Abstract
Dyskeratosis congenita is a rare inherited disorder characterized by abnormal skin manifestations. Morbidity and mortality from this disease is usually due to bone marrow failure, but idiopathic pulmonary fibrosis and an increased cancer predisposition also occur. Families with autosomal dominant dyskeratosis congenita display anticipation and have mutations in the telomerase RNA gene. We identified a three-generation pedigree with autosomal dominant dyskeratosis congenita, anticipation, and telomere shortening. We show that a null mutation in motif D of the reverse transcriptase domain of the protein component of telomerase, hTERT, is associated with this phenotype. This mutation leads to haploinsufficiency of telomerase, and telomere shortening occurs despite the presence of telomerase. This finding emphasizes the importance of telomere maintenance and telomerase dosage for maintaining tissue proliferative capacity and has relevance for understanding mechanisms of age-related changes.
Keywords: telomere, aplastic anemia, hTERT
Dyskeratosis congenita (DC) is a rare inherited disorder traditionally defined by a triad of mucocutaneous features: skin hyperpigmentation, oral leukoplakia, and nail dystrophy. The majority of patients suffer morbidity from progressive aplastic anemia (AA). In addition, pulmonary and liver fibrosis also occur in these patients. These latter phenotypes, although clinically asymptomatic, often appear first as fatal complications of treatment in the setting of bone marrow transplant and are related to hypersensitivity to preparative chemotherapy and radiation regimens (1, 2). The X-linked form of DC presents in the first decade, is most severe, and is a result of mutations in the DKC1 gene (3). The autosomal dominant (AD) form of DC is rare, has a later presentation, and is characterized by mutations in telomerase RNA (hTR) (4). Such families display anticipation, a worsening and earlier onset of symptoms with successive generations, which correlates with telomere shortening (5). However, among the families with AD DC, pedigrees are described with no skin manifestations but that nevertheless have hTR mutations, suggesting that these dermatologic findings are not critical to the genetic diagnosis and are not necessary for defining this disorder (6). Additionally, a small fraction of patients with apparently sporadic AA have been reported to have heterozygous mutations in hTR (7) and, more recently, in the catalytic component of telomerase, hTERT (8).
Telomerase is a specialized reverse transcriptase responsible for telomere length maintenance (9). Telomeres are required for chromosome stability and consist of tandem repeats of TTAGGG sequences bound by specific telomere-binding proteins. Telomerase adds telomeric repeats onto chromosome ends to offset the shortening that occurs during DNA replication. Telomerase consists of two essential components: the catalytic protein hTERT and the telomerase RNA (hTR) that specifies the repeat sequence added through an intrinsic template (10, 11). Telomere length limits the replicative capacity of primary fibroblasts and has been implicated in cellular aging (12, 13). In the presence of short telomeres, a DNA damage response is activated that leads to apoptosis (14-16). The shortest telomeres within a cell, not the average telomere length, are responsible for mediating this response that leads to cell death (17). Here, we show that mutations in the catalytic component of telomerase, hTERT can result in a complex phenotype of stem cell failure identical to AD DC. This phenotype shows anticipation; it presents earlier and more severely with successive generations. The anticipation is due to haploinsufficiency of telomerase that results in progressive shortening of telomeres.
Methods
Participants and Consent. The study was approved by the Johns Hopkins Hospital Institutional Review Board. Written informed consent was obtained from all participants.
Genotyping. Genomic DNA was extracted from peripheral blood pellets by using standard methods. Microsatellites were identified from the University of California Santa Cruz Genome Bioinformatics database at http://genome.ucsc.edu/. Genomic DNA was amplified with forward labeled fluorescent primers obtained from Qiagen (Valencia, CA). PCR products were analyzed on the ABI 3100 capillary electrophoresis instrument (Applied Biosystems). Pherograms were interpreted manually to determine allele size. hTERT sequencing was performed in both directions, and mutations were confirmed in three separately amplified PCR products. Primer sequences and PCR conditions are available upon request.
