Abstract
Ubiquitylation is an emerging mechanism implicated in a variety of nonproteolytic cellular functions. The attachment of a single ubiquitin (Ub) or poly-Ub (lysine 63) chains to proteins control gene transcription, DNA repair and replication, intracellular trafficking and virus budding. In these processes, protein ubiquitylation exhibits inducibility, reversibility and recognition by specialized domains, features similar to protein phosphorylation, which enable Ub to act as a signaling device. Here, we highlight several recent examples on how Ub regulates signaling and how signaling regulates ubiquitylation during physiological and pathological cellular processes.
Keywords: endocytosis, NF-κB, receptor tyrosine kinase, signal transduction, ubiquitin
Introduction
Ubiquitin (Ub) is a highly conserved protein of 8 kDa that becomes covalently attached to lysine (Lys) residues of target proteins. This occurs through a three-step process involving Ub-activating (E1), Ub-conjugating (E2) and Ub-ligating (E3) enzymes (Hershko and Ciechanover, 1998). The types of Ub modifications that can form are diverse. In the simplest form, a single Ub molecule is attached, which is defined as monoubiquitylation (Figure 1) (Hicke and Dunn, 2003). Alternatively, several Lys residues can be tagged with single Ub molecules, giving rise to multiple monoubiquitylation, also referred to as multiubiquitylation (Figure 1) (Haglund et al, 2003a). Since Ub contains seven Lys residues itself, Ub molecules can form different types of chains in an iterative process, known as polyubiquitylation (Pickart and Fushman, 2004). All seven Lys residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48 and Lys63) are possibly involved in chain formation in vivo, and Ub chains linked via Lys48 or 63 are the best characterized so far (Figure 1) (Hicke et al, 2005). It is clear that Lys48-linked poly-Ub (UbLys48) chains represent a signal for proteasomal degradation of modified substrates. This discovery was awarded the Nobel Prize in Chemistry 2004 and has been extensively reviewed (Hershko and Ciechanover, 1998). However, other types of Ub conjugates are involved in the regulation of different cellular processes, independently of proteolytic degradation (Haglund et al, 2003a; Hicke and Dunn, 2003; Krappmann and Scheidereit, 2005). Here, we focus on the novel role of Ub as a signaling mechanism to regulate signaling networks. We will discuss how different Ub modifications, in particular mono-Ub and UbLys63 chains, function as signaling-dependent devices for regulating cellular functions and establishing networks of protein–protein interactions. We will also highlight how ubiquitylation regulates signaling and vice versa.
Figure 1.
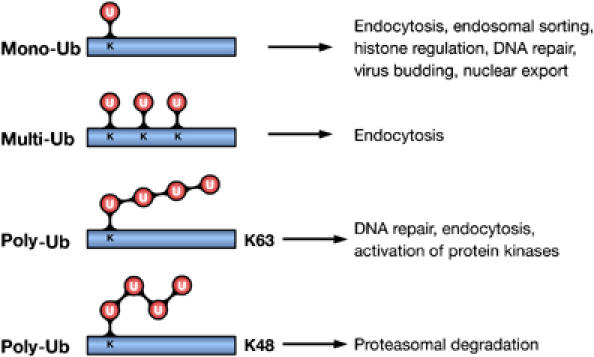
Ub modifications and their cellular functions. Attachment of a single Ub molecule to a single Lys (K) residue leads to protein monoubiquitylation (mono-Ub). Multiple monoubiquitylation (multi-Ub) results from the addition of several single Ub molecules to different Lys residues. These modifications are implicated in various nonproteolytic cellular functions, including endocytosis, endosomal sorting and DNA repair. Polyubiquitylation (poly-Ub) results from the attachment of a chain of Ub molecules to one or more Lys residues. Ub chains formed via Lys48 (K48) of Ub target-modified proteins for proteasomal degradation. Chains linked via Lys63 (K63) are implicated in DNA repair and activation of protein kinases.
Ub as an inducible and reversible signal
It is well established that protein ubiquitylation is induced by a vast variety of stimuli and upstream signaling events in cells. For example, various cell surface receptors become ubiquitylated upon stimulation with extracellular ligands (Hicke and Dunn, 2003). In addition, many cytoplasmic and nuclear proteins become ubiquitylated following their phosphorylation (Di Fiore et al, 2003; Muratani and Tansey, 2003). Moreover, the functions of Ub ligases are tightly regulated by signal-induced mechanisms such as compartmentalization, degradation, oligomerization and post-translational modifications (Thien and Langdon, 2001; Dikic et al, 2003). Regulation at this level is particularly important, since Ub ligases play a central role in substrate recognition and specificity. A second key feature of the ubiquitylation system is that Ub can be rapidly removed by deubiquitylating enzymes (DUBs), which serve to switch off the Ub signal or to shift between different modifications of the same Lys residue (Hershko and Ciechanover, 1998). Notably, ubiquitylation shares the two above-mentioned similarities with protein phosphorylation. In addition, both modifications are recognized by specific protein domains, providing a mechanism for translation of the Ub or phospho-specific signal to downstream effectors (Pawson et al, 2001; Hicke et al, 2005). Intriguingly, although it might not be universally required, phosphorylation is a signal that often precedes ubiquitylation of proteins, for example, phosphorylation of the Ub ligase (e.g. Cbl) or of the substrate protein (e.g. Eps15, Hrs and IκB), which indicates that the two modifications are in tight cooperation in cells. A major difference between the two systems is that Ub is a chemically more complex molecule than phosphate, since it has a larger surface to interact with other proteins. In addition, the fact that Ub can form chains increases the complexity even further. In fact, the UbLys63 and UbLys48 chains have different conformations, with the UbLys63 chain being much more extended than the one linked via Lys48, indicating that they likely have distinct targets and functions in the cell (Figure 1) (Pickart and Fushman, 2004). All these features allow the ubiquitylation system to integrate and synchronize protein networks all the way from the cellular membrane to the nucleus.
