Abstract
In toxigenic Vibrio cholerae, cholera toxin is encoded by the CTX prophage, which consists of a core region carrying ctxAB genes and genes required for CTXΦ morphogenesis, and an RS2 region encoding regulation, replication, and integration functions. Integrated CTXΦ is often flanked by another genetic element known as RS1 which carries all open reading frames (ORFs) found in RS2 and an additional ORF designated rstC. We identified a single-stranded circularized form of the RS1 element, in addition to the CTXΦ genome, in nucleic acids extracted from phage preparations of 32 out of 83 (38.5%) RS1-positive toxigenic V. cholerae strains analyzed. Subsequently, the corresponding double-stranded replicative form (RF) of the RS1 element was isolated from a representative strain and marked with a kanamycin resistance (Kmr) marker in an intergenic site to construct pRS1-Km. Restriction and PCR analysis of pRS1-Km and sequencing of a 300-bp region confirmed that this RF DNA was the excised RS1 element which formed a novel junction between ig1 and rstC. Introduction of pRS1-Km into a V. cholerae O1 classical biotype strain, O395, led to the production of extracellular Kmr transducing particles, which carried a single-stranded form of pRS1-Km, thus resembling the genome of a filamentous phage (RS1-KmΦ). Analysis of V. cholerae strains for susceptibility to RS1-KmΦ showed that classical biotype strains were more susceptible to the phage compared to El Tor and O139 strains. Nontoxigenic (CTX−) O1 and O139 strains which carried genes encoding the CTXΦ receptor toxin-coregulated pilus (TCP) were also more susceptible (>1,000-fold) to the phage compared to toxigenic El Tor or O139 strains. Like CTXΦ, the RS1Φ genome also integrated into the host chromosomes by using the attRS sequence. However, only transductants of RS1-KmΦ which also harbored the CTXΦ genome produced a detectable level of extracellular RS1-KmΦ. This suggested that the core genes of CTXΦ are also required for the morphogenesis of RS1Φ. The results of this study showed for the first time that RS1 element, which encodes a site-specific recombination system in V. cholerae, can propagate horizontally as a filamentous phage, exploiting the morphogenesis genes of CTXΦ.
Toxigenic Vibrio cholerae strains are lysogens of a filamentous bacteriophage designated CTXΦ (20), which carries the ctxAB genes encoding cholera toxin (CT). CTXΦ is unusual among filamentous phages because the phage genome encodes the functions necessary for a site-specific integration system and thus can integrate into the V. cholerae chromosome at a specific attachment site known as attRS, forming stable lysogens (7, 9, 20). A typical CTXΦ genome has two regions, the core and the RS2. The 4.6-kb core region encodes CT as well as the functions that are required for the virion morphogenesis, whereas the 2.5-kb RS2 region encodes the regulation, replication, and integration functions of the CTXΦ genome (21). In toxigenic V. cholerae, particularly in El Tor and O139 strains, the CTXΦ genome is integrated in the chromosome flanked by a 2.7-kb repetitive sequence originally referred to as the RS1 element, which is closely related to the RS2 region of CTXΦ (4, 5, 18, 21). DNA sequence analysis has shown that the RS2 region consists of three open reading frames (ORFs), rstR, rstA, and rstB, and two intergenic regions, ig1 and ig2 (21). The RS1 element is very similar to the RS2 region of CTXΦ in genetic and functional aspects but has an additional ORF termed rstC, which is absent in RS2 (4, 21). The origin of the RS1 element and its mechanism of transfer between V. cholerae strains are not clearly known.
The species V. cholerae contains a wide variety of strains and biotypes, receiving and transferring genes for virulence-associated factors as well as genes for other biochemical functions, including antibiotic resistance, capsular polysaccharides, and new surface antigens (7). Understanding the lateral or horizontal transfer of genes by phage, pathogenicity islands, and other accessory genetic elements can provide insights into how this bacterial pathogen emerges and evolves to become new strains. In previous studies we have shown the horizontal transfer of genes encoding CT by CTXΦ and the origination of new toxigenic strains from nontoxigenic progenitors (8, 9, 11, 20). In the present study we examined the transfer of RS1, a genetic element which encodes a site-specific recombination system (18). This study has also provided an understanding of the origin and evolution of unusual filamentous phages, such as CTXΦ, which have developed the ability to lysogenize their host.
MATERIALS AND METHODS
Bacterial strains.
The toxigenic V. cholerae strains analyzed in this study to investigate the production of RS1Φ included 69 clinical and 6 environmental strains belonging to O1 or O139 serogroups and 8 environmental strains belonging to non-O1 non-O139 serogroups. Clinical strains were isolated from patients who attended the treatment center of the International Centre for Diarrheal Disease Research, Bangladesh (ICDDR,B), located in Dhaka. The environmental strains were isolated from surface waters in Dhaka. Strains were stored either in lyophilized form or in sealed deep nutrient agar at room temperature. Before use, the identities of the V. cholerae cultures were confirmed by biochemical reaction and serology (23), and the presence of the RS1 and CTX elements was ascertained by using specific DNA probes as described below. Relevant characteristics of V. cholerae strains used as controls or as a recipient in phage transduction assays are listed in Table 1.
TABLE 1.
