Abstract
The secondary structures of metazoan mitochondrial (mt) tRNAsSer deviate markedly from the paradigm of the canonical cloverleaf structure; particularly, tRNASerGCU corresponding to the AGY codon (Y=U and C) is highly truncated and intrinsically missing the entire dihydrouridine arm. None of the mt serine isoacceptors possesses the elongated variable arm, which is the universal landmark for recognition by seryl-tRNA synthetase (SerRS). Here, we report the crystal structure of mammalian mt SerRS from Bos taurus in complex with seryl adenylate at an atomic resolution of 1.65 Å. Coupling structural information with a tRNA-docking model and the mutagenesis studies, we have unraveled the key elements that establish tRNA binding specificity, differ from all other known bacterial and eukaryotic systems, are the characteristic extensions in both extremities, as well as a few basic residues residing in the amino-terminal helical arm of mt SerRS. Our data further uncover an unprecedented mechanism of a dual-mode recognition employed to discriminate two distinct ‘bizarre' mt tRNAsSer by alternative combination of interaction sites.
Keywords: crystal structure, dual-mode recognition, mitochondria, seryl-tRNA synthetase, tRNA
Introduction
The fidelity of the protein synthesis is assured by the specific attachment of amino acids to the 3′-ends of their cognate tRNA species according to the algorithm of the genetic code (Ibba and Soll, 1999, 2000). This process is governed by a diverse family of enzymes called aminoacyl-tRNA synthetases (aaRS), each of which intrinsically discriminate with high selectivity among the structurally similar tRNAs and amino acids. The recognition of the amino acid is performed with precision in the active site of the catalytic domain, where the cognate amino acid is activated by adenosine triphosphate (ATP) to form an aminoacyl adenylate as a high-energy intermediate. After the complete accommodation of the CCA end of the cognate tRNA in the active site, the aminoacyl moiety is then transferred to the 2′- or 3′-OH of the terminal ribose of tRNA, with the release of AMP. The specificity for cognate tRNA(s) relies on the recognition (identity) elements spread on the surfaces of tRNA molecules (Giege et al, 1998). These identity elements are commonly localized in the anticodon loop, and in the acceptor stem, including the discriminator base located just upstream of the 3′ CCA end. However, an exception was found in the case of tRNA for serine (tRNASer), for which neither the anticodon loop nor the acceptor stem is involved in recognition (Himeno et al, 1990, 1997; Normanly et al, 1992; Achsel and Gross, 1993; Sampson and Saks, 1993). Owing to the absence of the conservation among the anticodons of the serine isoacceptors, the characteristic long variable arm of tRNASer is, therefore, used as a major determinant for specific recognition by seryl-tRNA synthetase (SerRS). Furthermore, recent biochemical studies show that a novel essential identity requirement for methanogenic archaea tRNAsSer additionally includes the discriminator base (G73) and a G30–C40 base pair in the anticodon stem (Korencic et al, 2004).
SerRS is a homodimeric enzyme belonging to the class II aaRS, characterized by a catalytic domain constructed from the antiparallel β-sheet architecture with three short conserved motifs: 1, 2 and 3 (Cusack et al, 1990; Eriani et al, 1990). Bacterial SerRSs consist of two domains: the globular C-terminal catalytic domain and the unique N-terminal antiparallel coiled-coil helical arm (Cusack et al, 1990; Fujinaga et al, 1993). The crystal structure of the SerRS–tRNASer complex from Thermus thermophilus supported the notion that identification of tRNASer mostly depends on specific recognition of the characteristic elongated variable arm of tRNASer exclusively through the sugar–phosphate backbone contacts by the N-terminal α-helical arm of SerRS (Biou et al, 1994; Cusack et al, 1996). Recent biochemical studies have also revealed that yeast and archaeal SerRSs employ a similar mechanism to discriminate their cognate tRNAs, thus suggesting that this idiosyncratic recognition of the variable arm of tRNASer by SerRS is evolutionarily conserved in all the three kingdoms of life (Lenhard et al, 1999; Bilokapic et al, 2004).
On the other hand, the findings of the extant translational machinery in metazoan mitochondrion have broken the paradigm for the tRNA structure. Many metazoan mitochondrial (mt) tRNAs have distinguished structural deviations from the canonical secondary cloverleaf shape of tRNAs (Sprinzl et al, 1998; Helm et al, 2000). Of our interest are two existing ‘bizarre' mt serine isoacceptors that are parts of the mammalian mt translation systems. Of these, tRNASerGCU corresponding to the AGY codons (Y=U and C) loses the entire D-arm (Steinberg et al, 1994), whereas tRNASerUGA recognizing the UCN codons (N=A, G, C and U) has an unusual cloverleaf configuration with an extended anticodon stem (Yokogawa et al, 1991; Watanabe et al, 1994) (Figure 1A). No three-dimensional structure is available for these mt tRNAsSer although the canonical L-shaped structure of tRNA has been unraveled by X-ray crystallographic studies three decades ago (Robertus et al, 1974). Nevertheless, intensive studies on the acceptor–anticodon interstem angles of several mt tRNAs using the method of transient electric birefringence render a view of an opened ‘L' shape with an obtuse interstem angle in the elbow region of mt tRNA (Frazer-Abel and Hagerman, 2004). Recently, different structural aspects of unusual tRNAs have been evoked from two crystal structures of tRNA bound to guanine transglycosylase (Ishitani et al, 2003), and of the tRNA-like domain of tmRNA complexed with SmpB (Gutmann et al, 2003). However, such variant RNA structures may only be possible through stabilization by extensive interactions with the protein surface.
Figure 1.
