Abstract
Current techniques in high-speed cell sorting are limited by the inherent coupling among three competing parameters of performance: throughput, purity, and rare cell recovery. Microfluidics provides an alternate strategy to decouple these parameters through the use of arrayed devices that operate in parallel. To efficiently isolate rare cells from complex mixtures, an electrokinetic sorting methodology was developed that exploits dielectrophoresis (DEP) in microfluidic channels. In this approach, the dielectrophoretic amplitude response of rare target cells is modulated by labeling cells with particles that differ in polarization response. Cell mixtures were interrogated in the DEP-activated cell sorter in a continuous-flow manner, wherein the electric fields were engineered to achieve efficient separation between the dielectrophoretically labeled and unlabeled cells. To demonstrate the efficiency of marker-specific cell separation, DEP-activated cell sorting (DACS) was applied for affinity-based enrichment of rare bacteria expressing a specific surface marker from an excess of nontarget bacteria that do not express this marker. Rare target cells were enriched by >200-fold in a single round of sorting at a single-channel throughput of 10,000 cells per second. DACS offers the potential for automated, surface marker-specific cell sorting in a disposable format that is capable of simultaneously achieving high throughput, purity, and rare cell recovery.
Keywords: cell sorting, microfluidics
Cell sorters are capable of separating a heterogeneous suspension of particles into purified fractions and thus have become an indispensable tool in biology and medicine. Emerging applications of cell sorting technology span a broad spectrum of pharmaceutical and biomedical fields that range from cancer diagnostics to cell-based therapies (1–3). The most widely used methodologies for cell separation are magnetic-activated cell sorting (MACS) and fluorescence-activated cell sorting (FACS). MACS is a selection technique that is capable of capturing a large number of target cells in parallel (4); however, the purity and recovery in MACS typically have large variances (5). In contrast, FACS relies upon serially screening each cell, yielding high performance in cell recovery and purity (6). However, because of the serial nature of its operation, FACS allows for a comparatively low throughput, typically in the range between 104 and 105 cells per second (7). Regardless of the mechanism, the performance of cell separation is typically characterized by three metrics. “Throughput” gauges how many cell characterization and sorting operations can be executed per unit of time, “purity” is the fraction of the target cells in the collection vessel, and “recovery” is the fraction of the input target cells successfully sorted into the collection vessel. Demands placed on cell sorting technologies continue to increase, because cell sorting applications are expanding and biological questions are becoming more complex (7). For example, rare cell sorting is particularly challenging, because target cells may occur at frequencies below one per million (8). Rare cell analysis and sorting have proven useful for low-abundance stem cell sorting (9), detection and isolation of rare circulating tumor cells (8), and screening of cell-based libraries (10).
Contemporary methodologies of cell sorting remain limited by the inherent coupling among the three competing performance parameters: throughput, purity, and rare cell recovery. Cell sorting technology that employs microfluidics provides an alternate strategy to decouple the three parameters through the use of arrayed devices that operate in parallel. In addition, it offers the potential to provide a disposable solution, which will eliminate sample cross-contamination. However, the reported performances of microfabricated cell sorters based on miniaturized MACS or FACS approaches lag significantly behind those of their macroscopic counterparts (11–14). An alternative potential mechanism for separation is dielectrophoresis (DEP), the translational motion of charge-neutral matter caused by polarization effects in nonuniform electric fields (15). Because of the relatively facile engineering of the electric fields and interface to integrated electronics, DEP provides an especially attractive force field for on-chip cell manipulation (16, 17).
