP with the Alexa 594 image. However, we did not observe any correlation between synaptic loss and the position of the rod-like structure (Fig. 5).
Fig. 5.
Effect of cofilin expression on the number of synaptic varicosities. (A) Overall cell morphology was not affected by cofilin(wt)-GFP expression. Cell morphology of 24 h after microinjection was compared with initial morphology. (B) Varicosity number is reduced in cofilin-GFP-expressing neurons, whereas it is not altered in either cofilin(S3E)-GFP-expressing neuron or buffer control neuron. Arrowheads and arrows indicate individual varicosities and rod-like structures, respectively. (C) Histogram shows significant reduction in varicosity number in the cofilin(wt)-GFP-expressing neurons as compared with those in the cofilin(S3E)-GFP-expressing neurons and buffer-injected neurons. The height of each bar corresponds to the mean ± SEM of change in varicosity number (*, P < 0.05). (Scale bar: 20 μm.)
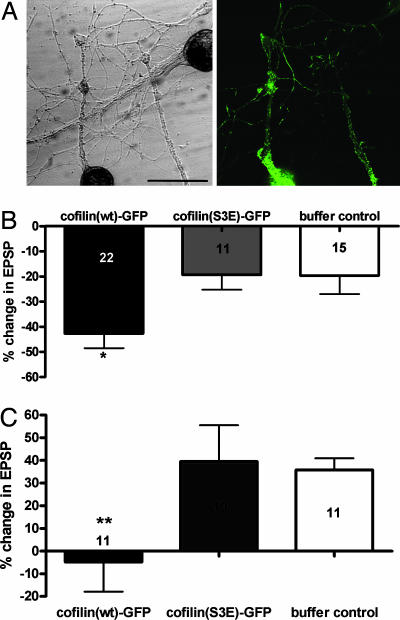
The Overexpression of Cofilin(wt)-GFP Impairs Both Basal Synaptic Transmission and Long-Term Synaptic Plasticity. Given that cofilin-(wt)-GFP expression affected the presynaptic structure, we next examined whether cofilin expression affects synaptic strength or synaptic plasticity. We performed the electrophysiological measurement of EPSPs in cofilin-GFP-expressing neurons. We measured the EPSP (1st EPSP) and then injected pNEXδ-cofilin-GFP DNA into presynaptic sensory neuron. The 2nd EPSP was measured 24 h after the microinjection. Expression of cofilin(wt)-GFP resulted in a significant decrease in EPSP amplitude (-42.7 ± 5.9%, n = 22), and this decrease is much bigger than that in cofilin(S3E)-GFP (-19.4 ± 5.9%, n = 11) and in buffer-injected control (-19.7 ± 7.3%, n = 15; one-way ANOVA: F = 4.64, df = 2, P < 0.02) (Fig. 6B), suggesting that Aplysia cofilin decreases the synaptic strength.
Fig. 6.
Effect of cofilin expression on the basal synaptic transmission. (A) Cofilin(wt)-GFP was expressed in sensory neurons synapsed with motor neurons, and EPSPs were measured before (0 h) and 24 h after microinjection. (Right) GFP fluorescence image of cultured neurons in Left. S, sensory neuron; M, motor neuron. (Scale bar: 50 μm.) (B) Cofilin(wt)-GFP expression in presynaptic sensory neurons showed a significant reduction in EPSP size as compared with cofilin(S3E)-GFP-expressing and buffer-injected neurons (*, P < 0.02). (C) Five pulses of 5-HT failed to increase the EPSP amplitude in cofilin(wt)-GFP-expressing neurons as compared with cofilin(S3E)-GFP-expressing neurons and buffer-injected control neurons (**, P < 0.01).
Next, we examined whether cofilin overexpression affects long-term synaptic facilitation induced by repeated pulses of 5-HT. Cofilin(wt)-GFP-expressing neurons did not produce an increase in EPSP (-4.9 ± 13.0%, n = 11) by five pulses of 5-HT treatment. In contrast, cofilin(S3E)-GFP-expressing neurons and buffer-injected control neurons showed a normal long-term facilitation (39.5 ± 16.1%, n = 10 and 35.8 ± 5.1%, n = 11, respectively; one-way ANOVA: F = 4.24, df = 2, P < 0.01) (Fig. 6C). Therefore, besides a decrease in the number of synaptic varicosities, cofilin-(wt)-GFP expression weakens synaptic strength and impairs long-term facilitation.