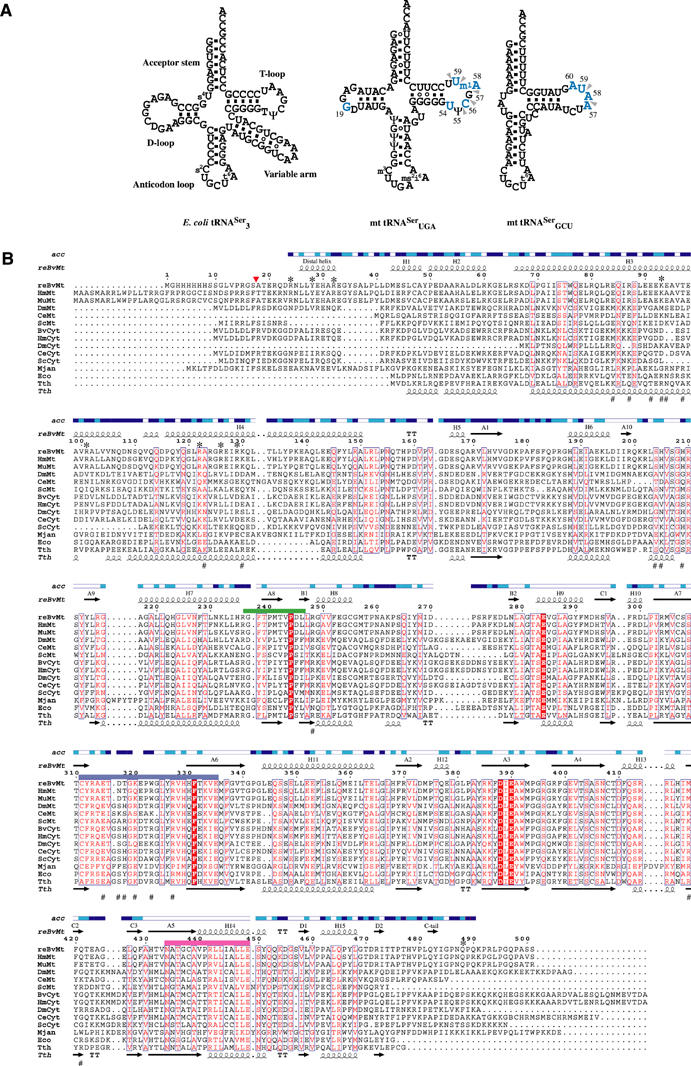
Unique structural characteristics of the tRNA/synthetase system for serine in mammalian mitochondria. (A) Secondary-structure diagrams of typical and atypical tRNAsSer are depicted in cloverleaf representations. The left panel shows the canonical serine isoacceptor from E. coli possessing an elongated variable arm as a discriminative tRNA identity for recognition by SerRS. This identity element disappears in both mammalian mt tRNAsSer, as shown in the middle and right panels for B. taurus mt tRNASerUGA and tRNASerGCU, respectively. Identity elements shifting to the T-loop of mt tRNAsSer as previously reported (Shimada et al, 2001) are illustrated in blue characters for the tRNA mutagenesis studies, and indicated by arrowheads for tRNA-footprinting experiments. (B) The amino-acid sequence alignments of SerRS from three domains of life. The organisms referred to in the sequences alignment and corresponding accession numbers of the Swiss Protein Data Base are as follows. The recombinant mt SerRS from B. taurus used for structure determination (reBvMt) having a hexahistidine tag in the N-terminus is in the top row and is numbered. A red arrowhead indicates the N-terminus of the mature protein in mitochondria. The secondary structure elements as defined by the crystal structure are shown above the sequence and labeled according to bacterial SerRS (Cusack et al, 1990). Solvent accessibility (acc) is shown in blue, cyan and white for accessible, intermediate and buried residues, respectively. Another five mt SerRSs from Homo sapiens (HmMt; Q9NP81), Mus musculus (MuMt; Q9JJL8), Drosophila melanogaster (DmMt), Caenorhabditis elegans (CeMt) and Saccharomyces cerevisiae (ScMt; P38705) are shown, followed by five eukaryotic cytoplasmic from B. taurus (BvCyt), H. sapiens (HmCyt; P49591), D. melanogaster (DmCyt), C. elegans (CeCyt; Q18678) and S. cerevisiae (ScCyt; P07284), then follows one archaeal from Methanococcus jannaschii (Mjan; Q58477), and two bacterial from E. coli (Eco; P09156) and T. thermophilus (Tth; P34945). The secondary structure elements of T. thermophilus are also shown below the sequence (Fujinaga et al, 1993). Conserved amino acids are in white in red-filled rectangles. Similar residues are in red, surrounded by blue lines. Class II synthetase motifs 1, 2 and 3 are underlined in green, purple and pink, respectively. Residues involved in tRNA recognition are indicated with asterisks for mt SerRS and with sharps for T. thermophilus SerRS, as suggested by mutational (in this study) and crystallographic studies (Biou et al, 1994), respectively.
Another striking feature of both mammalian mt tRNAsSer is the absence of the elongated variable arm which is a principal identity element of tRNASer shared among prokaryotic, eukaryotic and archaeal systems. Interestingly, we recently reported that a single mammalian mt SerRS from Bos taurus is capable of recognizing and charging each of the two isoacceptors, sharing no identical sequences or structural topologies, by serine to comparable degrees (Yokogawa et al, 2000). Further tRNA footprinting and mutagenesis experiments revealed that mt SerRS recognizes the nucleotides in the TΨC loop of each isoacceptor as an alternative, but the D–TΨC loop interaction between G19 and C56 is additionally required for mt tRNASerUGA recognition (Figure 1A), thus suggesting distinct mechanisms for the two substrate tRNAsSer (Shimada et al, 2001). Moreover, the fact that sequence similarity among the bacterial and eukaryotic counterparts is significantly lower in the N-terminal domain involved in tRNA binding strongly indicates a novel mechanism of tRNA recognition unique to mt systems (Figure 1B). As human mt tRNASer genes appear to be a hot spot for mutations associated with nonsyndromic deafness (Toompuu et al, 2002; Li et al, 2004), elucidation of the recognition mechanism is an essential step for understanding the pathogenesis of mitochondria diseases.
To provide an atomic basis for the mechanism of noncanonical tRNASer recognition, we cocrystallized the full-length mt SerRS from B. taurus complexed with seryl adenylate and determined the crystal structure to a resolution of 1.65 Å. To date, this is the first reported structure of an aaRS from organelles with the highest resolution of other known structures of full-length aaRS. The structure features two auxiliary extensions in both N- and C-termini of the enzyme that unexpectedly overlap the tRNA-binding site in our tRNA-docking model. This clash, combined together with the mutational analysis, provides the detail of a distinct mechanism for recognition of two unusual mt tRNAsSer by three separate regions of the mt enzyme. Both of the characteristic extensions are essential elements in recognition, but needed to be coupled with a defined set of residues in a small positively charged patch on the N-terminal helical arm. Notably, different combinations of recognition sites are exploited to recognize each mt isoacceptor, demonstrating a dual mode of recognition of two different ‘bizarre' mt tRNAsSer by a single enzyme. It is also fascinating to look through the evolutionary story of the recognition mechanism of the tRNASer/SerRS system from bacteria to mitochondria, which should reflect the functional complementarity taking place during the evolution from the RNA to the RNP worlds.
Results
Overall structure
The structure was solved by the method of molecular replacement using the T. thermophilus SerRS (Fujinaga et al, 1993) as an initial search model. The refined model, comprised of residues 24–490, yielded a crystallographic R factor of 21.0%, with a free R factor of 22.6% (see Table I and Supplementary Figure 1 online). Two molecules in the asymmetric unit form a homodimer as a shared feature of class II synthetases, burying 14% (3272 Å2) of the total surface area in the dimer interface. The overall monomeric structure is superficially similar to known bacterial SerRSs (Cusack et al, 1990; Fujinaga et al, 1993), as revealed by the root mean square deviation value (r.m.s.d.) of 1.27 Å over 375 Cα atoms compared with T. thermophilus SerRS and of 1.24 Å over 319 Cα atoms for Escherichia coli SerRS (Figure 2). Contrary to the low conservation in the amino-acid sequences, the N-terminal domain of mt SerRS also consists of a long α-helical hairpin arm, as observed in the bacterial counterparts. The α-helical arm is one or two turns longer and differs in orientation compared with those of the bacterial enzymes (Figure 3A). The solvent-exposed helical arm is approximately 65 Å long and makes an angle of about 120° between the two monomers in a dimer, which is slightly larger than that of bacterial SerRS (∼90°). Interestingly, there is a mitochondria-unique insertion forming a loop at the tip of the α-helical arm, which is mostly composed of polar residues, particularly three glutamine and two asparagine residues. Although the function of this region remains controversial, it is notable that the crystal packing is stabilized through the intermolecular contacts, taking place around the active site with this tip loop from the symmetry-related dimer (Supplementary Figure 3 online). This interaction concomitantly disorders the Lys320 and Glu321 residues in the motif 2 loop of one monomer and, in turn, disrupts a part of the helical configuration of the α-hairpin arm of the second monomer in the crystal structure.