Logarithm of Odds (LOD) Score Calculation. LOD score calculation was performed by using genetic analysis software genehunter 2.1) and based on single point linkage analysis assuming an autosomal dominant model and full penetrance (18).
FISH. Telomere FISH was performed on metaphases from stimulated primary T lymphocytes as described (19). Fifteen metaphases were analyzed. Primary fibroblasts were also available from the proband and II2 (Fig. 1) and were stained with a peptide nucleic acid-labeled telomere probe. Thirty interphase nuclei were analyzed. Quantitation was done by using image j, available at http://rsb.info.nih.gov/ij/.
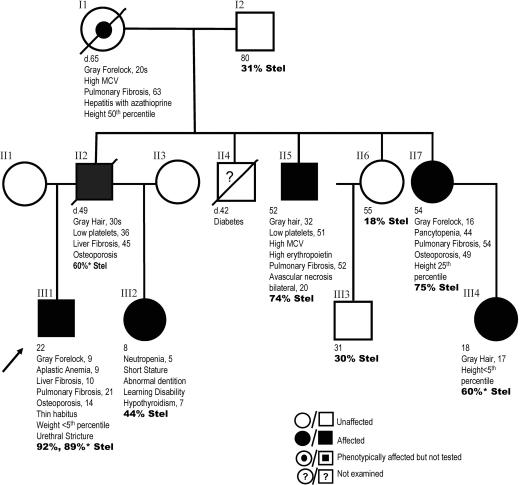
Fig. 1.
Pedigree with clinical features and age of onset. Individuals carrying the K902N mutation are indicated as affected. The pedigree displays anticipation in the onset of gray forelock, liver and lung fibrosis, and aplastic anemia. The onset and severity of symptoms correlates with the percentage of short telomeres in bold (% Stel). Short telomeres were defined as the number of telomeres having telomere fluorescent units <15 (Fig. 2). Quantitative FISH studies were done on primary lymphocytes. *, Telomeres measured in fibroblasts.
Mutagenesis of hTERT. Point mutations in the hTERT gene were generated essentially by using an overlap extension PCR strategy as described (20). Briefly, segments of the hTERT gene were PCR amplified by using combinations of an upstream hTERT-BamHI-F forward primer (5′-CATCAGGGGCAAGTCCTACGTC-3′), a downstream hTERT-NotI-R reverse primer (5′-GAGTGCGGCCGCCCGGGTCGAC-3′), and the internal primer pairs that contained desired mutant sequences K902A, K902N, and K902Q, in the middle of the primer. The amplified fragments were PCR-assembled, and the full-length PCR products were gel-purified, digested with BamHI and NotI, and cloned into the BamHI-NotI-digested phTERT-HA2 plasmid DNA. This plasmid encodes hTERT protein containing a C-terminal hemagglutinin (HA) tag for immunopurification of in vitro reconstituted telomerase enzyme. All mutant TERT genes were sequenced to confirm the presence of only the intended mutations.
In Vitro Reconstitution of Telomerase. Human telomerase complexes were reconstituted in vitro as described (21, 22). Briefly, epitope-tagged human TERT proteins were expressed from phTERT-HA2 (23) by using the TnT transcription/translation system (Promega), and assembled with purified T7 in vitro transcribed hTR. In vitro reconstituted telomerase complexes were immunoaffinity purified by using anti-HA F7 agarose beads (Santa Cruz Biotechnology) in the presence of 1× immunoprecipitation (IP) buffer (10 mM Hepes, pH 7.5/100 mM potassium glutamate/1 mM MgCl2/1 mM DTT/10% glycerol) and assayed for telomerase activity by using the direct telomerase assay protocol.
Telomere Repeat Amplification Protocol (TRAP) Assay. A TRAP assay was used to detect minimal activity of mutant telomerase containing TERT mutations K902A, K902N, and K902Q. In vitro reconstituted telomerase was diluted to 1/5×, 1/25×, and 1/125× and subjected to TRAP assay using TRAPeze telomerase detection kit according to the vender's instruction (Chemicon).