Ub mediates protein–protein interaction
Protein ubiquitylation influences the ability of a target protein to interact with other proteins and this is one of the keys to understanding how Ub regulates cellular processes. Nine specialized Ub-binding domains (UBDs) have been identified so far (Table I) (Hicke et al, 2005). These different UBDs might have evolved to allow a wide variety of proteins to interact directly with Ub or ubiquitylated proteins during various cellular processes. Interestingly, the UBDs show diverse structural folds, but they all contact the hydrophobic Ile44 patch of Ub, although with slight variation in the interaction surface. The affinity of the interaction is generally quite low and higher for poly-Ub chains than for mono-Ub, and there is a wide range of binding affinities for each type of domain (Table I). In cells, the affinity and specificity of the interaction to mono-Ub might be increased by additional interactions with the modified protein or formation of multimeric complexes of Ub-binding proteins (Di Fiore et al, 2003; Haglund et al, 2003a). Structural differences between different types of Ub chains might also provide specificity of the interaction in vivo (Kanayama et al, 2004). Dynamic interactions between ubiquitylated and Ub-binding proteins allow Ub networks to be formed in the cell. This concept has been suggested for several Ub-binding endocytic regulators that are involved in endosomal sorting of ubiquitylated cargo (Hicke and Dunn, 2003). Since UBDs are found in various proteins, Ub–UBD interactions might play important roles for the vast variety of cellular functions in which ubiquitylation is implicated.
Table 1.
Ub-binding domains and their characteristics
| UBD | Structure and mode of interaction with Ub | Kd (μM) | Motif length (aa) | Proteins containing UBD |
|---|---|---|---|---|
| CUE | Three-helix bundle with hydrophobic interaction surface. Conserved MFP and LL motifs required for Ub binding | ∼2–160 (mono-Ub) | 42–43 | Vps9p, Tollip |
| GAT | Three-helix bundle with hydrophobic and acidic interaction surface on helices α1 and α2 | ∼180 (mono-Ub) | 135 | GGA1, TOM1, TOM1L1 |
| GLUE | Not known | ∼430–490 (mono-Ub) | ∼135 | Eap45 |
| NZF | Four antiparallel β-strands. Zinc-finger with single zinc ion coordinated by four conserved cysteine residues. TF/Φ motif required for Ub binding | ∼100–400 (mono-Ub) | ∼35 | Np14, TAB2/3, Vps36p |
| PAZ | Not known | Not known | ∼58 | HDAC6 |
| UBA | Three-helix bundle with hydrophobic interaction surface. Conserved leucine residue required for Ub binding | ∼10–500 (mono-Ub) ∼30 nM (poly-Ub) | ∼45–55 | Rad23, Ede1p |
| UEV | Four α-helices and four β-strands. Homology to the catalytic subunit of Ubcs; lacking the catalytic cysteine residue | ∼100–500 (mono-Ub) | ∼145 | TSG101, Vps27p |
| UIM | Short α-helix. Two conserved serine residues required for Ub binding | ∼100–400 (mono- or poly-Ub) | ∼20 | Eps15, Epsin1/2, Ent1/2 Hrs/Vps27p |
| VHS |
Not known |
Not known |
150 |
STAM |
| Structural features and affinities of interactions with Ub are listed along with examples of proteins containing the identified UBDs. Although the domains have different structural folds, all of them contact the hydrophobic patch including Ile44 with slightly different interaction surfaces. aa=amino acids; CUE=coupling of Ub conjugation enzyme-like; Eps15=epidermal growth factor receptor substrate 15; Epsin=Eps15-interacting protein; GAT=GGA and TOM1; GGA=Golgi-localized, gamma-ear-containing, Arf (ADP-ribosylation factor)-binding protein; GLUE=Gram-like Ub-binding in Eap45; HDAC=histone deacetylase; HHM=Hidden Markow Model; Hrs=hepatocyte growth factor-regulated tyrosine kinase substrate; NZF=Np14 zinc-finger; PAZ=poly-Ub-associated zinc-finger; STAM=signal-transducing adaptor molecule; TAB=TAK1-binding protein; TOM1=target of Myb1; TSG101=tumor susceptibility gene 101; Ub=ubiquitin; UBA=Ub-associated; UEV=Ub E2 variant; UIM=Ub-interacting motif; VHS=Vps27, Hrs, STAM; Vps=vacuolar protein sorting; MFP=methionine, phenylalanine, proline; LL=dileucine; TF=threonine, phenylalanine; Φ=hydrophobic amino acid. | ||||
Regulatory role of mono-Ub
Monoubiquitylation might have several regulatory roles for the targeted protein, such as changes in subcellular localization, conformation, activity and protein interactions (Haglund et al, 2003a; Hicke and Dunn, 2003). Several UBD-containing proteins are monoubiquitylated themselves, and importantly, their UBDs are required for their own monoubiquitylation (Polo et al, 2002; Shih et al, 2003). A possible explanation for this is that the UBD recruits a Ub-loaded E2-conjugating enzyme and thus promotes its own monoubiquitylation (Di Fiore et al, 2003). Subsequent to monoubiquitylation, intramolecular Ub–UBD interactions might regulate the activity/Ub-binding ability of the UBD-containing protein to other ubiquitylated proteins or unconjugated Ub in the cell. There is an interesting example of this type of autoinhibition along the endocytic/biosynthetic pathways.