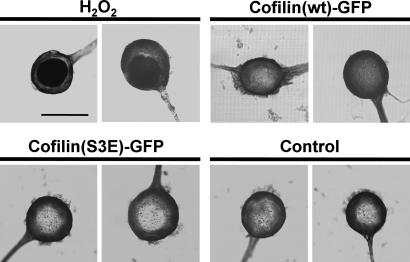
Synapse Loss by Cofilin-Actin Rod Formation Is Not Caused by Neuronal Cell Death. Because it was reported that cofilin is involved in an early step during the apoptosis induction (31), it is possible that synapse loss in cofilin-overexpressing neurons accompanies the process of cell death. Hence, we next examined whether cofilin expression induced apoptosis by the TUNEL assay. Endonucleolysis is known as an indicative process observed in apoptotic neurons (25). However, we could not detect the endonucleolytic process in cofilin-overexpressing neurons as shown by TUNEL staining 24 h after microinjection (n = 7) (Fig. 7). Positive control neurons that were treated with 1 mM hydrogen peroxide (22) were TUNEL-positive (n = 8), and cofilin(S3E)-GFP-expressing neurons (n = 4) and buffer-injected control neurons (n = 5) were TUNEL-negative. These data showed that nucleolytic apoptosis was not induced by cofilin overexpression, and the effects of cofilin overexpression on synaptic strength and plasticity are unlikely to be due to apoptotic processes.
Fig. 7.
TUNEL staining of cofilin-overexpressing neurons. Cofilin(wt)-GFP-expressing sensory neurons showed TUNEL-negative staining (five of five cells), whereas neurons treated with 1 mM hydrogen peroxide for 1 h showed TUNEL-positive staining (eight of eight cells). Cofilin(S3E)-GFP-expressing neurons (four of four cells) and control neurons (five of five cells) showed TUNEL-negative staining. Two neurons in each group are shown in this figure. (Scale bar: 50 μm.)
Discussion
In the present study, we found that cofilin overexpression induced rod-like structures in cultured Aplysia neuron, and the rod-like structures were composed of cofilin and actin. Cofilin overexpression disrupted synaptic structure and function without affecting neuronal survival.
Rod-like structures found in cofilin-overexpressing neurons were stained with anti-actin antibody, and these colocalization patterns of actin and cofilin-GFP are similar to those previously reported for ADF/cofilin-actin rods that are found in cells subjected to some stressful stimuli, such as heat shock; in such a case, rods are found in the nucleus or cytoplasm and are not co-stained with phalloidin (13). The rod-like structure in Aplysia neuron was not stained with phalloidin and disappeared after cytochalasin D treatment, indicating that it might be a cofilin-saturated F-actin structure.
ADF/cofilin-actin rods are also found in cultured rat hippocampal neurons treated with neurodegenerative stimuli (11). In this case, the neurodegenerative stimuli (e.g., glutamate treatment) appear to induce ADF/cofilin overactivation and lead to the formation of ADF/cofilin rods. Overexpression of wild-type (wt) or active mutant (A3) of Xenopus ADF/cofilin (Xac), which can mimic cofilin overactivation, also induced rod formation. However, cofilin overexpression does not always induce cofilin-actin rod formation. For example, Xac(wt) or Xac(A3) overexpression in cultured rat cortical neurons did not induce rod structures (11), but in this case, an increase in neurite outgrowth was observed. Differences in these results might be due to the difference in cell types used in which ADF/cofilin might be differently regulated or a difference in expression level between their experimental systems.
