Abstract
An 11-residue peptide with the sequence DSLEFIASKLA was identified from a genomic library of Bacillus subtilis by phage display as an efficient substrate for Sfp phosphopantetheinyl transferase-catalyzed protein labeling by small molecule-CoA conjugates. We name this peptide the “ybbR tag,” because part of its sequence is derived from the ybbR ORF in the B. subtilis genome. The site of Sfp-catalyzed ybbR tag labeling was mapped to the underlined Ser residue, and the ybbR tag was found to have a strong tendency for adopting an α-helical conformation in solution. Here we demonstrate that the ybbR tag can be fused to the N or C termini of target proteins or inserted in a flexible loop in the middle of a target protein for site-specific protein labeling by Sfp. The short size of the ybbR tag and its compatibility with various target proteins, the broad substrate specificity of Sfp for labeling the ybbR tag with small-molecule probes of diverse structures, and the high specificity and efficiency of the labeling reaction make Sfp-catalyzed ybbR tag labeling an attractive tool for expanding protein structural and functional diversities by posttranslational modification.
Keywords: coenzyme A, posttranslational modification, Bacillus subtillis, phage display
Nature expands the diversity of protein structure and function by posttranslational modification (1). To name a few examples, protein phosphorylation creates docking sites for partner proteins in kinase-dependent signaling pathways; protein glycosylation provides handles for selective receptor recognition; protein myristoylation and palmitoylation anchor the modified proteins to the membrane microenvironment; and protein lipoylation and phosphopantetheinylation install “swinging arms” responsible for substrate channeling during coupled enzymatic transformations.
Learning from nature, several investigators have described methods for site-specifically modifying proteins with small synthetic molecules so that affinity, fluorescent, and photocrosslinking probes are posttranslationally attached to the proteins of interest to study their biological functions in the cell proteome (2, 3). For example, short peptide sequences with a tetracysteine motif were found to react with biarsenical, allowing proteins fused to the peptide tag to be labeled with biarsenical fluorophores in living cells (4). As another example, the biotin ligase, BirA, was used to biotinylate a lysine side chain within a 15-residue acceptor peptide (AP) (5), and cell surface receptors fused with exposed AP peptide tag were labeled by biotin with BirA and subsequently labeled with fluorophores by the binding of streptavidin-fluorophore conjugates (6). Specific protein labeling has also been achieved by expressing the target protein as a fusion to O6-alkylguanine-DNA alkyltransferase (AGT), which catalyzes the transfer of the small-molecule label from O6-guanine to a reactive cysteine residue of AGT (7). The current methods often suffer drawbacks, such as high labeling background [tetracysteine tag (8)], indirect protein labeling [biotin ligase labeling with the use of streptavidin to attach the fluorophores in a second step (6)], and the large size of the AGT tag (207 residues) with the required use of AGT-deficient cell lines or inhibitors of wild-type AGT during the labeling process (9).
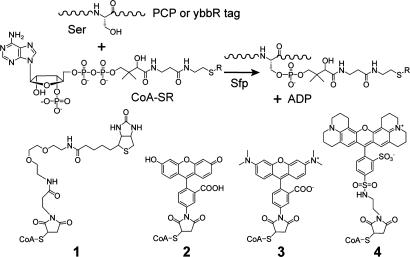
We recently reported the use of peptide carrier proteins (PCPs) for site-specific protein labeling with small molecules by Sfp phosphopantetheinyl transferase (10). PCPs are 80- to 120-residue domains or subunits distributed along the assembly line of multimodular biosynthetic enzyme nonribosomal peptide synthetase (NRPS) (11). In Bacillus subtilis, Sfp transfers the 4′-phosphopantetheinyl (Ppant) group of CoA onto a conserved serine residue of PCP in NRPS or acyl carrier protein (ACP) in polyketide synthase (PKS) (12) (Fig. 1). This posttranslational modification converts PCPs and ACPs from their inactive apo forms into active holo forms in which the Ppant group acts as a swinging arm for the transport of the reactive intermediates between spatially distinct reaction centers on NRPS and PKS. In addition to CoA, Sfp was found to covalently transfer small-molecule-Ppant conjugates to a PCP when the small molecules were attached to CoA via thioester, thioether, or disulfide linkages (13, 14). Thus, PCP was used as a tag for protein labeling by expressing the target proteins as fusions to an 80-residue PCP from a NRPS module, GrsA (10). The Sfp-catalyzed PCP-labeling reaction has been shown to proceed with high efficiency and specificity in cell lysates or on the surfaces of phages and live cells (10, 15, 16). Small-molecule probes, including biotin, Alexa Fluor dye, porphyrin, crosslinked peptides, and sugars, can all be site-specifically attached to PCP fused to the target protein in one simple step within 10-20 min. By using Sfp-catalyzed PCP labeling, we recently imaged the formation and endocytosis of the transferrin-transferrin receptor 1 complex during transferrin-mediated iron uptake by FRET (16). It has also been reported that α-agglutinin receptor and G protein-coupled receptor neurokinin-1 were fused to an ACP from Escherichia coli and specifically labeled with fluorophores by E. coli phosphopantetheinyl transferase (17).
Fig. 1.
Sfp-catalyzed PCP or ybbR tag modification at a specific Ser residue by various small-molecule-CoA conjugates (CoA-SR): 1, biotin-CoA; 2, fluorescein-CoA; 3, tetramethylrhodamine-CoA; and 4, Texas red-CoA.