The time-averaged dielectrophoretic force on a homogeneous sphere of radius rp, ignoring higher order effects of polarization, can be approximated as (18)
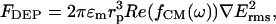 |
[1] |
where Erms is the electric field strength, εm is the permittivity of the suspending medium, ω is the angular frequency, and Re(fCM(ω)) is the real part of the dipolar Clausius–Mossotti (CM) factor. The CM factor is bound by the limits -0.5 < Re(fCM(ω)) < 1 and describes the relative polarization of the particle versus that of the surrounding medium given by 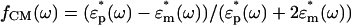 , where
, where 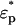 and
and 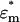 are the complex permittivities of the particle and medium, respectively. The term negative DEP (nDEP) corresponds to the phenomenon wherein the real part of the CM factor is negative (Re(fCM(ω)) < 0). In this region, the particles are physically repelled from the areas of higher electric field gradients into the weaker field region. In contrast, when Re(fCM(ω)) > 0, the phenomenon is called positive DEP, which corresponds to the effect of the particle being attracted to the region of higher electric field gradient (15).
are the complex permittivities of the particle and medium, respectively. The term negative DEP (nDEP) corresponds to the phenomenon wherein the real part of the CM factor is negative (Re(fCM(ω)) < 0). In this region, the particles are physically repelled from the areas of higher electric field gradients into the weaker field region. In contrast, when Re(fCM(ω)) > 0, the phenomenon is called positive DEP, which corresponds to the effect of the particle being attracted to the region of higher electric field gradient (15).
Previously, the native dielectrophoretic response of different cell types has been exploited for cell separation (18–20). However, the utility of this approach is limited to the separation of cells that possess significantly different dielectrophoretic response from that of other cells. In many applications, the target and nontarget cells exhibit similar responses, thereby precluding sorting based on the intrinsic dielectrophoretic phenotypes. To circumvent this limitation, in the present study, cells were labeled with polymeric beads to achieve significant differences in dielectrophoretic amplitude response between the cells bound to beads and the unlabeled background. Heterogeneous cell mixtures were then interrogated in a microfluidic device in a continuous-flow manner, wherein the electric fields were engineered to achieve efficient separation. To assess the efficiency of dielectrophoretic labeling and subsequent selection, we demonstrate affinity-based enrichment of rare Escherichia coli that display a specific surface marker from an excess of nontarget bacteria of the same species. To our knowledge, this study offers the first demonstration of enrichment of rare cells in a surface marker-specific manner by using DEP. By analogy with MACS and FACS, we term this process DEP-activated cell sorting (DACS).
Materials and Methods
Strains and Reagents. The bacterial strains used here display peptides as insertional fusions into the second extracellular loop of outer membrane protein OmpX of E. coli. PCR was used to generate the peptide inserts, which were cloned into the ompX gene at restriction sites inserted into the coding sequence after residue serine 53 of the mature protein and after the stop codon. A T7·tag epitope-containing clone was constructed by the insertion of amino acids MASMTGGQQMG flanked by linkers GQSGQ and GGS. All constructs were expressed in E. coli strain MC1061 (21) from the arabinose inducible promoter of plasmid pBAD33 (22) using the native ompX ribosome-binding site. Streptavidin R-phycoerythrin was obtained from Molecular Probes, and the biotinylated anti-T7·tag antibody was obtained from Novagen.
Fabrication of the DACS Device. The quadrupole electrodes that generate the electric fields for the dielectrophoretic separation were fabricated by e-beam evaporation of 300-nm Au/20-nm Ti on glass substrates and a liftoff process. Photosensitive polyimide HD4010 (HD MicroSystems, Santa Clara, CA) was used as the polymer spacer for the microchannel. Polyimide is chosen for the channel material because of its hydrolytic stability, high breakdown voltage, and inertness to most chemicals and solvents (23). The polyimide was spun onto the bottom substrate, and microfluidic channels of 20-μm depth were defined by photolithography. After dicing and creating microfluidic vias in the top plate, the two substrates were aligned and bonded in N2 atmosphere. Microfluidic inlets and outlets were manually fixed to the device with epoxy.