Direct Telomerase Assay. Activity of immunopurified telomerase was assayed directly without amplification (22). The reaction mixture (20 μl) contained 1× telomerase assay buffer (50 mM Tris·HCl, pH 8.0/50 mM KCl/1 mM MgCl2/5 mM 2-mercaptoethanol/1 mM spermidine), 1.0 μM telomere primer (TTAGGG3), 0.5 mM dATP, 0.5 mM dTTP, 2 μM dGTP, and 1.25 μM [α-32P]dGTP (800 Ci/mmol) (1 Ci = 37 GBq) with 6 μl of immunopurified telomerase complex. The reaction was incubated at 30°C for 1 h. The products were subjected to phenol-chloroform extraction and ethanol precipitation, followed by polyacrylamide electrophoresis. The gels were dried and exposed to Fuji phosphorimager screen. Repeat addition processivity was calculated by comparing the normalized intensity of the major product in the first and second repeats.
Results
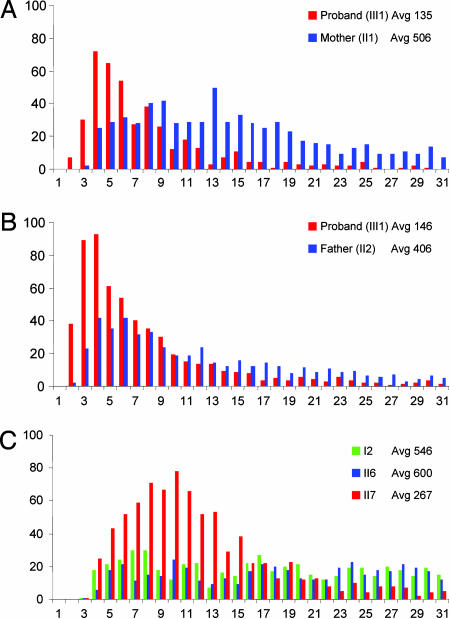
We identified a three-generation kindred with features of autosomal dominant DC but no skin manifestations (Fig. 1). The proband is a 22-yr-old male who presented with easy bruising and a gray forelock at age 9 (Fig. 1, III1). He was found to have AA, which has not required treatment. He has since been shown to have liver fibrosis, pulmonary fibrosis, osteoporosis, and a urethral stricture, all features described in DC patients. His family history displays a pattern of dominant inheritance of premature graying, AA, and fibrosis of the lung and liver (Fig. 1). There is anticipation in all four features. Because the anticipation suggested that telomere shortening may play a role in this disease, we examined telomeres directly by using quantitative FISH (Q-FISH). We found that the frequency of short telomeres was increased in the proband compared with both his parents (Fig. 2 A and B). This pattern of progressive telomere shortening with generations was also present in an aunt (Fig. 1, II7), who is less severely affected than the proband, when compared with the telomeres of her unaffected sister and father (Fig. 2C). In examining the entire pedigree, we found the severity of onset of the clinical phenotype correlated not only with a decrease in the average telomere length, but also with an increase in the shortest group of telomeres (Figs. 1 and 2). The shortest telomeres, not the average telomere length, are also responsible for the phenotypes seen in the telomerase knockout mouse where anticipation due to telomere shortening was first described (24).
Fig. 2.
Telomere length distributions of family members. Quantitative FISH shows the proband with shorter average telomere length than either parent in both primary fibroblasts (A) and lymphocytes (B). (C) The proportion of shortest telomeres (lymphocytes) is increased in an affected individual (II7) compared with both an unaffected sibling of comparable age (II6) and an unaffected parent (I2). The percentage of short telomeres in the proband (B) is increased compared with a less severely affected aunt (C, II7) and correlates with anticipation. Average telomere length (Avg) is indicated with individual identifiers shown in Fig. 1.
