Abstract
How organ identity is determined is a fundamental question in developmental biology. In Drosophila, field-specific selector genes, such as eyeless (ey) for eyes and vestigial (vg) for wings, participate in the determination of imaginal disc-specific identity. We performed gain-of-function screening and identified a gene named winged eye (wge), which encodes a bromo-adjacent homology domain protein that localizes at specific sites on chromosomes in a bromo-adjacent homology domain-dependent manner. Overexpression of wge-induced ectopic wings with antero-posterior and dorso-ventral axes in the eye field in a region-specific Hox gene-(Antennapedia) independent manner. Overexpression of wge was sufficient for ectopic expression of vg in eye discs. A context-dependent requirement of wge was demonstrated for vg expression in wing discs and for expression of eyes absent (eya), a control gene for eye development downstream of ey, in eye discs. In contrast to vg, however, overexpression of wge inhibited EY-mediated expression of eya. Consistent with colocalization on polytene chromosomes of WGE and Posterior sex combs (PSC), a Polycomb group gene product, we demonstrated an antagonistic genetic interaction between wge and Psc. These findings suggest that wge functions in the determination of disc-specific identity, downstream of Hox genes.
Keywords: determination, imaginal disc, organ identity, epigenetic regulation
To generate complex organs such as compound eyes, wings, and antennae, the correct cell types must be formed and correctly organized in three-dimensional dimensions, i.e., the specific organ identity must be determined in some specific morphogenetic field during development. Drosophila imaginal discs, the primordia of adult appendages and trunk, are valuable for investigating how specific identity is determined and maintained during development. The disc-specific determination state is established during embryogenesis or at early larval stages and is maintained until the discs differentiate into adult structures during metamorphosis. Some genes, such as eyeless (ey), vestigial (vg), and Distal-less (Dll), so-called master control genes or field-specific selector genes, have crucial roles in these functions, together with a cohort of subordinate transcription factors (1–4). For example, ey is required for eye development, and ectopic eyes can be induced on wings, legs, and antennae together with downstream genes, eyes absent (eya), sine oculis, and dachshund (1, 5–8). The ectopic expression of vg with its cofactor Scalloped (SD) induces ectopic wing-like structures on parts of the body other than the thorax (2, 9). Therefore, investigation of the regulatory mechanisms of upstream control of gene expression will advance this field. The region-specific Hox selector genes, such as Antennapedia (Antp) and Ultrabithorax, and the intracellular signaling pathways generating positional information within a morphogenetic field, such as Notch, Wingless (WG), and Decapentaplegic (DPP) signaling, are thought to regulate the expression of field-specific selector genes (4, 10–13).
We previously reported that, under the control of the eye-specific enhancer of ey, forced activation of Notch signaling induces ectopic structures, such as eyes, wings, legs, and antennae, in the eye-antennal field of the head, and respective control genes, ey, vg, and Dll, in eye-antennal discs in a context-dependent manner (10). For example, activation of Notch signaling induces ectopic expression of ey in antennal discs and ectopic eyes at the rostral membrane of the head, which is derived from the antennal disc. In combination with the expression of Antp, which is a Hox selector gene for the second thoracic segment, activation of Notch signaling induces vg and Dll in eye discs and ectopic wings and legs on the head. Notch signaling is not required for ey expression in eye discs (14), therefore the overexpression experiments might not reflect normal development in some aspects. The system does, however, provide a unique genetic screen that is useful for identifying genes capable of changing disc-specific identity. In this paper, we used a gain-of-function screen with 9,710 Gene Search (GS) lines (15) and identified a gene named winged eye (wge) that encodes a bromo-adjacent homology (BAH) domain protein, the overexpression of which induced ectopic wings on the head in an Antp-independent manner.
The BAH domain is frequently associated with other domains in proteins that are suggested to be involved in epigenetic regulation of gene expression such as bromodomains, plant homeodomain (PHD) fingers, and Suppressor of variegation 3-9, Enhancer of zeste, Trithorax (SET) domains (16). For example, in Drosophila, the BAH domain is present in ASH1 (absent, small, or homeotic discs 1) protein, which contains a SET domain and a PHD finger and belongs to the trithorax group (trxG) of activators (17). ASH1 is required for maintaining transcription of region-specific Hox selector genes such as Ultrabithorax and is thought to function by modulating chromatin structure with its histone methyl-transferase activity (18, 19). Here, we describe that wge is a chromatin-associated protein that is involved in the determination of imaginal disc identity in a context-dependent manner.
Materials and Methods
Fly Stocks. We performed two gain-of-function screenings. For the prescreening, ey-GAL4 females were crossed with the GS strains (15). For the screening, ey-GAL4/CyO p{w+, Act-GFP};UAS-Nact/TM3 p{w+, Act-GFP} females were crossed with the GS strains. UAS-ey, UAS-vg, vgBE-lacZ, dpp-GAL4, ey-lacZ, UAS-Antp, UAS-Nact, UAS-eya, eya-lacZ, dpp-lacZ, UAS-p35, and Psc1 were described in ref. 1, 2, 8, 10, and 20–23. The other transformants were generated by P element-mediated transformation. The wge40 strain, which had a 2,215-bp sequence deletion including the wge CDS, was established by retransposing the P element in the wgeGS15923 strain. Oregon R was used as the wild-type strain.
