Abstract
Fimbriae of Porphyromonas gingivalis, a periodontopathogen, play an important role in its adhesion to and invasion of host cells. The fimA genes encoding fimbrillin (FimA), a subunit protein of fimbriae, have been classified into five types, types I to V, based on nucleotide sequences. We previously reported that P. gingivalis with type II fimA was strongly associated with adult periodontitis. In the present study, we compared the abilities of recombinant FimA (rFimA) types I to V to adhere to and invade human gingival fibroblasts (HGF) and a human epithelial cell line (HEp-2 cells) by using rFimA-conjugated microspheres (rFimA-MS). There were no significant differences in the abilities of the rFimA-MS to adhere to HGF; however, the adhesion of type II rFimA-MS to HEp-2 cells was significantly greater than those of other types of rFimA-MS. We also observed that type II rFimA-MS invaded epithelial cells and accumulated around the nuclei. These adhesion and invasion characteristics were eliminated by the addition of antibodies to type II rFimA and α5β1-integrin. In contrast, Arg-Gly-Asp-Ser peptide and a synthetic peptide of proline-rich protein C had negligible inhibitory effects. Furthermore, P. gingivalis strain HW24D1 with type II fimA adhered to cells and invaded them more than strains with other fimA genotypes. These results suggest that type II FimA can bind to epithelial cells most efficiently through specific host receptors.
The initial event in most infectious diseases involves adhesion of pathogens to host tissues and subsequent invasion by the pathogens. Epithelial cells, which form a layer on the mucosal surface, are spontaneously exposed to bacterial attack and prevent the invasion of deeper tissues by bacteria. It has been postulated that in the oral cavity the innate host defense system limits the spread of oral bacteria by maintaining an intact epithelial barrier (13). Porphyromonas gingivalis, a gram-negative black-pigmented anaerobe, has been found to adhere to various oral surfaces in periodontitis patients (9, 39) and has also been detected within gingival tissues in vivo (31). It has also been shown that this organism invades oral epithelial cells in vitro (22, 32, 33). The abilities of P. gingivalis to adhere and invade have been strongly implicated in the periodontal pathogenicity of this organism.
P. gingivalis expresses a number of potential virulence factors which may contribute to the pathogenesis of periodontitis, and fimbriae of P. gingivalis are recognized as a major virulence factor influencing disease initiation and progression (14). Fimbriae are filamentous components on the cell surface, and their subunit protein, fimbrillin (FimA), reportedly mediates bacterial interaction with host tissues, which mediates bacterial adhesion and colonization at targeted sites. Several studies have shown that bacteria with fimbriae are capable of binding specifically to human salivary components as commensal bacteria (1, 3, 24, 29), as well as to a variety of host cells, including macrophages (37), epithelial cells (22), and fibroblasts (19). P. gingivalis fimbriae have also been shown to be critically important in bacterial invasion of human epithelial cells in studies performed with an fimA-deficient mutant (30, 40). These findings suggest that FimA plays a central role in host-bacterium interactions.
We previously demonstrated that P. gingivalis fimA genes can be classified into five types (types I to V) on the basis of their nucleotide sequences (4, 28). Recently, a sensitive PCR assay using fimA type-specific primer sets was developed to differentiate the five types of fimA genes found in the organisms in saliva and dental plaque samples collected from periodontitis patients (4, 28). The clonal distribution of specific fimA types was studied by examining P. gingivalis harbored by periodontitis patients and periodontally healthy adults with this PCR assay (5). A majority of the periodontitis patients were found to harbor type II fimA organisms, and the next most prevalent type was type IV. In contrast, in the healthy adults the most prevalent P. gingivalis fimA type was type I. These findings indicated that there may be disease-associated and non-disease-associated P. gingivalis strains. Thus, it is possible that clonal variations in fimbriae are related to bacterial infectious traits that influence periodontal disease development. Various investigators have obtained evidence that has been used to characterize P. gingivalis fimbriae based on their structural, genomic, functional, and immunological features (14). However, most studies of P. gingivalis fimbriae have been performed with organisms having type I FimA. Although some data regarding the purification and antigenic heterogeneity of FimA variants are available (23, 25, 27, 28), only a limited number of studies have been carried out to differentiate the functional traits of clonal FimA variants.
In this study, we generated five recombinant FimAs (rFimAs) corresponding to the clonal variants and characterized the abilities of the purified rFimA proteins to adhere to and invade host cells. To do this, we developed a new quantitative analysis method using rFimA-conjugated fluorescent microspheres (MS) and a confocal microscope and evaluated the abilities of FimA variants to bind to human epithelial cells and fibroblasts.
MATERIALS AND METHODS
Bacterial strains.
P. gingivalis strains ATCC 3277 (fimA type I), HW24D1 (fimA type II), 6/26 (fimA type III), HG564 (fimA type IV), and HNA-99 (fimA type V) (28) were selected from our culture collections. These organisms were grown in GAM broth (Nissui, Tokyo, Japan) supplemented with 5 μg of hemin per ml and 1 μg of menadione per ml anaerobically (80% N2, 10% H2, 10% CO2) at 37°C. Escherichia coli BL21 (Amersham Pharmacia Biotech, Uppsala, Sweden) was cultured in Luria-Bertani (LB) medium or on an LB agar plate supplemented with 100 μg of ampicillin per ml for expression of recombinant proteins.
Cell cultures.
Human pharyngeal carcinoma epithelial HEp-2 cells (CCL-23) were obtained from the American Type Culture Collection. Human gingival fibroblasts (HGF) were isolated from healthy human gingiva as described by Takada et al. (36) and were maintained in the logarithmic growth phase in plastic culture dishes for fewer than eight passages. These cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen Life Technologies, Carlsbad, Calif.) supplemented with 10% fetal calf serum, 20 μg of gentamicin per ml, and 4 mM l-glutamine at 37°C in the presence of 5% CO2.
Expression of P. gingivalis FimA in E. coli and purification of rFimA.