Because of the association of AD DC with mutations in hTR, we examined the hTR gene and locus. Sequencing of hTR revealed no abnormalities. Both blood and buccal mucosa were examined in the proband to exclude mosaicism. The karyotype was normal and revealed no visible deletions. To exclude a microdeletion of one of the hTR loci at 3q26.2, we performed FISH using a 6.8-kb probe spanning hTR and found two intact alleles (data not shown). To assess linkage to this region, we analyzed microsatellite markers covering 2.7 megabases around the hTR locus at D3S1614, D3S1282, D3S3523, and D3S3656. Under the assumption of dominant inheritance, we found LOD scores of 0.65, -2.3, -1,000, and -2.3, respectively. The negative LOD scores definitively excluded the 3q26.2 region, including hTR and its regulatory elements, from being implicated in this pedigree.
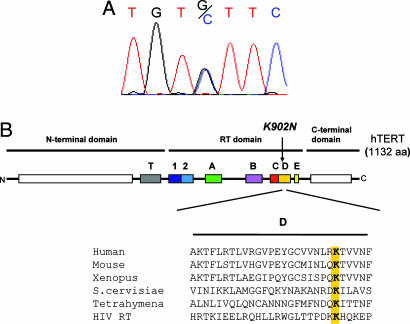
The absence of mutations in hTR, together with progressive telomere shortening, suggested that a mutation in other genes involved in telomerase or telomere maintenance may be present. We thus examined the coding sequences of hTERT in the proband. We identified a G to C missense mutation in exon 11 that results in a lysine to asparagine substitution in residue 902 (K902N) (Fig. 3A). We examined the segregation of the K902N mutation in the three-generation kindred and found that the mutation segregated with the phenotype in all affected members and was absent in the unaffected family members (Fig. 1). hTERT mutations were also recently reported in AA patients. This K902N change was not present as a polymorphism or mutation in 528 healthy controls in that study (8). The lysine residue at position 902 defines motif D, one of seven reverse transcriptase consensus motifs present in hTERT that is conserved in all reverse transcriptase families (Fig. 3B). Mutations of this highly conserved residue in HIV RT result in a severe reduction in activity and specifically affect the selection of the correct nucleotide in the catalytic site (25).
Fig. 3.
Point mutation in hTERT.(A) Chromatogram showing the presence of a heterozygous mutation G to C in the proband in exon 11 that results in a lysine to asparagine substitution in residue 902. (B) Schematic of hTERT protein with conserved reverse transcriptase domains shown in color. The K902N mutation lies within Motif D, which is conserved among telomerase proteins and across reverse transcriptase families, including HIV reverse transcriptase.
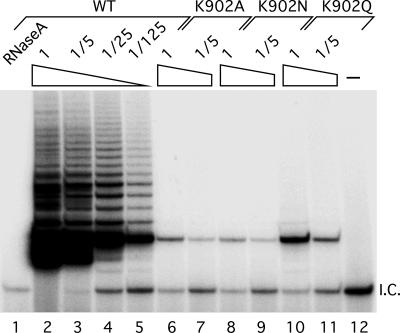
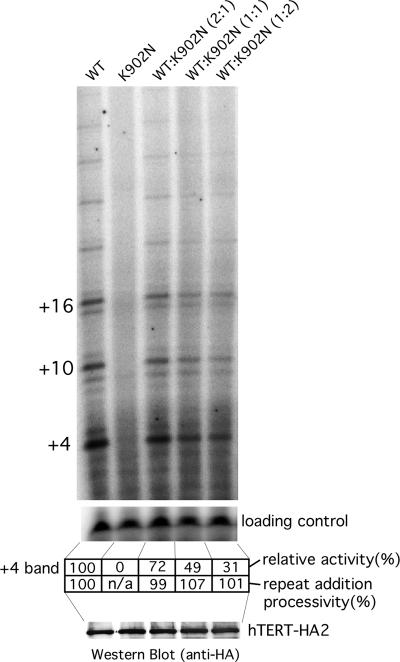
Based on the HIV reverse transcriptase studies, the K902N mutation was predicted to severely decrease enzyme activity of hTERT. To determine whether there were low levels of activity present, we used a telomere repeat amplification protocol (TRAP) assay that amplifies the products of the telomerase reaction (26). We generated three different mutations at position 902 and examined their effect on telomerase activity in vitro and found that all three mutations K902A, K902N, and K902Q had essentially no telomerase activity (<0.8%) by this very sensitive assay (Fig. 4). The dominant inheritance pattern of the phenotypes suggests that the K902N substitution results in either haploinsufficiency or a dominant negative mutation. To examine these possibilities, we mixed the K902N mutant TERT with wild-type TERT in different ratios and assayed activity in vitro. We found that activity decreased linearly with decreasing wild-type enzyme, indicating that the K902N allele does not inhibit the wild-type enzyme function (Fig. 5). Additionally, we quantitated the pattern of repeat addition in this direct assay and found that the K902N mutant did not affect the processivity of the wild-type enzyme. Thus, the K902N mutation likely causes haploinsufficiency for functional telomerase in this family.