Although ACPs and PCPs are of relatively small size (75-80 residues) compared with GFP (229 residues) (18), AGT (207 residues) (7), and mini-intein (154 residues) (19) that have been used for fusion protein construction and subsequent protein labeling, they are still significantly larger in size than the AP peptide (15 residue) for BirA-catalyzed biotin labeling (5) and many of the commonly used affinity peptide tags such as 6× histidine tag (20), myc tag (10 residues) (21), or flag tag (8 residues) (22). To overcome the structural bulk of ACPs or PCPs as peptide tags for protein labeling, we report here the development of a peptide tag known as the ybbR tag that can be as short as 11 residues in length and efficiently modified by Sfp with small-molecule probes of diverse structures. The ybbR tag was identified as an efficient substrate of Sfp from a genomic library of B. subtilis by phage display, and the N-terminal half of the tag was derived from an ORF of unknown function named ybbR in the B. subtilis genome. We have shown that the ybbR tag can be fused to either the N or C terminus or inserted within a flexible loop region of the target protein, and the ybbR-tagged proteins can be labeled by Sfp with small-molecule probes of diverse structures with high efficiency and specificity.
Materials and Methods
Detailed experimental procedures for the synthesis of CoA conjugates 1-4, cloning, Fourier-transform MS, circular dichroism (CD) measurements, and protein labeling are in supporting information, which is published on the PNAS web site.
Peptide-Labeling Kinetics. To test whether a specific peptide was the substrate of Sfp-catalyzed biotin-CoA modification, 200 μM biotin-CoA and 1 μM Sfp were incubated with 100 μM peptide in a 100-μl solution of 10 mM MgCl2/50 mM Hepes, pH 7.5, for 30 min at 37°C. Control reactions were also run in parallel with either Sfp or biotin-CoA excluded from the reaction. Reactions were then quenched by adding 30 μl of 4% trifluoroacetic acid (TFA) and analyzed by analytical HPLC with a reverse-phase C18 column by using a gradient of 0-60% CH3CN in 0.1% TFA/H2O over 30 min and monitored at 220 nm. Peptide-labeling reactions were also carried out at various pHs ranging from 5.0 to 8.5 with various buffering reagents (sodium acetate, 50 mM, pH 5.0; Mes, 50 mM, pH 6.0; Hepes, 50 mM, pH 7.0; Hepes, 50 mM, pH 8.0; Tris·HCl, 50 mM, pH 8.5) to test the effect of pH on the rate of Sfp-catalyzed peptide labeling. For the determination of kinetic parameters for Sfp-catalyzed peptide labeling at a saturating concentration of biotin-CoA or fluorescein-CoA, Sfp was added to a final concentration of 1 μM in 10 mM MgCl2/50 mM Hepes, pH 7.5, buffer with varying concentrations of the peptide ranging from 2 to 500 μM while holding the concentrations of the biotin-CoA or fluorescein-CoA conjugate constant at 150 μM. For the determination of kinetic parameters at saturating concentration of the peptide, the peptide concentration was held at 500 μM and the biotin-CoA or fluorescein-CoA concentration was varied from 2 to 200 μM. The reaction was allowed to proceed at 37°C for 5 min and was quenched and analyzed by HPLC as described above. HPLC peak areas were integrated, and the product concentration was calculated as a percent of the total peak area. Initial velocity data were fit to the Michaelis-Menten equation by the computer software kaleida-graph (Synergy Software, Reading, PA). The kinetic parameters for the Sfp-catalyzed PCP labeling were carried out in the same buffer (10 mM MgCl2/50 mM Hepes, pH 7.5) in the presence of 0.1 μM Sfp by either varying the concentration of PCP from 0.5 to 50 μM at a constant biotin-CoA concentration of 150 μM or varying the biotin-CoA concentration from 2 to 200 μM at a constant PCP concentration of 20 μM. The reaction was allowed to proceed at 37°C for 5 min before quenching by the addition of 30 μlof4%TFA to a 100-μl reaction mixture. The reaction was then analyzed by analytical HPLC with a reverse-phase C18 column by using a gradient of 30-50% CH3CN in 0.1% TFA/H2O over 30 min and was monitored at 280 nm.
Results
Identification of Truncated ybbR Proteins as Substrates of Sfp. We previously showed that PCP displayed on the surface of M13 phages can be specifically labeled with biotin by Sfp-catalyzed biotin-CoA transfer (15). As part of our effort to identify proteins subjected to Sfp-catalyzed posttranslational modification in the B. subtilis proteome, a genomic library of B. subtilis was displayed on the surface of M13 phages as pIII fusion proteins and selected for Sfp-catalyzed biotin-Ppant modification. Phages displaying proteins recognized by Sfp for posttranslational modification were covalently labeled with the biotin-Ppant group and selected by binding to immobilized streptavidin. The proteins subjected to Sfp-catalyzed posttranslational modification were then identified by sequencing the selected phage clones. The detailed phage selection results will be published elsewhere.
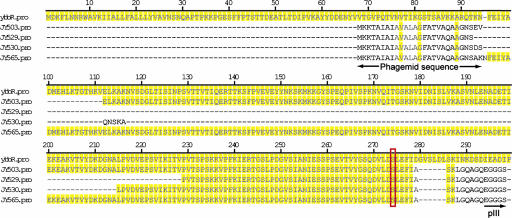
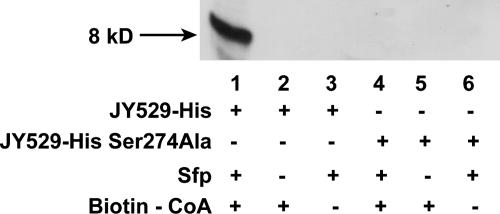
Besides known ACPs and PCPs, we found that truncated forms of the predicted ybbR protein (23) (residues 1-484 for full length) corresponding to residues 95-278 (JY565), 111-278 (JY503), 214-278 (JY530), and 229-278 (JY529) were selected multiple times (Fig. 2), and Sfp-catalyzed biotin labeling of phage-displayed ybbR truncates was confirmed by ELISA. JY529, the shortest ybbR-truncated protein selected by phage display, was cloned into the pET21b expression vector, and the 49-residue fragment was expressed as a fusion to a C-terminal 6× histidine tag (JY529-His). The purified protein was incubated with Sfp and biotin-CoA, and the biotin labeling of JY529-His was confirmed by Western blot analysis probed with a streptavidin-horseradish peroxidase conjugate, whereas control reactions with either no Sfp or no biotin-CoA added showed no biotin labeling on the Western blot (Fig. 3). This suggests that truncated ybbR proteins were selected by phage display due to their posttranslational modification by Sfp-catalyzed biotin-CoA labeling. The site of Ppant modification in JY529-His was mapped by Fourier-transform MS (FTMS) to Ser-274 (following the numbering in the full length ybbR) (see supporting information), which was then mutated to Ala by site-directed mutagenesis, and the Ser274Ala mutant of JY529-His could not be labeled with biotin after incubation with Sfp and biotin-CoA, as shown by Western blot (Fig. 3), confirming the assignment of FTMS.