Vps9 (mammalian Rabex-5) functions as a guanine exchange factor for Vps21 (Rab5) that regulates endosomal fusion during cargo transport both from the endocytic and biosynthetic pathways. Recent findings show that Vps9 contains a CUE domain, which binds to Ub (Donaldson et al, 2003; Shih et al, 2003). Importantly, Vps9 is required for sorting of Ste2p-Ub chimera to the vacuole. Vacuolar sorting of Ste2p-Ub chimera by Vps9 is blocked if the interaction of Vps9 with Ub is impaired by an Ile44 mutation. Moreover, yeast strains lacking Vps9 are defective in vacuolar sorting of Ste2p-Ub chimera (Donaldson et al, 2003). A possible scenario is that the CUE domain can engage in intramolecular interactions with covalently attached mono-Ub in the same molecule and would gain sorting activity for ubiquitylated cargo when this interaction is released.
Ub signaling networks in receptor endocytosis
Emerging evidence indicates that monoubiquitylation is an important signal during several steps of receptor endocytosis. Monoubiquitylation of various cell surface receptors functions as an endosomal sorting signal that targets them for lysosomal degradation (Hicke and Dunn, 2003). Receptors may also recycle back to the cell surface and loss of receptor ubiquitylation has indeed been correlated with enhanced recycling (Levkowitz et al, 1998). The requirement of ubiquitylation for the early steps of receptor endocytosis is still a matter of controversy based on the fact that Ub is sufficient to facilitate the removal of receptors from the cell membrane, but is also dispensable for internalization of many transmembrane receptors in vivo (Holler and Dikic, 2004). On the other hand, the importance of monoubiquitylation for endocytic sorting of receptors into the inner vesicles of the forming multivesicular body (MVB), which are destined for lysosomal degradation is firmly established (Katzmann et al, 2002; Hicke and Dunn, 2003). The Ub ligase Cbl promotes multiple monoubiquitylation of activated receptor tyrosine kinases (RTKs), ensuring their efficient lysosomal targeting (Haglund et al, 2003b). In addition, several endocytic regulators harbor UBDs that interact with mono-Ub and are required for endosomal sorting of ubiquitylated cargo (Table II). These proteins include Eps15, epsins, Hrs and tumor susceptibility gene 101 (TSG101), which are localized in specific endocytic compartments and can direct the endosomal sorting of the ubiquitylated cargo (Raiborg et al, 2002; Hicke and Dunn, 2003). More recently, the adaptor proteins GGA and TOM1 have also been implicated in sorting of ubiquitylated proteins (Puertollano and Bonifacino, 2004; Scott et al, 2004).
Table 2.
Ub-binding endocytic regulators and their functions
| Protein |
UBD | Protein function | |
|---|---|---|---|
| Yeast | Mammals | ||
| Ede1p | Eps15 | UBA/UIM | Endocytic adaptor protein at the plasma membrane and sorting endosome. Endocytosis of ubiquitylated EGFRs and endosomal sorting in mammalian cells |
| Ent1p/2p | Epsin1/2 | UIM | Endocytic adaptor protein at the plasma membrane. Internalization of Ste2p in yeast and endocytosis of ubiquitylated EGFRs in mammalian cells |
| Gga | GGA | GAT | Monomeric clathrin adaptor protein. Ub-dependent sorting of Gap1 from the TGN to endosomes and EGFRs from early endosomes to the lysosome |
| Hse1p | STAM1/2 | UIM and VHS | Endocytic adaptor protein at the sorting endosome. Sorting of vacuolar markers into the vacuole in yeast and of ubiquitylated cargo, including EGFRs, into the MVB in mammalian cells |
| — | TOM1 | GAT | Adaptor protein with VHS and GAT domains recruited to early endosomes by Tollip. Recruitment of ubiquitylated proteins to early endosomes |
| Vps9p | Rabex-5 | CUE/? | GEF for Vps21p/Rab5 at the sorting endosome. Sorting of Ste2p into the vacuole in yeast |
| Vps23p | TSG101 | UEV | ESCRT-I component. Sorting of monoubiquitylated CPS into the MVB in yeast and of ubiquitylated EGFRs for lysosomal degradation in mammalian cells |
| Vps27p | Hrs | UIM | Endocytic adaptor protein at the sorting endosome. Sorting of ubiquitylated cargo into the vacuole in yeast and of ubiquitylated EGFRs and TfR-Ub chimera into the MVB in mammalian cells |
| Vps36p |
Eap45 |
NZF/GLUE |
ESCRT-II component. Sorting of ubiquitylated cargo into the vacuole/MVB |