Five types of rFimAs were expressed in E. coli BL21 as fusion proteins with glutathione S-transferase (GST) by using a pGEX-6P1 vector (Amersham Pharmacia Biotech). Genomic DNA from P. gingivalis strains were isolated and purified with a DNA isolation kit (Gentra Systems, Minneapolis, Minn.) for PCR amplification and cloning of fimA gene fragments. Representative strains with the five fimA types were selected as mentioned above. The primers used were as follows: for type I fimA, sense primer 5′-TAGGATCCGCTTTTGGAGTTGGCGATGACGAA and antisense primer 5′-TAGAATTCTTACCAAGTAGCATTCTGACCAAC; for type II fimA, sense primer 5′-TAGGATCCGCTTTTGGAGAAGACGAATCAAAG and antisense primer 5′-TAGAATTCTTACCAAGTAGCATTCTGACCAAC; for type III fimA, sense primer 5′-TAGGATCCGCTTTTGGAAATGCGGGAGACGAA and antisense primer 5′-GCGAATTCTTACCAAATAACATTTTGTACAAC; for type IV fimA, sense primer 5′-TAGGATCCGCTGTAGGCGATGGCCTTGCAGAT and antisense primer 5′-TAGCCCGGGTTACCAAGTAGCAGCCTGATTAAC; and for type V fimA, sense primer 5′-TAGGATCCGAAACAGAACCCAACAGTCTCCTT and antisense primer 5′-GCGAATTCTTAATTCCAAATGGCACTTTGGTT. These primers were designed for amplification of the fimA genes encoding whole mature proteins without a leader sequence by using the GenBank database (accession numbers: type I fimA, D17795; type II fimA, D17797; type III fimA, D17801; type IV fimA, D17802; and type V fimA, AB027294), and each primer contained an EcoRI, BamHI, or SmaI site (underlined) for in-frame cloning into the expression vector. PCR amplification was performed in a 50-μl (total volume) reaction mixture containing each primer at a concentration of 0.2 μM, 100 ng of template DNA, and 2.5 U of ExTaq (Takara Shuzo, Otsu, Japan) according to the manufacturer’s instructions. The PCR products were separated by agarose gel electrophoresis, and the amplified fimA genes (about 1.2 kb) were extracted with QIAEX II (Qiagen, Dusserdorf, Germany). The gene fragments were digested with BamHI and EcoRI (types I, II, III, and V) or with BamHI and SmaI (type IV) and then cloned in frame downstream from the GST-incorporated PreScission protease (Amersham Pharmacia Biotech) cleavage site of the pGEX6P-1 expression vector (Amersham Pharmacia Biotech). The nucleotide sequences of the cloned genes were determined by performing a dye terminator reaction with a model 310 genetic analyzer (PE Applied Biosystems, Foster City, Calif.).
For expression of the recombinant proteins, GST-rFimA expression vectors were transfected into E. coli BL21. The recombinant strains were grown in LB broth containing 100 μg of ampicillin per ml at 30°C with shaking until the mid-log phase, and expression of GST-rFimAs was induced by adding 1 mM isopropyl-β-d-galactopyranoside (IPTG). After 3 h of incubation, the cells were harvested by centrifugation, resuspended in ice-cold phosphate-buffered saline (PBS), and then disrupted by sonication. Since a majority of GST-rFimA was found to be solubilized in the supernatant following removal of cellular debris by centrifugation, proteins in the supernatant were subjected to affinity chromatography (Glutathione Sepharose 4B; Amersham Pharmacia Biotech) as recommended by the manufacturer. The purified recombinant fusion proteins were treated with PreScission protease (Amersham Pharmacia Biotech), and the cleavage products were passed twice over an affinity chromatography column. Cleaved target proteins were recovered in the flowthrough fraction and then stored in 50 mM Tris HCl (pH 8.5)-1 mM EDTA at −20°C. Protein concentrations were determined with a bicinchoninic acid protein assay kit (Pierce, Rockfort, Ill.). The purity of the rFimAs was determined by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Coomassie brilliant blue staining.
Covalent coupling of rFimA or BSA to fluorescent MS.
Approximately 2.1 × 1010 carboxylate-modified fluorescent MS (diameter, 0.5 μm; Molecular Probes, Eugene, Oreg.) were incubated with 0.5 mg (0.2 mg/ml) of purified rFimA or with 0. 5 mg of bovine serum albumin (BSA) (Sigma, St. Louis, Mo.) in 50 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 6.0) at room temperature for 15 min to create protein-MS complexes. 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide was added to a final concentration of 0.4 mg/ml, and the pH was adjusted to 6.5 with 0.1 M NaOH. The reaction mixture was then incubated on a rocking shaker for 2 h at room temperature. To quench the reaction, glycine was added to a final concentration of 0.1 M, and then the mixture was incubated for 30 min at room temperature. The protein-MS complexes were then washed extensively with PBS (pH 7.4) five times and stored at 4°C in PBS containing 1% BSA. The numbers of protein-MS complexes were counted with a FACScan (Becton Dickinson, Franklin Lakes, N.J.) for 5 min (flow rate, 300 μl/min), and the concentration was adjusted. To determine coupling efficiency, the amounts of bound protein were measured with a bicinchoninic acid protein assay kit (Pierce) before blocking with PBS containing BSA.
Fluorescence analysis of adhesion and invasion of MS covalently coupled to rFimA or BSA.
Approximately 2 × 104 HEp-2 cells or fibroblasts were seeded onto eight-well LabTek II chamber slides (Nalge-Nunc International, Rochester, N.Y.) and grown for 2 days. The cells were then incubated with rFimA-MS or BSA-MS (1 × 107 to 1 × 108 MS per chamber) at 37°C in the presence of 5% CO2. After appropriate incubation periods, the cells were washed extensively with PBS five times to remove the nonadherent MS and then fixed with 4% paraformaldehyde in PBS for 20 min at room temperature. After the cells were washed with PBS, they were incubated with Oregon Green 488-conjugated phalloidin (Molecular Probes) to stain for filamentous F-actin.
To analyze and quantify the adhesion and invasion of rFimA-MS or BSA-MS, a laser scanning confocal microscope (model LSM510; Carl Zeiss, Thornwood, N.Y.) was used. The excitation wavelength and detection filter settings were 505 and 550 nm (band pass), respectively, for Oregon Green 488 and 560 and 615 nm (band pass) for FluoSpheres (red fluorescence). The pinhole size was set to an airy unit of 1.0 to 1.2 depending on optimal trade-off between the z resolution and the signal/noise ratio of the images in each experiment. Fluorescent images were collected at a magnification of ×400, while the laser power and irradiation time were minimized to avoid photobleaching and possible photodynamic effects. The images were analyzed with LSM510 software (Carl Zeiss). To quantify MS that adhered to or invaded cultured cells, the sizes of the red images were calculated with the histogram tool of the LSM510 software. The software was used to calculate the area of a red image in a ×400-magnification field (0.053 mm2) by setting the threshold low value to 100 in order to avoid contamination by the excitation wavelength of Oregon Green 488. The total numbers of fibroblasts and HEp-2 cells in a 0.053-mm2 field were 35 ± 3 and 110 ± 5, respectively. The efficiency of adhesion or invasion of MS was expressed as the average area in the fields of images collected, and the data were expressed as means ± standard deviations based on three independent experiments. To analyze the distribution of the MS that invaded cells, optical sections were obtained along the z axis at 0.15-μm intervals (60 sections; thickness, 9 μm), and the images of x-z and y-z planes were reconstructed with the orthogonal section tool of the LSM510 software.
Adhesion and invasion assay.