Clonal Analysis. Mutant clones for wge in a Minute background were generated by flp-mediated mitotic recombination (24) and marked by the absence of Ubi-GFP expression. To induce recombination, yw hs-flp/yw;FRT82B wge40/FRT82B Ubi-GFP Rps3Plac92 larvae were heat-shocked (37°C for 1 h) 48 or 72 h after egg deposition (AED). GAL4-expressing clones, based on the excision of the FRT cassette (25), were induced by heat shock (37°C for 1 h) 48 h AED to the following flies: yw hsFLP, Ay-GAL4 UAS-GFP/+ (or eya-lacZ or ey-lacZ), and UAS-wge/+.
The molecular cloning, RT-PCR, histochemistry procedures, and the primers used are published as Supporting Text, which is published as supporting information on the PNAS web site.
Results
Identification of Genes Capable of Inducing Ectopic Appendages in the Eye Field. We previously reported that artificial activation of Notch signaling induces various ectopic appendages, such as ectopic eyes, antennae, wings, and legs, in the eye field in a context-dependent manner (10). In this system, the ey enhancer-dependent activation of Notch signaling and gene expression was crucial for transdetermination within a limited time window and for shutting off the forced expression once the transdetermination was induced. To identify the genes capable of changing disc-specific identity, we used a system with a P element-based GS vector (15). For a pilot experiment, 106 lines harboring the GS vector (GS lines) were crossed with flies carrying the ey enhancer-GAL4 (ey-GAL4) and a construct for the constitutively active Notch receptor under an upstream-activating sequence for GAL4 (UAS-Nact). Ectopic structures were induced in the eye field in combination with Notch signaling activation in five lines. For example, ectopic wings were induced in GS 1068 in which ey-GAL4 drives the expression of PGRP-LE, CG8509, and sd, encoding a cofactor of VG (26). In the absence of Notch signaling activation, all five lines had reduced eye phenotypes. Therefore, to increase the screening efficiency, we introduced a prescreening step in which GS lines were crossed with the ey-GAL4 driver, and the resulting lines with reduced eye phenotypes were crossed with the ey-GAL4, UAS-Nact line. To confirm the effects of the prescreening, we crossed 74 negative GS lines with normal eyes in a prescreening with an ey-GAL4, UAS-Nact line. None of the lines had ectopic structures in the eye field, indicating that the prescreening was effective (data not shown). In the prescreening, 8,486 lines had normal eyes and 1,202 lines had reduced eye phenotypes. In 9 of 9,710 lines, ectopic structures were induced in the eye field, e.g., ectopic wings in the GS 15923, even in the absence of Notch signaling activation. We then crossed the resulting 729 lines with reduced eye phenotypes or ectopic structures in the eye field in the prescreening and 22 lines that were selected without prescreening with the ey-GAL4, UAS-Nact line, and ectopic structures were induced in 45 lines in combination with Notch signaling activation (wing, 3 lines; antenna, 26 lines; leg, 16 lines). The results of these screenings are summarized in Table 1, which is published as supporting information on the PNAS web site, and the data including the phenotype images are available on the Drosophila Gene Search Project web site (http://gsdb.biol.metro-u.ac.jp/%7Edclust).
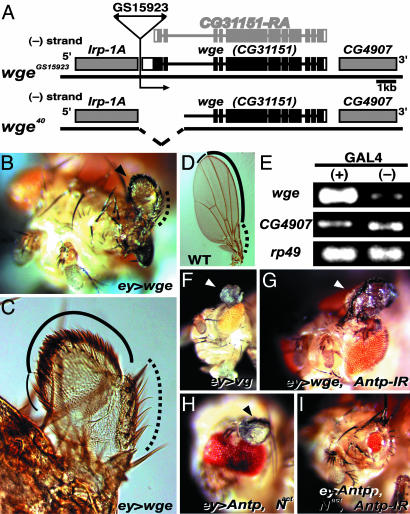
Overexpression of winged eye Induces Ectopic Wings with Antero-Posterior and Dorso-Ventral Axes in an Antp-Independent Manner. In this paper, we focus on the GS15923 line, because well organized wings with antero-posterior and dorso-ventral axes were induced in the eye field when GS15923 was crossed with ey-GAL4. Using the inverse PCR method, we determined the insertion site of the GS vector in GS15923 and found a predicted gene, CG31151, adjacent to the insertion site (Fig. 1A). CG31151 was the only gene expressed in a GAL4-dependent manner in GS15923 (Fig. 1E). After cloning cDNAs corresponding to CG31151, we identified an ORF of a previously uncharacterized gene that we named winged eye (wge), which has a 345-bp extension at the 5′ end from the estimated translation initiation site of expressed sequence tag (EST) clones corresponding to CG31151 (RA, Fig. 1A). The sequence of an EST clone, LP24488, overlapped by 693 bp with the 5′ end of the cloned cDNA and 20 bp of the 5′ end of the RA transcript, confirming the identified ORF. Wge encodes a 1,658-aa protein with two Gln-rich, one Ala-rich, and one Ser-rich domain at the N-terminal half and a bipartite nuclear localization signal and a BAH domain in the C-terminal half, implying that WGE is involved in epigenetic regulation of gene expression (Fig. 5A, which is published as supporting information on the PNAS web site). This prediction is consistent with the specific localization of WGE on polytene chromosomes as described below (Fig. 4A). Ectopic wing induction by forced wge expression was confirmed by using UAS-wge transgenic flies (Fig. 1 B and C; see also Table 2, which is published as supporting information on the PNAS web site). The WGE-mediated ectopic wings had the costa with spine bristles, the triple row of bristles, and the double row of bristles that are formed at the anterior-proximal part of the wing, at the anterior wing margin, and at the distal wing margin in wild-type wing, respectively (Fig. 1 C and D), indicating that WGE-mediated ectopic wings are correctly organized along the antero-posterior and dorso-ventral axes. In contrast, as reported before (2), VG-mediated ectopic wings in the eye field were outgrowths with wing hair that did not have any wing margins, suggesting that WGE is involved in the wing formation upstream of VG (Fig. 1F).