Cell Sorting Using DACS. For cell labeling, 50 μl of cells (2 × 109 cells per ml) were harvested and mixed with biotinylated T7·tag monoclonal antibody (Novagen and EMD Biosciences, San Diego) at a final concentration of 100 nM. After incubation on ice for 1 h with gentle agitation, both antibody-labeled and unlabeled cells were pelleted by centrifugation at 2,650 × g for 5 min and resuspended in 100 μl of sterile-filtered 1× PBS (pH 7.4). The cells were washed again by centrifugation at 2,650 × g for 5 min. Streptavidin-coated polystyrene beads (5 μl, 1 × 108 beads per ml, Bangs Laboratories, Carmel, IN) were added to the cells, which were resuspended in 1× PBS (100 μl) at a final concentration of 109 cells per ml. The mixture was incubated on ice for 1 h, washed twice in PBS (1 ml), and resuspended in 0.1× PBS (600 μl) supplemented with 1% BSA (Fraction V, Sigma-Aldrich). To prevent settling during DACS screening, the density of the solution was adjusted to that of polystyrene beads (1.06 g/ml) by including glycerol at a final concentration of 20% (vol/vol).
For DACS experiments, tygon tubing (inner diameter of 0.02 inches, Fisher Scientific) was attached to the inlets and outlets of the device. The device was placed beneath the objective of an epifluorescent microscope for visualization. To allow easy access to the objective lens, the device was inverted with all of the tubing facing away from the lens of the microscope. The electrodes were connected through two card-edge connectors to a function generator (AFG320, Tektronix). The frequency and the amplitude of the applied voltage were monitored by a digital oscilloscope (54622A, Agilent Technologies, Palo Alto, CA). A dual-track programmable syringe pump setup (Ph.D. 2000, Harvard Apparatus) delivered both the cell mixture and the sorting buffer into the device at a constant flow rate. The syringes were placed on ice to minimize cell growth during the sorting. The device and the tubing were filled with sorting buffer (0.1× PBS/20% glycerol/1% BSA) to drive out air bubbles before pumping. The volumetric flow rate during sorting was 50–200 μl/h. When the velocity of the fluid flow stabilized, the voltage (sine wave, 20-V peak-to-peak voltage at 500 kHz) was turned on. The flow of the beads in the microchannel was monitored through a charge-coupled device camera.
The enriched cell solution and waste were collected separately by using 1.5-ml centrifuge tubes. The collected enriched cells were grown in LB medium overnight to amplify the selected population. A second round of induction, labeling, DACS, and growth was performed for improved target cell purity. After each use, the DACS device was sterilized by infusing bleach (2%) into the microchannels, soaking for 10 min, and repeating. The device was then flushed with sterilized deionized water (2 ml) followed by 1 ml of ethanol [70% (vol/vol)].
Analysis by conventional FACS (FACSAria, BD Biosciences) was carried out by growing, inducing, and labeling the population of the cells with biotinylated T7·tag monoclonal antibody as described previously. The cells then were washed twice and incubated on ice with streptavidin-phycoerythrin (20 nM) for 45–60 min. Cells were washed once and resuspended in cold PBS at a final concentration of ≈106 cells per ml and immediately analyzed by flow cytometry.
Results and Discussion
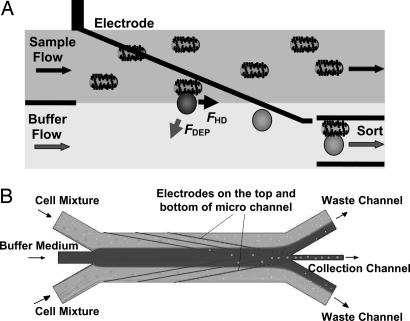
Dielectrophoretic Labeling and Electrokinetics. The DACS device was designed and constructed to exploit the differences in dielectrophoretic response between unlabeled and bead-labeled cells. A schematic view of the device design is shown in Fig. 1, where the matching electrodes on the top and bottom walls of the microchannel establish an electric field with the highest field gradient occurring close to the electrodes. The electrodes were fabricated at an angle of 15° to the direction of the fluid flow to reduce the nDEP force required for the deflection. As the mixture enters this region, the dielectrophoretically labeled cells are selectively deflected by nDEP. As a result, target cells can be electrokinetically funneled into the collection channel while the unlabeled cells are rejected into the waste channel.