TABLE1.Characteristics of control bacterial strains and plasmids used in the study
| Strain or plasmid | Relevant characteristic | Source or reference |
|---|---|---|
| Plasmids | ||
| pCTX-Km | RF of genetically marked CTXΦ genome derived from strain SM44 | 20 |
| pRS1-Km | Extrachromosomal RF of RS1 element carrying an intergenic kanamycin resistance (Kmr) marker | This study |
| pIG-Km | Derivative of the RF DNA of a wild-type CTXΦ carrying an intergenic Kmr marker | 16 |
| Strains | ||
| P-27459 | Toxigenic V. cholerae O1 El Tor strain isolated from a patient in Bangladesh | 18 |
| Bang-2 | Derivative of P27459 in which the core and RS sequences (including attRS) were deleted | 18 |
| Bah-1 | Derivative of El Tor strain E7946 in which the core region of the CTX element were deleted | 18 |
| Bah-2 | Derivative of E7946 in which the core and RS sequences (including attRS) were deleted | 18 |
| AL-11089 | Clinical toxigenic V. cholerae O139 which produces both CTXΦ and RS1Φ in the culture supernatant | This study |
| MO10 | Toxigenic V. cholerae O139 strain isolated from a patient in India | 22 |
| Bengal-2 | Derivative of MO10 in which both the core and RS sequences (including attRS) were deleted | 22 |
| RV-508 | Derivative of classical biotype strain 569B which constitutively expresses CT, TCP, and other ToxR-regulated gene products | 20 |
| Env-002 | Environmental nontoxigenic V. cholerae O1 El Tor strain | This study |
| SA-317 | Clinical nontoxigenic V. cholerae O1 El Tor strain | 11 |
| AOE-12/39 | Clinical nontoxigenic V. cholerae O139 strain | 11 |
| Env-99 | Environmental nontoxigenic V. cholerae O139 strain | 11 |
Probes and hybridization.
The RS1Φ genome was detected by its positive hybridization with the rstR and rstC gene probes but negative reactions with probes corresponding to the core region of CTXΦ. The probes used in this study included a 0.5-kb EcoRI fragment of pCVD27 (13), encoding the ctxA gene, and a 2.1-kb SphI-XbaI fragment of pCTX-Km containing the entire zot and ace genes and part of orfU (9, 20). The El Tor and classical biotype-specific rstR probes were SacI-XbaI fragments of pHK1 and pHK2, respectively (15). The rstC probe was either a synthetic oligonucleotide or a PCR amplicon with primers derived from the sequence of rstC (GenBank accession no. U83795). PCR amplicons corresponding to different other regions of RS1 element or the CTXΦ genome (Fig. 1) were also used as probes in hybridization assays whenever appropriate. In addition, strand-specific oligonucleotide probes (Table 2) corresponding to the plus and minus strands of CTX and RS1 elements were used to detect the presence of single-stranded phage genomes. Colony blots or Southern blots were prepared by using nylon filters (Hybond; Amersham International, Plc., Ayelesbury, United Kingdom) and processed by standard methods (17, 19). The polynucleotide probes were labeled by random priming (12) using a Random Primers DNA Labeling kit (Bethesda Research Laboratories, Gaithersburg, Md.) and [α-32P]dCTP (3,000 Ci/mmol; Amersham), and oligonucleotide probes were labeled by 3′-tailing by using terminal deoxynucleotide transferase and [α-32P]dCTP (Amersham). Southern blots and colony blots were hybridized with the labeled probes and autoradiographed as described previously (8, 9, 11).
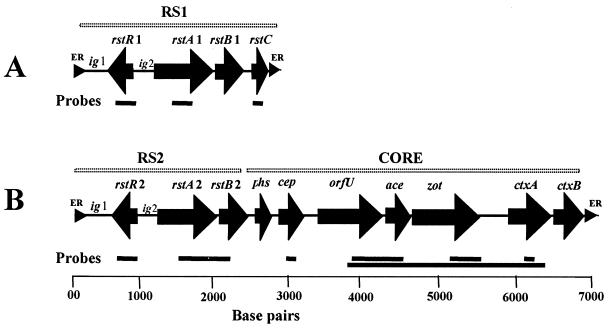
FIG. 1.
Schematic representation of the genetic organization of an RS1 element (A) and a CTX prophage comprising an RS2 region and a core region (B). Solid arrows represent ORFs, whereas small triangles represent ER sequences. Horizontal bars represent the DNA probes used in this study.
TABLE 2.
Sequence of oligonucleotide probes and primers used to analyze the RS1Φ genome
| Primer | Sequence of probe or primer | Description and/or specificity | Source or reference |
|---|---|---|---|
| 1 | 5′-TCTATCTCTGTAGCCCCTATTACG | Specific for the plus strand of ctxA gene and hence the single-stranded genome of CTXΦ | 8 |
| 2 | 5′-CTCAGACGGGATTTGTTAGGCACG | Specific for minus strand of ctxA gene | 8 |
| 3a | 5′-ATGAGTTTGAAACCATACACTTT | Forward primer for rstC and specific for the minus strand of the RS1 element | This study |
| 3b | 5′-TTACAGTGATGGATCAGTCAAT | Reverse primer for rstC and specific for the plus strand of the RS1 element | This study |
| 4a | 5′-GCCTAAATCCGCAACTCAAGG | Forward primer for rstC and specific for the minus strand of the RS1 element | This study |
| 4b | 5′-CAATGGTACTGACGACTGCCCC | Reverse primer for ig1 and specific for the minus strand of the RS1 element | This study |
PCR assays.
All oligonucleotides used either as probes or PCR primers were synthesized commercially by Oswel DNA Service (University of Edinburgh, Edinburgh, United Kingdom), and PCR reagents and kits were purchased from the Perkin-Elmer Corporation (Norwalk, Conn.). The presence of tcpA genes specific for the classical and El Tor biotypes was determined by using a multiplex PCR assay (14). PCR assays for different regions of the RS1 element were performed with primers based on the published sequence of RS1 as described in Table 2. The expected size of the PCR amplicons was ascertained by electrophoresis in agarose gels. The identities of all PCR products were further verified with specific oligonucleotide probes.
Induction of lysogens.