Fig. 1.
Identification of winged eye, the overexpression of which induces ectopic wings in the eye field and its Antp-independent function on ectopic wing induction. (A) Schematic representation of the genomic regions of wge. The black boxes represent the wge ORF. The direction of GS vector-mediated transcription, the insertion site of the P element GS15923, and previously estimated transcript of CG31151 (RA) from EST clones are indicated. The broken line represents the deletion in wge40. (B) WGE-mediated induction of ectopic wings in ey-GAL4, UAS-wge fly. Arrowhead indicates an ectopic wing. (C) Higher magnification of B. WGE-mediated ectopic wings have the costa with spine bristles (dotted lines), the triple row of bristles (bold line), the double row of bristles (narrow line), and the posterior rows of hairs (the remaining wing margin). (D) These structures are formed at the anterior-proximal part of wing (dotted line), anterior wing margin (bold line), distal wing margin (narrow line), and posterior wing margin in the wild-type wing, respectively. (E) GAL4-dependent induction of wge. RT-PCR was performed with hs-GAL4, GS15923 larvae (+), and GS15923 (-). rp49 was used as an internal control. (F) VG-mediated wing-like outgrowth without margin bristles formed in the eye field of the ey-GAL4, UAS-vg fly. Arrowhead indicates ectopic outgrowth. (G) Antp-independent induction of ectopic wings in the ey-GAL4, UAS-wge, UAS-Antp-IR fly. Arrowhead indicates ectopic wing with margin bristles. (H) Induction of ectopic wing in the ey-GAL4, UAS-Nact, UAS-Antp fly. Arrowhead indicates an ectopic wing with margin bristles. (I) Eye field of the ey-GAL4, UAS-Nact, UAS-Antp, UAS-Antp-IR fly.
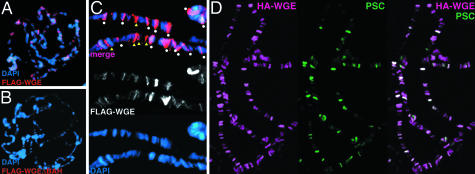
Fig. 4.
BAH domain-dependent localization of WGE and colocalization with PSC at specific sites of polytene chromosomes. (A and B) FLAG-tagged wild-type WGE and FLAG-tagged mutant protein (ΔBAH) lacking the BAH domain were expressed in a salivary gland (nonspecific expression of target genes are induced in a salivary gland by the GAL4/UAS system). The polytene chromosomes of ey-GAL4, UAS-FLAG-wge larvae (A) and ey-GAL4, UAS-FLAG-wgeΔBAH larvae (B) were stained with an anti-FLAG antibody. Immunostaining (red) and DAPI staining (DNA, blue) is merged. (C) The binding sites of WGE (red color is changed to white) were analyzed with DAPI staining (blue) in higher magnifications. Some WGE signals are overlapped with DAPI staining (circles), and others do not overlap with DAPI staining (arrowheads) in a merged picture. (D) Colocalization of PSC with WGE on polytene chromosomes. The polytene chromosomes of ey-GAL4, UAS-HA-wge larvae were stained with anti-HA antibody (red) and anti-PSC monoclonal antibody (green). Almost all PSC binding sites are coincident with some of WGE binding sites.