Adhesion of and invasion by bacteria were quantified basically by using a standard antibiotic protection assay as described previously (22). Briefly, five strains of P. gingivalis with different fimA genotypes were incubated with 0.1 mCi of [methyl-3H]thymidine for 24 h, and then the bacterial cells were harvested and washed in prereduced sterile PBS. The number of bacteria in each suspension was estimated by determining the optical density at 600 nm and extrapolating from a standard curve, as described previously (21). 3H-labeled P. gingivalis (1 × 107 cells) was added to monolayers of HEp-2 cells (1 × 105 cells/well) in a 24-well culture plate and incubated in the presence of 5% CO2 at 37°C for 90 min. External, nonadherent bacteria were removed by washing the preparation three times with PBS, and then the cells were disrupted by adding 100 μl of distilled water and incubating the preparation for 20 min. The numbers of adherent and invading organisms were counted with a scintillation counter, and adhesion and invasion values were calculated from the amounts of 3H recovered from infected cells as percentages of the total amount of 3H obtained from P. gingivalis. To measure the amount of invading bacteria, P. gingivalis-infected HEp-2 cells were incubated for an additional 1 h with Dulbecco’s modified Eagle’s medium containing 300 μg of gentamicin per ml and 200 μg of metroimidazole per ml. The cells were washed three times with PBS, and the amounts of internalized bacteria were measured as described above.
Inhibition studies.
For inhibition studies, cell monolayers were preincubated for 30 min with rabbit anti-rFimA (type II) antibody (26), RGD peptide (Peptide Institute, Osaka, Japan), GRGD peptide (Peptide Institute), and proline-rich protein C (PRP-C) peptide (2) along with mouse monoclonal anti-α5β1-integrin (Chemicon, Temecula, Calif.) or mouse monoclonal anti-αVβ3-integrin antibodies (Chemicon) 30 min before incubation. Normal rabbit serum (Life Technologies) was added at a dilution of 1:50 as a negative control. Purified mouse immunoglobulin G (Southern Biotechnology Associates, Inc., Birmingham, Ala.) was added at a final concentration of 5 μg/ml. At this concentration, the inhibitors had no effect on cell viability as determined by Alamer blue staining (BioSource International, Camarillo, Calif.).
Statistical analyses.
All data are expressed below as means ± standard deviations. Statistical analyses were performed with an unpaired Student’s t test. Multiple comparisons were performed by one-way analysis of variance and Sheffe’s test using STAT View software (SAS Institute Inc., Cary, N.C.).
RESULTS
Purification of type I to V rFimA proteins and conjugation with fluorescent MS.
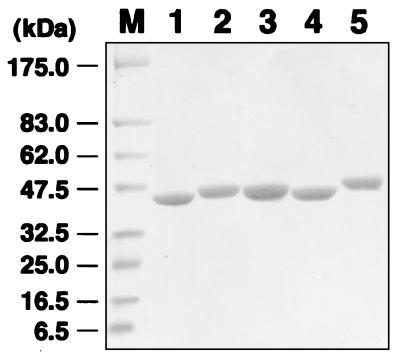
Five variants of rFimA were purified from recombinant E. coli BL21 extracts and were detected as a single band at an apparent molecular mass of 41 to 45 kDa by SDS-PAGE, as expected (Fig. 1). To determine the abilities of the rFimAs to adhere to and invade host cells, these purified proteins and BSA (as a control) were covalently coupled to 2.1 × 1010 MS. Similar amounts of the rFimA proteins and BSA were found to be coupled to the MS, as follows: 337.0 μg of type I rFimA, 319.5 μg of type II rFimA, 368.4 μg of type III rFimA, 343.8 μg of type IV rFimA, 354.0 μg of type V rFimA, and 379.5 μg of BSA.
FIG. 1.
SDS-PAGE of purified rFimAs. The purity of each rFimA was determined by Coomassie brilliant blue staining. An aliquot (20 μg) of each protein was loaded onto a lane. Lane M, prestained molecular weight markers (New England Biolabs); lane 1, type I rFimA; lane 2, type II rFimA; lane 3, type III rFimA; lane 4, type IV rFimA; lane 5, type V rFimA.
Adhesion of rFimA-MS to HGF and invasion.
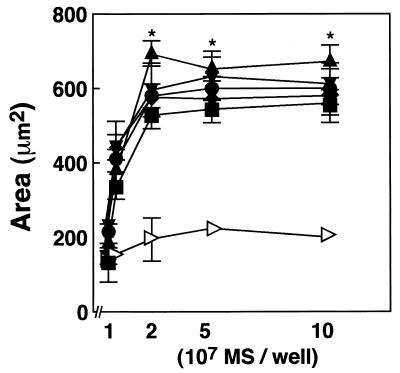
We first evaluated the abilities of rFimA-MS to adhere to and invade HGF. To determine a suitable ratio of rFimA-MS to HGF, a quantitative assay was performed. Confluent HGF (2 × 104 cells) were incubated with different numbers (107 to 108 MS/well) of rFimA-MS or BSA-MS for 1 h. The amounts of MS bound to HGF were determined by image analysis as described in Materials and Methods (Fig. 2). The levels of adhesion and invasion of each rFimA-MS preparation reached plateaus (area of MS bound to HGF, 500 to 700 μm2) when 2 × 107 MS were added to a well containing 2 × 104 cells. BSA-MS exhibited negligible abilities to adhere to and invade HGF (area, ca. 200 μm2). Therefore, we decided to use a cell/MS ratio of 1:1,000 in subsequent experiments.
FIG. 2.
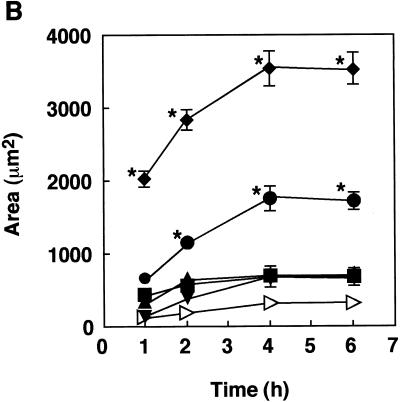
Determination of cell/MS ratio for quantitative analysis. HGF were cultured on LabTekII chamber slides (eight wells; 2 × 104 cells per well) for 2 days at 37°C and then incubated with various concentrations of rFimA-MS (type I [▪], type II [⧫], type III [•], type IV [▴], or type V [▾]) or BSA-MS (◃) for 1 h. The amounts of rFimA-MS and BSA-MS that adhered are expressed as the total areas of the red images in the field of a confocal microscope (magnification, ×400), and at least five fields were analyzed for each experiment. The values are means and standard errors of the means based on three individual experiments. *, P < 0.01.