Fig. 4.
Telomerase activity of TERT mutants. In vitro reconstituted telomerase with TERT mutants K902A, K902N, and K902Q were assayed for telomerase activity by using TRAP. Diluted (1/5×, 1/25×, or 1/125×) telomerases were subjected to TRAP assay. A negative control with RNase A-inactivated telomerase was included for the wild-type and mutant enzymes. The labeled product seen in lanes 6-11 represents a very low level of telomerase activity detected by this sensitive PCR-based assay. The activity in these lanes is <1/125 of that of the wild-type enzyme or <0.8%.
Fig. 5.
Effect of mutant TERT K902N on wild-type telomerase activity. Telomerase reconstituted with different ratios (2:1, 1:1, and 1:2) of wild-type and K902N mutant TERT was analyzed by direct telomerase activity assay to determine whether the mutant TERT would inhibit TERT function. The relative activity of each reaction was measured by the intensity of the +4 band. Levels of TERT proteins in each reaction were measured by Western blotting probed with anti-HA antibody. Repeat addition processivity was calculated by comparing the normalized intensity of the major product in the first and second repeats.
Discussion
This family study reports the association of a null mutation in hTERT with the phenotype of AD DC in a three-generation pedigree that displays anticipation. The phenotype is complex and manifests as organ failure in the bone marrow, as well as the lungs, liver, and bones. This mutation results in haploinsufficiency of telomerase that leads to telomere shortening across generations. The severity of phenotypes correlates with the number of short telomeres inherited. The earlier onset of phenotypes in later generations implicates telomere length in limiting proliferative capacity in both the bone marrow as well as other solid tissues. In particular, the pattern of anticipation in idiopathic pulmonary fibrosis suggests that this disorder, like aplastic anemia, might represent stem cell failure within the lung. The complex phenotype seen in this family, in contrast to isolated hematologic abnormalities described recently in patients with AA, may be related to a shorter telomere starting set point in our family, or observing the effects of telomere shortening in later generations. The absence of telomerase activity seen with the K902N mutant allele may also contribute to a higher rate of telomere loss compared with hTERT mutations that are only hypomorphic.
Our findings underscore the fact that telomere shortening can occur in the presence of telomerase and that this shortening can lead to disease phenotypes. Haploinsufficiency for telomerase that results in progressive telomere shortening with anticipation has been described in families with mutations in the telomerase RNA component, and this study shows that hTERT dosage is also critical for maintaining telomere function. The dosage of both hTR and hTERT has previously been shown to be critical for telomere maintenance in mouse models (27, 28), and recently stem cell failure phenotypes were also seen in wild-derived mice heterozygous for telomerase RNA (L.-Y. Hao, M.A., M. A. Strong, B. Karim, D. M. Feldser, D. Huso, and C.W.G., unpublished data). Insufficient levels of telomerase leads to telomere shortening, and the shortest telomeres limit tissue renewal capacity most prominently in tissues with high turnover. This effect is particularly relevant given the heterogeneity of telomere length in the population and the possibility that certain diseases or exposures can lead to high turnover states. It may also shed light on epidemiologic phenomena of telomere shortening being linked to mortality due to age-associated diseases (29).