Toxigenic V. cholerae strains were grown in Luria broth (LB) at 30°C up to an absorbance at 540 nm (A540) of 0.2. The cells were collected by centrifugation, washed, and resuspended in fresh LB. The suspension was divided into aliquots to which mitomycin C (Sigma Chemical Company, St. Louis, Mo.) was added to a final concentration of 20 ng/ml and incubated for 6 h at 30°C. Strains grown in a similar way but without mitomycin C were used as controls. The culture supernatants were analyzed for extracellular phages carrying the CTX and RS1 elements by previously described methods (8). Briefly, supernatant fluids were sterilized by filtration through 0.22-μm (pore-size) filters (Millipore Corp., Bedford, Mass.). To confirm that the filtrates did not contain any bacterial cells, aliquots of the filtrates were streaked on Luria agar (LA) plates and incubated overnight at 37°C. The sterile filtrates were mixed with 1/4 volumes of a solution containing 20% polyethylene glycol (PEG 6000) and 10% NaCl and then centrifuged at 12,000 × g to precipitate the phage particles. The precipitate was dissolved in a solution containing 20 mM Tris-Cl (pH 7.5), 60 mM KCl, 10 mM MgCl, and 10 mM NaCl and then digested with pancreatic DNase I (100 U/ml) and RNase A (50 μg/ml) at 37°C for 2 h to remove possible nucleic acids carried over from lysed bacterial cells. The solution was extracted with phenol-chloroform to disrupt possible phage particles, and the total nucleic acids were precipitated with ethanol. To determine the presence of single-stranded phage genome, the nucleic acid preparations were analyzed by agarose gel electrophoresis and Southern blot hybridization. Aliquots of nucleic acid preparations digested with the single-strand-specific mung bean nuclease (BRL), and aliquots of the undigested preparations were tested in adjacent lanes. Mung bean nuclease-treated double-stranded pCTX-Km DNA was used as a control to test the stability of double-stranded DNA in this assay. Preparations containing CTXΦ and RS1 DNA were identified by hybridizing Southern blots derived from the agarose gels with appropriate probes.
Analysis of pRS1.
To analyze the extrachromosomal replicative form (RF) of RS1 element (pRS1), plasmid DNA was prepared by a standard method (17), purified by using microcentrifuge filter units (Ultrafree-Probind, Sigma), and analyzed by Southern blot hybridization for the presence of RS1-specific sequences and the absence of CTXΦ-specific core genes. The pRS1 DNA was purified by electroelution from an agarose gel and was labeled by inserting a kanamycin resistance (Kmr) marker into an intergenic NotI site. This was done by digesting pRS1 with NotI and ligating the linearized plasmid with a Kmr determinant derived from NotI-cleaved pIG-Km (16). The ligated DNA was used to electroporate a recipient V. cholerae strain and plated on LA plates containing kanamycin (50 μg/ml). Kmr colonies were selected, and pRS1-Km isolated from these colonies was examined by restriction analysis, PCR assays, and nucleotide sequencing of the probable junction of circularization of the RS1 element.
DNA sequencing.
Nucleotide sequencing was performed with an Applied Biosystems automated DNA sequencing system (ABI Prism 310; PE Applied Biosystems, Foster City, Calif.) by using a BigDye Terminator Cycle Sequencing Ready Reaction kit (PE Applied Biosystems). The nucleotide sequences of both the strands were determined with primers designed at several positions of RS1 (Table 2) and finally processed through the Sequencher alignment program, version 4.0 (Gene Codes Corporation, Inc.). Comparative analysis of the 300-bp region of pRS1 was performed with the Sequencher program, while the published sequence of RS1 (GenBank accession no. U83795) was used as the reference.
Transduction assays.
The susceptibility of different recipient strains to RS1-KmΦ was assayed with phage prepared from the culture supernatant fluids of strain O395 (pRS-Km) and by using the strain RV508 as a control recipient. The assays were done both in the adult rabbit ileal loops and under in vitro laboratory conditions by using modifications of previously described methods (10, 11). Briefly, 10-ml aliquots of filtered sterile supernatant fluids were used to precipitate phage particles, and the pellet was suspended in 100 μl of TES buffer (20 mM Tris-HCl, pH 7.5; 10 mM NaCl; 0.1 mM Na2EDTA). The recipient strain was grown in LB at 37°C, and the cells were precipitated by centrifugation and resuspended in fresh LB. Approximately 105 bacterial cells mixed with 10 μl of the phage preparation in a final volume of 100 μl were inoculated into the ileal loops of adult rabbits obtained from the breeding facilities of the Animal Resources Branch of the ICDDR,B and were prepared as described previously (10). For each recipient strain, at least five loops were inoculated. After 16 h, the rabbits were sacrificed and contents of the ileal loops were collected. The inside of the loops was washed out with 1 ml of 10 mM phosphate-buffered saline (pH 7.4) and collected in the same tube. Dilutions of the ileal loop fluids were analyzed by plating on LA plates containing kanamycin (50 μg/ml) and on plates without kanamycin. The ratio of Kmr transduced colonies to the total number of colonies derived from the recipient strain was calculated and is expressed as the percentage of recipient cells infected. For in vitro assays, mixtures of phage and recipient cells as described above were prepared. Each mixture was inoculated into 5 ml of LB and incubated for 16 h at 30°C, and aliquots of the culture were plated on LA plates containing kanamycin and incubated overnight at 30°C.
Analysis of infected cells.
Representative infected colonies were grown overnight in LB containing kanamycin (50 μg/ml), and cells were precipitated by centrifugation. The supernatant fluids of the cultures were titrated for the presence of RS1-KmΦ particles with strain RV508 as the recipient. Integration of the phage genome into the chromosome of the recipient cells was studied by comparative Southern blot analysis of total DNA and plasmid preparations from the phage-infected and the corresponding native strains as described previously (8, 9, 10).
Electron microcopy of phage particles.
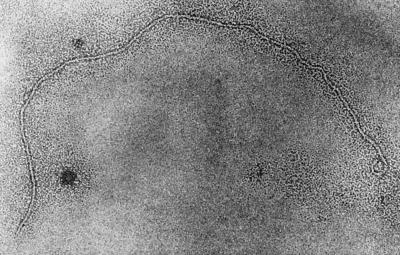
Phage prepared from the supernatant of strain O395 (pRS1-Km) and the corresponding native strain (negative control) were concentrated by precipitation, and the crude preparation was used to determine the morphology of the phage as described previously (2). The phage particles were negatively stained with 2% uranyl acetate and were examined under a Philips transmission electron microscope (model 420T).