Table 1.
Summary of crystallographic data and refinement statistics
| Seryl adenylate complex | |
|---|---|
| Data set | |
| Wavelength (Å) | 0.8123 |
| Resolution (Å) | 99–1.65 |
| Space group | C2221 |
| Unit-cell parameters (Å) | a=79.9, b=230.4, c=135.6 |
| Observed total/unique reflections | 917392/145408 |
| Completeness (%) | 96.6 (96.9)a |
| Rmergeb (%) | 5.0 (37.2) |
| I/σ (I) | 30.7 (1.8) |
| Redundancy | 6.3 (4.3) |
| Structure refinement | |
| Resolution (Å) | 10–1.65 |
| Number of reflections (working/free) | 130099/14531 |
| Rworkc/Rfree (%) | 21.0/22.6 |
| Protein/ligand/water atoms | 7437/58/611 |
| Ramachandrand (%) | 91.1/8.1/0.9/0.0 |
| R.m.s.d. bonds/angles (Å/deg) | 0.005/1.35 |
| Average B value (Å2) | 28.53 |
| aValues in the parentheses refer to the highest resolution shell (1.69–1.65 Å). | |
| bRmerge=∑j∣Ij−〈Ij〉∣/∑jIj, where 〈Ij〉 is the average intensity of reflection j for its symmetry equivalents. | |
| cRwork=∑∣Fobs∣−k∣Fcalc∣/∑∣Fobs∣. In all, 9.7% of randomly chosen reflections were used for calculation of Rfree. | |
| dFractions of residues in the most favored/allowed/generously allowed/disallowed regions of the Ramachandran plot. | |
Figure 2.
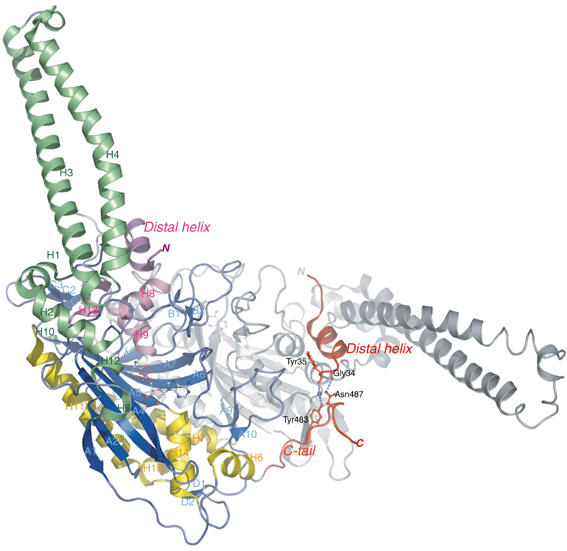
Ribbon representation of the three-dimensional crystal structure of B. taurus mt SerRS at 1.65 Å resolution, depicted in different colors for three α-helical bundles and β-sheet strands. The mitochondria-unique N-terminal distal helix (of monomer 2) and C-tail (of monomer 1) are colored in red, with the stick representations showing the mutual interaction sites. The second monomer is drawn as gray ribbons for clarity. Secondary structure elements are denominated corresponding to bacterial SerRS (Cusack et al, 1990). This figure and subsequent figures were composed using PyMol (http://pymol.sourceforge.net/).
Figure 3.
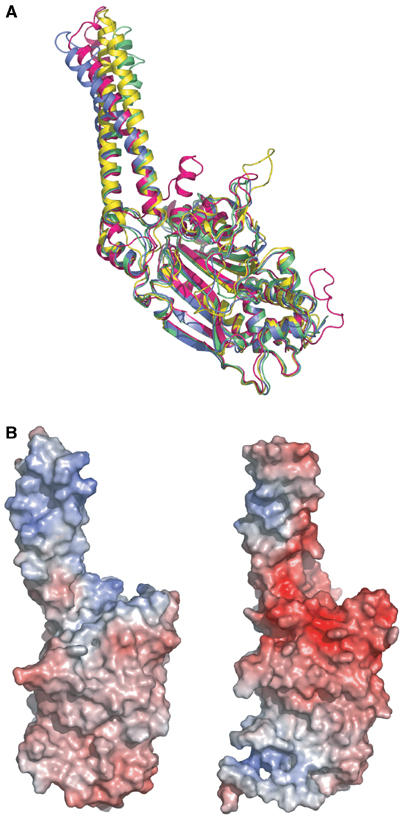
Distinct features of B. taurus mt SerRS compared with bacterial SerRS. (A) Superposition of Cα traces of mt SerRS with the known structures of bacterial SerRSs reveals characteristic insertions in both N- and C-terminus. B. taurus mt SerRS is colored in magenta, whereas SerRS from T. thermophilus and E. coli are in blue and yellow, respectively. The second monomer of T. thermophilus SerRS in the tRNASer complex (green) reveals the alteration of the orientation and curvature of the helical arm during tRNA recognition (Biou et al, 1994). (B) Solvent-accessible surfaces of T. thermophilus (left panel) and mt (right panel) SerRS are represented in the same orientation corresponding to Figure 5A, and colored according to electrostatic potential (red for negative and blue for positive), suggesting the interaction sites with the phosphate backbone of tRNA, which differ substantially between the two molecules.
The active site C-terminal globular domain is built around an eight-stranded β-sheet, encompassed by three helical bundles. Unbiased difference electron densities of the natural intermediate, that is, seryl adenylate, are found in the active sites of both monomers surrounded by nine water molecules, but no density for pyrophosphate or magnesium is observed in the current structure (Supplementary Figure 2 online). Specific recognition of seryl adenylate by the mt synthetase is almost identical to the previous report for two intermediate analogs bound to T. thermophilus SerRS (Belrhali et al, 1994), with the exception of a direct polar interaction (2.84 Å) between the side chain of the invariant Ser404 and the phosphate O3 oxygen atom.