| The functions of endocytic Ub-binding proteins and the role of their UBDs in cellular processes are indicated. Some proteins have different UBDs in the yeast or mammalian homologs. In these cases, both domains are shown in the table with the yeast domain first and the mammalian second (i.e. UBA (yeast)/UIM (mammalian)). In the case of STAM1, both the UÌM and the VHS domain have been implicated in Ub binding. A UBD of Rabex-5 has not yet been published. CUE=coupling of Ub conjugation enzyme-like; EGFR=epidermal growth factor receptor; Eps15=epidermal growth factor receptor substrate 15; Epsin=Eps15-interacting protein; Gap1=general amino-acid permease; GAT=GGA and TOM1; GGA=Golgi-localized, gamma-ear-containing, Arf (ADP-ribosylation factor)-binding protein; GEF=guanine nucleotide exchange factor; GLUE=Gram-like Ub-binding in Eap45; ESCRT=endosomal sorting complex required for transport; Hrs=hepatocyte growth factor-regulated tyrosine kinase substrate; MVB=multivesicular body; NZF=Np14 zinc-finger; RIP=TNF receptor-interacting protein; STAM=signal-transducing adaptor molecule; TfR=transferrin receptor; TGN=trans-Golgi network; TOM1=target of Myb1; Ub=ubiquitin; TSG101=tumor susceptibility gene 101; UBA=Ub associated; UBD=Ub-binding domain; UEV=Ub E2 variant; UIM=Ub-interacting motif; VHS=Vps27, Hrs, STAM; Vps=vacuolar protein sorting. | |||
It is becoming apparent that numerous receptors can transmit signals from endocytic compartments and that these might be qualitatively different from those initiated at the plasma membrane (Miaczynska et al, 2004). It is clear that signaling via neurotrophin (NT) receptors occurs after their internalization. For instance, the receptor tyrosine kinase TrkA promotes NGF-induced cell survival when at the cell surface, whereas it induces differentiation when internalized (Zhang et al, 2000). Importantly, NT-3 binding to TrkA induces internalization and recycling and regulates neurite outgrowth, while NGF induces endocytosis of TrkA, retrograde signaling and transport, and sympathetic neuron survival (Kuruvilla et al, 2004). In addition, several lines of evidence indicate that signaling via mitogen receptors, such as the EGFR, also can occur subsequent to their endocytosis. For instance, inhibition of clathrin-dependent receptor endocytosis significantly blocks activation of MAPK in response to EGF (Vieira et al, 1996). Moreover, many adaptor and signaling molecules (including Grb2, Crk and Ras) translocate to endosomes with activated EGF and TrkA receptors. The signaling from endosomes seems to be physiologically relevant, because signal transduction of EGFRs in endosomes is required for cell survival (Sorkin and Von Zastrow, 2002). Different endocytic routes taken by the receptor might influence its signaling properties (Sigismund et al, 2005). Similarly, in the case of the transforming growth factor (TGF)-β receptor, clathrin-mediated receptor endocytosis promotes signaling, whereas lipid-raft/caveolar internalization is required for receptor turnover (Di Guglielmo et al, 2003). For Notch receptors, which regulate several developmental decisions, monoubiquitylation and endocytosis are prerequisites for efficient receptor activation and signaling (Gupta-Rossi et al, 2004). Moreover, the Notch receptor ligands undergo ubiquitylation, which is required for ligand endocytosis in the signal-sending cell and Notch signaling in the signal-receiving cell (Lai et al, 2005).
Thus, ubiquitylation is an important signal for directing the subcellular localization and intracellular traffic of receptors as well as to regulate receptor signaling by modulating the quality, strength and duration of the signal.
Ub networks in the regulation of NF-κB signaling
The NF-κB transcription factor family members control various biological processes such as apoptosis and proliferation and are among others activated by the proinflammatory cytokines tumor necrosis factor α (TNFα) and interleukin-1β (IL-1β) (Silverman and Maniatis, 2001). In resting cells, NF-κB is kept inactive in the cytoplasm by association with the inhibitor IκBα. The IκB kinase (IKK), consisting of two catalytic subunits (IKKα and IKKβ) and the regulatory subunit NEMO (NF-κB essential modulator)/IKKγ, induces phosphorylation of IκBα, which targets it for Lys48-linked polyubiquitylation and proteasomal degradation (Karin and Ben-Neriah, 2000). In this way, NF-κB gets released and activated, and translocates into the nucleus where it triggers the expression of its target genes (Figure 2A). Here, we will briefly discuss the evidence for the regulation of NF-κB signaling by ubiquitylation. For a complete overview of the topic, we refer to a recent review (Krappmann and Scheidereit, 2005).
Figure 2.