RESULTS AND DISCUSSION
Detection of RS1Φ genome.
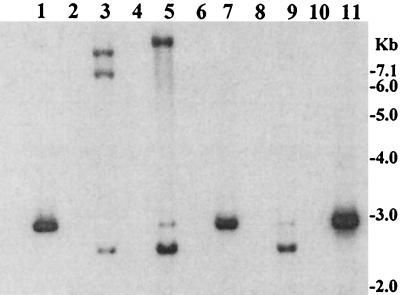
Nucleic acids extracted from phage particles derived from the culture supernatant fluids of toxigenic V. cholerae strains were found to contain two kinds of single-stranded DNA. This included the CTXΦ genome and a smaller genome (∼2.7 kb) which contained the RS1 sequence but none of the core genes of CTXΦ. Probes corresponding to the core regions of the CTXΦ genome did not hybridize with the 2.7-kb DNA, whereas a probe specific for rstC hybridized with this 2.7-kb DNA and not with the CTXΦ genome. The 2.7-kb DNA was also positive with probes for rstR, rstA, and rstB genes. Of 83 RS1-positive toxigenic strains tested, 32 (38.5%) showed the presence of both CTX- and RS1-specific DNA in the phage nucleic acid preparations (Table 3). As evidenced by the intensities of the respective phage DNA bands (Fig. 2B), most of these strains produced CTX- and RS1-specific phage DNA with similar efficiency. Only the positive strands of the respective nucleic acids were present in the phage DNA preparations. This was evidenced by the finding that DNA extracted from the particles hybridized with the positive-strand-specific oligonucleotide probes but were negative with the negative-strand-specific probes shown in Table 2.
TABLE 3.
Mitomycin C-induced production of CTXΦ and RS1Φ in culture supernatants of RS1-positive toxigenic V. cholerae strains as assayed by the presence of different phage-specific DNAs in total nucleic acids extracted from phage preparations
| V. cholerae strains | Yr of isolation | Source | No. of strains analyzed (n = 83) | No. of strains producing extracellular phage particles
|
|
|---|---|---|---|---|---|
| CTXΦ (n = 52) | RS1Φa (n = 32) | ||||
| O1, El Tor | 1969–2000 | Patient | 37 | 21 | 11 |
| O1, El Tor | 1998–2000 | Sea water | 6 | 2 | 2 |
| O139 | 1993–1999 | Patient | 32 | 27 | 19 |
| Non-O1 non-O139 | 1998–2001 | Sea water | 8 | 42 | 40 |
All strains producing RS1Φ also produced CTXΦ.
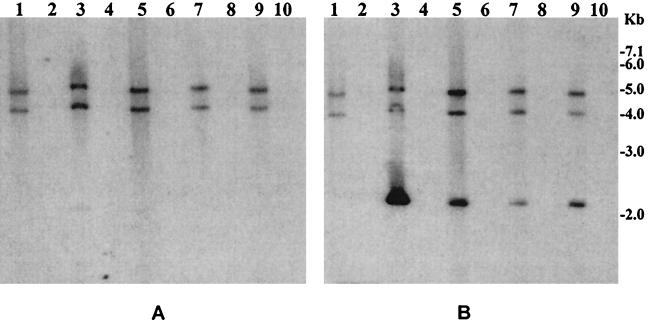
FIG. 2.
Supernatant fluids of toxigenic V. cholerae O1 or O139 strains grown in the presence of mytomycin C (20 ng/ml) were sterilized by filtration through 0.22-μm (pore-size) filter and were used to precipitate possible bacteriophage particles. The precipitates were dissolved in an appropriate buffer and treated with DNase I and RNase A to remove contaminating exogenous DNA or RNA. Total phage nucleic acids were isolated and analyzed by using the ctxA probe (A) and the rstR probe (B). Southern blot hybridization analysis of total phage nucleic acids derived from six different V. cholerae strains are shown in lanes 1, 3, 5, 7, and 9. The intensities of the phage DNA bands indicate that the titer of RS1 phage was comparable to that of CTX phage for most strains. Aliquots of phage DNA preparations digested with the single-strand-specific mung bean nuclease (BRL) were loaded in lanes 2, 4, 6, 8, and 10, which are adjacent to the lanes containing the corresponding intact phage DNA. Numbers indicating molecular sizes of bands correspond to supercoiled DNA ladder (Bethesda Research Laboratories).
The previously reported structure of RS1 showed the presence of four ORFs (rstR, rstA, rstB, and rstC) and two intergenic regions (ig1 and ig2). Whereas ig1 represents the upstream end of the element, rstC is located at the downstream end. In the present study, a PCR assay with a sense primer corresponding to the rstC region and an antisense primer corresponding to the ig1 region of RS1 amplified a short fragment of DNA spanning both of these regions, as would be expected for a circular structure for this 2.7-kb DNA. This also suggested the presence of a novel junction between igI and rstC, in the circularized form of RS1 element. This particle-derived nucleic acid, like the CTXΦ genome, was also resistant to restriction endonuclease digestion but sensitive to DNase I as well as the single-strand-specific nuclease, mung bean nuclease (Fig. 2). These findings confirmed that the 2.7-kb DNA was indeed a single-stranded circular DNA that corresponded to the positive strand of the RS1 element and resembled the genome of a possible filamentous phage. The preparation of plasmid DNA from strains positive for the single-stranded RS1 element and Southern hybridization analysis also provided evidence for the presence of an extrachromosomal RF of the RS1 element (data not shown).
Since the RS1 element of V. cholerae is known to be flanked by an end repeat (ER) sequence on either side, it was speculated previously that the RS1 element could possibly circularize off the chromosome by recombination of these ER sequences and thus form an extrachromosomal RF DNA (20). In the present study, we confirmed the presence of the excised RF of the RS1 element (pRS1), as well as the existence of a single-stranded form of the element. Moreover, the single-stranded RS1 element was present in particles, since it was protected from nuclease digestion during the phage preparation. These findings suggested the possibility of horizontal transfer of RS1 packaged in phage particles and raised questions regarding whether the RS1 element itself constituted the genome of a possible filamentous phage (RS1Φ).