Fig. 2.
Alignment of B. subtilis ybbR ORF amino acids 1-299 with the truncated ybbR clones JY503, JY529, JY530, and JY565 selected by phage display. Sequences matching that of ybbR were highlighted by yellow shadings. Truncated ybbR protein sequences in the phagemid are preceded by the sequence of a leader peptide and followed by the sequence of phage capsid protein pIII. The Ppant-modified Ser-274 (full length ybbR numbering) in the phagemid clones is boxed in red.
Fig. 3.
Western blot analysis of JY529-His labeled with biotin by Sfp-catalyzed biotin-CoA modification. The Ser274Ala mutant of JY529-His was used as the control, and the Western blot was probed with streptavidin-horseradish peroxidase.
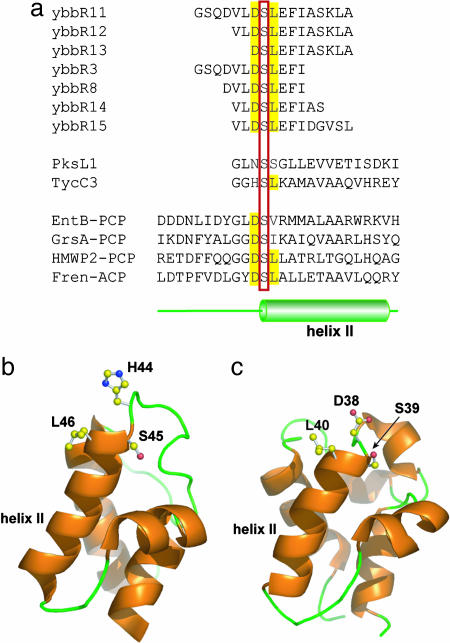
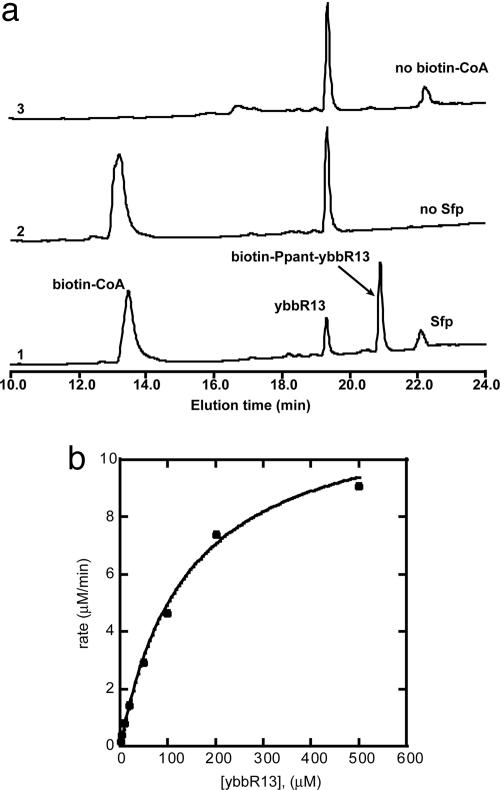
Characterization of Short ybbR Peptides as Substrate of Sfp-Catalyzed Ppant Modification. JY529-His showed no revealing sequence homology with ACPs or PCPs, the known substrates of Sfp. The site of Ppant modification in JY529-His, Ser-274, was very close to the C terminus of the protein, in contrast to Ppant-modified Ser in ACPs or PCPs, which is in the middle of the 80- to 90-residue protein with 40-45 residues to either end of the protein. We thus tested whether the C-terminal peptide of JY529-His encompassing the Ser-274 residue could be recognized by Sfp as a substrate. Short peptides corresponding to the flanking sequence of Ser-274 in JY529-His were synthesized (Fig. 4a) and incubated with Sfp and biotin-CoA, followed by HPLC analysis. Upon incubation with Sfp and biotin-CoA for 30 min at 37°C, >75% of peptide ybbR13 (DSLEFIASKLA) was found to be modified by biotin-CoA, as shown by the HPLC trace 1 with a product peak at 21 min (Fig. 5a). The product formation depends on the presence of both biotin-CoA and Sfp, because no product was formed with either biotin-CoA or Sfp excluded from the labeling reaction (traces 2 and 3, Fig. 5a). MALDI confirmed that the product of Sfp-catalyzed ybbR13 modification by biotin-CoA had the same mass as biotin-Ppant-conjugated ybbR13 ([M+H]+, calculated 2,059.0, observed 2,059.2). Similarly, peptides of 17 and 13 residues in length, ybbR11 (GSQDVLDSLEFIASKLA) and ybbR12 (VLDSLEFIASKLA), with nine-residue LEFIASKLA extending beyond Ppant-modified Ser-274 (underlined), can both be recognized by Sfp for biotin-CoA loading as shown by HPLC and MALDI. However, in contrast, peptides with three to five C-terminal residues missing from the sequence LEFIASKLA beyond Ser-274, ybbR3 (GSQDVLDSLEFI), ybbR8 (DVLDSLEFI), and ybbR14 (VLDSLEFIAS) were not the substrates of Sfp, denoting the importance of the C-terminal residues in ybbR11-13 for Sfp recognition.