Fig. 1.
Operational principle of DACS. (A) The DACS concept: Cells entering in the sample stream are only deflected into the collection stream if they are labeled with a dielectrophoretically responsive label. (B) Schematic view of the electrode region of the microchannels with sample and buffer inlets, as well as waste and collection outlets.
For the given electrode geometry shown in Fig. 1B, the DEP force near the electrode pair can be approximated by (24)
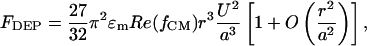 |
[2] |
where U is the applied rms voltage and a is the channel height. Because of the small thickness of the electrodes compared with the channel height, the perturbation of the electrodes on the flow of the fluid was negligible. Both polystyrene beads and E. coli cells experience nDEP force with Re(fCM) ≈ -0.5 at 500 kHz, at a medium conductivity between 100 and 200 mS/m. Simultaneously, the beads and cells also experience a viscous drag force. Approximating the cells and particles as rigid spheres in a low Reynold's number flow, the viscous drag force was estimated by using Stokes' equation, FHD = 6πηvr, where η is the surrounding fluid viscosity and v is the difference in velocity between the cell or particle and the surrounding flow. Therefore, deflection was expected to occur when 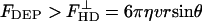 , where θ is the angle between the electrodes and the direction of fluid flow. Thus, the maximum flow velocity for cell deflection (25) is given by
, where θ is the angle between the electrodes and the direction of fluid flow. Thus, the maximum flow velocity for cell deflection (25) is given by
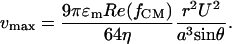 |
[3] |
Because the device was operated such that the flow velocity did not exceed vmax, the approaching, labeled E. coli were deflected into the collection channel, whereas the unlabeled clones were not deflected and continued their flow into the waste channels. Assuming εm = 80ε0, U = 7.4 V, and a = 20 μm, the DEP forces on the labeled and unlabeled E. coli were calculated to be ≈388 and 4.6 pN, respectively. At a total volumetric flow rate of 300 μl/h per microchannel with η ≈ 0.002 kg·m-1·s-1 for 20% glycerol (26), the fluid velocity is ≈v = 3 mm/s near the electrodes. As a result, the viscous drag forces on the labeled and unlabeled E. coli cells were ≈368 and 57 pN, respectively. Thus, using these operating conditions in conjunction with shallow angles between the fluid velocity and electrodes, the DEP force was designed to be insufficient in deflecting the unlabeled cells but large enough to selectively deflect the labeled cells.