Comparison with the known bacterial structure
The most notable features of mt SerRS compared to bacterial SerRS are two amino-acid extensions found in both termini of the enzyme (Figure 3A). The N-terminal extension consists of a short helix (residue 25–33) connected to the first helix H1 in a helix–loop–helix motif, with an extended loop embracing the coiled-coil helical arm. This terminal helix, hereafter designated as the ‘distal helix', is unique to and highly conserved among mammalian mitochondria. The distal helix is capped by two invariant arginine residues (Arg24 and Arg32), and forms an α-helix bundle together with the adjacent helices H8, H9 and H13 at the shoulder of the molecule (Figure 2). The other remarkable prolongation observed in the carboxyl terminus, designated as the ‘C-tail', is an over 40 Å long flexible loop stretching away from the body of the monomer. The C-tail lies on the backside of the molecular surface of the other subunit and makes extensive intermonomer interactions contributing to the dimer interface. In particular, the short internal β-strand (residues 482–483) forms an antiparallel sheet with strand C1 of the related counterpart. This interfacial interaction is peculiar to mt SerRS and functionally anchors the flexible C-tail in the dimeric molecule. In the crystal structure, the C-tail is apparently in close proximity to the distal helix of the other subunit, and makes a hydrogen bond between the side chain of Asn487 and the main-chain carbonyl oxygen of Gly34, as well as a water-mediated hydrogen bond that links residue Tyr483 with the main chain of Tyr35 (Figure 2). Based on sequence alignments, the eukaryotic (cytoplasmic) SerRSs also have the long C-terminal extension; however, the amino-acid sequences are substantially different from those of the mt enzymes. Interestingly, it has been reported, for yeast cytoplasmic SerRS, that the C-terminal extension plays a role in stabilizing the dimeric structure, and deletion of this region significantly attenuates the aminoacylation activity of the enzyme (Lenhard et al, 1998).
In a search for clues to the tRNA-recognition mechanism, we further evaluated the surface electrostatic potential of mt SerRS (Figure 3B). In contrast to the bacterial T. thermophilus SerRS, which have a wide range of positively charged patches spread over the helical arm and play an important role in the interaction with the phosphate backbone of tRNASer, the monomeric surface of mt SerRS reveals two exclusive positively charged patches localized on one side of the helical arm composed of four basic residues: Lys93, Arg122, Arg126 and Arg129 and, unexpectedly, in the cleft formed by the C-tail and a loop (residues Arg199–His205) between strand A9 and A10. These limited positive patches essentially reveal the distinct property of mt SerRS regardless of the similarity in shape with bacterial SerRS, and are conceivably candidates for the interaction sites with the backbone of mt tRNASer.
Docking model with yeast tRNAPhe
A tRNA-docking model was generated by superposing the active-site domain of mt SerRS onto that of T. thermophilus SerRS in complex with cognate tRNASer (Biou et al, 1994). As one-third of tRNASer in the complex structure is disordered and mt tRNAsSer do not possess the long variable arm, the crystal structure of yeast tRNAPhe (PDB code: 1EVV) was further superimposed onto tRNASer in the complex structure, finally yielding a model of mt SerRS complexed with canonical yeast tRNAPhe (Figure 4). As mt tRNASerGCU do not have the D-arm, while mt tRNASerUGA is thought to possess an altered cloverleaf structure (Yokogawa et al, 1991; Watanabe et al, 1994), we dare not to consider this docking model to be sufficiently accurate to discuss the exact mechanism of recognition (i.e. protein side chain–RNA base interaction) of mt tRNASer; however, the docking model could provide some aspects of interactions between the protein and tRNA, which is quite useful in prediction of potentially important sites of interaction for further mutational experiments. The most striking feature of the docking model is the severe steric clashes seen in the elbow region of tRNA by the distal helix and the C-tail (of the second monomer), which reaches deep into the body of the modeled tRNA. This is consistent with our earlier report showing that the T-loop of each mt tRNASer is an identity element for mt SerRS (Shimada et al, 2001). It is likely that the accommodation of the substrate tRNASer induces the substantial conformational alterations in the N-terminal domain, including a movement of the helical arm toward the incoming tRNA, as observed for T. thermophilus SerRS, and a simultaneous repositioning of three short helices in the N-terminus (i.e. the distal helix, H1 and H2 helices). Nevertheless, the current model clearly suggests the recruitment of the characteristic distal helix and C-tail in mt tRNAsSer-specific recognition.
Figure 4.
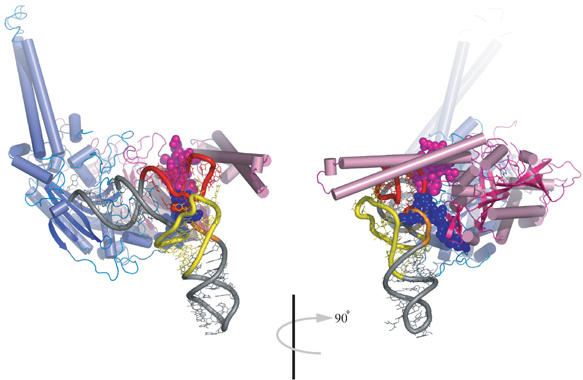
Docking model of yeast tRNAPhe onto mt SerRS. Overall view with the protein shown in ribbon representation is colored in blue and pink for monomers 1 and 2, respectively. The intact tRNA is depicted in gray, except for the D-arm (yellow), variable arm (orange) and the T-arm (red). Seryl adenylates in the active sites are rendered in stick. Mitochondria-specific insertions in the N-terminus of monomer 1 and C-terminus of monomer 2 are depicted in spacefill representations. The right panel is a 90° rotated view clearly revealing steric clashes with the elbow region of tRNA.
Mutational analysis
In order to obtain further insight into the mechanism of tRNA recognition, a series of mt SerRS variants were constructed with mutations in the distal helix, the helical arm and the C-tail (Figure 5A), and subsequently investigated by an in vitro aminoacylation assay with both of the native mt tRNAsSer purified from bovine liver to homogeneity. The results evidently revealed a crucial effect on the aminoacylation capacity of both deletion mutants of the N-terminal distal helix (residues Arg24–Arg32) and the C-tail (residues His478–Ser501) for either mt tRNASerUGA or tRNASerGCU. Inside the distal helix, Asn25 and Glu29 did not affect the aminoacylation capacity. In contrast, two basic residues Arg24 and Arg32 at both ends of the distal helix are likely to be involved in interactions with both mt tRNASerUGA and tRNASerGCU, while the mutation of Tyr28 affected the aminoacylation of tRNASerUGA exclusively. As none of the single mutations could completely extinguish the aminoacylation ability of mt SerRS, the recognition of tRNA may be achieved in a concert of multiple residues in the distal helix. This is reminiscent of the helix–loop–helix motif found in transcription factors such as MyoD (Ma et al, 1994), where it mediates sequence-specific DNA–protein interaction. On the opposite site, the ΔC-10 (deletion of the last 10 residues from the C-terminus) mutant did not affect serylation activity, whereas the ΔC-11 mutant (i.e. deletion from Lys491) significantly decreased the serylation activity for both substrate tRNAs. It is surprising that the cloverleaf-shaped mt tRNASerUGA is more susceptible to the length of the C-tail, as it requires Gln488 (ΔC-14 mutant) for reasonable serylation ability (the ΔC-16 mutant can still charge tRNASerGCU by serine to some extent). Therefore, the C-tail is likely not compensating for the D-arm of tRNA, and probably recognizes some inherent structural features of each mt tRNAsSer. The fact that the C-tail forms a part of the dimer interface raised the possibility of an alternative interpretation that the deletion in this region might weaken the internal contacts and destabilize the local structure of the enzyme. However, we could not detect any differences in dimerization between these deletion mutants compared to the wild-type (WT) protein assessed by the gel filtration analysis (data not shown). Intriguingly, the kinetic parameters determination of the ΔC-15 mutant showed a 28-fold higher Km value than the WT enzyme for tRNASerGCU (0.78 and 21.92 μM for the WT and the ΔC-15 mutant, respectively), while the kcat remained substantially unchanged. Conversely, the ΔC-15 mutant decreased the kcat by nine-fold (12.1 × 10−3 and 1.3 × 10−3 s−1 for the WT and the ΔC-15 mutant, respectively), whereas it barely affected the Km for tRNASerUGA. These unexpected contradictory results suggested that the C-tail may recognize each of serine isoacceptors in a distinct manner, and clearly showed that the loss of serylation activity was not caused by the destabilization of the dimeric structure of the enzyme.