Fig. 4.
Sequence analysis of the ybbR peptides. (a) Alignment of ybbR peptides with peptide sequences flanking the Ppant-modified Ser (in red box) in known PCPs and ACPs. The conserved Asp and Leu residues at the site of Ppant modification were highlighted by yellow shadings. The position of helix II based on the NMR structures of TycC3-PCP and FrenN-ACP is also shown. (b) NMR structures of TycC3-PCP [Protein Data Bank (PDB) ID code 1DNY] and (c) FrenN-ACP (PDB ID code 1OR5). Ser-45 in TycC-PCP and Ser-39 in FrenN-ACP at the tip of helix II were posttranslationally modified by Sfp.
Fig. 5.
Kinetic analysis of Sfp-catalyzed peptide ybbR13 modification by biotin CoA. (a) HPLC traces of the ybbR13 peptide-labeling reaction with biotin CoA and Sfp added (trace 1) and the control reactions with either Sfp (trace 2) or biotin-CoA (trace 3) excluded from the reaction mixture. A 100-μl solution of 100 μM peptide in 10 mM MgCl2/50 mM Hepes, pH 7.5, was incubated with 200 μM biotin-CoA/1 μM Sfp at 37°C for 30 min before HPLC analysis. (b) Michaelis-Menten plot for the measurement of kinetic parameters of Sfp-catalyzed ybbR13 labeling at a saturating concentration of biotin-CoA (150 μM).
Interestingly, the sequence of the last five residues, ASKLA at the C termini of the Sfp active peptides ybbR11-13, is not encoded by the ybbR ORF in B. subtilis but is part of the sequence between the last residue of the truncated ybbR protein Ile-278 and the 6× histidine tag in JY529-His (Fig. 2). Similarly, in the selected phage clones, the peptide sequence ASKLG was part of the linker between residue Ile-278 and phage capsid protein pIII (Fig. 2). Peptide ybbR15 (VLDSLEFIDGVSL), which has the original ybbR sequence flanking the Ppant-modified Ser-274 and the same number of residues beyond Ser-274 as in Sfp active peptides ybbR11-13, failed to be the substrate of Sfp (data not shown), suggesting that the full length ybbR protein in the B. subtilis proteome may not be modified by Sfp.
The activities of Sfp with peptide ybbR13 and biotin-CoA as substrates were determined at various pHs ranging from 5.0 to 8.5, and it was found that Sfp showed the highest activity between pH 7.0 and 8.0, thus all subsequent kinetic measurements were done at pH 7.5. Fig. 5b shows a typical Michaelis-Menten plot for the Sfp-catalyzed ybbR peptide modification reaction. Detailed kinetic analysis was carried out for the Sfp-catalyzed modification of peptides ybbR11-13 by biotin-CoA and fluorescein-CoA (Table 1). Peptides ybbR11-13 have the same nine residues (LEFIASKLA) C-terminal to Ppant-modified Ser-274 but have different numbers of residues N-terminal to Ser-274: seven for ybbR11, three for ybbR12, and only one for ybbR13. They all showed similar Km (123-142 μM) and kcat (9.3-12.1 min-1) values for Sfp-catalyzed peptide modification at saturating biotin-CoA concentration (150 μM) (Table 1). This suggests that the N-terminal sequence of the ybbR peptides does not play a significant role for Sfp recognition. Comparison with the GrsA-PCP with a Km of 4.1 μM and a kcat of 10.3 min-1 for Sfp-catalyzed biotin-CoA loading shows that peptides ybbR11-13 have a 30-fold higher Km and a similar kcat, which may be due to the larger surface area of PCP that can interact with Sfp. Similar values of Km and kcat for biotin-CoA were found at saturating concentrations of ybbR13 peptide (500 μM) and PCP (20 μM), respectively, suggesting that the binding of either PCP or peptide as substrates to Sfp did not affect the binding and turnover of biotin-CoA. YbbR13 peptide was modified by fluorescein-CoA at a similar rate as biotin-CoA with a Km of 69.9 μM and a kcat of 19.1 min-1 (Table 1), suggesting that Sfp retains its substrate promiscuity for the modification of the short-peptide substrates instead of PCP and does not differentiate various small-molecule probes conjugated to CoA, which would be a very desirable property for site-specific protein labeling using the ybbR tags.
Table 1. Kinetic parameters of Sfp-catalyzed ybbR tag and PCP modification.