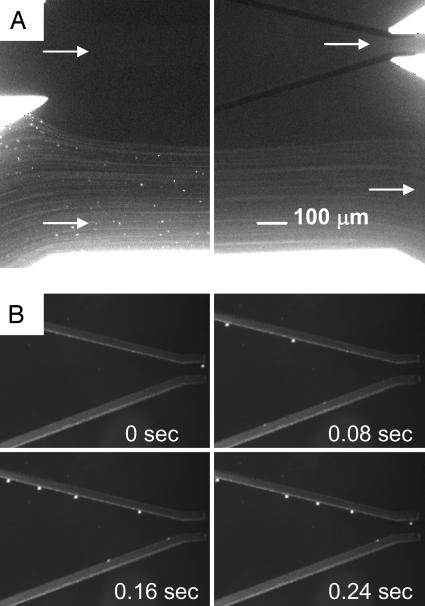
Microfluidics. The DACS device operates at low Reynold's numbers, in the range of 0.1 < Re = ρvL/η < 1, where v denotes the characteristic flow velocity and L denotes the characteristic length determined by the channel geometry. The fluid density and dynamic viscosity are given by ρ and η, respectively. To maximize the purity performance at the collection channels, we introduce the concept of a “buffer flow.” The idea is analogous to having a “sheath flow” in FACS; however, the buffer flow serves a different function. In FACS, the sheath flow surrounds the cell mixture and serves to lower the shear stress on the cells and to align the cells in single file (i.e., hydrodynamic focusing). The buffer flow in the DACS geometry is inverted such that the cell mixture flanks the buffer flow (Fig. 1). The cell mixture was introduced from the side channels while buffer solution of the same density and conductivity was introduced through the central inlet channel, creating an initial cell concentration profile that is devoid of any cells in the buffer stream at the collection channel. In the absence of an electric field, all DEP-responsive particles followed the streamlines and entered the waste channel (Fig. 2A). When the electrodes were energized, the DEP particles were selectively deflected into the buffer stream (Fig. 2B). Particles moved from the sample stream into the buffer stream because of nDEP deflection near the edges of the electrodes where the gradient of the electric field was maximal. Similarly, unlabeled E. coli were unable to enter the collection channel, because Brownian diffusion across the streamline that separates the cell mixture and the buffer stream was negligible. The average lateral diffusion length for cells in the microchannel is roughly a few microns and thus insufficient for the unlabeled cells to cross the streams and enter the collection channel. As a result, the unlabeled cells remain in the cell mixture and flow through the waste channel. Conversely, using mixtures of DEP-labeled and unlabeled cells, only the labeled cells were collected effectively, and thus high purity and rare cell recovery were achieved without compromising the throughput in each microchannel. Several design parameters influencing the DACS performance could be optimized for a particular volumetric throughput. In particular, these parameters include (i) the ratio of stream velocities, (ii) the angle of electrodes, (iii) the width of the collection channel, and (iv) the separation distance between the sample and buffer stream interface and the collection channel centerline.
Fig. 2.
Optical micrographs of the stabilized flow at the inlet and outlet channels. (A) The unlabeled cells follow the streamline and enter the waste channels, and the diffusion of the cells into the buffer stream is minimal. (B) Sequentially captured images of polystyrene beads moving under the influence of DEP deflection. Quadrupole electrodes guide the beads to the center of the microchannel. Total volume flow rate is 240 μl/h. Applied voltage is 20 V peak-to-peak at 500 kHz.
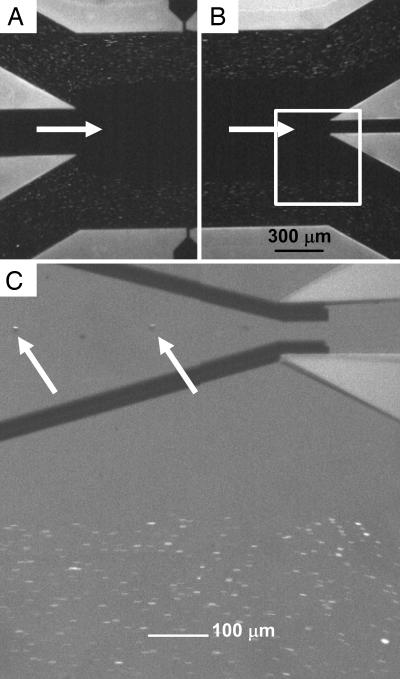
DACS Enrichment of Rare Bacterial Cells. Unlabeled fluorescent E. coli cells introduced at the device inlet (Fig. 3A) followed the streamlines and passed into the waste stream at the outlet (Fig. 3B). In the absence of dielectrophoretic labeling of the target cells, >99.9% of cells followed the streamlines into the waste channel as verified through flow cytometry analysis (not shown). In contrast, DEP-labeled cells effectively crossed the streamlines into the buffer flow and entered the collection channel (Fig. 3C). At a total flow rate of 300 μl·h-1 per microchannel, ≈95% of the labeled cells were recovered, as determined by flow cytometry (not shown).