Figure 5.
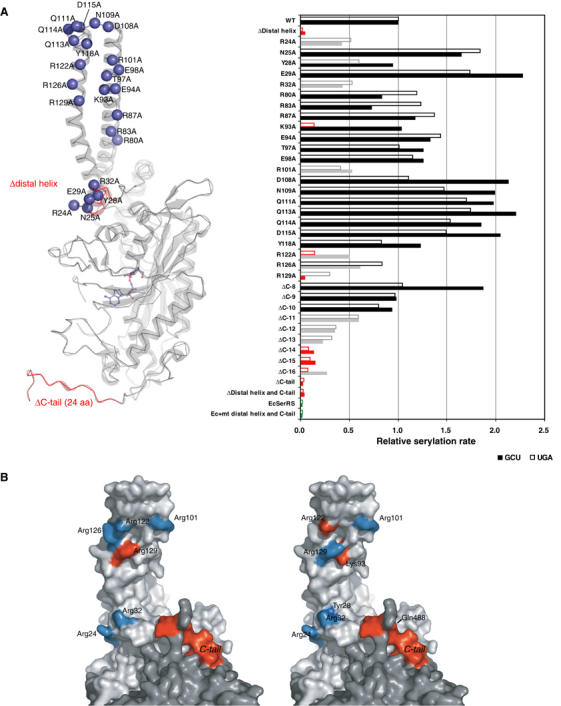
Mutagenesis studies of SerRS variants. (A) In all, 12 deletion and 23 alanine-substituted mutants of mt SerRS illustrated in the left panel, and a single mutant of E. coli SerRS with the mt N-terminal distal helix and C-tail inserted, were assayed by in vitro aminoacylation (shown in the right panel) with two native mt serine isoacceptors: tRNAsSerUGA (empty bar) and tRNAsSerGCU (filled bar). The results indicate the initial rate of serylation relative to the WT mt enzyme. Mutations retaining the serylation ability comparable to the WT are colored in black, whereas mutations that lower the serylation rate to <60% of the WT are shown in gray, and mutants that lose the serylation ability (<20%) are in red. (B) Residues involved in recognition of mt tRNASerGCU (left panel) and tRNASerUGA (right panel) are mapped onto the surface of the dimeric structure (light and dark gray for each monomer). The effective sites are colored in cyan, while the critical residues are depicted in red.
Another 18 alanine-substituted mutants were introduced into both sides of the helical arm and into the inherent tip loop. All mutations in the tip loop did not reduce the aminoacylation ability, but slightly stimulated serylation for both cases of mt tRNAsSer. Thus, we conclude that the tip loop is not used as a mean to recognize the tRNA, although the amino acids in this region are conserved among the mammalian mt SerRS. On the other hand, five residues (i.e. Lys93, Arg101, Arg122, Arg126 and Arg129), which constitute a positive patch on the helical arm, clearly showed depletion of enzyme activity, with remarkable differences between the two substrate tRNAsSer. The K93A harboring enzyme failed to serylate tRNASerUGA efficiently, but could serylate tRNASerGCU comparable to the WT protein. Similar decrement in the serylation ability for either tRNASerUGA or tRNASerGCU was observed with the R101A and R126A mutations. The R122A mutant significantly reduced the serylation activity for tRNASerGCU, but even more dramatically for tRNASerUGA. The most severe effect on serylation of tRNASerGCU was observed in the R129A mutant, which completely failed to serylate tRNASerGCU, as did the distal helix deletion mutant.
Discussion
Combining the crystal structure with the mutagenesis analysis, we have identified the amino-acid residues that are crucial for serylation of mt tRNASer residing in three separated regions: the distal helix, the C-tail and a narrow positively charged patch on the N-terminal helical arm of the mt SerRS molecule. As the distal helix and the C-tail are found only in mt SerRS, the mechanism for recognition of tRNA should be unique to the mt enzyme. However, it has been proven that recognition of tRNA relies on the synergetic action between both extensions and the helical arm, as suggested by the fact that an E. coli SerRS variant with the insertion of the distal helix and the C-tail from mt SerRS could not serylate mt tRNAsSer (Figure 5A). As the distal helix and the C-tail of mt SerRS obscure the landing site for the T-arm of tRNA in our docking model, we hypothesize that binding to the cognate tRNASer stimulates the movement of the long helical arm of the enzyme and leads to structural rearrangement of the N-distal helix, as well as the interactive C-tail. Here, we propose a model for mt tRNASer recognition as an RNA-sandwich model in which the distal helix and the C-tail of mt SerRS make contacts with the major groove side of the acceptor helix of mt tRNASer, while the positive patch on the helical arm simultaneously approaches tRNA at the opposite face of the acceptor helix, thus locking up the T-loop of tRNA (Figure 6B). By contrast, the bacterial, and possibly the eukaryotic, SerRS exploit a distinct protein-sandwich recognition in which the variable arm and the T-loop of tRNASer flank the helical arm of SerRS (Figure 6A). Moreover, the results clearly illustrate the distinct set of amino-acid residues responsible for recognition of each cognate mt tRNASer (Figure 5B). For tRNASerUGA, three residues, Arg24, Tyr28 and Arg32, in the distal helix and the basic residues Lys93 and Arg122 on the helical arm are crucial, whereas Arg24 and Arg32 flanking the distal helix, and Arg129 on the helical arm are indispensable for tRNASerGCU recognition. The 11 amino acids at the C-tail (His478–Gln488) are essential for both serine isoacceptors. However, the C-tail is also likely to play a role in the dual-mode recognition, although the precise mechanism remains unclear, suggested by significant differences between two isoacceptors in the kinetic parameters and the sensitivity to the deletion mutants. As supported by our earlier work on tRNA (Shimada et al, 2001), we have established clear evidence for a dual-mode mechanism for recognition of two distinct mt tRNAsSer in mammalian mitochondria. Due to the absence of analogy of the primary and secondary structures between the two mt tRNAsSer, mt SerRS adopt an alternative combination of the recognition sites for each cognate tRNASer, and plausibly performs discrimination by scanning through the phosphate–sugar backbone of tRNASer using the basic residues residing on the helical arm, as previously reported for the case of bacterial SerRS (Biou et al, 1994). Therefore, mt SerRS recognizes the distinctive shape of each cognate tRNASer by an indirect readout mechanism (Hauenstein et al, 2004). Residue Lys93 on the helical arm is a good candidate obligated to monitor the critical D–TΨC loop tertiary interaction of the G19−C56 base pair in mt tRNASerUGA, as proposed by the kinetic studies on tRNA variants (Shimada et al, 2001). Indeed, in our docking model, Lys93 is in close proximity to make contact with the G19−C56 base pair. Interestingly, the corresponding base pair of G19–C56 in bacterial tRNASer is also recognized through a hydrogen bond with Ala55 and base stacking with Pro59 in the T. thermophilus SerRS−tRNASer complex structure (these residues are not conserved in mt SerRS). To our knowledge, this is the first report of an aaRS that uses a different mechanism to recognize its distinct isoacceptor tRNAs, which coincidentally possess discrete individual identity elements.