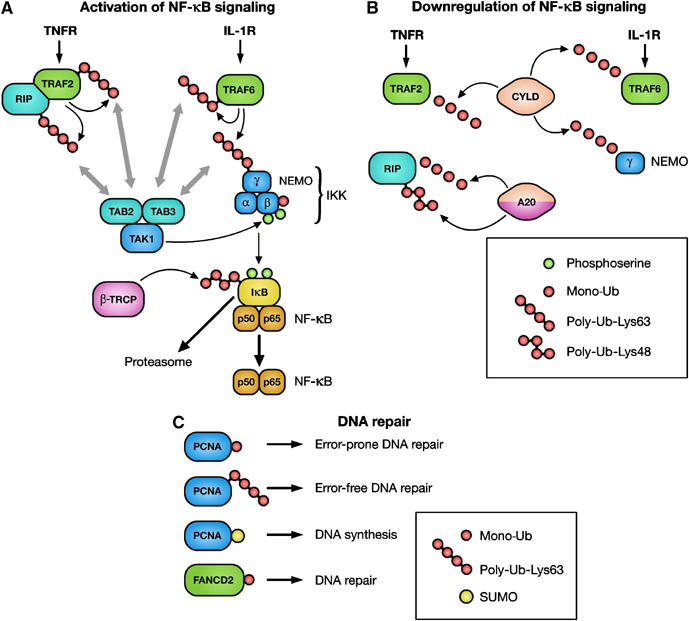
Ub in the regulation of NF-κB signaling and DNA repair. (A) Activation of the Ub ligases TRAF2 and TRAF6 leads to the modification of TRAF2, TRAF6, RIP and NEMO with UbLys63 chains. TAK1 is thought to be recruited to these proteins via TAB2/3, which harbor UBDs. Binding of TAB2/3 to Ub is required for the activation of TAK1, which in turn activates IKK. Serine phosphorylation of IKKβ triggers its monoubiquitylation. Once activated, IKK mediates serine phosphorylation of IκB, which triggers ubiquitylation by the Ub ligase β-TRCP, followed by proteasomal IκB degradation. When NF-κB is released, it translocates to the nucleus and activates transcription. IκB, inhibitor-κB; IKK, IκB kinase; IL-1R, interleukin-1 receptor; NEMO, NF-κB essential modulator; NF-κB, nuclear factor-κB; RIP, receptor-interacting protein; TAB, TAK1-binding protein; TAK1, TGF-β-activated kinase 1; TNFR, tumor necrosis factor receptor; TRAF, TNFR-associated factor; β-TRCP, transducin-repeat-containing protein. (B) Deubiquitylation of TRAF2, TRAF6 and NEMO by the DUB CYLD leads to inhibition of NF-κB activation. Moreover, deubiquitylation of RIP by A20 is followed by A20-mediated Lys48-linked polyubiquitylation of RIP. This modification targets RIP for proteasomal degradation. CYLD, cylindromatosis. (C) During DNA damage, monoubiquitylation and Lys63-linked polyubiquitylation regulates switching between translesion and high-fidelity polymerases, leading to error-prone or error-free DNA repair, respectively. Sumolyation of PCNA is involved in DNA synthesis. Monoubiquitylation of FANCD2 is associated with DNA repair.
The activation of both IKK and the kinase that activates IKK, TGF-β-activated kinase (TAK1), requires UbLys63 chains synthesized by the Ub ligase TRAF6 (Figure 2A) (Krappmann and Scheidereit, 2005). In the case of IKK, modification of the regulatory subunit NEMO with UbLys63 leads to the activation of IKK and thus of NF-κB (Zhou et al, 2004). Moreover, activation of IKK requires monoubiquitylation of IKKβ, which is induced by its serine phosphorylation (Carter et al, 2003). TAK1 is found in complexes with the TAK1-binding proteins (TAB1, TAB2 and TAB3), which stimulate its kinase activity. Recent findings show that TAB2 and TAB3 harbor a novel type of Ub-binding zinc-finger domain that preferentially binds UbLys63 chains. Importantly, this domain is required for the ability of TAB2 and TAB3 to activate TAK1 (Kanayama et al, 2004). Possibly, binding of TAB proteins to UbLys63-modified NEMO would bring TAK1 close to IKK and thereby facilitate TAK1-mediated activation of the IKK/NF-κB pathway. These findings illustrate how UbLys63 chains may function as a scaffold to assemble signaling complexes.
An essential component of the TNF receptor 1 signaling complex, RIP (receptor-interacting protein), becomes modified by the Ub ligase TRAF2 with UbLys63 chains upon TNF-α stimulation (Figure 2B) (Silverman and Maniatis, 2001). A20, a potent feedback inhibitor of NF-κB signaling, acts both as a DUB that specifically removes UbLys63 chains from RIP and as a Ub ligase that subsequently attaches UbLys48 chains, thereby promoting proteasomal degradation of RIP (Wertz et al, 2004). Interestingly, deubiquitylation of RIP is a prerequisite for ubiquitylation by A20, and therefore Lys63-linked polyubiquitylation of RIP seems to protect it from degradation and to ensure activation of NF-κB. Another DUB, the tumor suppressor cylindromatosis (CYLD), removes UbLys63 chains from TRAF2, TRAF6 and NEMO, also leading to the downregulation of NF-κB signaling (Brummelkamp et al, 2003). In conclusion, modification of IKKβ with mono-Ub and NEMO, TRAFs and RIP with UbLys63 chains are critical events for activating NF-κB, whereas Ub removal seems to be a common theme for downregulation of NF-κB signaling.
Ub signaling networks in the nucleus
Many findings have established that monoubiquitylation plays a pivotal role for transcriptional regulation by modification of histones and transcription factors. More details on Ub networks during transcription are found in a recent review (Muratani and Tansey, 2003). Here, we focus on Ub signaling networks during DNA repair.