Previously, we have demonstrated that, in combination with activation of Notch signaling, expression of Antp by ey-GAL4 induces ectopic wings with antero-posterior and dorso-ventral axes, which are similar to WGE-mediated ectopic wings (Fig. 1H and ref. 10). To investigate the requirement of ANTP for the WGE-mediated induction of ectopic wings, we generated transgenic flies possessing an inverted repeat (IR) expression construct of Antp cDNA that specifically inhibits ANTP expression in a GAL4-dependent manner due to RNA interference. Antp-IR expression inhibited the formation of ectopic wings induced by the expression of Antp and Notch signaling activation, indicating that the construct works as a specific inhibitor of ANTP expression (Fig. 1I and Table 2). Antp-IR expression, however, did not inhibit the formation of ectopic wings when coexpressed with wge (Fig. 1G and Table 2). Consistent with this result, forced expression of wge did not induce ectopic expression of ANTP in eye discs of ey-GAL4;UAS-wge larvae (Fig. 2 A and B). These results indicated that wge overexpression induces ectopic wings in an Antp-independent manner. Moreover, wge-IR expression suppressed the formation of ectopic wings induced by the expression of Antp and Notch signaling activation (Table 2). These results indicate that WGE acts downstream of Antp and Notch signaling in the ectopic wing induction.
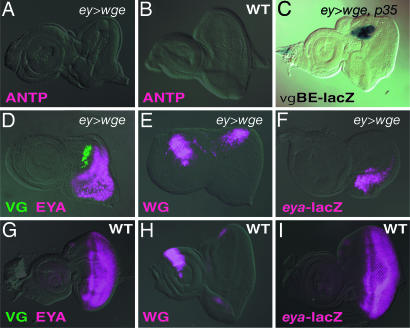
Fig. 2.
WGE-mediated induction of VG and WG and WGE-mediated suppression of EYA in eye discs. (A and D–F) Immunostaining of eye discs of ey-GAL4, UAS-wge larvae with antibodies against ANTP (A), VG (green) and EYA (red purple) (D), WG (E), and β-galactosidase (F). (F) An eye disc of an ey-GAL4, UAS-wge, eya-lacZ larva. (C) X-gal staining (blue) of an eye disc of an ey-GAL4, UAS-wge, vgBE-lacZ, UAS-p35 larva. To avoid vgBE-lacZ-mediated cell death, p35, a cell death inhibitor (22), was coexpressed. (B, G–I) Immunostaining of eye discs of wild-type larvae with antibodies against ANTP (B), VG (green) and EYA (red purple) (G), WG (H), and β-galactosidase (I). (I) An eye disc of an eya-lacZ transgenic larva. Staining is merged with the bright field image (except C). In all images, posterior is to the right and dorsal is up.
Overexpression of wge Induces Ectopic Expression of vg and wg and Represses EY-Mediated Expression of eya. Ectopic expression of vg in various imaginal discs induces ectopic wing-like outgrowth (2). In combination with WG signaling, however, it induces wings with wing margins (27). WG signaling is also suggested to participate in the determination of wing discs (13). We investigated whether wge overexpression induces ectopic expression of VG and WG in eye imaginal discs when it induces ectopic wings with wing margins. In wild type, VG is expressed in the wing discs but not in eye discs (Fig. 2G; see also Fig. 6F, which is published as supporting information on the PNAS web site), whereas, in ey-GAL4;UAS-wge, there was significant expression of VG in the eye discs (Fig. 2D). The forced expression of wge also activated vg D/V boundary enhancer-lacZ (vgBE-lacZ) (Fig. 2C). These results indicate that wge overexpression is sufficient to induce the ectopic expression of vg, a control gene for wing formation. In third-instar larvae, WG was expressed at the peripheral edge of the dorsal and ventral sides of wild-type eye discs (Fig. 2H). Overexpression of wge by ey-GAL4 enhanced the dorsal expression of WG and suppressed the ventral expression of WG in eye discs (Fig. 2E). These results were consistent with the findings that wge overexpression induced ectopic wings at the dorsal part of the eye field. Therefore, WGE is suggested to induce ectopic wings upstream of VG and WG.
We then investigated the effects of wge overexpression on the expression of genes involved in the determination of eye identity, such as ey and eya. Overexpression of wge-repressed EYA expression at the dorsal side of the eye discs, in contrast to WG expression, which was up-regulated at the dorsal side by WGE (Fig. 2 D and G). Similar results were obtained with eya-lacZ transgenic larvae (Fig. 2 F and I). The enhancer trap line of eya-lacZ reflects endogenous expression of EYA (Fig. 2 G and I and refs. 8 and 20). Double staining revealed that the ectopic induction of VG did not overlap with EYA expression when wge was overexpressed in eye discs (Fig. 2D). These results suggest that WGE-mediated down-regulation of eya is involved in the ectopic induction of vg. We could not examine the effects of eya overexpression on WGE-mediated ectopic vg expression, however, because eya overexpression in eye discs inhibits eye disc development. Further analysis is required to determine the relationship between the down-regulation of eya and the ectopic induction of vg.