Finally, our results emphasize the importance of recognizing family clusters of hematologic disorders with organ fibroses, premature hair graying, and other features of DC. Patients with AA who have a family history of hematologic disorders have traditionally been thought of as having constitutional forms of AA. Unlike patients who present with idiopathic AA, constitutional AA patients do not respond to immunosuppression. Our pedigree study suggests that, similarly, patients who present with AA and have a family history of premature graying or organ fibroses may also not respond to immunosuppressive therapy and, like other DC patients, may be exquisitely sensitive to bone marrow transplant preparative regimens. The same may be true for patients with idiopathic pulmonary fibrosis where similar family history patterns may predict a lack of response to immunosuppression. Identifying these pedigrees, however, can be obscured by anticipation due to subtle phenotypes in earlier generations. Additionally, first presentations in individuals within a single family can be heterogeneous and may depend on environmental exposures in less severely affected generations. The absence of skin findings in a large proportion of AD DC families suggests that considering these disorders more broadly as inherited syndromes of telomere shortening may facilitate their diagnosis.
Acknowledgments
We thank members of the family and their clinicians. We also thank Dr. David Valle and Dr. Harry C. Dietz for helpful discussions. This work was supported by an American Association for Cancer Research-Amgen, Inc. Fellowship in Cancer Research (to M.A.) and an Institute for Cellular Engineering-Molecular and Cellular Engineering Pilot Program grant (to C.W.G.).
Author contributions: M.A., J.-L.C., and C.W.G. designed research; M.A., J.-L.C., Y.-P.C.C., A. Hawkins, and C.A.G. performed research; M.A., J.-L.C., Y.-P.C.C., J.R.E., A.C., A. Hamosh, and C.W.G. analyzed data; R.A.B. and A.R.C. identified the proband; and M.A. and C.W.G. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: AA, aplastic anemia; AD, autosomal dominant; DC, dyskeratosis congenita; hTERT, human telomerase reverse transcriptase; LOD, logarithm of odds; HA, hemagglutinin; TRAP, telomere repeat amplification protocol.
References
- 1.Yabe, M., Yabe, H., Hattori, K., Morimoto, T., Hinohara, T., Takakura, I., Shimizu, T., Shimamura, K., Tang, X. & Kato, S. (1997) Bone Marrow Transplant. 19, 389-392. [DOI] [PubMed] [Google Scholar]
- 2.Rocha, V., Devergie, A., Socie, G., Ribaud, P., Esperou, H., Parquet, N. & Gluckman, E. (1998) Br. J. Haematol. 103, 243-248. [DOI] [PubMed] [Google Scholar]
- 3.Knight, S. W., Heiss, N. S., Vulliamy, T. J., Greschner, S., Stavrides, G., Pai, G. S., Lestringant, G., Varma, N., Mason, P. J., Dokal, I. & Poustka, A. (1999) Am. J. Hum. Genet. 65, 50-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vulliamy, T., Marrone, A., Goldman, F., Dearlove, A., Bessler, M., Mason, P. J. & Dokal, I. (2001) Nature 413, 432-435. [DOI] [PubMed] [Google Scholar]
- 5.Vulliamy, T., Marrone, A., Szydlo, R., Walne, A., Mason, P. J. & Dokal, I. (2004) Nat. Genet. 36, 447-449. [DOI] [PubMed] [Google Scholar]
- 6.Fogarty, P. F., Yamaguchi, H., Wiestner, A., Baerlocher, G. M., Sloand, E., Zeng, W. S., Read, E. J., Lansdorp, P. M. & Young, N. S. (2003) Lancet 362, 1628-1630. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi, H., Baerlocher, G. M., Lansdorp, P. M., Chanock, S. J., Nunez, O., Sloand, E. & Young, N. S. (2003) Blood 102, 916-918. [DOI] [PubMed] [Google Scholar]