| Biotin-CoA with 500 μM peptide or 20 μM PCP
|
Peptide or PCP with 150 μM biotin CoA
|
Fluorescein-CoA with 500 μM peptide
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Peptide sequence | kcat, min–1 | Km, μM | kcat/Km, μM–1·min–1 | kcat, min–1 | Km, μM | kcat/Km, μM–1·min–1 | kcat, min–1 | Km, μM | kcat/Km, μM–1·min–1 | |
| ybbR11 | GSQDVLDSLEFIASKLA | 12.1 | 142 | 0.085 | ||||||
| ybbR12 | VLDSLEFIASKLA | 9.3 | 128 | 0.073 | ||||||
| ybbR13 | DSLEFIASKLA | 14.7 | 60.8 | 0.242 | 11.2 | 123 | 0.091 | 19.1 | 69.9 | 0.273 |
| GrsA-PCP | ...DNFYALGGDSIKAIQVAAR... | 10.3 | 32.4 | 0.318 | 10.3 | 4.1 | 2.51 | |||
CD Characterization of the ybbR Peptides. When Ser-274 in Sfp active peptides ybbR11-13 were aligned with Ppant-modified Ser in PCPs or ACPs from known NRPS and PKS modules, PksL (23), TycC3 (24), EntB (25), GrsA (26), HMWP2 (27), and FrenN (28), a conserved DSL tripeptide sequence motif was identified at the Ppant-modified Ser (underlined) (Fig. 4a). Previous NMR structural studies on TycC3-PCP (24) and FrenN-ACP (28) suggested that PCP and ACP adopted an antiparallel four-helix bundle fold with the Ppant-modified Ser at the end of a long flexible loop, immediately followed by helix II (Fig. 4 b and c). Because the nine-residue sequence LEFIASKLA following the Ppant-modified Ser-274 in ybbR11-13 was found to be the key element for Sfp recognition and could be mapped to helix II in the known structures of PCP and ACP, we tested whether the ybbR peptides could form an α-helix in solution to be recognized by Sfp as substrates. The ybbR peptides were dissolved in 5 mM potassium phosphate buffer, pH 7.5, with 30% 2,2,2-trifluoroethanol (TFE), and the peptide conformation was measured by CD spectroscopy (see supporting information). The CD spectra of Sfp active peptides ybbR11, ybbR12, and ybbR13 all showed two minima at 208 and 222 nm and an isodichroic point close to 200 nm, which are characteristics of an α-helical conformation (29). The α-helix content of these peptides in aqueous TFE were estimated to be 57.3%, 36.0%, and 35.1% for ybbR11, ybbR12, and ybbR13, respectively (30). In contrast, the Sfp inactive peptides ybbR3, ybbR8, ybbR14, and ybbR15 exhibited a lesser extent of α-helical conformation (16.6%, 11.9%, 16.6%, and 7.7%, respectively), suggesting C-terminal residues ASKLA in peptides ybbR11-13 were important for the propensity of these peptides to adopt α-helical conformation in solution.
Because peptides ybbR11-13 all had a strong tendency for α-helix formation in solution and were good substrates of Sfp, we tested whether short peptides with sequences flanking the Ppant-modified Ser and encompassing helix II in known PCPs and ACPs could be the substrates of Sfp as well. Based on the sequences of TycC3-PCP (24) and PksL-ACP (23), two peptides, TycC3 (GGHSLKAMAVAAQVHREY) and PksL1 (GLNSSGLLEVVETISDKI), were synthesized with the Ppant-modified Ser (underlined) at the fourth residue followed by 11 residues involved in helix II, the same as the arrangements in ybbR12 (Fig. 4a). Although the CD spectra of peptides TycC3 and PksL1 both showed significant α-helical content (22.49% and 36.91%), no biotin-Ppant-modified peptides were identified by HPLC or MALDI after overnight incubation of the peptides with Sfp and biotin-CoA at various pHs ranging from 5.0 to 8.5. Therefore, peptides with sequences encompassing helix II in TycC3-PCP and PksL-ACP were not substrates of Sfp.
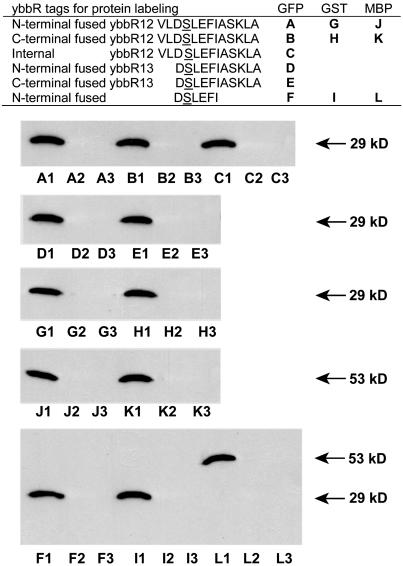
YbbR Peptides as Tags for Site-Specific Protein Labeling by Sfp. To test whether Sfp-active ybbR peptides could be used as tags for site-specific protein labeling, 13-residue ybbR12 peptide (VLDSLEFIASKLA) was fused to the N or C termini of enhanced GFP (EGFP), GST, and maltose-binding protein (MBP), respectively, and the purified fusion proteins were found to be labeled with biotin Ppant in the presence of Sfp and biotin-CoA, as shown by Western blot and ELISA (data not shown). Furthermore, N- or C-terminal ybbR12-tagged EGFP, GST, and MBP in cell lysates can all be labeled by biotin in the presence of Sfp and biotin-CoA, as shown by Western blot probed with streptavidin-horseradish peroxidase, and the control reactions with either biotin-CoA or Sfp excluded did not give any biotin labeling (Fig. 6) nor did the control reactions in which the target protein without the ybbR tag was incubated with biotin-CoA and Sfp (data not shown), suggesting the labeling reaction strictly depends on Sfp-catalyzed biotin Ppant transfer onto the ybbR tag.
Fig. 6.
Western blot analysis of biotin-labeled ybbR fusions A-L, as shown in the table. Labeling reactions were carried out in cell lysates in which both biotin-CoA and Sfp were added (1) or only biotin-CoA (2) or Sfp (3) was added as control.