Clonal activation of WG signaling represses eya expression in eye discs (28). To examine the effects of clonal induction of wge overexpression on eya and wg expression, we applied a cell lineage tracer technique by using a combination of the flp/FRT and GAL4/UAS recombinase systems (25). In this system, in which wge-overexpressing cells are labeled with GFP, eya-lacZ expression was repressed by clonal induction of wge overexpression in a cell-autonomous manner (Fig. 6A), whereas WG expression was not induced when the wge-expressing clone was induced, even on the dorsal side of the eye discs (Fig. 6C). These results indicate that overexpression of wge represses expression of eya in eye discs in a cell-autonomous manner, independent of the up-regulation of wg. Overexpression of wge-inhibited expression of eya, a gene downstream of ey; however, ey-lacZ expression in eye discs was not repressed by clonal induction of wge overexpression (Fig. 6B). Consistent with these results, EY-mediated ectopic induction of eya was inhibited by coexpression of wge. Ectopic induction of ey under the control of dpp-GAL4 induced ectopic induction of eya-lacZ in the leg (Fig. 6I), wing, and antennal discs (8), but the ectopic expression of eya-lacZ was totally suppressed by the coexpression of wge (Fig. 6J). Coexpression of GFP did not suppress the EY-mediated induction of eya, indicating a specific effect of wge overexpression on eya expression (data not shown). These results suggest that overexpression of wge suppresses the eye development program downstream of ey. EYA repression occurs only on the dorsal side of the eye disc when wge is overexpressed with ey-GAL4, although clonally overexpressed wge also represses EYA on the ventral side. Further analysis is required to explain these phenomena.
Although ey-GAL4-dependent overexpression of wge-induced WG expression on the dorsal side of the eye discs, clonal induction of wge overexpression did not induce WG expression in eye discs. Consistent with these results, clonal induction of wge overexpression did not induce either ectopic expression of VG in eye discs (Fig. 6D) or ectopic formation of wings in the eye field on the head (data not shown), suggesting that overexpression of wge in a relatively large field is required for the transformation of eye to wing. Repression of eya but absence of vg expression in clonal overexpression of wge might represent an intermediate step toward wing transformation, which has to be analyzed further. In wing discs, clonal induction of wge overexpression did not affect expression of either VG or WG (Fig. 6 E and G), indicating a specific effect of clonal induction of wge overexpression on eya expression in eye discs.
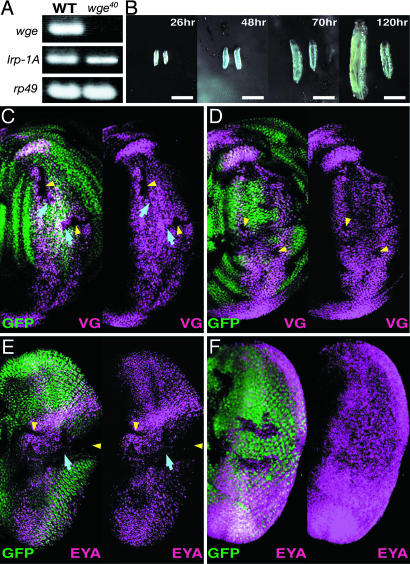
Context-Dependent Requirement of wge for vg Expression in Wing Discs. To investigate the requirement of wge for wing development, we generated a wge-deficient mutant by mobilizing the P element. The deletion of wge was screened by using genomic PCR in 126 excision lines. In one line, wge40, sequencing analysis after genomic PCR revealed that a 2,215-bp sequence, including the wge first exon, was deleted (Fig. 1 A). In wge40, there was no wge expression, and the expression of a gene neighboring wge, Irp-1A, was not affected, indicating that wge40 is a wge null mutant (Fig. 3A). The embryogenesis of wge40 was quite normal, but the development of wge40 gradually stopped after the first-instar larval stage, suggesting crucial roles of wge in larval development or growth (Fig. 3B). A rare rescue (3%) of larval lethality was observed by wge overexpression by using a heat-shock promoter, reflecting the context-dependent requirement of wge for development as described below. Rescue was never observed, however, in the absence of heat shock (29°C for 2 h every 24 h). We then introduced wge mutant clones by using the flp/FRT system with the Minute technique (24). When the wge mutant clones were introduced in wing discs 48 h AED by heat shock, no VG expression was observed in many cells within the clones, but some cells in the clones still expressed VG (Fig. 3C). The expression of dpp-lacZ was observed in the wge mutant clones, indicating a specific effect of the wge mutation on vg expression (Fig. 7C, which is published as supporting information on the PNAS web site). As described below, the size of these clones was relatively small, whereas VG expression was not affected in all but a few small-sized clones when the wge mutant clones were introduced 72 h AED (Fig. 3D). These results indicate that wge is required for vg induction in wing discs in a context-dependent manner. Compared with the wge mutant clones that were introduced 72 h AED, the wge mutant clones that were introduced 48 h AED were small in size, suggesting stage-specific involvement of wge on the growth of disc cells.
Fig. 3.