Our data showed that cofilin overexpression disrupted synaptic structure and function but did not affect neuronal survival in Aplysia neuron. Given these results, the rod-like structures observed in our study seem to be similar in some aspects to the protein inclusions such as Hirano bodies, because Hirano bodies are also mainly composed of actin and actin-associated proteins, including ADF and cofilin. Furthermore, Hirano bodies are not thought to affect cell survival because surviving neurons in neurodegenerative diseases have intact cell bodies despite synapse loss (11); and the induced formation of Hirano bodies in Dictyostelium and some other cell lines is not associated with cell death and does not significantly affect cell growth (32). The cause of Hirano-body formation has not been determined, but it appears to be related to the abnormal regulation of the actin cytoskeleton (11, 32). Taken together, the ADF/cofilin-actin rod has been suggested to be a precursor of the Hirano body (11).
How are the rod-like structures formed by the overexpression or overactivation of cofilin? Cofilin is known to accelerate actin bundling (13, 33), which may lead to aggregation formation. Another possibility is that cofilin may bind to F-actin bundles and shear it, thereby producing discrete rod structures (28). However, this may not be our case because we could not find such prominent F-actin bundle structures in Aplysia neuron (Fig. 3 B and C). It is also possible that cofilin may induce F-actin bundling (34), and this F-actin bundle is subsequently sheared to form rod-like structures. In this aspect, relatively long filament-like structures could be regarded as F-actin bundles induced by cofilin overexpression. In either case, cofilin overexpression might convert monomeric actins to the insoluble forms, thereby leading to depletion in the monomeric actin pool.
In this manner, cofilin-actin rods may sequester actin monomers (32), thereby reducing the available G-actin sources, and eventually resulting in the collapse of the actin cytoskeleton required for maintaining synaptic structures and trafficking synaptic vesicles. Another possible mechanism for the synapse loss by cofilin-actin rods is the disruption of the microtubule structure by the rod formation; this may lead to the malfunction in axonal transport in a cell-wide manner and eventually result in disruption of synaptic function as suggested by Minamide et al. (11). Furthermore, because we observed the structural change in the varicosity that did not contain the rod-like structure, the disruptive effect of cofilin overexpression is not synapse-specific but cell-wide.
Synaptic plasticity requires the modulation of the actin cytoskeleton (35-38) in which actin is a key downstream molecule that regulates synaptic growth. F-actin reorganization is important in regulating spine morphogenesis during long-term potentiation in mammals (35-37). Similarly, the 5-HT signal activates molecules that reorganize the presynaptic actin network in Aplysia neurons (38). Therefore, the impairment of long-term facilitation could result from disturbing the actin reorganization by cofilin overexpression. Besides destabilizing the existing synaptic structures, a shortage of required molecules, including actin, caused by cofilin-actin-rod formation might interfere with synaptic growth induced by repeated pulses of 5-HT.
Cofilin(wt)-GFP expression induced rod-like structure formation, but the rod structures appeared only a given amount of time after microinjection. The overexpressed cofilin(wt)-GFP appears to be regulated by endogenous kinase(s) at the initial stage of overexpression when GFP fluorescence showed a diffuse pattern. However, at a later stage, the overexpressed cofilin might overwhelm the ability of the endogenous kinase(s), possibly leading to an increase in active forms (unphosphorylated wild-type proteins), thus mimicking cofilin overactivation and inducing rod-like structure formation. Although we overexpressed cofilin to mimic overactivation in our study, the activity of endogenous cofilin might be regulated by posttranslational regulation in neurodegenerative disease. This idea is supported by the finding that there is no difference in the expression levels of both ADF and cofilin between the hippocampal tissues from normal individuals and Alzheimer's disease patients (39).
In conclusion, we showed that abnormal function of cofilin could induce rod-like structures and disrupt both synaptic structure and synaptic function in Aplysia sensory-to-motor synapse. These results could provide a clue for revealing the cellular and molecular mechanisms underlying memory and cognitive dysfunction in neurodegenerative diseases.
Acknowledgments
This work was supported by the Korea Ministry of Science and Technology under the National Research Laboratory Program (M1-0104-00-0140) and the BRC-Frontier Program (M103KV010012 03K2201 01210), and by the Marine and Extreme Genome Research Center Program, Ministry of Marine Affairs and Fisheries, Republic of Korea. D.-H.J., J.-H.H., Sue-Hyun Lee, and H.K. were supported by a BK21 Research Fellowship from the Korea Ministry of Education and Human Resources Development.