Shorter ybbR tags were fused to the target proteins, and it was found when 11-residue peptide tag ybbR13 (DSLEFIASKLA) was fused to either the N or C terminus of EGFP, the fusion protein could still be labeled by Sfp-catalyzed biotin-CoA transfer (Fig. 6). When a six-residue peptide DSLEFI was fused to the C termini of EGFP, GST, or maltose-binding protein, none of the fusion proteins were labeled with biotin in the presence of Sfp and biotin CoA. In contrast, when the same peptide sequence was fused to the N termini of the target proteins, all of the fusion proteins were labeled with biotin by Sfp-catalyzed biotin-CoA transfer (Fig. 6). This suggests that residues ASKLA at the C terminus of ybbR13 (DSLEFIASKLA) were crucial for the formation of α-helical conformation and subsequent Sfp recognition when the ybbR tag was at the C terminus of the protein. In contrast, when the ybbR tag was at the N terminus, the presence of residues ASKLA in the ybbR tag might not be necessary, because the tag was followed by the N-terminal residues of the target proteins, which might facilitate α-helical formation of the N-terminal ybbR tag to be recognized by Sfp.
Because the truncated ybbR protein was selected by phage display in which Sfp-catalyzed biotin labeling of the ybbR tag VLDSLEFIASKL was flanked by the truncated ybbR protein and phage capsid protein pIII at the N and C termini, respectively, we rationalized that the ybbR tag might still function as the substrate of Sfp when the tag was inserted into a flexible loop region in the middle of the target protein. To test this idea, ybbR tag with flanking glycine residues (GGGTVLDSLEFIASKLAGGG, 20 residues) was inserted in place of Tyr-145 of EGFP to give the construct EGFP-ybbR145, because it was previously reported that calmodulin or zinc finger proteins inserted at the same position in EGFP did not affect protein fluorescence (31). Biotin labeling of EGFP-ybbR145 by Sfp in cell lysates was confirmed by Western blot analysis (Fig. 6), and the EGFP-ybbR was also labeled site specifically with tetramethylrhodamine and Texas red upon Sfp-catalyzed fluorophore-CoA transfer onto the internal ybbR tag in EGFP-ybbR145 to give dual-colored GFP (see supporting information).
To quantify the yield of the protein-labeling reaction, various EGFP protein constructs in Fig. 6 with the ybbR tags fused to the N or C terminus or inserted in the middle of the protein were labeled with biotin after incubation with Sfp and biotin-CoA at 37°C for 30 minutes. Streptavidin-coated agarose beads were then added, and >80% of various ybbR-tagged EGFP proteins can be immobilized on the streptavidin beads, suggesting the high efficiency of the labeling reaction (see supporting information).
Discussion
Sfp phosphopantetheinyl transferase activates NRPS and PKS clusters in B. subtilis by posttranslational transfer of the Ppant group from CoA to a conserved Ser residue in each of the PCPs and ACPs embedded in the biosynthetic NRPS and PKS assembly lines (12). It was previously noted that the Ppant-modified Ser (underlined) in the PCPs is located in a conserved motif with the sequence DxFFxxLGG(H/D)S(L/I) (“x” denotes any of the 20 proteinogenic amino acids, and “H/D” denotes either H or D at the same position) (32). However, a 19-residue peptide with the sequence GVTDNFFMIGGHSLKAMMM from the B. subtilis SrfB1-PCP encompassing the conserved motif was previously tested for Sfp-catalyzed Ppant modification and was found not to be a substrate of Sfp (33). Here we report the identification from a phage-displayed library of the B. subtilis genome, a short 11-residue peptide tag ybbR13 (DSLEFIASKLA) with part of the sequence derived from the B. subtilis ORF ybbR, to be an efficient substrate of Sfp. The ybbR tag has the conserved DSL tripeptide sequence at the N terminus (Fig. 4a), matching the conserved sequence motif of a PCP, although it is missing the DxFFxxLGG sequence at its N terminus, which is conserved in PCPs. In fact, N-terminal extensions on the ybbR peptide (ybbR11 and ybbR12) did not affect the activity of the ybbR peptide as a substrate of Sfp, suggesting the N terminus of the ybbR tag plays a minor role for interaction with Sfp. In contrast, when ybbR12 was truncated by three or five residues from the C terminus (ybbR14 and ybbR8), both peptides failed to be a substrate of Sfp, revealing the importance of the C-terminal sequence of the ybbR tag for Sfp recognition.
We also found that the activity of the ybbR tag as the substrate of Sfp-catalyzed Ppant modification was coupled to its tendency to adopt α-helical conformation in solution. All Sfp active peptides ybbR11-13 showed high contents of α-helical conformation in 30% 2,2,2-trifluoroethanol, as measured by CD spectra, whereas C-terminal-truncated peptides ybbR3, ybbR8, and ybbR14 showed poor α-helical formation and were not substrates of Sfp modification. Because in PCP and ACP domains, the Ppant-modified Ser is immediately followed by helix II, which has been reported as a key element for Sfp recognition (34-36), it is plausible to propose that the Sfp active ybbR peptides adopt an α-helical conformation for the peptide sequence SLEFIASKLA with the Ppant-modified Ser at the tip of the α-helix upon its binding to Sfp. Following this notion, peptides TycC3 and PksL1 encompassing the Ppant-modified Ser and the helix II region of the known PCP and ACP were tested for Sfp-catalyzed biotin-CoA modification but were found not to be substrates of Sfp. Although these peptides also had a tendency to form α-helices, as shown by their CD spectra, that they were not recognized by Sfp suggests the peptide sequence flanking the Ppant-modified Ser in PCP or ACP needs to be presented to Sfp as part of the intact PCP or ACP domain, whereas the ybbR peptide tag selected by phage display has some unique yet unidentified features in addition to a tendency for α-helix formation that facilitates binding between the ybbR tag and Sfp.