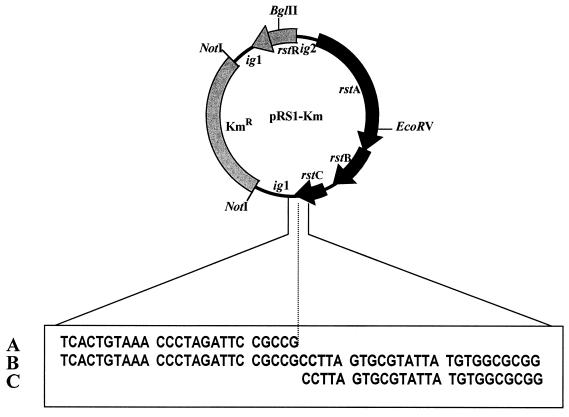
To begin to investigate this issue, we isolated pRS1 from the O139 strain AL11089, and introduced a Kmr marker in an intergenic region (ig1) to construct pRS1-Km. This allowed us to prepare large amounts of the RF DNA and to perform detailed molecular analysis. The restriction endonuclease cleavage pattern of this DNA completely agreed with that derived from the published sequence of RS1 element (GenBank accession no. U83795). PCR analysis with primers corresponding to different regions of the ORFs and intergenic sequences also agreed with the expected size of the amplicons (Fig. 3). Finally, we determined the nucleotide sequence of a 300-bp region, including the possible junction of circularization, by using primers corresponding to the ig1 and rstC regions (Table 2). The derived nucleotide sequence agreed entirely with the published sequence and showed that the circularized RS1 element formed a novel junction between ig1 and rstC in the process of its excision from the bacterial chromosome (Fig. 4).
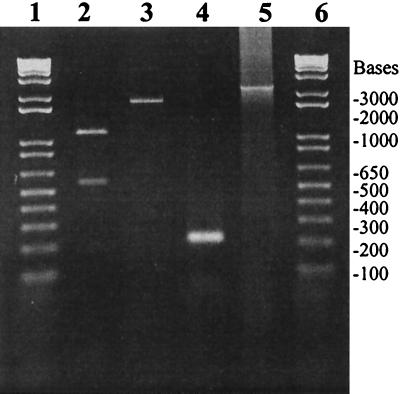
FIG. 3.
PCR analyses of the genomic organization of RS1Φ genome. The identities of the PCR amplicons are as follows: lane 5, rstR/rstB; lane 4, junction of rstC/ig1; lane 3, rstr/rstC; and lane 2, PCR product shown in lane 3 digested with BglII. Lanes 1 and 6 contain molecular size markers.
FIG. 4.
Schematic representation of pRS1-Km and the nucleotide sequence of the novel junction. The published nucleotide sequences of the 5′ end of ig1 (A) and the 3′ end of rstC (C) of the RS1 element have been aligned against the relevant sequence derived from pRS1-Km (B) by using primers corresponding to the rstC and ig1 regions (see the text for details).
The introduction of the Kmr marker into pRS1 also allowed us to conveniently monitor the generation of possible phage particles from the RF DNA. Introduction of pRS1-Km into a classical biotype strain, O395, by electroporation led to the production of Kmr transducing particles in the culture supernatant of the transformed strain. This was demonstrated by incubating a number of potential recipient strains (Table 4) with the cell-free culture supernatant (sterilized by filtration through 0.22-μm filters) derived from O395(pRS1-Km). It was found that the Kmr phenotype was transmitted to the recipient strains by the supernatant fluid of O395(pRS1-Km), and some of the Kmr transductants in turn produced new Kmr transducing particles in the culture supernatants (Table 5). We concentrated these particles by precipitation with a PEG-NaCl solution as described above and analyzed their stability, infectious activity, and nucleic acid contents. The particles were heat- and chloroform-labile but resistant to DNase (DNase I) and RNase. Analysis of nucleic acids extracted from the particles showed the presence of single positive-strand form of pRS1-Km, as evidenced by hybridization with single-strand-specific probes. These findings suggested that the transducing particle was most likely a filamentous bacteriophage that we designated RS1Φ. We further examined the morphology of these particles by electron microscopy. Electron microscopic examination of phage preparations from supernatant fluids derived from strain O395 carrying pRS1-Km showed a typical filamentous structure (Fig. 5) that was absent in similar preparations obtained from the corresponding native strain.
TABLE 4.
Susceptibility of different V. cholerae strains to a genetically marked derivative of RS1Φ
| Strain | Description | Presence of CTX- and TCP-specific genes and attachment sequencea
|
Susceptibility (%) to RS1-KmΦc
|
Integration of phage genomed | |||||
|---|---|---|---|---|---|---|---|---|---|
| rstRb | ctxA | zot | tcpA | attRS | In vitro | In rabbit | |||
| AK-31047 | O1, El Tor | + | + | + | + | + | 1.6 × 10−3 | 2.9 × 10−3 | − |
| P-27459 | O1, El Tor | + | + | + | + | + | 2.0 × 10−4 | 1.8 × 10−2 | − |
| AF-1471 | O1, El Tor | + | + | + | + | + | 2.3 × 10−3 | 7.2 × 10−3 | − |
| Bang-2 | O1, El Tor | − | − | − | + | − | 3.1 | 8.6 | − |
| Bah-2 | O1, El Tor | − | − | − | + | − | 4.1 | 7.2 | − |
| Env-002 | O1,El Tor | − | − | − | + | + | 2.7 | 18.2 | + |
| SA-317 | O1, El Tor | − | − | − | + | + | 3.9 | 10.2 | + |
| RV508 | O1, classical | + | + | + | + | + | 70.6 | 72.9 | − |
| O395 | O1, classical | + | + | + | + | + | 25.4 | 65.2 | − |
| TCP-2 | O1, classical | + | − | + | − | + | 0 | 0 | − |
| AL-11089 | O139 | + | + | + | + | + | 3.0 × 10−4 | 7.5 × 10−3 | − |
| MO10 | O139 | + | + | + | + | + | 2.6 × 10−3 | 6.2 × 10−3 | − |
| Bengal-2 | O139 | − | − | − | + | − | 1.5 | 5.5 | − |
| Env-99 | O139 | − | − | − | + | + | 7.5 | 19.2 | + |
| AOE-12/39 | O139 | − | − | − | + | + | 3.2 | 12.8 | + |
| AM19226 | Non-O1, non-O139 | − | − | − | − | + | 0 | 0 | |
The presence of different genes was detected by using DNA probes and PCR assays.