Fig. 3.
Flow pattern of fluorescent cells in the DACS device. (A and B) Optical micrographs of streaming E. coli cells at the inlet (A) and outlet (B) channels. (C) An expanded view using optical microscopy, showing DEP particles entering the collection channel after being focused into the center of the stream. The arrows identify two beads carrying bound cells. Total volume flow rate is 300 μl/h, and the applied voltage is 20 V peak-to-peak at 500 kHz.
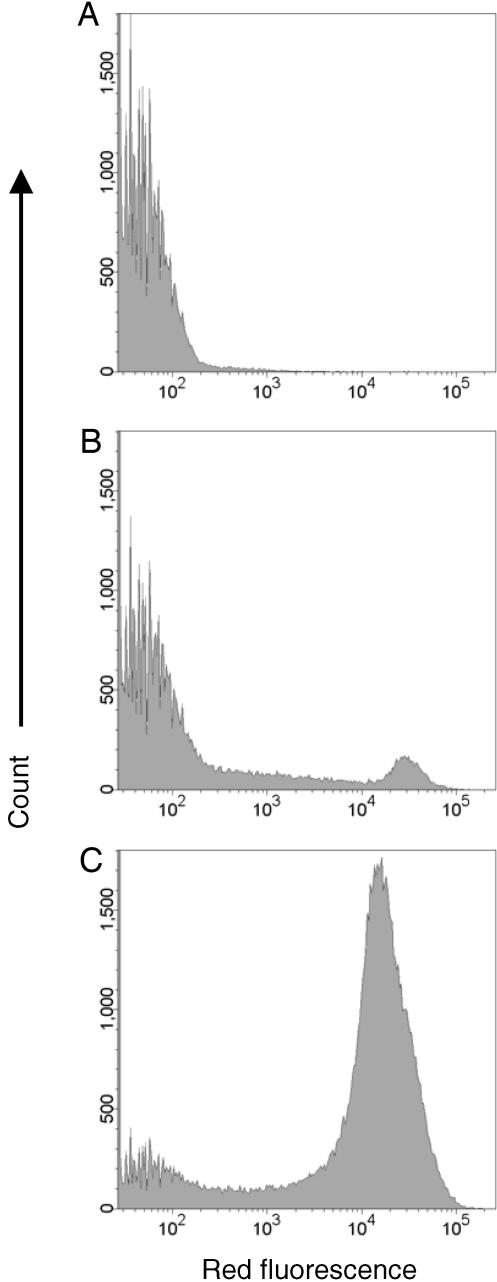
The utility of DACS for marker-specific separation of rare target cells was investigated by using E. coli that display on their outer surface different peptide antigens. In particular, the rare target cells were E. coli that display a peptide recognized by a monoclonal antibody. Background, nontarget cells consisted of a bacterial display library of random peptide insertions into outer membrane protein OmpX, analogous to that described in ref. 27. To facilitate quantification of the enrichment obtained using DACS, the bacterial display library (nontarget cells) was spiked with bacteria (at a ratio of 1:5,000) that display the T7·tag epitope (MASMTGGQQMG) recognized by a T7·tag-specific monoclonal antibody. The initial frequency of target cells was accurately measured with conventional flow cytometry after labeling with biotinylated T7·tag antibody and then streptavidin-phycoerythyrin. The threshold for the detection of the marker-specific cells with flow cytometry was ≈1 per 100,000 (data not shown). Before DACS, the frequency of target cells (i.e., those displaying T7·tag and binding to the anti-T7·tag antibody) was 0.02%, as determined by flow cytometry (Fig. 4A).
Fig. 4.
Enrichment of T7·tag mAb-binding clones as measured by flow cytometry. Induced cells were labeled with 20 nM biotin-T7 mAb and 20 nM streptavidin-phycoerythrin. (A) Unselected population. (B) After one round of DACS. (C) After two rounds of DACS.