A helical wheel representation of the ybbR tag sequence SLEFIASKLA revealed that the proposed α-helix adopted by the ybbR tag has an amphiphilic distribution of the side chains with nonpolar residues Ile, Leu, and Ala on one side of the helix and charged or polar residues Lys, Glu, and Ser on the opposite side (see supporting information). The corresponding helical wheel plots of the helix II region of the TycC3-PCP and FrenN-ACP with known NMR structures showed no significant similarities with the ybbR tag in terms of distribution or alignment of certain type of residues on a specific side of the helix. We thus expect that structural studies of ybbR peptide substrates bound to Sfp will be required to decipher the recognition rules.
The shortest Sfp active peptide ybbR13 (DSLEFIASKLA) is only 11 residues long with a similar kcat for Sfp-catalyzed biotin-CoA labeling as that of the full length PCP and 30-fold higher Km than that of PCP. Thirty-minute incubation of the ybbR-tagged proteins with Sfp and biotin-CoA results in >80% of the protein being labeled with biotin. Sfp retained its substrate promiscuity for the ybbR tag modification with small-molecule probes conjugated to CoA and catalyzed fluorescein-CoA transfer to the ybbR tag at a similar rate as that of biotin-CoA. The ybbR peptide tag has been shown to be recognized by Sfp when fused to the N and C termini of various proteins, and the site-specific labeling reaction of the ybbR tagged proteins by Sfp can be carried out in cell lysates within a short time (10-20 min). Interestingly, we also showed that the ybbR tag inserted in a flexible loop in the middle of EGFP could still be labeled by Sfp with various fluorophore-CoA conjugates.
The phage selection on the B. subtilis proteome turns up a peptide substrate with part of the sequence from the ybbR ORF of an unknown function. It is yet to be tested whether full length ybbR is an authentic substrate for posttranslational phosphopantetheinylation in B. subtilis cells, although the fusion of ybbR fragments to phage capsid protein pIII created a hybrid peptide that can be recognized by Sfp. This highlights the power of phage selection for the discovery of a novel class of peptide tags for protein labeling. Other bacterial genomes may yield additional examples on selection.
Conclusion
Sfp-catalyzed ybbR tag posttranslational modification has the following advantages for versatile protein labeling:
The size of the ybbR tag (11 residues) is much smaller compared with the size of PCP or ACP (75-80 residues).
The ybbR tag has an excellent portability for fusion to various proteins for protein labeling: the ybbR tag can be attached to the N or C terminus of the target protein or inserted into a flexible loop in the middle of the target protein and labeled site specifically by Sfp-catalyzed small-molecule CoA modification.
Sfp has broad substrate specificities with respect to the small-molecule probes conjugated to CoA, including sugars, affinity probes such as biotin, glutathione, fluorescent probes such as fluorescein, Alexa Fluor dyes, and redox probes such as porphyrin. Furthermore, Sfp-catalyzed ybbR tag labeling by various small-molecule probes is highly specific and efficient and can be accomplished in one step.
We thus expect Sfp-catalyzed ybbR tag posttranslational modification would have broad applications for expanding the structural and functional diversity of ybbR-tagged recombinant proteins in vitro and in vivo. At present, the in vivo use is limited by the inability of CoA derivatives to cross biological membranes, but a recent report on the uptake of a fluorescent pantetheine derivative by E. coli, in vivo conversion to the fluorescent CoA, and phosphopantetheinyl transfer to the VibB PCP indicates a way forward for intracellular labeling of ybbR-tagged proteins (37).
Supplementary Material
Acknowledgments
We thank Stephen C. Blacklow and Cheryll Sanchez-Irizarry for helping with the CD measurements. P.D.S. was supported by a National Science Foundation Postdoctoral Fellowship in Microbiology Biology (DBI-0200307). This work was supported by Grants GM20011 (to C.T.W.), GM58213 (to R.K.), and HL32854 (to D.E.G.) from the National Institutes of Health.
Author contributions: J.Y. and C.T.W. designed research; J.Y., P.D.S., S.M.M., Z.Z., and A.J.L. performed research; J.Y. contributed new reagents/analytic tools; J.Y., D.E.G., N.L.K., R.K., and C.T.W. analyzed data; and J.Y. and C.T.W. wrote the paper.
Abbreviations: CD, circular dichroism; EGFP, enhanced GFP; AGT, O6-alkylguanine-DNA alkyltransferase; PCPs, peptide carrier proteins; NRPS, nonribosomal peptide synthetase; ACP, acyl carrier protein; PKS, polyketide synthase; Ppant, 4′-phosphopantetheinyl.
References
- 1.Walsh, C. T. (2005) Posttranslational Modification of Proteins: Expanding Nature's Inventory (Roberts, Englewood, CO).