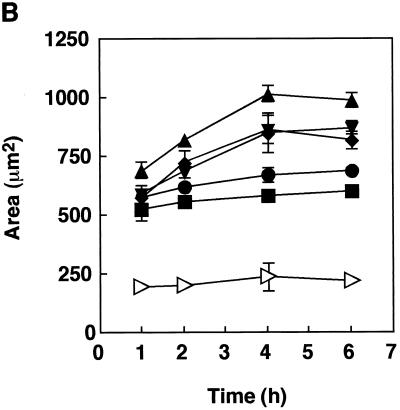
We then evaluated the adhesion of rFimA-MS to fibroblasts and the invasion of fibroblasts by rFimA-MS for various incubation periods (Fig. 3). All of the rFimA-MS exhibited significant abilities to adhere to and invade HGF compared to the abilities of BSA-MS (Fig. 3A). The levels of adhesion and invasion for type II, IV, and V rFimA-MS reached plateaus after 4 h of incubation, whereas additional incubation did not markedly enhance the binding of type I and III rFimA-MS (Fig. 3B). Although significant differences among the abilities of the five rFimA variants to adhere to and invade HGF were not observed, more type II, IV, and V rFimA-MS than other types of rFimA-MS bound to HGF.
FIG. 3.
Quantitative analysis of adhesion of rFimA-MS to HGF. (A) Confocal microscopic analysis of adhesion of rFimA-MS. HGF were cultured on a LabTekII chamber slide (eight wells; 2 × 104 cells per well) for 2 days at 37°C. rFimA-MS were added to each well and incubated for 4 h. After incubation, the cells were extensively washed and fixed. Images (red, rFimA-MS) were collected and analyzed by using a confocal microscope. Magnification, ×400. Bar, 50 μm. (B) Time course of adhesion of rFimA-MS (type I [▪], type II [⧫], type III [•], type IV [▴], or type V [▾]) or BSA-MS (◃ ) to fibroblasts. The amounts of rFimA-MS and BSA-MS that adhered are expressed as the total areas of the red images in the field of a confocal microscope (magnification, ×400), and at least five fields were analyzed in each experiment. The values are means and standard errors of the means based on three individual experiments.
Increased adhesion of type II rFimA to epithelial cells and increased invasion of epithelial cells by type II rFimA.
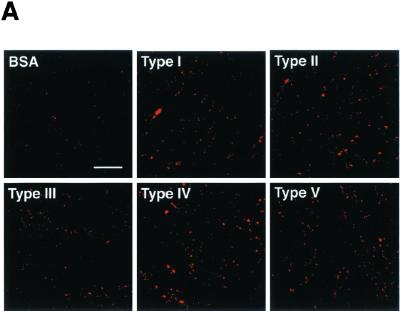
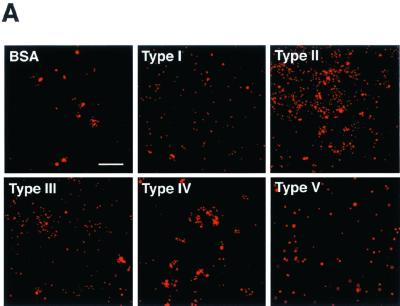
Since oral epithelial cells are thought to mediate the initial anchoring of P. gingivalis, we examined the abilities of various rFimAs to adhere to HEp-2 cells and to invade these cells over the course of 6 h. Different types of rFimA-MS (2 × 1010 MS) were incubated with 1 × 104 HEp-2 cells. All types clearly bound to the cells (Fig. 4A), and the adhesion and invasion activities reached maximum levels after 4 h of incubation (Fig. 4B). Apparently, of the various types of rFimA-MS tested, type II rFimA-MS adhered most prominently to HEp-2 cells. Furthermore, type III rFimA-MS had greater ability to adhere than type I, IV, or V rFimA-MS; however, the ability of type III rFimA-MS to adhere was less than the ability of type II rFimA-MS. There were no significant differences in the abilities of type I, IV, and V rFimA-MS and BSA-MS to adhere to and invade HEp-2 cells. These findings clearly indicate that only P. gingivalis type II rFimA had such abilities.
FIG. 4.
Quantitative analysis of adhesion of rFimA-MS to epithelial cells. (A) Confocal microscopic analysis of adhesion of rFimA-MS. HEp-2 cells were cultured on LabTekII chamber slides (eight wells; 2 × 104 cells per well) for 2 days at 37°C. rFimA-MS were added to each well and incubated for 4 h. After incubation, the cells were extensively washed and fixed. Images (red, rFimA-MS) were collected and analyzed with a confocal microscope. Magnification, ×400. Bar, 50 μm. (B) Time course of adhesion of rFimA-MS (type I [▪], type II [⧫], type III [•], type IV [▴], or type V [▾]) or BSA-MS (◃) to HGF. The amounts of rFimA-MS and BSA-MS that adhered are expressed as the total areas of the red images in the field of a confocal microscope (magnification, ×400), and at least five fields were analyzed for each experiment. The values are means and standard errors of the means based on three individual experiments. An asterisk indicates that a P value is <0.01.
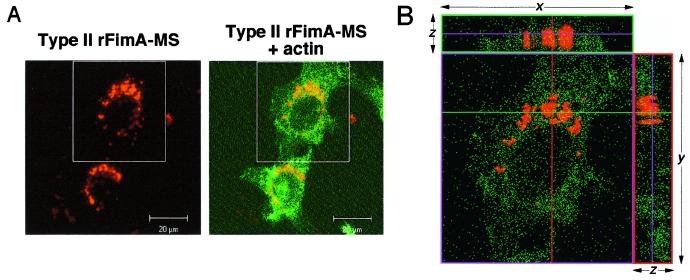
It is suspected that P. gingivalis fimbriae mediate invasion of epithelial cells by the organism. To determine whether rFimA alone is sufficient for invasion of mammalian cells, we examined whether rFimA-MS could be internalized in epithelial cells. A total of 1 × 104 HEp-2 cells were incubated with 2 × 1010 type II rFimA-MS for 1 to 6 h. As shown in Fig. 5A, type II rFimA-MS were found to be internalized in HEp-2 cells after 6 h of incubation and accumulated uniformly around the nuclei. Optical sectioning of a typical cell (Fig. 5B) in the x-z and y-z planes confirmed that the type II rFimA-MS were inside the HEp-2 cell and localized in the perinuclear area. Similar results were obtained with other types of rFimA-MS; however, the numbers of invading MS were significantly lower than the numbers of type II rFimA-MS, and no invading MS alone or BSA-MS were observed in HEp-2 cells (data not shown).
FIG. 5.
(A) Confocal microscopic analysis of type II rFimA-MS invasion. HEp-2 cells were incubated with type II rFimA-MS for 6 h. After the cells were washed, they were fixed and stained with Oregon Green 488-conjugated phalloidin. Images (red, rFimA-MS; green, actin) were collected (thickness, 0.15 μm) and analyzed with a confocal microscope (magnification, ×1,000). The box indicates an area that was sectioned along the vertical axis (see panel B). (B) Intracellular localization of type II rFimA-MS in HEp-2 cells. A cell was optically sectioned along the vertical (z) axis, yielding horizontal sections (x-y planes) at 0.15-μm intervals, and the x-z and y-z planes were constructed with LSM510 software.