After a single round of sorting using the DACS device, the frequency of target cells reached 1:20 (5%) at a single-channel throughput of 104 s-1, a 250-fold enrichment (Fig. 4B). A second round of DACS further enriched the target cells to ≈65% of the population (Fig. 4C). Each step enabled sorting of 2–3 × 107 cells in 1 h, a rate comparable to conventional cell sorters. Remarkably, a repeat sort wherein all bead wash steps were completely eliminated from the protocol yielded an equivalent enrichment after two rounds (data not shown), indicating that, unlike MACS, wash steps are not essential for high purity sorting using DACS.
The use of DACS methodology may be extendable to the separation of a wide spectrum of biological species, including molecules, viruses, bacteria, and mammalian cells. Because the DACS microfabrication process is relatively versatile, the device geometry and characteristic lengths within the device can be tailored to specific cell types. In particular, the current implementation may prove to be useful in screening of bacterial and yeast display libraries to develop peptide and antibody affinity reagents where the selection process is typically based on a single surface marker. However, a sequential label-stripping and relabeling will enable the extension to multiple surface markers for other applications. Finally, in the current configuration, the limits of throughput performance are governed by the balance between the DEP force relative to the hydrodynamic forces on the labeled cells. However, it is expected that the integration of parallel sorting stages within the microfluidic chip will greatly augment the throughput capability and may enable the prospect of high-performance, automated cell sorting in a disposable format.
Acknowledgments
We thank Dr. Sang-Ho Lee and Marin Sigurdson for helpful discussions. H.T.S. thanks Dr. Ira Skurnick for continued encouragement. This work was supported by the Office of Naval Research Young Investigator Program (N000140410456), the Army Research Office Institute for Collaborative Biotechnologies (DAAD1903D004), University of California Biotechnology Research and Education Program graduate student training Grant 2004-30, and the Samsung Advanced Institute of Technology. Microfabrication was carried out in the Nanofabrication Facility at University of California, Santa Barbara.
Author contributions: X.H., P.H.B., C.D.M., P.S.D., and H.T.S. designed research; X.H., P.H.B., and J.Q. performed research; X.H., P.H.B., J.Q., P.S.D., and H.T.S. contributed new reagents/analytic tools; X.H., P.H.B., P.S.D., and H.T.S. analyzed data; and X.H., P.H.B., P.S.D., and H.T.S. wrote the paper.
Abbreviations: DEP, dielectrophoresis; DACS, DEP-activated cell sorting; MACS, magnetic-activated cell sorting; FACS, fluorescence-activated cell sorting; CM, Clausius–Mossotti; nDEP, negative DEP.
References
- 1.Gee, A. P. & Durett, A. G. (2002) Cytotherapy 4, 91-92. [DOI] [PubMed] [Google Scholar]
- 2.Thiel, A., Scheffold, A. & Radbruch, A. (1998) Immunotechnology 4, 89-96. [DOI] [PubMed] [Google Scholar]
- 3.Wachtel, S. S., Shulman, L. P. & Sammons, D. (2001) Clin. Genet. 59, 74-79. [DOI] [PubMed] [Google Scholar]