- 2.Hahn, M. E. & Muir, T. W. (2005) Trends Biochem. Sci. 30, 26-34. [DOI] [PubMed] [Google Scholar]
- 3.Prescher, J. A. & Bertozzi, C. R. (2005) Nat. Chem. Biol. 1, 13-21. [DOI] [PubMed] [Google Scholar]
- 4.Griffin, B. A., Adams, S. R. & Tsien, R. Y. (1998) Science 281, 269-272. [DOI] [PubMed] [Google Scholar]
- 5.Beckett, D., Kovaleva, E. & Schatz, P. J. (1999) Protein Sci. 8, 921-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, I., Howarth, M., Lin, W. & Ting, A. (2005) Nat. Methods 2, 99-104. [DOI] [PubMed] [Google Scholar]
- 7.Keppler, A., Gendreizig, S., Gronemeyer, T., Pick, H., Vogel, H. & Johnsson, K. (2003) Nat. Biotechnol. 21, 86-89. [DOI] [PubMed] [Google Scholar]
- 8.Adams, S. R., Campbell, R. E., Gross, L. A., Martin, B. R., Walkup, G. K., Yao, Y., Llopis, J. & Tsien, R. Y. (2002) J. Am. Chem. Soc. 124, 6063-6076. [DOI] [PubMed] [Google Scholar]
- 9.Juillerat, A., Heinis, C., Sielaff, I., Barnikow, J., Jaccard, H., Kunz, B., Terskikh, A. & Johnsson, K. (2005) ChemBioChem 6, 1263-1269. [DOI] [PubMed] [Google Scholar]
- 10.Yin, J., Liu, F., Li, X. & Walsh, C. T. (2004) J. Am. Chem. Soc. 126, 7754-7755. [DOI] [PubMed] [Google Scholar]
- 11.Stachelhaus, T., Huser, A. & Marahiel, M. A. (1996) Chem. Biol. 3, 913-921. [DOI] [PubMed] [Google Scholar]
- 12.Lambalot, R. H., Gehring, A. M., Flugel, R. S., Zuber, P., LaCelle, M., Marahiel, M. A., Reid, R., Khosla, C. & Walsh, C. T. (1996) Chem. Biol. 3, 923-936. [DOI] [PubMed] [Google Scholar]
- 13.Belshaw, P. J., Walsh, C. T. & Stachelhaus, T. (1999) Science 284, 486-489. [DOI] [PubMed] [Google Scholar]
- 14.La Clair, J. J., Foley, T. L., Schegg, T. R., Regan, C. M. & Burkart, M. D. (2004) Chem. Biol. 11, 195-201. [DOI] [PubMed] [Google Scholar]
- 15.Yin, J., Liu, F., Schinke, M., Daly, C. & Walsh, C. T. (2004) J. Am. Chem. Soc. 126, 13570-13571. [DOI] [PubMed] [Google Scholar]
- 16.Yin, J., Lin, A. J., Buckett, P. D., Wessling-Resnick, M., Golan, D. E. & Walsh, C. T. (2005) Chem. Biol. 12, 999-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.George, N., Pick, H., Vogel, H., Johnsson, N. & Johnsson, K. (2004) J. Am. Chem. Soc. 126, 8896-8897. [DOI] [PubMed] [Google Scholar]
- 18.Tsien, R. Y. (1998) Annu. Rev. Biochem. 67, 509-544. [DOI] [PubMed] [Google Scholar]
- 19.Lesaicherre, M. L., Lue, R. Y., Chen, G. Y., Zhu, Q. & Yao, S. Q. (2002) J. Am. Chem. Soc. 124, 8768-8769. [DOI] [PubMed] [Google Scholar]
- 20.Hochuli, E., Dobeli, H. & Schacher, A. (1987) J. Chromatogr. 411, 177-184. [DOI] [PubMed] [Google Scholar]
- 21.Evan, G. I., Lewis, G. K., Ramsay, G. & Bishop, J. M. (1985) Mol. Cell. Biol. 5, 3610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hopp, T. P., Pricket, K. S., Price, V. L., Libby, R. T., March, C. J., Ceretti, D. P., Urdal, D. L. & Conlon, P. J. (1988) Bio/Technology 6, 1204-1210. [Google Scholar]
- 23.Kunst, F., Ogasawara, N., Moszer, I., Albertini, A. M., Alloni, G., Azevedo, V., Bertero, M. G., Bessieres, P., Bolotin, A., Borchert, S., et al. (1997) Nature 390, 249-256. [DOI] [PubMed] [Google Scholar]
- 24.Weber, T., Baumgartner, R., Renner, C., Marahiel, M. A. & Holak, T. A. (2000) Struct. Fold. Des. 8, 407-418. [DOI] [PubMed] [Google Scholar]
- 25.Gehring, A. M., Bradley, K. A. & Walsh, C. T. (1997) Biochemistry 36, 8495-8503. [DOI] [PubMed] [Google Scholar]
- 26.Stachelhaus, T. & Walsh, C. T. (2000) Biochemistry 39, 5775-5787. [DOI] [PubMed] [Google Scholar]
- 27.Keating, T. A., Miller, D. A. & Walsh, C. T. (2000) Biochemistry 39, 4729-4739. [DOI] [PubMed] [Google Scholar]
- 28.Li, Q., Khosla, C., Puglisi, J. D. & Liu, C. W. (2003) Biochemistry 42, 4648-4657. [DOI] [PubMed] [Google Scholar]
- 29.Fasman, G. D., ed. (1996) Circular Dichroism and the Conformational Analysis of Biomolecules (Plenum, New York).
- 30.Greenfield, N. & Fasman, G. D. (1969) Biochemistry 8, 4108-4116. [DOI] [PubMed] [Google Scholar]
- 31.Baird, G. S., Zacharias, D. A. & Tsien, R. Y. (1999) Proc. Natl. Acad. Sci. USA 96, 11241-11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marahiel, M. A., Stachelhaus, T. & Mootz, H. D. (1997) Chem. Rev. 97, 2651-2674. [DOI] [PubMed] [Google Scholar]
- 33.Quadri, L. E., Weinreb, P. H., Lei, M., Nakano, M. M., Zuber, P. & Walsh, C. T. (1998) Biochemistry 37, 1585-1595. [DOI] [PubMed] [Google Scholar]
- 34.Finking, R., Mofid, M. R. & Marahiel, M. A. (2004) Biochemistry 43, 8946-8956. [DOI] [PubMed] [Google Scholar]
- 35.Mofid, M. R., Finking, R., Essen, L. O. & Marahiel, M. A. (2004) Biochemistry 43, 4128-4136. [DOI] [PubMed] [Google Scholar]
- 36.Mofid, M. R., Finking, R. & Marahiel, M. A. (2002) J. Biol. Chem. 277, 17023-17031. [DOI] [PubMed] [Google Scholar]
- 37.Clarke, K. M., Mercer, A. C., La Clair, J. J. & Burkart, M. D. (2005) J. Am. Chem. Soc. 127, 11234-11235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.