The El Tor and O139 strains carried El Tor type rstR, whereas the classical strains carried the classical type rstR gene.
Susceptibility to the phage was determined by using a genetically marked phage. See the text for details.
Integration of the phage genome into the host chromosome was determined by Southern blot analysis of genomic and plasmid DNA isolated from native and infected strains.
TABLE 5.
Production of cell-free phage particles by V. cholerae strains carrying a genetically marked derivative of RS1Φ
| Strain | Description | Titer of RS1-KmΦ in culture supernatant (particles/ml)a |
|---|---|---|
| AF-1471(pRS1-Km) | Toxigenic V. cholerae O1 El Tor strain carrying pRS1-Km | 0.5 × 102 |
| AL-11089(pRS1-Km) | Toxigenic V. cholerae O139 carrying pRS1-Km | 1.2 × 102 |
| P-27459(pRS1-Km) | Toxigenic V. cholerae O1 El Tor strain carrying pRS1-Km | 1.8 × 102 |
| Bang-2(pRS1-Km) | Nontoxigenic (ΔCTX) V. cholerae O1 El Tor strain carrying pRS1-KmΦ | 0 |
| Bah-2(pRS1-Km) | Nontoxigenic (ΔCTX) V.cholerae O1 El Tor strain carrying pRS1-KmΦ | 0 |
| MO-10(pRS1-Km) | Toxigenic V. cholerae O139 strain carrying pRS1-Km | 2.1 × 102 |
| Bengal-2(pRS1-Km) | Nontoxigenic (ΔCTX) V. cholerae O139 strain carrying pRS1-Km | 0 |
| Env-002(RS1-Km) | Environmental CTXΦ−V. cholerae O1 El Tor strain lysogenized with pRS1-KmΦ | 0 |
| SA-317 (RS1-Km) | Nontoxigenic (CTXΦ−) V. cholerae O1 El Tor strain lysogenized with RS1-KmΦ | 0 |
| RV508(pRS1-Km) | Classical V. cholerae O1 strain carrying pRS1-Km | 4.5 × 104 |
| O395(pRS1-Km) | Classical V. cholerae O1 strain carrying pRS1-Km | 3.6 × 104 |
| Env-99(pRS1-Km) | Environmental CTXΦ−V. cholerae O139 strain carrying pRS1-Km | 0 |
| AOE-12/39(RS1-Km) | Environmental CTXΦ−V. cholerae O139 strain lysogenized with RS1-KmΦ | 0 |
Values are averages of three independent assays, using strain RV508 as the recipient.
FIG. 5.
Morphology of an RS1-Km phage particle isolated from an O395 strain (pRS1-Km). Note the typical filamentous structure. Magnification, ×180,000
To determine whether these Kmr phage particles simultaneously contained both RS1 and CTXΦ DNA, we used Kmr phage particles prepared from toxigenic El Tor and O139 strains containing pRS1-Km to infect the classical recipient strain RV508. Nucleic acids extracted from Kmr phage particles derived from transduced RV508 were reexamined with appropriate probes. These nucleic acids hybridized with an RS1-specific probe (rstC) but did not hybridize with probes for the core genes (zot, ace, and orfU) of CTXΦ, thus ruling out the possibility of the presence of both RS1 and CTX genomes in the same particle.
Infection by RS1Φ and analysis of recipient strains.
We tested the susceptibility of recipient strains both under in vitro laboratory conditions and inside the ileal loops of adult rabbits. The Kmr marker present in the genome of the phage allowed us to conveniently detect and analyze infected cells. Of a total of 16 strains exposed to RS1-KmΦ, 14 strains (87.5%) that were positive for the CTXΦ receptor TCP were infected by RS1Φ, although the level of susceptibility varied (Table 4). The two TCP-negative strains included in the study were completely resistant, and no kanamycin-resistant transductant was detected in these strains when assayed under identical conditions. Previous studies with CTXΦ have shown that the efficiency of transduction was considerably higher in vivo, since the phage receptor TCP is known to express more efficiently under in vivo conditions (8, 9, 10, 20). In the present study, infection with RS1Φ also occurred more efficiently in the ileal loops of rabbits than under in vitro conditions (Table 4). The classical biotype strains which are known to express more TCP were also more susceptible to the phage compared to the El Tor and O139 strains (Table 4).
The susceptibility of toxigenic El Tor and O139 strains to RS1-KmΦ was significantly lower (between 2 and 26 Kmr colonies were recovered, with mean percent susceptibilities of between 2 × 10−4 and 2.3 × 10−3) compared to that of the nontoxigenic (CTXΦ−) V. cholerae O1 and O139 strains. The mean susceptibility of these nontoxigenic strains varied between 3.1 and 7.7% in vitro and between 8.6 and 19.2% in the in vivo assays. Previous studies have described heteroimmunity among CTX phages (15). Toxigenic classical strains of V. cholerae O1 are known to be infected by CTXΦ isolated from El Tor biotype strains, whereas toxigenic El Tor strains are resistant to further infection by the same phage. The molecular basis for this heteroimmunity and DNA sequences of the classical and El Tor CTXΦ repressors have also been studied (3, 15). Recent reports have described the existence of at least three widely diverse rstR genes carried by different CTX phages: CTXETΦ, CTXclassΦ, and CTXcalcΦ (3, 6). The RS1-KmΦ used in our study carried an El Tor type rstR gene, and the susceptibility of nontoxigenic El Tor and O139 strains was at least ∼1,000-fold higher compared to that of toxigenic El Tor and O139 strains which already carried an El Tor rstR as part of the CTX prophage. Hence, this was consistent with the heteroimmunity previously described for CTX phages (15).