- 4.Miltenyi, S., Muller, W., Weichel, W. & Radbruch, A. (1990) Cytometry 11, 231-238. [DOI] [PubMed] [Google Scholar]
- 5.Chalmers, J. J., Zborowski, M., Sun, L. P. & Moore, L. (1998) Biotechnol. Progr. 14, 141-148. [DOI] [PubMed] [Google Scholar]
- 6.Ashcroft, R. G. & Lopez, P. A. (2000) J. Immunol. Methods 243, 13-24. [DOI] [PubMed] [Google Scholar]
- 7.Ibrahim, S. F. & van den Engh, G. (2003) Curr. Opin. Biotechnol. 14, 5-12. [DOI] [PubMed] [Google Scholar]
- 8.Gross, H. J., Verwer, B., Houck, D., Hoffman, R. A. & Recktenwald, D. (1995) Proc. Natl. Acad. Sci. USA 92, 537-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shizuru, J. A., Negrin, R. S. & Weissman, I. L. (2005) Annu. Rev. Med. 56, 509-538. [DOI] [PubMed] [Google Scholar]
- 10.Georgiou, G., Stathopoulos, C., Daugherty, P. S., Nayak, A. R., Iverson, B. L. & Curtiss, R. (1997) Nat. Biotechnol. 15, 29-34. [DOI] [PubMed] [Google Scholar]
- 11.Fu, A. Y., Spence, C., Scherer, A., Arnold, F. H. & Quake, S. R. (1999) Nat. Biotechnol. 17, 1109-1111. [DOI] [PubMed] [Google Scholar]
- 12.Kruger, J., Singh, K., O'Neill, A., Jackson, C., Morrison, A. & O'Brien, P. (2002) J. Micromech. Microeng. 12, 486-494. [Google Scholar]
- 13.Chen, C. C., Zappe, S., Sahin, O., Zhang, X. J., Fish, M., Scott, M. & Solgaard, O. (2004) Sens. Actuators B Chem. 102, 59-66. [Google Scholar]
- 14.Wang, M. M., Tu, E., Raymond, D. E., Yang, J. M., Zhang, H. C., Hagen, N., Dees, B., Mercer, E. M., Forster, A. H., Kariv, I., et al. (2005) Nat. Biotechnol. 23, 83-87. [DOI] [PubMed] [Google Scholar]
- 15.Pohl, H. A. (1978) Dielectrophoresis: The Behavior of Neutral Matter in Nonuniform Electric Fields (Cambridge Univ. Press, Cambridge, U.K.).
- 16.Muller, T., Pfennig, A., Klein, P., Gradl, G., Jager, M. & Schnelle, T. (2003) IEEE Eng. Med. Biol. Mag. 22, 51-61. [DOI] [PubMed] [Google Scholar]
- 17.Seger, U., Gawad, S., Johann, R., Bertsch, A. & Renaud, P. (2004) Lab Chip 4, 148-151. [DOI] [PubMed] [Google Scholar]
- 18.Gascoyne, P. R. C. & Vykoukal, J. (2002) Electrophoresis 23, 1973-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker, F. F., Wang, X. B., Huang, Y., Pethig, R., Vykoukal, J. & Gascoyne, P. R. (1995) Proc. Natl. Acad. Sci. USA 92, 860-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng, J., Sheldon, E. L., Wu, L., Heller, M. J. & O'Connell, J. P. (1998) Anal. Chem. 70, 2321-2326. [DOI] [PubMed] [Google Scholar]
- 21.Casadaban, M. J. & Cohen, S. N. (1980) J. Mol. Biol. 138, 179-207. [DOI] [PubMed] [Google Scholar]
- 22.Guzman, L. M., Belin, D., Carson, M. J. & Beckwith, J. (1995) J. Bacteriol. 177, 4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mittal, K. L. (2001) Polyimides and Ohter High Temperature Polymers: Synthesis, Characterization, and Applications (Utrecht, Boston).
- 24.Durr, M., Kentsch, J., Muller, T., Schnelle, T. & Stelzle, M. (2003) Electrophoresis 24, 722-731. [DOI] [PubMed] [Google Scholar]
- 25.Fiedler, S., Shirley, S. G., Schnelle, T. & Fuhr, G. (1998) Anal. Chem. 70, 1909-1915. [DOI] [PubMed] [Google Scholar]
- 26.Uribe, S. & Sampedro, J. G. (2003) Biol. Proced. Online 5, 108-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bessette, P. H., Rice, J. J. & Daugherty, P. S. (2004) Protein Eng. Des. Sel. 17, 731-739. [DOI] [PubMed] [Google Scholar]