Adhesion of P. gingivalis with different fimA genotypes to epithelial cells and invasion of epithelial cells by these P. gingivalis variants.
Although type II rFimA-MS bound most prominently to HEp-2 cells, it was not clear whether the different fimA types could affect the ability of P. gingivalis cells to adhere to and invade epithelial cells. Therefore, we examined the abilities of five P. gingivalis strains with different fimA genotypes to adhere to and invade HEp-2 cells. As shown in Table 1, P. gingivalis strain HW24D1 (type II fimA) invaded significantly more than the other strains tested; almost 30% of the bacteria invaded HEp-2 cells. Compared with organisms with the other types of fimA, the type III organism (strain 6/26) exhibited weak efficiency, and strain ATCC 33277 (type I fimA) exhibited marked abilities to adhere to and invade HEp-2 cells. These data indicate that the P. gingivalis strain with type II fimA had the greatest abilities to adhere to and invade epithelial cells, as demonstrated in the rFimA experiments.
TABLE 1.
Adhesion to and invasion of HEp-2 cells by P. gingivalis strains with five different fimA genotypes
| Strain | fimA type | % Adhesion + invasiona | % Invasiona |
|---|---|---|---|
| ATCC 33277 | I | 35.1 ± 0.5b | 13.8 ± 1.4b |
| HW24D1 | II | 80.1 ± 4.9bc | 27.9 ± 5.2bc |
| 6/26 | III | 3.3 ± 0.3c | 3.0 ± 0.4c |
| HG564 | IV | 12.0 ± 1.3c | 9.8 ± 2.3c |
| HNA-99 | V | 7.1 ± 0.1c | 4.1 ± 0.3c |
The values are means ± standard deviations (n = 4).
Significantly different (P < 0.01) as determined by Sheffe’s test.
Significantly different (P < 0.001) as determined by Sheffe’s test.
Effects of anti-type II rFimA antibodies or salivary component on type II rFimA-MS invasion.
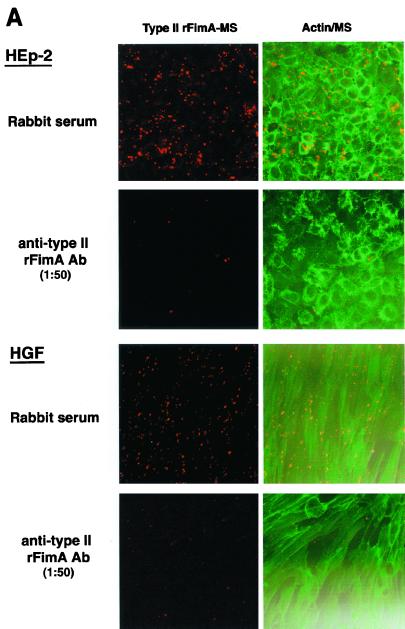
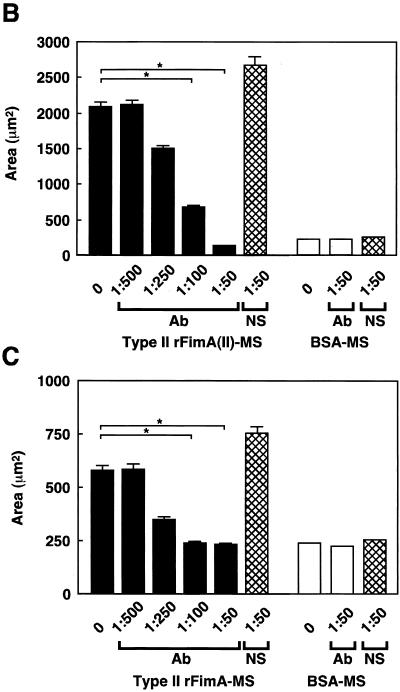
Anti-type II rFimA antibodies were added to a reaction mixture to confirm the specificity of type II rFimA-MS adhesion to and invasion of HEp-2 cells. Adhesion of type II rFimA-MS to HEp-2 cells and invasion were almost completely inhibited by the antibodies (Fig. 6A), while adhesion to and invasion of HGF were significantly inhibited by the antibodies (Fig. 6A). Normal rabbit serum did not have any effect on the adhesion of and invasion by type II rFimA-MS. Furthermore, the inhibitory effect of the antibodies was shown to be dose dependent in both cell lines, indicating the specific nature of the binding of the type II rFimA-MS to the host cells (Fig. 6B and C). In addition, the synthetic peptide of the C-terminal region of salivary PRP-C did not affect the binding of type II rFimA-MS to HEp-2 cells or HGF (data not shown), whereas this peptide effectively inhibited the binding of an organism with type II FimA to saliva (1). These results indicate that type II rFimA binds to cells through mechanisms different from the mechanisms of binding to salivary molecules.
FIG. 6.
Inhibition of adhesion or invasion of type II rFimA-MS by addition of anti-type II rFimA specific antibody. (A) Confocal microscopic analysis of adhesion of rFimA-MS to HEp-2 cells and fibroblasts. Cells were preincubated with anti-type II rFimA specific antibody for 30 min and then incubated with type II rFimA-MS for 1 h. Magnification, ×400. Bar, 50 μm. (B) Effects of anti-type II antibody on adhesion to HEp-2 cells by type II rFimA-MS. (C) Effects of anti-type II antibody on adhesion to HGF cells by type II rFimA-MS. Different concentrations of antibody were added 30 min before the addition of rFimA-MS. The amounts of MS that adhered to or invaded cells are expressed as the areas in the field of a confocal microscope (magnification, ×400), and at least five fields were analyzed for each experiment. The values are means and standard errors of the means based on three individual experiments. An asterisk indicates that a P value is <0.01. Ab, antibody; NS, nonimmunized serum.
Involvement of α5β1-integrins in fimbrial adhesion.
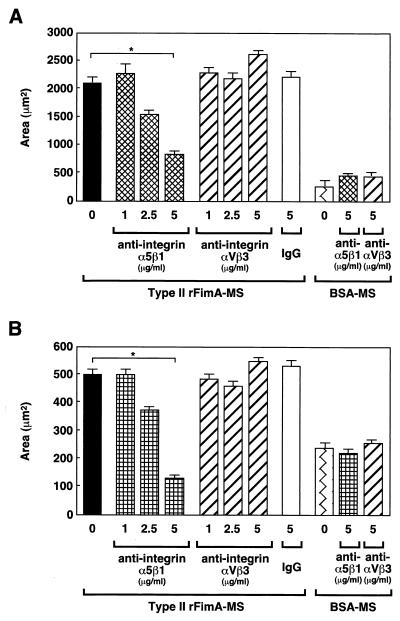
P. gingivalis fimbriae are also known to specifically interact with integrin expressed on macrophages (37). Thus, we examined whether fimbrial adhesion is mediated by integrin moieties. The effects of anti-integrin antibodies and synthetic RGD peptide on the adhesion of rFimA-MS to host cells were evaluated. Addition of anti-α5β1-integrin antibodies inhibited adhesion of type II rFimA-MS to both HEp-2 cells and HGF in a dose-dependent manner, and a dose of 5 μg/ml resulted in significantly decreased adhesion (Fig. 7). Addition of anti-αVβ3-integrin or purified mouse immunoglobulin G had little effect on fimbrial adhesion.