A context-dependent requirement of wge for vg expression in wing discs and for eya expression in eye discs. (A) Lack of wge transcript in wge40 mutant. RT-PCR was performed with first- and second-larval stage wild-type (WT) and wge40. rp49 was used as an internal control. (B) Developmental defect of wge40. Wild-type larvae (left side) and wge40 larvae (right side) at the indicated times AED are represented. (Scale bars: 1 mm.) (C–F) wge mutant clones were introduced in wing discs (C and D) and eye discs (E and F) at 48 h (C and E) and 72 h AED (D and F). Immunostaining (red purple) with antibody against VG (C and D) and EYA (E and F) is merged with GFP (green). wge mutant clones lack GFP signals. Yellow arrowheads indicate cells with no VG (C) or EYA expression (E) within the clones. Blue arrows indicate cells expressing VG (C) or EYA (E) within the clones. In D, yellow arrowheads indicate wge mutant clones with no VG expression. In C and D, dorsal is to the left and anterior is up. In E and F, posterior is to the right and dorsal is up.
wge Is Expressed Ubiquitously and Is Required for the Formation of Various Appendages. Clonal induction of wge overexpression represses eya-lacZ expression in eye discs. We investigated derepression of eya in the wge mutant clones. Contrary to our prediction, there was no misexpression of EYA in wge mutant clones that were introduced at both 48 and 72 h AED in imaginal discs such as antennal, leg (data not shown), and wing discs (Fig. 7 A and B). In eye discs, EYA was not expressed in many cells within the wge mutant clones but was expressed in some cells when the clones were introduced 48 h AED (Fig. 3E), whereas the expression of EYA was not affected in the clones when the wge mutant clones were introduced 72 h AED (Fig. 3F). These results indicate that wge is required for eya expression in eye discs in a context-dependent manner, which is similar to the function of wge in the regulation of vg expression in wing discs. Consistent with the requirement of wge for the function of vg and eya, adult structures, such as eyes, wings, and legs, were malformed when wge mutant clones were introduced 72 h AED (Fig. 7 D–H). The mutant clones were distinguished by the absence of Ubi-GFP (Fig. 7E). Pupal lethality was induced when wge mutant clones were introduced 48 h AED. The participation of wge in the development and growth of various appendages confirmed the ubiquitous expression of wge (Fig. 8, which is published as supporting information on the PNAS web site). RT-PCR findings revealed constitutive expression of wge throughout the larval and pupal developmental stages and ubiquitous expression of wge in larval tissues and imaginal discs (Fig. 8 A and B). The ubiquitous expression of wge in various tissues was confirmed by in situ hybridization experiments (Fig. 8 C–J).
WGE Localizes at Specific Chromatin Sites in a BAH Domain-Dependent Manner. WGE has a BAH domain that is frequently found in proteins participating in the epigenetic regulation of gene expression. To investigate the nuclear localization and chromatin association of WGE, we expressed FLAG-tagged wild-type WGE and FLAG-tagged mutant protein (ΔBAH) lacking the BAH domain and stained salivary glands with anti-FLAG antibody (Figs. 4 and 5). Both wild-type WGE and ΔBAH were localized in the nuclei of the salivary glands (Fig. 5C), whereas only wild-type protein, and not ΔBAH, localized at specific sites on polytene chromosomes (Fig. 4 A and B). These results indicate that WGE associates with chromatin in a BAH domain-dependent manner. The association of WGE with chromatin seems to be crucial for WGE function, because ΔBAH did not induce ectopic wings in the eye field when it was expressed by ey-GAL4 (Table 2).
The binding sites of WGE were analyzed with DAPI staining (DNA) under higher magnification. Some signals overlapped with DAPI staining and others did not overlap with DAPI staining, suggesting that WGE localizes in both bands and interbands of polytene chromosomes (Fig. 4C). We then compared the WGE binding sites on polytene chromosomes to that of Posterior sex combs (PSC). Almost all PSC binding sites were coincident with some WGE binding sites, suggesting that some WGE function is related to PSC function (Fig. 4D). The genetic interaction between wge and Psc was investigated with wge40 and Psc1 (Table 3, which is published as supporting information on the PNAS web site). The extra sex comb phenotype of Psc1 was suppressed by the loss of one dose of wge, suggesting a wge function similar to that of trithorax-group (trxG) genes, which antagonize PcG genes. There was a maternal effect in wge40 single heterozygotes, as indicated by the appearance of an additional sex comb on the second tarsomere of the first leg. Such a transformation of the second to the first tarsomere of the leg occurs in some PcG mutants such as multi sex combs and cramped (29, 30). The additional sex comb phenotype of wge40 was suppressed by a partial loss-of-function of Psc. In both wge40 and Psc1 single heterozygotes, the number of sex comb teeth on the first tarsomere of the first legs was increased, whereas there was no significant modification of the number of sex comb teeth in double heterozygotes. These results indicate that wge and Psc have antagonistic roles in both transformation from the second thoracic legs to the first thoracic legs and transformation from the second tarsomere to the first tarsomere of the first leg but not in the increase in the number of sex comb teeth.
Database analysis revealed a genome sequence of Anopheles gambiae, ENSANGP00000005615, with striking similarity to wge; for example, there was a 77% amino acid identity in the BAH domain. Comparison of the two sequences led to the identification of a previously uncharacterized protein domain named highly corresponding region (HCR: 126 aa, amino acids 1130–1255) with similarity with the A. gambiae gene product (57% amino acid identity), KIAA1447 human protein (41%), CAGL79 human protein (33%), and BC060615 mouse protein (42%) (Fig. 5B). These results suggest that wge is evolutionarily conserved in insects and mammals.
