Abstract
The Frizzled (Fz) receptors contain seven transmembrane helices and an amino-terminal cysteine-rich domain (CRD) that is sufficient and necessary for binding of the ligands, the Wnts. Recent genetic experiments have suggested, however, that the CRD is dispensable for signaling. We engineered fz CRD mutant transgenes and tested them for Wg signaling activity. None of the mutants was functional in cell culture or could fully replace fz in vivo. We also show that replacing the CRD with a structurally distinct Wnt-binding domain, the Wnt inhibitory factor, reconstitutes a functional Wg receptor. We therefore hypothesized that the function of the CRD is to bring Wg in close proximity with the membrane portion of the receptor. We tested this model by substituting Wg itself for the CRD, a manipulation that results in a constitutively active receptor. We propose that Fz activates signaling in two steps: Fz uses its CRD to capture Wg, and once bound Wg interacts with the membrane portion of the receptor to initiate signaling.
Keywords: Frizzled, signaling, Wingless, Wnt
Introduction
Owing to their fundamental role in development and disease, it is of great interest to elucidate how Wnt proteins activate intracellular signaling. Recently, much progress has been made in identifying cytoplasmic signaling components that act downstream of the Wnt receptor Frizzled (Fz), which is responsible for regulating the stability of β-catenin, the main effector of Wnt signaling. In comparison, relatively little is known about the initial events that occur at the membrane, including which structural features of Fz are required to activate the cytoplasmic signaling machinery (Veeman et al, 2003a; Logan and Nusse, 2004).
In Drosophila, Wg signaling through Armadillo (Arm, Drosophila β-catenin) is required for patterning embryos (reviewed in Logan and Nusse, 2004; Tolwinski and Wieschaus, 2004; Bejsovec, 2005). The embryonic epidermis secretes a cuticle with groups of cells that produce hair-like projections, called denticles, separated by those that produce smooth or ‘naked' cuticle. wg signaling promotes the formation of naked cuticle by promoting the post-translational stabilization of the transcriptional coactivator, Arm. As Arm protein accumulates in the cytoplasm and nucleus, it interacts with the TCF/LEF transcription factor Pangolin (Pan) to regulate target genes responsible for cell fate changes. In Drosophila, fz acts redundantly with another member of the Fz family, frizzled2 (fz2) as a receptor for Wg. Either fz or fz2 (Bhanot et al, 1996; Kennerdell and Carthew, 1998; Bhanot et al, 1999; Chen and Struhl, 1999), together with the membrane component, arrow (arr), form a receptor complex for the Wg protein (Wehrli et al, 2000). Embryos lacking wg, arr or both fz and fz2 cannot initiate Arm signaling and, thus, are completely covered by denticles.
All Fz receptors contain within their extracellular portion a region called the cysteine-rich domain (CRD), named for its invariant pattern of 10 cysteine residues. Furthermore, a domain with homology to the CRD is present on a receptor involved in Hedgehog signaling, Smoothened (Smo; Alcedo et al, 1996; van den Heuvel and Ingham, 1996) and in the receptor tyrosine kinase Ror2, which is proposed to activate Arm/β-catenin-independent (noncanonical) Wnt signaling (Oishi et al, 2003). The CRD of Fz has been crystallized (Dann et al, 2001) and binds Wnt proteins with nanomolar affinity (Hsieh et al, 1999b; Wu and Nusse, 2002). Several mutations in a fz CRD have been engineered that affect Wnt binding (Hsieh et al, 1999b). Given that CRD domains confer Wnt binding, it was unexpected that fz transgenes lacking the CRD were reported to respond normally to Wg and activate Arm signaling in vivo (Chen et al, 2004). What is the function of this highly conserved portion of fz? We addressed this question by testing a set of CRD variants for Arm signaling in cell culture and in vivo.
Results
Generating fz transgenes to test for CRD requirements
Studies in different experimental systems have addressed the role for the CRD in Wnt signaling, but with conflicting outcomes. It was initially found that the CRD of the fz receptor is necessary for Wnt binding (Bhanot et al, 1996; Hsieh et al, 1999b; Dann et al, 2001; Wu and Nusse, 2002). In addition, expression of the CRD alone acts as a dominant negative, indicating that this portion of fz can nonproductively bind Wnt ligands, limiting the available pool for endogenous receptors (Cadigan et al, 1998). However, the role of the CRD as the sole determinant of Wnt binding has been recently challenged by experiments showing that Fz transgenes lacking a CRD are still capable of functioning in Arm signaling (Chen et al, 2004).