FIG. 7.
Inhibition of adhesion or invasion of type II rFimA-MS by anti-integrin antibody. (A) Effects of anti-integrin antibodies on adhesion of type II rFimA-MS to HEp-2 cells. (B) Effects of anti-integrin antibodies on adhesion of type II rFimA-MS to HGF. Different concentrations of antibodies were added 30 min before the addition of rFimA-MS. The amounts of MS that adhered to or invaded cells are expressed as the areas in the field of a confocal microscope (magnification, ×400), and at least five fields were analyzed in each experiment. The values are means and standard errors of the means based on three individual experiments. An asterisk indicates that a P value is <0.01. IgG, immunoglobulin G.
Furthermore, synthetic RGD peptides or GRGD peptides did not affect the adhesion of rFimA-MS to HEp-2 and HGF and invasion (data not shown). These results indicate that type II rFimA binding may be mediated by the surface α5β1-integrin, although the mechanism may not depend on the RGD motif.
DISCUSSION
Heterogeneities among various P. gingivalis strains based on their immunological and genetic properties and virulence potentials have been investigated previously (16, 26). Results of a study performed with an infection model using rodents suggested that P. gingivalis can be divided into organisms with invasive traits and organisms with noninvasive traits depending on the capsular structure of the organism (7, 39). However, little information regarding the predominant types of P. gingivalis involved in chronic periodontitis is available. Our recent studies have clearly shown that P. gingivalis with a specific fimA genotype is strongly associated with the destructive progression of periodontitis (4, 5, 28). The findings suggest that FimA clonal variation may be related to the periodontopathogenicity of P. gingivalis. Thus, in the present study we investigated the functional differences in adhesion to and invasion of host cells.
The quantitative method employed in this study was useful for characterizing the abilities of rFimAs to mediate bacterial adhesion to and invasion of host tissues. In the assay performed with HGF, it was difficult to distinguish adhesion of rFimA-MS from invasion (Fig. 3). However, the abilities of different rFimA-MS to be internalized in epithelial cells were successfully evaluated (Fig. 5B). All types of rFimA-MS markedly interacted with host cells, and compared to the other types, type II rFimA-MS exhibited noticeably greater abilities to adhere and invade. We also noted that the invading MS accumulated uniformly around the nuclei (Fig. 5), especially when the MS were coupled with type II rFimA. A similar accumulation of P. gingivalis whole cells in the perinuclear region was reported in a study of the invasion of epithelial cells (6). These results suggest that fimbriae are an essential component of P. gingivalis invasion of epithelial cells.
The involvement of fimbriae in the adhesion of P. gingivalis to fibroblasts and epithelial cells and in invasion of these cells has been examined with a number of cell lines, including oral epithelial and endothelial cells (8, 10, 11, 40). All of the studies demonstrated that fimbriae play a critical role in the interaction of P. gingivalis with host cells, as did a study in which a fimA-deficient mutant was used (30). However, in all these investigations the researchers used P. gingivalis strains 381 and ATCC 33277, which are genetically identical and possess the type I fimA gene. In other studies, several strains of P. gingivalis with different fimA types were compared to examine their abilities to invade endothelial and epithelial cells; however, the results revealed no clear relationship between invasiveness and specific fimA types (12). In this study, P. gingivalis with type II fimA (strain HW24D1) adhered to and invaded HEp-2 cells significantly more than other P. gingivalis strains with different fimA genotypes. The ability of strain HW24D1 to adhere to and invade cells was correlated with the ability of type II rFimA-MS to adhere to and invade cells. Thus, the data strongly suggest that type II FimA has the greatest capacity to adhere to and invade host cells, and type II fimA-harboring organisms play a central role in the development of periodontitis. Although type III rFimA-MS effectively bound to HEp-2 cells at a level similar to that of type II rFimA-MS, P. gingivalis strain 6/26 (type III fimA) was the strain that was least able to adhere to and invade cells. This contradiction between the ability of intact cells to adhere to and invade cells and the ability of rFimA-MS to adhere to and invade cells may be due to additional factors, such as different levels of encapsulation and protease expression. In fact, binding of P. gingivalis fimbriae to HGF was enhanced by an Arg-specific protease due to proteolytic exposure of hidden receptors (20). Thus, further studies are needed to analyze the effect of other factors likely to differentiate the pathogenicities of P. gingivalis strains.
The adhesion and invasion of type II rFimA-MS were inhibited by anti-α5β1-integrin antibodies but not by RGD peptides. Binding of fibronectin to various bacterial cell surface components has been demonstrated for a number of pathogens, including Staphylococcus aureus, Streptococcus pyogenes, Candida albicans, and Treponema pallidum (11, 15, 18, 38). In particular, the fibronectin binding protein on the cell surface of S. pyogenes can interact with fibronectin through its repeated domain, and the fibronectin binding protein-fibronectin complex recognizes the RGD motif of cell surface molecules, resulting in adhesion followed by invasion of host cells (17). Our findings suggest that α5β1-integrin is a key molecule for facilitating P. gingivalis invasion. However, it is not clear whether P. gingivalis type II fimA interacts directly with α5β1-integrin. In addition, adhesion of type II rFimA-MS to fibroblasts was completely inhibited by anti-α5β1-integrin antibodies. Scragg et al. (34) reported that P. gingivalis cells and β1-integrin were colocalized on the cell surface of fibroblasts, which may support our conclusion. However, the binding of rFimA-MS to epithelial cells was not completely inhibited (a reduction of approximately 70% was observed) by the addition of anti-α5β1-integrin antibodies, suggesting that there was an unknown receptor of type II FimA in the epithelial cells. The involvement of fibronectin in the interaction with the host cells should be studied further.