Figure 6.
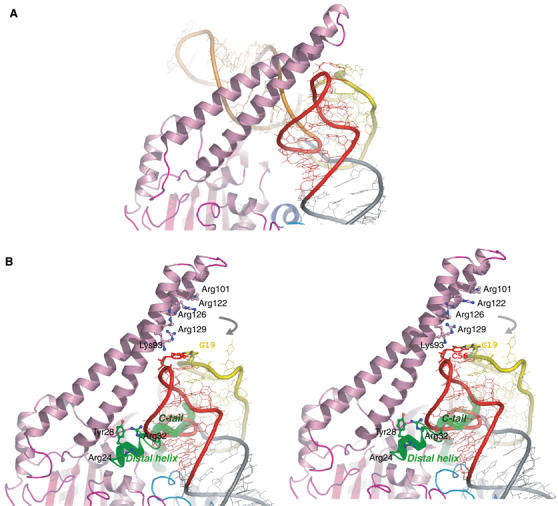
Proposed discriminating model of the SerRS–tRNASer complex. (A) illustrates the protein-sandwich model based on the crystal structure of T. thermophilus SerRS–tRNASer (Biou et al, 1994; Cusack et al, 1996). Proteins and tRNAs are colored corresponding to Figure 4. (B) Stereo view of the mt discriminating complex suggests a dissimilar RNA-sandwich model in which three separate parts of the protein are encompassing the T-loop of the modeled tRNA. The mitochondria-unique extensions are shown in green ribbon and tube for the distal helix and the C-tail, respectively. Residues involved in RNA–protein interactions are drawn in stick representations. Gray arrows indicate a likely movement of the helical arm upon tRNA binding to make interactions with the T-loop of tRNA.
The canonical tRNASer is characterized by a long variable arm (∼20 bases) between the anticodon stem and the T-arm, which SerRS employs as a tRNA identity to discriminate its cognate tRNAs from all other species. This elongated variable arm is well conserved throughout the evolutionary process from prokaryotes to eukaryotes, with the only exception being the mitochondria. The shortening of the long variable arm of mt tRNAsSer may be evolutionarily compensated by the acquisition of the distal helix and the C-tail of mt SerRS, which consequently forces an alteration of the identity requirement to be the T-loop of mt tRNAsSer. Supposing that mt tRNASerUGA possesses a pseudo-cloverleaf structure similar to the other mt tRNAs, it would have been a challenge for mt SerRS to discriminate its cognate tRNASerUGA from other species in a highly specific manner, thus driving the introduction of some additional recognition sites to ensure specificity. We believe that this resulted in the dual-mode tRNA-recognition mechanism of mt SerRS. Our results also provide an example of the RNA–protein coevolution and complementarity in the mitochondrion that may mirror the transition process of the RNA to the RNP worlds. Further crystallographic works on the mt SerRS–tRNASer complex structures will allow us a comprehensive interpretation of the recognition mechanism, as well as the evolutionary pathways of the tRNA/synthetase system.
Materials and methods
Structure determination and refinement
The mature B. taurus mt SerRS gene (the precise N-terminus of the native protein has been previously determined on SerRS isolated from bovine mitochondria) was cloned into the expression vector pET-19b (Novagen) with a hexahistidine tag followed by a thrombin-cleavage site at the N-terminus, and crystals diffracting to 1.6 Å were grown from PEG 8000 and lithium sulfate in the presence of ATP and L-Ser as described (Chimnaronk et al, 2004). The phase problem was solved by molecular replacement in a resolution range of 15–3.5 Å using AMoRe (Navaza, 2001), with T. thermophilus SerRS (33% identity to mt SerRS) as a search model (PDB code: 1SRY). E. coli SerRS was neglected for the trial in molecular replacement, due to the higher divergence of the amino-acid sequences (28% identity to mt SerRS). Two related solutions were obtained, providing R factors of 53.0 and 53.9, respectively. Initial phases were improved by averaging, solvent flattening and histogram matching with DM in the CCP4i suite (Collaborative Computational Project, 1994), yielding an interpretable electron density map. At this point, the clear density of homodimeric mt SerRS was visible and the main chains were then automatically traced by running 15 cycles of ARP/wARP (Perrakis et al, 1999). The resulting peptide fragments were subjected to the newly released program Lafire (Local-correlation-coefficient-based Automatic Fitting for Refinement; Lafire version 1.0 is available at http://altair.sci.hokudai.ac.jp/g6/Research/Lafire_English.html), which allowed the automatic building of most of the side chains into the model. In the ∣Fo−Fc∣ electron density map, we found an unbiased density of seryl adenylate in the active site. The model of the adenylate intermediate was built using TURBO (Roussel and Cambillau, 1991), and the full model was subsequently refined straightforward by Lafire running with CNS (Brunger et al, 1998). Briefly, refinement was carried out by a single round of rigid-body refinement followed by several rounds of conjugate gradient minimization and individual B-factor refinement without the noncrystallographic symmetry restraint due to the high-resolution data. At the last stage, several rounds of the water pick and water delete procedures alternating with positional and B-factor refinement were performed until the free R factor (Rfree) converged, yielding the model including 611 water molecules. At this point, the crystallographic R factor dropped to 21.7% (Rfree=23.8%) and further manual refinement, followed by a single round of Refmac (Murshudov, 1997), energy minimization and B-factor positional refinement in CNS, yielded the final R factor of 21.0% and Rfree of 22.6%. The side chain flipping for His, Asn and Gln residues was checked by the hydrogen bond analysis feature from the program WHAT IF (Vriend, 1990). The final structure shows a good stereochemistry and geometry, as validated by the program PROCHECK (Laskowski et al, 1993): 91.1% of the residues are in the most favored regions of the Ramachandran plot, with none in the disallowed regions. In all, 23 amino-acid residues including the histidine tag from the N-terminus and 11 from the C-terminus could not be modeled. Due to the crystal packing interactions, two residues, Lys320 and Glu321, in the motif 2 loop of monomer 1 are also disordered in the refined structure.