After infection, the RS1Φ genome integrated into the chromosome of most of the nontoxigenic strains that carried the attRS sequence, as demonstrated by Southern blot analysis of plasmid and chromosomal DNA preparations from representative infected cells (Fig. 6). In strains that did not contain attRS, the phage genome was maintained as a plasmid when cells were grown in the presence of kanamycin. In toxigenic El Tor and O139 strains in which the attRS sequence was blocked by a resident CTX prophage, as well as in the classical biotype strains, the RS1 genome was also maintained as a plasmid. To analyze strains infected with RS1-KmΦ, chromosomal DNA was digested with BglIl to cleave within the rstR gene, and an rstR probe was used to determine the number of fragments produced. Since there is one BglII site within the rstR gene, one copy of integrated RS1 element produced two bands, whereas unintegrated RF DNA produced one band after linearization of the circular form. Thus, hybridization of BglII-digested genomic DNA from infected strains with the rstR probe and interpretation of the resulting restriction patterns confirmed that the phage genome integrated into the chromosome of some of the recipient cells (Fig. 6). Although plasmid preparations from the freshly infected cells showed the presence of the phage genome, the lysogens spontaneously lost the RF of the phage, as confirmed by subsequent analysis of plasmid preparations (data not shown).
FIG. 6.
Southern hybridization analysis of genomic DNA isolated from V. cholerae O1 and O139 strains infected with RS1-KmΦ and the corresponding native strains. Total DNA or plasmids were digested with BglII and probed with the rstR probe. The strain number and description of the strains shown in different lanes are as follows: lane 1, pRS1-Km linearized with BglII; lane 2, El Tor strain SA-317 (native); lane 3, SA317(pRS1-Km); lane 4, El Tor strain Env-002 (native); lane 5, Env-002(pRS1-Km); lane 6, Bah-2; lane 7, Bah-2(pRS1-Km); lane 8, Env-99 (native); lane 9, Env-99(pRS1-Km); lane 10, Bengal-2 (native); lane 11, Bengal-2(pRS1-Km), The integration of RS1-KmΦ DNA into the chromosome of recipient strains SA-317 and Env-002 is shown, whereas in strains Bah-2 and Bengal-2 the phage genome is shown to be present in the RF form. Both of these strains lack the attRS sequence. Numbers indicating the molecular sizes of bands correspond to a 1-kb DNA ladder (BRL).
Morphogenesis of RS1Φ.
We studied the production of infectious RS1-KmΦ particles by strains that were transduced to Kmr by the genetically marked phage. While most strains were susceptible to phage prepared from strain O395(pRS1-Km), only transduced strains which had a resident CTXΦ genome produced new Kmr phage particles in their supernatant fluids. The structure of RS1 element is known to be similar to that of RS2, which constitutes part of the CTX prophage and encodes functions related to regulation, integration, and replication of the phage genome (21). RS1 apparently encodes four polypeptides, two of which, RstR and RstA, are identical to the corresponding polypeptides encoded by RS2. The rstB of RS1 is also nearly identical to that of RS2 (21). The RS1 element carries an additional ORF not present in RS2. This ORF, termed rstC, is predicted to encode a 74-amino-acid polypeptide. The function of rstC is not clear, but it seems unlikely that rstC encodes all of the functions required for morphogenesis of the RS1 phage. This is supported by the observation that, in the present study, only CTXΦ-positive strains infected by RS1-KmΦ consistently produced a detectable level of phage particles (Table 5), whereas the nontoxigenic (CTX−) strains did not produce a detectable level of the phage, even though these strains harbored the RS1 element either in its integrated form or as an RF. This indicated that RS1 is possibly packaged in a phage particle, the morphogenesis of which at least partially depends on the core genes of CTXΦ.
Origin of CTXΦ.
Whereas in the CTX prophage the core region of the prophage resides downstream and adjacent to the rstB gene of RS2, in RS1 rstB is followed by rstC, an additional ORF (21). Our results showed that RS1 can propagate as a filamentous phage aided by core genes of CTXΦ. It appears that in CTXΦ rstC is substituted by the core genes that are involved in the morphogenesis of CTXΦ particles. Thus, CTXΦ may have evolved from an ancestral RS1 element by substitution of rstC and acquisition of additional ORFs and thus become a complete phage genome. RS1 encodes a site-specific recombination system, as well as a repressor protein, RstR, which has significant similarity to repressor proteins of both gram-negative and gram-positive bacteria (1, 21). Similar repressors include Salmonella enterica serovar Typhimurium phage P22 protein c2, Lactococcus lactis phage Tuc 20009 repressor, coli phage 434 repressor, and Pseudomonous aeruginosa phage D3 repressor (1, 21). Thus, many of these repressor homologues of RstR are phage encoded and play critical roles in the maintenance of lysogeny. However, CTXΦ is unusual among filamentous phages since it has the ability to integrate into the host chromosome and reside there as a prophage. This property of CTXΦ can possibly be attributed to its having an RS1 origin.
RS1 is found in most epidemic strains, along with CTX prophages, despite the fact that RS1 carries the rstR gene, which is supposed to provide immunity to infection by CTXΦ carrying the same rstR gene. Moreover, the presence of tandemly integrated RS1-CTX arrays or at least two CTX prophages in tandem is required for the production of CTXΦ particles (3, 5). Since the same strains can simultaneously produce CTXΦ and RS1Φ, both of which use the same receptor (TCP) to infect a recipient strain (as shown in the present study), it appears that in the natural habitat the recipient strains are possibly infected with both phages simultaneously and that subsequently these phage genomes integrate in the chromosome, producing the observed RS1-CTX arrays.