Anti-αvβ3-integrin (vitronectin receptor) antibodies did not inhibit the binding of type II rFimA-MS to the cell surface (Fig. 7). It has also been demonstrated that there is a lack of colocalization of αvβ3-integrin and P. gingivalis components in P. gingivalis-infected fibroblasts (34). P. gingivalis fimbriae may not have a specific ability to bind to αvβ3-integrin on the cell surface. However, our previous study indicated that P. gingivalis fimbriae could bind to vitronectin with significant specific affinity. Furthermore, αvβ3-integrin-overexpressing CHO cells interacted with P. gingivalis cells at significantly greater levels than mock CHO cells interacted T. Nakamura, A. Amano, I. Nakagawa, N. Okahashi, and S. Hamada, unpublished data). Thus, it is possible that the anti-αvβ3-integrin antibodies in the present study did not saturate the binding sites for type II rFimA on the αvβ3-integrin molecules.
Salivary components, such as proline-rich protein, proline-rich glycoprotein, and statherin, are known to specifically bind to P. gingivalis fimbriae and are thought to promote colonization of P. gingivalis in the oral cavity (1, 29). The C-terminal peptide region of proline-rich protein or proline-rich glycoprotein is recognized as a receptor for fimbriae, and the PRP-C peptide effectively inhibits fimbrial binding to whole saliva (3). However, addition of PRP-C peptide did not affect the profile of binding of type II rFimA-MS to the host cells, indicating that unknown mechanisms are involved in fimbrial binding to cell surface molecules.
The functional region of type I FimA for binding to epithelial cells has been described by Sojar et al. (35). Antibodies against peptides 49 to 68 (VVMANTAGAMELVGKTLAEVK) and 69 to 90 (ALTTELTAENQEAAGLIMTAEP) of type I FimA polypeptide markedly inhibited the adhesion of P. gingivalis cells to KB cells. In our study, type II rFimA-MS bound to and invaded host cells at a level that was fivefold greater than that of type I rFimA-MS (Fig. 5). However, peptides 49 to 68 of type I and II FimAs were identical except for three residues (VVMANTGRMKLAGKTLAEVK, residues in boldface type), and the sequence of peptides 69 to 90 is completely conserved in these two types of FimA. These findings suggest that other binding sites involved in the interaction with the host cell may be present in type II FimA. In fact, the N-terminal region (corresponding to amino acid residues 1 to 90) and the C-terminal region (corresponding to amino acid residues 251 to 341) are highly conserved in type I and type II FimAs; however, other FimA regions exhibit variable levels of similarity. Characterization of the active sites for binding of type II FimA to epithelial cells with an FimA truncated mutant is now under way in our laboratories.
In summary, we suggest that the abilities of P. gingivalis to adhere to and invade epithelial cells depend on FimA type specificity. Type II rFimA-MS exhibited a significantly greater ability to adhere to and invade epithelial cells than the other types of rFimA-MS. The distinction was especially evident in the interaction with epithelial cells and less evident in the interaction with fibroblasts. There were not significant differences in the abilities of type I, IV, and V rFimA-MS to adhere to and invade both epithelial cells and fibroblasts. Together, our findings indicate that type II FimA may be an important virulence trait for colonization and destruction of periodontal tissues by P. gingivalis and a critical determinant for associating the organisms possessing type II fimA with periodontitis. Since P. gingivalis fimbriae are thought to be a major adhesin involved in adherence to and colonization of oral surfaces, we will investigate whether different fimA types influence these events at the beginning of the infection process.
Acknowledgments
We thank S. Morishima for his generous gift of the type II rFimA-specific antibody.
This work was supported by grants-in-aid for scientific research B-13557181 and C-12671994 from the Ministry of Education, Science, Sports, and Culture of Japan.
Editor: V. J. DiRita
REFERENCES
- 1.Amano, A., H. T. Sojar, J. Y. Lee, A. Sharma., M. J. Levine, and R. J. Genco. 1994. Salivary receptors for recombinant fimbrillin of Porphyromonas gingivalis. Infect. Immun. 62: 3372–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amano, A., T. Fujiwara, H. Nagata, M. Kuboniwa, A. Sharma, H. T. Sojar, R. J. Genco, S. Hamada, and S. Shizukuishi. 1997. Porphyromonas gingivalis fimbriae mediate coaggregation with Streptococcus oralis through specific domains. J. Dent. Res. 76: 852–857. [DOI] [PubMed] [Google Scholar]
- 3.Amano, A., S. Shizukuishi, H. Horie, S. Kimura, I. Morisaki, and S. Hamada. 1998. Binding of Porphyromonas gingivalis fimbriae to proline-rich glycoproteins in parotid saliva via a domain shared by major salivary components. Infect. Immun. 66: 2072–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amano, A., I. Nakagawa, K. Kataoka, I. Morisaki, and S. Hamada. 1999. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J. Clin. Microbiol. 37: 1426–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amano, A., M. Kuboniwa, I. Nakagawa, S. Akiyama, I. Morisaki, and S. Hamada. 2000. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J. Dent. Res. 79: 1664–1668. [DOI] [PubMed] [Google Scholar]
- 6.Belton, C. M., K. T. Izutsu, P. C. Goodwin, Y. Park, and R. J. Lamont. 1999. Fluorescence image analysis of the association between Porphyromonas gingivalis and gingival epithelial cells. Cell. Microbiol. 1: 215–224. [DOI] [PubMed] [Google Scholar]
- 7.Chen, P. B., M. E. Neider, S. J. Millar, H. S. Reynold, and J. J. Zambon. 1987. Effect of immunization on experimental Bacteroides gingivalis infection in a murine model. Infect. Immun. 55: 2534–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu, B. 1999. Multiple infections in carotid atherosclerotic plaques. Am. Heart J. 138: S534–S536. [DOI] [PubMed] [Google Scholar]
- 9.Christersson, L. A., C. L. Fransson, R. G. Dunford, and J. J. Zambon. 1992. Subgingival distribution of periodontal pathogenic microorganisms in adult periodontitis. J. Periodontol. 63: 418–425. [DOI] [PubMed] [Google Scholar]
- 10.Despande, R. G., M. B. Khan, and C. A. Genco. 1998. Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis. Infect. Immun. 66: 5337–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorn, B. R., W. A. Dunn, Jr., and A. Progulke-Fox. 1999. Invasion of human coronary artery cells by periodontal pathogens. Infect. Immun. 67: 5792–5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorn, B. R., J. N. Burks, K. N. Seifert, and A. Progulske-Fox. 2000. Invasion of endothelial and epithelial cells by strains of Porphyromonas gingivalis. FEMS Microbiol. Lett. 187: 139–144. [DOI] [PubMed] [Google Scholar]