Author contributions: D.-H.J., J.-H.H., Seung-Hee Lee, and B.-K.K. designed research; D.-H.J., J.-H.H., Seung-Hee Lee, Sue-Hyun Lee, and H.K. performed research; Y.-S.L. and H.P. contributed new reagents/analytic tools; D.-H.J., J.-H.H., Seung-Hee Lee, Y.-S.L., H.P., and B.-K.K. analyzed data; and D.-H.J. and B.-K.K. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: ADF, actin-depolymerizing factor; EPSP, excitatory postsynaptic potential; 5-HT, 5-hydroxytryptamine.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ094143).
References
- 1.Moon, A. & Drubin, D. G. (1995) Mol. Biol. Cell 6, 1423-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maciver, S. K. & Hussey, P. J. (2002) Genome Biol. 3, reviews3007. [DOI] [PMC free article] [PubMed]
- 3.Galkin, V. E., Orlova, A., Lukoyanova, N., Wriggers, W. & Egelman, E. H. (2001) J. Cell Biol. 153, 75-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGough, A., Pope, B., Chiu, W. & Weeds, A. (1997) J. Cell Biol. 138, 771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohashi, K., Hosoya, T., Takahashi, K., Hing, H. & Mizuno, K. (2000) Biochem. Biophys. Res. Commun. 276, 1178-1185. [DOI] [PubMed] [Google Scholar]
- 6.Sarmiere, P. D. & Bamburg, J. R. (2004) J. Neurobiol. 58, 103-117. [DOI] [PubMed] [Google Scholar]
- 7.Tojima, T. & Ito, E. (2004) Prog. Neurobiol. 72, 183-193. [DOI] [PubMed] [Google Scholar]
- 8.Gohla, A., Birkenfeld, J. & Bokoch, G. M. (2005) Nat. Cell Biol. 7, 21-29. [DOI] [PubMed] [Google Scholar]
- 9.Wiggan, O., Bernstein, B. W. & Bamburg, J. R. (2005) Nat. Cell Biol. 7, 8-9. [DOI] [PubMed] [Google Scholar]
- 10.Niwa, R., Nagata-Ohashi, K., Takeichi, M., Mizuno, K. & Uemura, T. (2002) Cell 108, 233-246. [DOI] [PubMed] [Google Scholar]
- 11.Minamide, L. S., Striegl, A. M., Boyle, J. A., Meberg, P. J. & Bamburg, J. R. (2000) Nat. Cell Biol. 2, 628-636. [DOI] [PubMed] [Google Scholar]
- 12.Kuhn, T. B., Meberg, P. J., Brown, M. D., Bernstein, B. W., Minamide, L. S., Jensen, J. R., Okada, K., Soda, E. A. & Bamburg, J. R. (2000) J. Neurobiol. 44, 126-144. [PubMed] [Google Scholar]
- 13.Nishida, E., Iida, K., Yonezawa, N., Koyasu, S., Yahara, I. & Sakai, H. (1987) Proc. Natl. Acad. Sci. USA 84, 5262-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson, P. H. & Tomlinson, B. E. (1977) J. Neurol. Sci. 33, 199-206. [DOI] [PubMed] [Google Scholar]
- 15.Mitake, S., Ojika, K. & Hirano, A. (1997) Kaohsiung J. Med. Sci. 13, 10-18. [PubMed] [Google Scholar]
- 16.Mori, H., Tomonaga, M., Baba, N. & Kanaya, K. (1986) Acta Neuropathol. (Berlin) 71, 32-37. [DOI] [PubMed] [Google Scholar]
- 17.Hirano, A., Dembitzer, H. M., Kurland, L. T. & Zimmerman, H. M. (1968) J. Neuropathol. Exp. Neurol. 27, 167-182. [PubMed] [Google Scholar]
- 18.Kaang, B. K. (1996) Neurosci. Lett. 221, 29-32. [DOI] [PubMed] [Google Scholar]