Waldor and coworkers (21) had speculated previously that RS1 may be capable of excision from the chromosome independently of CTXΦ due to the presence of ER sequences flanking both ends of RS1 element. In the present study, we confirmed not only the presence of the excised form of the RS1 element but also the existence of a single-stranded form of the RS1 element and that the RS1 element of V. cholerae can propagate as the genome of a filamentous phage, RS1Φ. We have also demonstrated the horizontal transfer of RS1 element to a variety of V. cholerae strains through lysogenic conversion by RS1Φ. Since RS1Φ carries the rstR gene encoding a phage repressor identical to the RS2-encoded repressor of CTXΦ, the use of RS1Φ may have practical applications in conferring immunity against CTXΦ infection in constructing potential vaccine strains.
Acknowledgments
We thank Matthew Waldor, New England Medical Center, Boston, Mass., for the rstR probes and Afjal Hossain for secretarial assistance.
This research was funded in part by the U.S. Agency for International Development under grant HRN-5986-A-00-6005-00 with the ICDDR,B, and by the U.S. National Institutes of Health under grant RO1 AI39129-01A1 with the Department of International Health, Johns Hopkins University, and the ICDDR,B. The ICDDR,B is in turn supported by countries and agencies which share its concern for the health problems of developing countries.
Editor: J. T. Barbieri
REFERENCES
- 1.Aggarwal, A. K., D. W. Rodgers, M. Drottar, M. Ptashne, and S. C. Harrison. 1998. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science 242: 899–907. [DOI] [PubMed] [Google Scholar]
- 2.Albert, M. J., N. A. Bhuyan, A. Rahman, A. N. Ghosh, K. Hultenby, A. Weintraub, S. Nahar, A. K. M. G. Kibriya, M. Ansaruzzaman, and T. Shimada. 1996. Phage specific for Vibrio cholerae O139 Bengal. J. Clin. Microbiol. 34: 1843–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, E. F., A. J. Heilpern, and M. K. Waldor. 2000. Molecular analysis of a putative CTXΦ precursor and evidence for independent acquisition of distinct CTXΦs by toxigenic Vibrio cholerae. J. Bacteriol. 182: 5530–5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campos, J., R. Fando, A. Silva, B. L. Rodriguez, and J. A. Benitez. 1998. Replication function of the RS1 element associated with Vibrio cholerae CTXΦ prophage. FEMS Microbiol. Lett. 164: 141–147. [DOI] [PubMed] [Google Scholar]
- 5.Davis, B. M., and M. K. Waldor. 2000. CTXΦ contains a hybrid genome derived from tandemly integrated elements. Proc. Natl. Acad. Sci. USA 97: 8572–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, B. M., K. E. Moyer, E. F. Boyd, and M. K. Waldor. 2000. CTX prophages in classical biotype of Vibrio cholerae: functional phage genes but dysfunctional phage genomes. J. Bacteriol. 182: 6992–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62: 1301–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faruque, S. M., Asadulghani, A. R. M. A. Alim, M. J. Albert, K. M. N. Islam, and J. J. Mekalanos. 1998. Induction of the lysogenic phage encoding cholera toxin in naturally occurring strains of toxigenic V. cholerae O1 and O139. Infect. Immun. 66: 3752–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faruque, S. M., Asadulghani, M. N. Saha, A. R. M. A. Alim, M. J. Albert, K. M. N. Islam, and J. J. Mekalanos. 1998. Analysis of clinical and environmental strains of nontoxigenic Vibrio cholerae for susceptibility to CTXΦ: molecular basis for the origination of new strains with epidemic potential. Infect. Immun. 66: 5819–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faruque, S. M., M. M. Rahman, Asadulghani, K. M. N. Islam, and J. J. Mekalanos. 1999. Lysogenic conversion of environmental Vibrio mimicus strains by CTXΦ. Infect. Immun. 67: 5723–5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faruque, S. M., Asadulghani, M. M. Rahman, M. K. Waldor, and D. A. Sack. 2000. Sunlight-induced propagation of the lysogenic phage encoding cholera toxin. Infect. Immun. 68: 4795–4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feinberg, A., and B. Volgelstein. 1984. A technique for radio labelling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 137: 266–267. [DOI] [PubMed] [Google Scholar]
- 13.Kaper, J. B., J. G. Morris, Jr., and M. Nishibuchi. 1988. DNA probes for pathogenic Vibrio species, p. 65–77. In F. C. Tenover (ed.), DNA probes for infectious disease. CRC Press, Inc., Boca Raton, Fla.
- 14.Keasler, S. P., and R. H. Hall. 1993. Detection and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet 341: 1661. [DOI] [PubMed] [Google Scholar]
- 15.Kimsey, H. H., and M. K. Waldor. 1998. CTXΦ immunity: application in the development of cholera vaccines. Proc. Natl. Acad. Sci. USA 95: 7035–7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazar, S., and M. K. Waldor. 1998. ToxR-independent expression of cholera toxin from the replicative form of CTXΦ. Infect. Immun. 66: 394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Pearson, G. D. N., A. Woods, S. L. Chiang, and J. J. Mekalanos. 1993. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc. Natl. Acad. Sci. USA 90: 3750–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98: 503–517. [DOI] [PubMed] [Google Scholar]
- 20.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272: 1910–1914. [DOI] [PubMed] [Google Scholar]
- 21.Waldor, M. K., E. J. Rubin, D. N. Gregory, H. H. Kimsey, and J. J. Mekalanos. 1997. Regulation, replication and integration functions of the Vibrio cholerae CTXΦ are encoded by region RS2. Mol. Microbiol. 24: 917–926. [DOI] [PubMed] [Google Scholar]
- 22.Waldor, M. K., and J. J. Mekalanos. 1994. Emergence of a new cholera pandemic: molecular analysis of virulence determinants in Vibrio cholerae O139 and development of a live vaccine prototype. J. Infect. Dis. 170: 278–283. [DOI] [PubMed] [Google Scholar]
- 23.World Health Organization. 1974. World Health Organization guidelines for the laboratory diagnosis of cholera. Bacterial Disease Unit, World Health Organization, Geneva, Switzerland.