Emerging data show that DNA damage and DNA repair are signaling responses that are tightly linked to ubiquitylation. Proliferating cell nuclear antigen (PCNA), the replicative processivity factor, forms a clamp that encircles DNA during DNA replication (Haracska et al, 2004). Two types of Ub modifications of PCNA determine which DNA polymerases will bind to PCNA and which type of DNA repair will take place. Monoubiquitylation of PCNA on Lys164 triggers error-prone DNA repair, whereas attachment of UbLys63 chains at the same site signals error-free DNA repair (Figure 2C) (Hoege et al, 2002; Stelter and Ulrich, 2003). Interestingly, the same Lys residue that undergoes both mono-Ub and UbLys63 poly-Ub is also sumoylated. SUMO (small Ub-like modifier) attachment to PCNA is involved in normal DNA synthesis during S phase (Hoege et al, 2002). Thus, the same Lys residue in PCNA undergoes three different mutually exclusive modifications. A model has been proposed where modification of PCNA with mono-Ub or UbLys63 chains regulates the engagement of low-fidelity lesion-bypass polymerases or high-fidelity replicases to the site of replication, respectively (Friedberg et al, 2005).
Interplay between the reversible SUMO and Ub modifications seems to be a common regulatory mechanism in nuclear Ub networks. NF-κB activation is triggered by two pathways, one that is initiated by TNF-α or IL-1β (Figure 2A) and one initiated by DNA damage (Huang et al, 2003). Upon DNA damage, the localization of NEMO, the NF-κB essential modulator, is controlled by sequential modifications with SUMO and Ub. First, sumoylation of NEMO leads to its accumulation in the nucleus. Thereafter, NEMO desumolyation and phosphorylation by the DNA response kinase ATM leads to ubiquitylation of NEMO on the same two Lys residues that were modified with SUMO (Huang et al, 2003). Importantly, NEMO ubiquitylation leads to its translocation to the cytoplasm where it can associate with and activate the IKK, leading to the activation of NF-κB.
DNA damage induces phosphorylation and monoubiquitylation of the FANCD2 protein (Fanconi anemia protein of subtype 2) (Figure 2C), which triggers interaction with the tumor suppressor protein breast cancer 1 (BRCA1) in chromatin-associated nuclear foci (D'Andrea and Grompe, 2003; Gregory et al, 2003; Vandenberg et al, 2003). These foci are implicated in activation of the S-phase checkpoint and DNA repair. Deubiquitylation of FANCD2 may play a critical role for the recycling of FANCD2 when cells recommence the cell cycle after DNA damage (Nijman et al, 2005). Together, these examples highlight the importance for ubiquitylation in orchestrating nuclear signaling networks during DNA repair.
Ub networks as a target for therapeutic interventions
Alterations of ubiquitylation pathways have been shown to contribute to the pathogenesis of several diseases, such as cancer and neurodegenerative disorders. In fact, mutations of Ub, ubiquitylation enzymes (E1s, E2s and E3s), DUBs and ubiquitylation substrates are all found in human disorders (Jiang and Beaudet, 2004). The fact that many Ub ligases are proto-oncogenes emphasizes the importance of ubiquitylation for maintenance of cellular homeostasis (Fang et al, 2003).
It is well-established that constitutive RTK signaling, resulting from receptor overexpression, autocrine growth factor loops and activating mutations, may lead to cell transformation and cancer (Blume-Jensen and Hunter, 2001). In addition, loss of negative regulation of RTKs is an important factor that contributes to enhanced receptor signaling (Dikic and Giordano, 2003). For instance, receptor mutations that lead to loss of the binding site for the Ub ligase Cbl as well as Cbl mutants lacking the Ub ligase activity cause defective receptor downregulation (Peschard and Park, 2003; Bache et al, 2004b). The ESCRT machinery recognizes ubiquitylated cargo at the sorting step into the inner vesicles of the MVB, targeting the cargo for lysosomal degradation. Importantly, two of the ESCRT-I subunits, TSG101 and hepatocellular carcinoma-related protein 1 (HCRP1), are linked to the development of tumors (Katzmann et al, 2002; Bache et al, 2004a). Thus, proper Ub-dependent lysosomal targeting of activated RTKs prevents constitutive receptor signaling and carcinogenesis. Therefore, new therapies not only aim to reduce receptor activity but also to induce RTK internalization and degradation (Rowinsky, 2004).
Monoclonal antibodies and small molecule kinase inhibitors have proven to act partially via Ub-dependent receptor degradation. For instance, monoclonal antibodies, used for the treatment of breast cancer tumors overexpressing ErbB2, enhance Cbl-mediated receptor ubiquitylation and downregulation (Klapper et al, 2000). Some types of cancer exhibit constitutive NF-κB activity, which promotes cell proliferation and inhibits apoptosis (Karin and Ben-Neriah, 2000). For this pathway, proteasomal inhibitors are explored as potential therapeutics blocking IκB degradation and thus NF-κB activation. The reversible proteasome inhibitor Bortezomib (Velcade™) has shown positive results for the treatment of relapsed and refractory multiple myeloma and is the first proteasome inhibitor approved in clinics (Adams and Kauffman, 2004). Moreover, more specific small-molecule inhibitors that interfere specifically with Ub ligase binding to tumor-suppressing proteins, such as IκB, might be promising. An additional example is VHL, an E3 ligase subunit associated with von Hippel-Lindau disease, a cause of familial kidney tumors (Fang et al, 2003). These examples show that detailed knowledge of the molecular mechanisms that are regulated by protein ubiquitylation in both biological and physiological processes is required to consider components of these pathways as targets for therapeutics.