The final 3D coordinates and structure factors have been deposited in the Protein Data Bank under the accession code 1WLE.
Preparation of native mt tRNASer from bovine liver
Fresh cow livers (5 kg) were gently homogenized in 10 l of the acidic buffer containing 50 mM CH3COONa, 10 mM (CH3COO)2Mg. The nucleic acid fractions were subsequently extracted by phenol/chloroform and the aqueous phase was diluted with milli Q water before being loaded onto a 4l DEAE-Sepharose Fast Flow anion-exchange column (Amersham Biosciences), pre-equilibrated in buffer containing 20 mM Hepes-NaOH, pH 7.5, 8 mM MgCl2 and 200 mM NaCl. The tRNA fraction was obtained with a 45 l linear gradient of MgCl2 and NaCl to final concentrations of 16 and 450 mM, respectively. The fractions enriched with mt tRNAs were monitored by dot hybridization using the complementary synthetic DNA probe. The tRNA fractions were pooled together and precipitated by iso-propanol. The total yield was 1700 mg RNA, as estimated by UV absorbance at 260 nm.
The RNA pellet was redissolved in a binding buffer (30 mM Hepes-NaOH, pH 7.5, 15 mM EDTA, 1.2 M NaCl) and passed through the modified immobilized DNA probe column, called the ‘chaplet column' (Suzuki et al, 2002), at 65°C. Briefly, the biotinylated DNA probe (∼30 bases) complementary to each mt tRNASer was immobilized on an avidin Sepharose column (Amersham Biosciences) connected in tandem and the tRNA mixture was circulated through the chaplet column using a peristaltic pump to entrap the target mt tRNAs. After washing out nonspecific tRNAs with the wash buffer (15 mM Hepes-NaOH, pH 7.5, 7.5 mM EDTA, 0.6 M NaCl), each individual mt tRNA was eluted separately with a low-salt buffer (0.5 mM Hepes-NaOH, pH 7.5, 0.25 mM EDTA, 20 mM NaCl) at 65°C. The purified mt tRNAsSer were completely activated by 3′ terminal CCA repair reaction using bovine mt CCA-adding enzyme (Nagaike et al, 2001) and further purified by 10% denaturing PAGE. The native mt tRNAs purified in this manner typically showed plateau aminoacylation levels of approximately 95 and 70% for tRNASerGCU and tRNASerUGA, respectively.
Mutational analysis
All of the mt SerRS variants were constructed using the QuikChange™ site-directed mutagenesis kit according to the manufacturer's protocol (Stratagene). Chimeric E. coli SerRS was generated by a stepwise procedure. First, the C-tail of mt SerRS was incorporated using the modified QuikChange™ protocol with two 69-base-long primers which have ∼20 bases complementary to each other at their 5′-ends, and to the insertion site on the template at their 3′-ends. The extracted plasmid DNA was verified by DNA sequencing and then used as the template for the next insertion step. The distal helix of mt SerRS was subsequently introduced half-by-half by two rounds of PCR reaction with long primers in a similar manner as mentioned above. Protein variants were overexpressed as described for the WT protein, and purified by TALON cobalt affinity resin (BD Bioscience) in a step-wise manner. The purified mutant proteins were more than 80% pure, as judged by SDS–PAGE. Aminoacylation assays were performed at 37°C in a reaction mixture containing 100 mM Hepes-NaOH, pH 7.6, 15 mM MgCl2, 5 mM DTT, 60 mM KCl, 1 mM spermine, 2 mM ATP, 0.1 mg/ml BSA, 32.2 μM 14C-serine, 400 nM enzyme and 2 μM pure tRNA. Assay conditions were adjusted to be sensitive enough to evaluate mutant activity. Reactions were initiated by addition of the enzyme to the reaction mixture. Aliquots were withdrawn at appropriate time intervals, spotted onto 3MM filter pads (Whatmann) and immediately rinsed in chilled 5% trichloroacetic acid (TCA). Filters were washed twice with 5% TCA and once more with 100% ethanol prior to measurement of radioactivities by scintillation counting.
Supplementary Material
Supplementary Information
Acknowledgments
We thank K Harata for kindly providing the workstation for data processing, and S Cusack for a gift of the refined coordinates of E. coli SerRS and T. thermophilus SerRS-tRNASer complex. We also thank M Yao and I Tanaka for numerous helpful advice and assistance in structure determination. This research was supported in part by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan (KW), and in part by a grant from the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science and Technology of Japan (TS). Research was further granted by NIH code GM32734-17 given to LL Spremulli as PI (JN and MGJ).