Discussion
WGE localizes at specific sites on polytene chromosomes in a BAH domain-dependent manner, and wge overexpression induces a gain-of-function transformation of eyes to wings. The extra sex comb phenotype of Psc1 is suppressed by the loss of one dose of wge. The characteristics of wge are similar to that of trxG genes. The trxG genes were identified as suppressors of the Polycomb phenotype and are implicated in the activation of Hox selector genes (31). Therefore, similar to loss-of-function mutations in the PcG genes, gain-of-function mutations in trxG genes cause ectopic expression of Hox selector genes and homeotic transformations. TrxG proteins also localize at specific sites on polytene chromosomes, and one of the trxG proteins, ASH1, has a BAH domain (16). There are several functional differences, however, between wge and trxG genes. One major functional difference is Hox selector gene independence on homeotic transformation. Mutations of the trxG genes cause homeotic transformations through the modulation of transcriptional regulation of the Hox selector genes. On the other hand, overexpression of wge induces ectopic wings in an Antp-independent manner. Although endogenous wing development is considered to be independent of Antp (32), Antp was the only Hox selector gene examined that induced eye-to-wing transformation in the system (F. Prince, T.K., S. Plaza, D. Resendez-Perez, M. Berry, S.K., and W.J.G., unpublished data). In addition, wge overexpression does not induce ectopic expression of Antp in eye discs. Therefore, wge induces eye-to-wing transformation in an independent Hox selector gene. Moreover, wge is required for the ectopic wing formation that is induced by the expression of Antp and the activation of Notch signaling. These results suggest that wge is involved in the regulation of field-specific selector gene expression but not in the regulation of region-specific Hox selector gene expression. There is probably a regulatory mechanism that determines the field-specific identity after determination of region-specific identity by Hox selector genes. Another difference between wge and trxG is that the trxG functions as activators, whereas wge overexpression also represses eya.
Wge is required for the expression of both vg in wing discs and eya in eye discs in a context-dependent manner. Overexpression of wge, however, induces ectopic expression of vg and represses eya expression in eye discs. In wing discs, wge overexpression does not induce either ectopic expression of vg or repression of vg. Therefore, wge regulates expression of vg and eya in a context-dependent manner. Consistent with the context-dependent function of wge, wge is expressed ubiquitously throughout larval to pupal development and in various tissues. The field-specific identity should be determined from an equivalent group of cells. This characteristic is observed not only in normal development but also in artificial situations of imaginal discs called transdetermination, in which, after regenerative cell growth, disc cells change their determined state to another determined state, e.g., a leg disc transdetermines to a wing disc (33, 34). Transdetermination is a polyclonal event and not the result of either differentiation of reserve cells or somatic mutations (34). Context-dependent regulation of gene expression by a ubiquitously expressed gene might explain how differences are created within a group of equivalent cells.
Supplementary Material
Acknowledgments
We thank B. Bello, D. Brower, S. Carroll, G. Halder, Y. Hiromi, T. Murata, the Bloomington Stock Center (Indiana University), Drosophila Genetic Resource Center in the Kyoto Institute of Technology (Kyoto), Genetic Strain Research Center of the National Institute of Genetics (Mishima, Japan), and Developmental Studies Hybridoma Bank (Iowa City, IA) for fly stocks and antibodies and S. Hirose and S. Hayashi for helpful advice. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, the Japan Society for the Promotion of Science, and the Program for the Promotion of Basic Research Activities for Innovative Biosciences. W.J.G. is supported by the Kantons of Basel and the Swiss National Science Foundation.
Author contributions: T.K., Y.O., W.J.G., T.A., and S.K. designed research; T.K., T.S., M.T., and S.K. performed research; T.K., W.J.G., and S.K. analyzed data; and S.K. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: AED, after egg deposition; BAH, bromo-adjacent homology; GS, gene search; IR, inverted repeat; PSC, Posterior sex combs.
Data deposition: The sequence reported in this paper has been deposited in the DDBJ/EMBL/GenBank databases (accession no. AB190214).