Conclusions and perspectives
With similarities to phosphorylation, ubiquitylation is emerging as a signal involved in the regulation of a variety of biological processes. In order to understand how these responses are controlled at the molecular level, much more needs to be understood about how Ub modification controls protein function, activity and localization and how the Ub signal is propagated and translated to regulate downstream cellular events. A key to unraveling these questions is the recognition of ubiquitylated substrates by Ub-binding proteins in vivo. This might give insights into how induction of protein ubiquitylation by several signaling processes allows the formation of networks of protein–protein interactions. Moreover, proteomic approaches to study the dynamics of Ub–UBD interactions upon ligand stimulation as well as live-imaging techniques to visualize Ub signaling networks in vivo will empower a more comprehensive analysis of cellular signaling events governed by ubiquitylation. The growing detailed knowledge about ubiquitylation signaling pathways during normal physiology will also help in understanding how their deregulation leads to disease.
Acknowledgments
We apologize to investigators whose important contributions were not included in this review due to space limitations. We thank Carl-Henrik Heldin and members of the Dikic laboratory for critical reading of the manuscript. ID gratefully acknowledges support from the Deutsche Forschungsgemeinschaft and the Boehringer Ingelheim Foundation.
References
- Adams J, Kauffman M (2004) Development of the proteasome inhibitor Velcade (Bortezomib). Cancer Invest 22: 304–311 [DOI] [PubMed] [Google Scholar]
- Bache KG, Slagsvold T, Cabezas A, Rosendal KR, Raiborg C, Stenmark H (2004a) The growth-regulatory protein HCRP1/hVps37A is a subunit of mammalian ESCRT-I and mediates receptor down-regulation. Mol Biol Cell 15: 4337–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bache KG, Slagsvold T, Stenmark H (2004b) Defective downregulation of receptor tyrosine kinases in cancer. EMBO J 23: 2707–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume-Jensen P, Hunter T (2001) Oncogenic kinase signalling. Nature 411: 355–365 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Nijman SM, Dirac AM, Bernards R (2003) Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 424: 797–801 [DOI] [PubMed] [Google Scholar]
- Carter RS, Pennington KN, Ungurait BJ, Arrate P, Ballard DW (2003) Signal-induced ubiquitination of I kappaB Kinase-beta. J Biol Chem 278: 48903–48906 [DOI] [PubMed] [Google Scholar]
- D'Andrea AD, Grompe M (2003) The Fanconi anaemia/BRCA pathway. Nat Rev Cancer 3: 23–34 [DOI] [PubMed] [Google Scholar]
- Di Fiore PP, Polo S, Hofmann K (2003) When ubiquitin meets ubiquitin receptors: a signalling connection. Nat Rev Mol Cell Biol 4: 491–497 [DOI] [PubMed] [Google Scholar]
- Di Guglielmo GM, Le Roy C, Goodfellow AF, Wrana JL (2003) Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nat Cell Biol 5: 410–421 [DOI] [PubMed] [Google Scholar]
- Dikic I, Giordano S (2003) Negative receptor signalling. Curr Opin Cell Biol 15: 128–135 [DOI] [PubMed] [Google Scholar]
- Dikic I, Szymkiewicz I, Soubeyran P (2003) Cbl signaling networks in the regulation of cell function. Cell Mol Life Sci 60: 1805–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson KM, Yin H, Gekakis N, Supek F, Joazeiro CA (2003) Ubiquitin signals protein trafficking via interaction with a novel ubiquitin binding domain in the membrane fusion regulator, Vps9p. Curr Biol 13: 258–262 [DOI] [PubMed] [Google Scholar]
- Fang S, Lorick KL, Jensen JP, Weissman AM (2003) RING finger ubiquitin protein ligases: implications for tumorigenesis, metastasis and for molecular targets in cancer. Semin Cancer Biol 13: 5–14 [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Lehmann AR, Fuchs RP (2005) Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell 18: 499–505 [DOI] [PubMed] [Google Scholar]
- Gregory RC, Taniguchi T, D'Andrea AD (2003) Regulation of the Fanconi anemia pathway by monoubiquitination. Semin Cancer Biol 13: 77–82 [DOI] [PubMed] [Google Scholar]
- Gupta-Rossi N, Six E, LeBail O, Logeat F, Chastagner P, Olry A, Israel A, Brou C (2004) Monoubiquitination and endocytosis direct gamma-secretase cleavage of activated Notch receptor. J Cell Biol 166: 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Di Fiore PP, Dikic I (2003a) Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem Sci 28: 598–603 [DOI] [PubMed] [Google Scholar]
- Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I (2003b) Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol 5: 461–466 [DOI] [PubMed] [Google Scholar]
- Haracska L, Torres-Ramos CA, Johnson RE, Prakash S, Prakash L (2004) Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol Cell Biol 24: 4267–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hicke L, Dunn R (2003) Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol 19: 141–172 [DOI] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP (2005) Ubiquitin-binding domains. Nat Rev Mol Cell Biol 6: 610–621 [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Holler D, Dikic I (2004) Receptor endocytosis via ubiquitin-dependent and -independent pathways. Biochem Pharmacol 67: 1013–1017 [DOI] [PubMed] [Google Scholar]
- Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S (2003) Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell 115: 565–576 [DOI] [PubMed] [Google Scholar]