- 19.Schacher, S. & Proshansky, E. (1983) J. Neurosci. 3, 2403-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaang, B. K., Pfaffinger, P. J., Grant, S. G., Kandel, E. R. & Furukawa, Y. (1992) Proc. Natl. Acad. Sci. USA 89, 1133-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaefer, A. W., Kabir, N. & Forscher, P. (2002) J. Cell Biol. 158, 139-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bershadsky, A. D., Gelfand, V. I., Svitkina, T. M. & Tint, I. S. (1980) Exp. Cell Res. 127, 421-429. [DOI] [PubMed] [Google Scholar]
- 23.Han, J. H., Lim, C. S., Lee, Y. S., Kandel, E. R. & Kaang, B. K. (2004) Learn. Mem. 11, 421-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huh, M., Han, J. H., Lim, C. S., Lee, S. H., Kim, S., Kim, E. & Kaang, B. K. (2003) J. Neurochem. 85, 282-285. [DOI] [PubMed] [Google Scholar]
- 25.Lim, C. S., Lee, J. C., Kim, S. D., Chang, D. J. & Kaang, B. K. (2002) Brain Res. 941, 137-145. [DOI] [PubMed] [Google Scholar]
- 26.Lappalainen, P., Kessels, M. M., Cope, M. J. & Drubin, D. G. (1998) Mol. Biol. Cell 9, 1951-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaw, A. E., Minamide, L. S., Bill, C. L., Funk, J. D., Maiti, S. & Bamburg, J. R. (2004) Electrophoresis 25, 2611-2620. [DOI] [PubMed] [Google Scholar]
- 28.Rathke, P. C., Seib, E., Weber, K., Osborn, M. & Franke, W. W. (1977) Exp. Cell Res. 105, 253-262. [DOI] [PubMed] [Google Scholar]
- 29.Iida, K. & Yahara, I. (1986) Exp. Cell Res. 164, 492-506. [DOI] [PubMed] [Google Scholar]
- 30.Cooper, J. A. (1987) J. Cell Biol. 105, 1473-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chua, B. T., Volbracht, C., Tan, K. O., Li, R., Yu, V. C. & Li, P. (2003) Nat. Cell Biol. 5, 1083-1089. [DOI] [PubMed] [Google Scholar]
- 32.Maselli, A., Furukawa, R., Thomson, S. A., Davis, R. C. & Fechheimer, M. (2003) Eukaryot. Cell 2, 778-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfannstiel, J., Cyrklaff, M., Habermann, A., Stoeva, S., Griffiths, G., Shoeman, R. & Faulstich, H. (2001) J. Biol. Chem. 276, 49476-49484. [DOI] [PubMed] [Google Scholar]
- 34.Aizawa, H., Sutoh, K. & Yahara, I. (1996) J. Cell Biol. 132, 335-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukazawa, Y., Saitoh, Y., Ozawa, F., Ohta, Y., Mizuno, K. & Inokuchi, K. (2003) Neuron 38, 447-460. [DOI] [PubMed] [Google Scholar]
- 36.Meng, Y., Zhang, Y., Tregoubov, V., Falls, D. L. & Jia, Z. (2003) Rev. Neurosci. 14, 233-240. [DOI] [PubMed] [Google Scholar]
- 37.Meng, Y., Zhang, Y., Tregoubov, V., Janus, C., Cruz, L., Jackson, M., Lu, W. Y., MacDonald, J. F., Wang, J. Y., Falls, D. L. & Jia, Z. (2002) Neuron 35, 121-133. [DOI] [PubMed] [Google Scholar]
- 38.Udo, H., Jin, I., Kim, J. H., Li, H. L., Youn, T., Hawkins, R. D., Kandel, E. R. & Bailey, C. H. (2005) Neuron 45, 887-901. [DOI] [PubMed] [Google Scholar]
- 39.Maciver, S. K. & Harrington, C. R. (1995) NeuroReport 6, 1985-1988. [DOI] [PubMed] [Google Scholar]