- 8.Yamaguchi, H., Calado, R. T., Ly, H., Kajigaya, S., Baerlocher, G. M., Chanock, S. J., Lansdorp, P. M. & Young, N. S. (2005) N. Engl. J. Med. 352, 1413-1424. [DOI] [PubMed] [Google Scholar]
- 9.Greider, C. W. & Blackburn, E. H. (1985) Cell 43, 405-413. [DOI] [PubMed] [Google Scholar]
- 10.Greider, C. W. & Blackburn, E. H. (1989) Nature 337, 331-337. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura, T. M., Morin, G. B., Chapman, K. B., Weinrich, S. L., Andrews, W. H., Lingner, J., Harley, C. B. & Cech, T. R. (1997) Science 277, 955-959. [DOI] [PubMed] [Google Scholar]
- 12.Harley, C. B., Futcher, A. B. & Greider, C. W. (1990) Nature 345, 458-460. [DOI] [PubMed] [Google Scholar]
- 13.Allsopp, R. C., Vaziri, H., Patterson, C., Goldstein, S., Younglai, E. V., Futcher, A. B., Greider, C. W. & Harley, C. B. (1992) Proc. Natl. Acad. Sci. USA 89, 10114-10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.d'Adda di Fagagna, F., Reaper, P. M., Clay-Farrace, L., Fiegler, H., Carr, P., Von Zglinicki, T., Saretzki, G., Carter, N. P. & Jackson, S. P. (2003) Nature 426, 194-198. [DOI] [PubMed] [Google Scholar]
- 15.Hao, L. Y., Strong, M. A. & Greider, C. W. (2004) J. Biol. Chem. 279, 45148-45154. [DOI] [PubMed] [Google Scholar]
- 16.Hemann, M. T., Rudolph, K. L., Strong, M. A., DePinho, R. A., Chin, L. & Greider, C. W. (2001) Mol. Biol. Cell 12, 2023-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemann, M. T., Strong, M. A., Hao, L. Y. & Greider, C. W. (2001) Cell 107, 67-77. [DOI] [PubMed] [Google Scholar]
- 18.Kruglyak, L., Daly, M. J., Reeve-Daly, M. P. & Lander, E. S. (1996) Am. J. Hum. Genet. 58, 1347-1363. [PMC free article] [PubMed] [Google Scholar]
- 19.Lansdorp, P. M., Verwoerd, N. P., van de Rijke, F. M., Dragowska, V., Little, M. T., Dirks, R. W., Raap, A. K. & Tanke, H. J. (1996) Hum. Mol. Genet. 5, 685-691. [DOI] [PubMed] [Google Scholar]
- 20.Ge, L. & Rudolph, P. (1997) BioTechniques 22, 28-30. [DOI] [PubMed] [Google Scholar]
- 21.Chen, J. L., Opperman, K. K. & Greider, C. W. (2002) Nucleic Acids Res. 30, 592-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen, J. L. & Greider, C. W. (2003) Genes Dev. 17, 2747-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen, J. L. & Greider, C. W. (2003) EMBO J. 22, 304-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blasco, M. A., Lee, H. W., Hande, M. P., Samper, E., Lansdorp, P. M., DePinho, R. A. & Greider, C. W. (1997) Cell 91, 25-34. [DOI] [PubMed] [Google Scholar]
- 25.Canard, B., Chowdhury, K., Sarfati, R., Doublie, S. & Richardson, C. C. (1999) J. Biol. Chem. 274, 35768-35776. [DOI] [PubMed] [Google Scholar]
- 26.Kim, N. W., Piatyszek, M. A., Prowse, K. R., Harley, C. B., West, M. D., Ho, P. L., Coviello, G. M., Wright, W. E., Weinrich, S. L. & Shay, J. W. (1994) Science 266, 2011-2015. [DOI] [PubMed] [Google Scholar]
- 27.Erdmann, N., Liu, Y. & Harrington, L. (2004) Proc. Natl. Acad. Sci. USA 101, 6080-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hathcock, K. S., Hemann, M. T., Opperman, K. K., Strong, M. A., Greider, C. W. & Hodes, R. J. (2002) Proc. Natl. Acad. Sci. USA 99, 3591-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cawthon, R. M., Smith, K. R., O'Brien, E., Sivatchenko, A. & Kerber, R. A. (2003) Lancet 361, 393-395. [DOI] [PubMed] [Google Scholar]