- 13.Gibbons, R. J. 1989. Bacterial adhesion to oral tissues: a model for infectious diseases. J. Dent. Res. 68: 750–760. [DOI] [PubMed] [Google Scholar]
- 14.Hamada, S., A. Amano, S. Kimura, I. Nakagawa, S. Kawabata, and I. Morisaki. 1998. The importance of fimbriae in the virulence and ecology of some oral bacteria. Oral Microbiol. Immunol. 13: 129–138. [DOI] [PubMed] [Google Scholar]
- 15.Hasty, D. L., and H. S. Courtney,. 1996. Group A streptococcal adhesion. All of the theories are correct. Adv. Exp. Med. Biol. 408: 81–94. [PubMed] [Google Scholar]
- 16.Holt, S. C., L. Kesavalu, S. Walker, and C. A. Genco. 1999. Virulence factors of Porphyromonas gingivalis. Periodontol. 2000 20: 168–238. [DOI] [PubMed] [Google Scholar]
- 17.Joh, D., E. R. Wann, B. Kreikemeyer, P. Speziale, and M. Hook. 1999. Role of fibronectin-binding MSCRAMMs in bacterial adhesion and entry into mammalian cells. Matrix Biol. 18: 211–223. [DOI] [PubMed] [Google Scholar]
- 18.Klotz, S. A. 1994. Plasma and extracellular matrix proteins mediate in the fate of Candida albicans in the human host. Med. Hypotheses 42: 328–334. [DOI] [PubMed] [Google Scholar]
- 19.Kontani, M., S. Kimura, I. Nakagawa, and S. Hamada. 1997. Adhesion of Porphyromonas gingivalis to matrix proteins via a fimbrial cryptic receptor exposed by its own arginine-specific protease. Mol. Microbiol. 24: 1179–1187. [DOI] [PubMed] [Google Scholar]
- 20.Kontani, M., H. Ono, H. Shibata, Y. Okamura, T. Tanaka, T. Fujiwara, S. Kimura, and S. Hamada. 1996. Cysteine protease of Porphyromonas gingivalis 381 enhances binding of fimbriae to cultured human fibroblasts and matrix proteins. Infect. Immun. 64: 756–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuboniwa, M., A. Amano, S. Shizukuishi, I. Nakagawa, and S. Hamada. 2001. Specific antibodies to Porphyromonas gingivalis Lys-gingipain by DNA vaccination inhibit bacterial binding to hemoglobin and protect mice from infection. Infect. Immun. 69: 2972–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamont, R. J., A. Chan, C. M. Belton, K. T. Izutsu, D. Vasel, and A. Weinberg. 1995. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 63: 3878–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, J. Y., H. T. Sojar, G. S. Bedi, and R. J. Genco. 1991. Porphyromonas (Bacteroides) gingivalis fimbrillin: size, amino-terminal sequence, and antigenic heterogeneity. Infect. Immun. 59: 383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, J. Y., H. T. Sojar, G. S. Bedi, and R. J. Genco. 1992. Synthetic peptides analogous to the fimbrillin sequence inhibit adhesion of Porphyromonas gingivalis. Infect. Immun. 60: 1662–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, J. Y., H. T. Sojar, A. Amano, and R. J. Genco. 1995. Purification of major fimbrial proteins of Porphyromonas gingivalis. Protein Expr. Purif. 6: 496–500. [DOI] [PubMed] [Google Scholar]
- 26.Mayrand, D., and S. C. Holt,. 1988. Biology of asaccharolytic black-pigmented Bacteroides species. Microbiol. Rev. 52: 134–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naito, Y., H. Tohda, K. Okuda, and I. Takazoe. 1993. Adhesion and hydrophobicity of invasive and noninvasive strains of Porphyromonas gingivalis. Oral Microbiol. Immunol. 8: 195–202. [DOI] [PubMed] [Google Scholar]
- 28.Nakagawa, I., A. Amano, R. K. Kimura, T. Nakamura, S. Kawabata, and S. Hamada. 2000. Distribution and molecular characterization of Porphyromonas gingivalis carrying a new type of fimA gene. J. Clin. Microbiol. 38: 1909–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura, T., A. Amano, I. Nakagawa, and S. Hamada. 1999. Specific interactions between Porphyromonas gingivalis fimbriae and human extracellular matrix proteins. FEMS Microbiol. Lett. 175: 267–272. [DOI] [PubMed] [Google Scholar]
- 30.Njoroge, T., R. J. Genco, H. T. Sojar, N. Hamada, and C. A. Genco. 1997. A role for fimbriae in Porphyromonas gingivalis invasion of oral epithelial cells. Infect. Immun. 65: 1980–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papapanou, P. N., J. Sandros, K. Lindberg, M. J. Duncun, R. Niederman, and U. Nannmark. 1994. Porphyromonas gingivalis may multiply and advance within stratified human junctional epithelium in vitro. J. Periodontal Res. 29: 374–375. [DOI] [PubMed] [Google Scholar]
- 32.Sandros, J., P. Papapanou, and G. Dahlen. 1993. Porphyromonas gingivalis invades oral epithelial cells in vitro. J. Periodontal Res. 28: 219–226. [DOI] [PubMed] [Google Scholar]
- 33.Sandros, J., P. Papapanou, U. Nannmark, and G. Dahlen. 1994. Porphyromonas gingivalis invades human pocket epithelium in vitro. J. Periodontal Res. 29: 62–69. [DOI] [PubMed] [Google Scholar]
- 34.Scragg, M. A., S. J. Cannon, M. Rangarajan, D. M. Williams, and M. A. Curtis. 1999. Targeted disruption of fibronectin-integrin interactions in human gingival fibroblasts by the RI protease of Porphyromonas gingivalis W50. Infect. Immun. 67: 1837–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sojar, H. T., Y. Han, N. Hamada, A. Sharma, and R. J. Genco. 1999. Role of the amino-terminal region of Porphyromonas gingivalis fimbriae in adhesion to epithelial cells. Infect. Immun. 67: 6173–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takada, H., J. Mihara, I. Morisaki, and S. Hamada. 1991. Induction of interleukin-1 and -6 in human gingivalis fibroblast cultures stimulated with Bacteroides lipopolysaccharide. Infect. Immun. 59: 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeshita, A., Y. Murakami, Y. Yamashita, M. Ishida, S. Fujisawa, S. Kitano, and S. Hanazawa. 1998. Porphyromonas gingivalis fimbriae use β2 integrin (CD11/CD18) on mouse peritoneal macrophages as a cellular receptor, and the CD18 β chain plays a functional role in fimbrial signaling. Infect. Immun. 66: 4056–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Umemoto, T., Y. Nakatani, Y. Nakamura, and I. Namikawa. 1993. Fibronectin-binding proteins of a human oral spirochete, Treponema denticola. Microbiol. Immunol. 37: 75–78. [DOI] [PubMed] [Google Scholar]
- 39.Van Steenbergen, T. J., F. G. Delemarre, F. Namavar, and J. De Graaff. 1987. Differences in virulence within the species Bacteroides gingivalis. Antonie Leeuwenhoek 53: 233–244. [DOI] [PubMed] [Google Scholar]
- 40.Weinberg, A., C. M. Belton, Y. Park, and R. J. Lamont. 1997. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 65: 313–316. [DOI] [PMC free article] [PubMed] [Google Scholar]