References
- Achsel T, Gross HJ (1993) Identity determinants of human tRNASer: sequence elements necessary for serylation and maturation of a tRNA with a long extra arm. EMBO J 12: 3333–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belrhali H, Yaremchuk A, Tukalo M, Larsen K, Berthet-Colominas C, Leberman R, Beijer B, Sproat B, Als-Nielsen J, Grubel G, Legrand JF, Lehmann M, Cusack S (1994) Crystal structures at 2.5 angstrom resolution of seryl-tRNA synthetase complexed with two analogs of seryl adenylate. Science 263: 1432–1436 [DOI] [PubMed] [Google Scholar]
- Bilokapic S, Korencic D, Soll D, Weygand-Durasevic I (2004) The unusual methanogenic seryl-tRNA synthetase recognizes tRNASer species from all three kingdoms of life. Eur J Biochem 271: 694–702 [DOI] [PubMed] [Google Scholar]
- Biou V, Yaremchuk A, Tukalo M, Cusack S (1994) The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science 263: 1404–1410 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54 (Part 5): 905–921 [DOI] [PubMed] [Google Scholar]
- Chimnaronk S, Jeppesen MG, Shimada N, Suzuki T, Nyborg J, Watanabe K (2004) Crystallization and preliminary X-ray diffraction study of mammalian mitochondrial seryl-tRNA synthetase. Acta Crystallogr D 60: 1319–1322 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Cusack S, Berthet-Colominas C, Hartlein M, Nassar N, Leberman R (1990) A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature 347: 249–255 [DOI] [PubMed] [Google Scholar]
- Cusack S, Yaremchuk A, Tukalo M (1996) The crystal structure of the ternary complex of T. thermophilus seryl-tRNA synthetase with tRNASer and a seryl-adenylate analogue reveals a conformational switch in the active site. EMBO J 15: 2834–2842 [PMC free article] [PubMed] [Google Scholar]
- Eriani G, Delarue M, Poch O, Gangloff J, Moras D (1990) Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 347: 203–206 [DOI] [PubMed] [Google Scholar]
- Frazer-Abel AA, Hagerman PJ (2004) Variation of the acceptor–anticodon interstem angles among mitochondrial and non-mitochondrial tRNAs. J Mol Biol 343: 313–325 [DOI] [PubMed] [Google Scholar]
- Fujinaga M, Berthet-Colominas C, Yaremchuk AD, Tukalo MA, Cusack S (1993) Refined crystal structure of the seryl-tRNA synthetase from Thermus thermophilus at 2.5 Å resolution. J Mol Biol 234: 222–233 [DOI] [PubMed] [Google Scholar]
- Giege R, Sissler M, Florentz C (1998) Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res 26: 5017–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann S, Haebel PW, Metzinger L, Sutter M, Felden B, Ban N (2003) Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB. Nature 424: 699–703 [DOI] [PubMed] [Google Scholar]
- Hauenstein S, Zhang CM, Hou YM, Perona JJ (2004) Shape-selective RNA recognition by cysteinyl-tRNA synthetase. Nat Struct Mol Biol 11: 1134–1141 [DOI] [PubMed] [Google Scholar]
- Helm M, Brule H, Friede D, Giege R, Putz D, Florentz C (2000) Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA 6: 1356–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H, Hasegawa T, Ueda T, Watanabe K, Shimizu M (1990) Conversion of aminoacylation specificity from tRNATyr to tRNASer in vitro. Nucleic Acids Res 18: 6815–6819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H, Yoshida S, Soma A, Nishikawa K (1997) Only one nucleotide insertion to the long variable arm confers an efficient serine acceptor activity upon Saccharomyces cerevisiae tRNALeu in vitro. J Mol Biol 268: 704–711 [DOI] [PubMed] [Google Scholar]
- Ibba M, Soll D (1999) Quality control mechanisms during translation. Science 286: 1893–1897 [DOI] [PubMed] [Google Scholar]
- Ibba M, Soll D (2000) Aminoacyl-tRNA synthesis. Annu Rev Biochem 69: 617–650 [DOI] [PubMed] [Google Scholar]
- Ishitani R, Nureki O, Nameki N, Okada N, Nishimura S, Yokoyama S (2003) Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell 113: 383–394 [DOI] [PubMed] [Google Scholar]
- Korencic D, Polycarpo C, Weygand-Durasevic I, Soll D (2004) Differential modes of transfer RNASer recognition in Methanosarcina barkeri. J Biol Chem 279: 48780–48786 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Lenhard B, Orellana O, Ibba M, Weygand-Durasevic I (1999) tRNA recognition and evolution of determinants in seryl-tRNA synthesis. Nucleic Acids Res 27: 721–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard B, Praetorius-Ibba M, Filipic S, Soll D, Weygand-Durasevic I (1998) C-terminal truncation of yeast SerRS is toxic for Saccharomyces cerevisiae due to altered mechanism of substrate recognition. FEBS Lett 439: 235–240 [DOI] [PubMed] [Google Scholar]
- Li X, Fischel-Ghodsian N, Schwartz F, Yan Q, Friedman RA, Guan MX (2004) Biochemical characterization of the mitochondrial tRNASer(UCN) T7511C mutation associated with nonsyndromic deafness. Nucleic Acids Res 32: 867–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma PC, Rould MA, Weintraub H, Pabo CO (1994) Crystal structure of MyoD bHLH domain–DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell 77: 451–459 [DOI] [PubMed] [Google Scholar]
- Murshudov GN (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nagaike T, Suzuki T, Tomari Y, Takemoto-Hori C, Negayama F, Watanabe K, Ueda T (2001) Identification and characterization of mammalian mitochondrial tRNA nucleotidyltransferases. J Biol Chem 276: 40041–40049 [DOI] [PubMed] [Google Scholar]
- Navaza J (2001) Implementation of molecular replacement in AMoRe. Acta Crystallogr D 57: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Normanly J, Ollick T, Abelson J (1992) Eight base changes are sufficient to convert a leucine-inserting tRNA into a serine-inserting tRNA. Proc Natl Acad Sci USA 89: 5680–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS (1999) Automated protein model building combined with iterative structure refinement. Nat Struct Biol 6: 458–463 [DOI] [PubMed] [Google Scholar]
- Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, Klug A (1974) Structure of yeast phenylalanine tRNA at 3 A resolution. Nature 250: 546–551 [DOI] [PubMed] [Google Scholar]
- Roussel A, Cambillau C (1991) Turbo Frodo: Silicon Graphics Geometry. Mountain View, CA: Silicon Graphics [Google Scholar]
- Sampson JR, Saks ME (1993) Contributions of discrete tRNASer domains to aminoacylation by E. coli seryl-tRNA synthetase: a kinetic analysis using model RNA substrates. Nucleic Acids Res 21: 4467–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada N, Suzuki T, Watanabe K (2001) Dual mode recognition of two isoacceptor tRNAs by mammalian mitochondrial seryl-tRNA synthetase. J Biol Chem 276: 46770–46778 [DOI] [PubMed] [Google Scholar]
- Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S (1998) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 26: 148–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S, Gautheret D, Cedergren R (1994) Fitting the structurally diverse animal mitochondrial tRNAsSer to common three-dimensional constraints. J Mol Biol 236: 982–989 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Wada T, Saigo K, Watanabe K (2002) Taurine as a constituent of mitochondrial tRNAs: new insights into the functions of taurine and human mitochondrial diseases. EMBO J 21: 6581–6589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toompuu M, Yasukawa T, Suzuki T, Hakkinen T, Spelbrink JN, Watanabe K, Jacobs HT (2002) The 7472insC mitochondrial DNA mutation impairs the synthesis and extent of aminoacylation of tRNASer(UCN) but not its structure or rate of turnover. J Biol Chem 277: 22240–22250 [DOI] [PubMed] [Google Scholar]
- Vriend G (1990) WHAT IF: a molecular modeling and drug design program. J Mol Graph 8: 29, 52–56 [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Kawai G, Yokogawa T, Hayashi N, Kumazawa Y, Ueda T, Nishikawa K, Hirao I, Miura K, Watanabe K (1994) Higher-order structure of bovine mitochondrial tRNA(SerUGA): chemical modification and computer modeling. Nucleic Acids Res 22: 5378–5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokogawa T, Shimada N, Takeuchi N, Benkowski L, Suzuki T, Omori A, Ueda T, Nishikawa K, Spremulli LL, Watanabe K (2000) Characterization and tRNA recognition of mammalian mitochondrial seryl-tRNA synthetase. J Biol Chem 275: 19913–19920 [DOI] [PubMed] [Google Scholar]
- Yokogawa T, Watanabe Y, Kumazawa Y, Ueda T, Hirao I, Miura K, Watanabe K (1991) A novel cloverleaf structure found in mammalian mitochondrial tRNA(Ser) (UCN). Nucleic Acids Res 19: 6101–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