References
- 1.Halder, G., Callaerts, P. & Gehring, W. J. (1995) Science 267, 1788-1792. [DOI] [PubMed] [Google Scholar]
- 2.Kim, J., Sebring, A., Esch, J. J., Kraus, M. E., Vorwerk, K., Magee, J. & Carroll, S. B. (1996) Nature 382, 133-138. [DOI] [PubMed] [Google Scholar]
- 3.Gorfinkiel, N., Morata, G. & Guerrero, I. (1997) Genes Dev. 11, 2259-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Affolter, M. & Mann, R. (2001) Science 292, 1080-1081. [DOI] [PubMed] [Google Scholar]
- 5.Bonini, N. M., Bui, Q. T., Gray-Board, G. L. & Warrick, J. M. (1997) Development (Cambridge, U.K.) 124, 4819-4826. [DOI] [PubMed] [Google Scholar]
- 6.Pignoni, F., Hu, B., Zavitz, K. H., Xiao, J., Garrity, P. A. & Zipursky, S. L. (1997) Cell 91, 881-891. [DOI] [PubMed] [Google Scholar]
- 7.Chen, R., Amoui, M., Zhang, Z. & Mardon, G. (1997) Cell 91, 893-903. [DOI] [PubMed] [Google Scholar]
- 8.Halder, G., Callaerts, P., Flister, S., Walldorf, U., Kloter, U. & Gehring, W. J. (1998) Development (Cambridge, U.K.) 125, 2181-2191. [DOI] [PubMed] [Google Scholar]
- 9.Simmonds, A. J., Liu, X., Soanes, K. H., Krause, H. M., Irvine, K. D. & Bell, J. B. (1998) Genes Dev. 12, 3815-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurata, S., Go, M. J., Artavanis-Tsakonas, S. & Gehring, W. J. (2000) Proc. Natl. Acad. Sci. USA 97, 2117-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar, J. P. & Moses, K. (2001) Cell 104, 687-697. [DOI] [PubMed] [Google Scholar]
- 12.Curtiss, J., Halder, G. & Mlodzik, M. (2002) Nat. Cell Biol. 4, E48-E51. [DOI] [PubMed] [Google Scholar]
- 13.Maves, L. & Schubiger, G. (2003) Curr. Opin. Genet. Dev. 13, 472-479. [DOI] [PubMed] [Google Scholar]
- 14.Kenyon, K. L., Ranade, S. S., Curtiss, J., Mlodzik, M. & Pignoni, F. (2003) Dev. Cell 5, 403-414. [DOI] [PubMed] [Google Scholar]
- 15.Toba, G., Ohsako, T., Miyata, N., Ohtsuka, T., Seong, K. H. & Aigaki, T. (1999) Genetics 151, 725-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Callebaut, I., Courvalin, J. C. & Mornon, J. P. (1999) FEBS Lett. 446, 189-193. [DOI] [PubMed] [Google Scholar]
- 17.Tripoulas, N., LaJeunesse, D., Gildea, J. & Shearn, A. (1996) Genetics 143, 913-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beisel, C., Imhof, A., Greene, J., Kremmer, E. & Sauer, F. (2002) Nature 419, 857-862. [DOI] [PubMed] [Google Scholar]
- 19.Byrd, K. N. & Shearn, A. (2003) Proc. Natl. Acad. Sci. USA 100, 11535-11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonini, N. M., Leiserson, W. M. & Benzer, S. (1993) Cell 72, 379-395. [DOI] [PubMed] [Google Scholar]
- 21.Sanicola, M., Sekelsky, J., Elson, S. & Gelbart, W. M. (1995) Genetics 139, 745-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hawkins, C. J., Wang, S. L. & Hay, B. A. (1999) Proc. Natl. Acad. Sci. USA 96, 2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adler, P. N., Charlton, J. & Brunk, B. (1989) Dev. Genet. 10, 249-260. [DOI] [PubMed] [Google Scholar]
- 24.Xu, T. & Rubin, G. M. (1993) Development (Cambridge, U.K.) 117, 1223-1237. [DOI] [PubMed] [Google Scholar]
- 25.Ito, K., Awano, W., Suzuki, K., Hiromi, Y. & Yamamoto, D. (1997) Development (Cambridge, U.K.) 124, 761-771. [DOI] [PubMed] [Google Scholar]
- 26.Takehana, A., Katsuyama, T., Yano, T., Oshima, Y., Takada, H., Aigaki, T. & Kurata, S. (2002) Proc. Natl. Acad. Sci. USA 99, 13705-13710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baena-Lopez, L. A. & Garcia-Bellido, A. (2003) Development (Cambridge, U.K.) 130, 197-208. [DOI] [PubMed] [Google Scholar]
- 28.Baonza, A. & Freeman, M. (2002) Development (Cambridge, U.K.) 129, 5313-5322. [DOI] [PubMed] [Google Scholar]
- 29.Santamaría, P. & Randsholt, N. B. (1995) Mol. Gen. Genet. 246, 282-290. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto, Y. Girard, F. Bello, B. Affolter, M. & Gehring, W. J. (1997) Development (Cambridge, U.K.) 124, 3385-3394. [DOI] [PubMed] [Google Scholar]
- 31.Orlando, V. (2003) Cell 112, 599-606. [DOI] [PubMed] [Google Scholar]
- 32.Carroll, S. B., Weatherbee, S. D. & Langeland, J. A. (1995) Nature 375, 58-61. [DOI] [PubMed] [Google Scholar]
- 33.Hadorn, E. (1965) in Genetic Control of Differentiation: Brookhaven Symposia in Biology No. 18 (Brookhaven Natl. Lab., Upton, NY) pp. 148-159. [Google Scholar]
- 34.Gehring, W. (1972) in Results and Problems in Cell Differentiation, Vol. 5, eds. Ursprung, H. & Nöthiger, R. (Springer, Berlin) pp. 36-58. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.