- Jiang YH, Beaudet AL (2004) Human disorders of ubiquitination and proteasomal degradation. Curr Opin Pediatr 16: 419–426 [DOI] [PubMed] [Google Scholar]
- Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, Chiu YH, Deng L, Chen ZJ (2004) TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell 15: 535–548 [DOI] [PubMed] [Google Scholar]
- Karin M, Ben-Neriah Y (2000) Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol 18: 621–663 [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD (2002) Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol 3: 893–905 [DOI] [PubMed] [Google Scholar]
- Klapper LN, Waterman H, Sela M, Yarden Y (2000) Tumor-inhibitory antibodies to HER-2/ErbB-2 may act by recruiting c-Cbl and enhancing ubiquitination of HER-2. Cancer Res 60: 3384–3388 [PubMed] [Google Scholar]
- Krappmann D, Scheidereit C (2005) A pervasive role of ubiquitin conjugation in activation and termination of IkappaB kinase pathways. EMBO Rep 6: 321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuruvilla R, Zweifel LS, Glebova NO, Lonze BE, Valdez G, Ye H, Ginty DD (2004) A neurotrophin signaling cascade coordinates sympathetic neuron development through differential control of TrkA trafficking and retrograde signaling. Cell 118: 243–255 [DOI] [PubMed] [Google Scholar]
- Lai EC, Roegiers F, Qin X, Jan YN, Rubin GM (2005) The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development 132: 2319–2332 [DOI] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon WY, Beguinot L, Geiger B, Yarden Y (1998) c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev 12: 3663–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miaczynska M, Pelkmans L, Zerial M (2004) Not just a sink: endosomes in control of signal transduction. Curr Opin Cell Biol 16: 400–406 [DOI] [PubMed] [Google Scholar]
- Muratani M, Tansey WP (2003) How the ubiquitin–proteasome system controls transcription. Nat Rev Mol Cell Biol 4: 192–201 [DOI] [PubMed] [Google Scholar]
- Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, Bernards R (2005) The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell 17: 331–339 [DOI] [PubMed] [Google Scholar]
- Pawson T, Gish GD, Nash P (2001) SH2 domains, interaction modules and cellular wiring. Trends Cell Biol 11: 504–511 [DOI] [PubMed] [Google Scholar]
- Peschard P, Park M (2003) Escape from Cbl-mediated downregulation: a recurrent theme for oncogenic deregulation of receptor tyrosine kinases. Cancer Cell 3: 519–523 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Fushman D (2004) Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol 8: 610–616 [DOI] [PubMed] [Google Scholar]
- Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP (2002) A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416: 451–455 [DOI] [PubMed] [Google Scholar]
- Puertollano R, Bonifacino JS (2004) Interactions of GGA3 with the ubiquitin sorting machinery. Nat Cell Biol 6: 244–251 [DOI] [PubMed] [Google Scholar]
- Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H (2002) Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol 4: 394–398 [DOI] [PubMed] [Google Scholar]
- Rowinsky EK (2004) The erbB family: targets for therapeutic development against cancer and therapeutic strategies using monoclonal antibodies and tyrosine kinase inhibitors. Annu Rev Med 55: 433–457 [DOI] [PubMed] [Google Scholar]
- Scott PM, Bilodeau PS, Zhdankina O, Winistorfer SC, Hauglund MJ, Allaman MM, Kearney WR, Robertson AD, Boman AL, Piper RC (2004) GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat Cell Biol 6: 252–259 [DOI] [PubMed] [Google Scholar]
- Shih SC, Prag G, Francis SA, Sutanto MA, Hurley JH, Hicke L (2003) A ubiquitin-binding motif required for intramolecular monoubiquitylation, the CUE domain. EMBO J 22: 1273–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigismund S, Woelk T, Puri C, Maspero E, Tacchetti C, Transidico P, Di Fiore PP, Polo S (2005) Clathrin-independent endocytosis of ubiquitinated cargos. Proc Natl Acad Sci USA 102: 2760–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman N, Maniatis T (2001) NF-kappaB signaling pathways in mammalian and insect innate immunity. Genes Dev 15: 2321–2342 [DOI] [PubMed] [Google Scholar]
- Sorkin A, Von Zastrow M (2002) Signal transduction and endocytosis: close encounters of many kinds. Nat Rev Mol Cell Biol 3: 600–614 [DOI] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD (2003) Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191 [DOI] [PubMed] [Google Scholar]
- Thien CB, Langdon WY (2001) Cbl: many adaptations to regulate protein tyrosine kinases. Nat Rev Mol Cell Biol 2: 294–307 [DOI] [PubMed] [Google Scholar]
- Vandenberg CJ, Gergely F, Ong CY, Pace P, Mallery DL, Hiom K, Patel KJ (2003) BRCA1-independent ubiquitination of FANCD2. Mol Cell 12: 247–254 [DOI] [PubMed] [Google Scholar]
- Vieira AV, Lamaze C, Schmid SL (1996) Control of EGF receptor signaling by clathrin-mediated endocytosis. Science 274: 2086–2089 [DOI] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM (2004) De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430: 694–699 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Moheban DB, Conway BR, Bhattacharyya A, Segal RA (2000) Cell surface Trk receptors mediate NGF-induced survival while internalized receptors regulate NGF-induced differentiation. J Neurosci 20: 5671–5678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Wertz I, O'Rourke K, Ultsch M, Seshagiri S, Eby M, Xiao W, Dixit VM (2004) Bcl10 activates the NF-kappaB pathway through ubiquitination of NEMO. Nature 427: 167–171 [DOI] [PubMed] [Google Scholar]