To address the function of the CRD, we made a series of modified fz transgenes (Figure 1A). We engineered two mutations into the CRD of fz that are predicted to disrupt specifically Wnt binding, but otherwise leave the protein intact (Hsieh et al, 1999b). These mutations (fz57 and fz81) insert three amino acids (GSG) (Figure 1B) that map to the surface of the CRD folded structure (Dann et al, 2001). We also constructed a form of fz in which the entire CRD was deleted (fzΔCRD1) and two forms where it was replaced by another domain. In one case, we exchanged the fz CRD with the CRD of Drosophila smoothened (fzSMO), a domain that does not bind Wg (Wu and Nusse, 2002). In the other case, we substituted the Wnt inhibitory factor (WIF) domain of the human WIF (fzWIF), a secreted molecule that can bind to Wnt proteins, including Wg (Hsieh et al, 1999b). In addition, we generated transgenes where the Drosophila Wnt genes, wg and wntD (CG8458 formerly wnt8) (Ganguly et al, 2005; Gordon et al, 2005), are substituted for the CRD (wg∷fzΔCRD and wntD∷fzΔCRD). Lastly, we obtained two previously described CRD deletions of fz2 and fz (fz2ΔCRD and fzΔCRD2) (Chen et al, 2004). The fzΔCRD2 transgene differs from the version we engineered (fzΔCRD1) in three ways: fzΔCRD2 uses the signal sequence of wg, it is tagged with three copies of the Flu epitope and leaves eight amino acids of the CRD including the 10th conserved cysteine residue. For comparison, we made a modified form of fzΔCRD2 that eliminates the eight amino acids, but preserves the wg signal sequence and Flu epitopes (fzΔCRD3) (Figure 1B).
Figure 1.
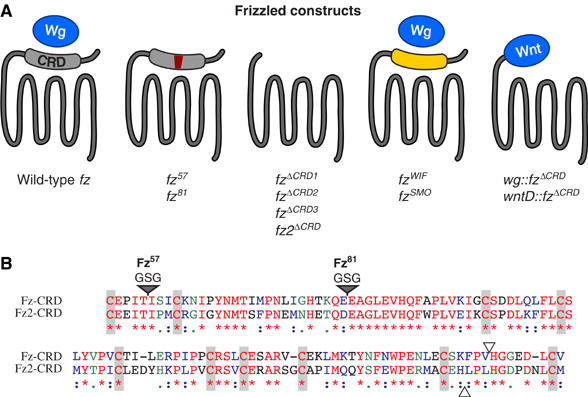
Design fz CRD variants. (A) To test the role of the CRD in Arm signaling, we engineered the following fz constructs into a vector for constitutive expression in Drosophila via the tubulin-α1 promoter (pTub): Wild-type fz (581 amino acids). fz57 and fz81 insert three amino acids (GSG) between residues in the CRD and are predicted to interfere with Wg binding (Hsieh et al, 1999b). fzΔCRD1 specifically deletes the CRD in-frame (amino acids 53–164). This leaves the remainder of the receptor intact, including the native signal sequence and the entire extracellular ‘hinge' region lying between the CRD and the first transmembrane domain. fzΔCRD2 and fz2ΔCRD replace the entire N-terminal portions (up to the open triangles shown in panel B), including the native signal sequences of fz and fz2 with the wg signal sequence and three Flu epitopes (Chen et al, 2004). fzΔCRD3 specifically removes the eight amino acids of the CRD present in fzΔCRD2 but absent in fzΔCRD1, fzWIF and fzSMO specifically exchange the CRD with the WIF domain of hWIF (Hsieh et al, 1999a) or the CRD of Drosophila smoothened (Alcedo et al, 1996; van den Heuvel and Ingham, 1996). wg∷fzΔCRD and wntD∷fzΔCRD fusions join the full-length Wg or WntD protein in-frame with the region of Fz C-terminal to the CRD in fzΔCRD1 (amino acids 165–581). Wnt-Fz fusions use the signal sequence native to their respective Wnt protein. (B) Alignment of the Drosophila Fz and Fz2 amino-acid sequence within the CRD. The 10 cysteine residues defining this domain are indicated by shaded vertical bars. Identical amino acids are indicated in red (*). Amino acids with conserved properties are indicated in purple (:) and green (.). Two mutations previously characterized to disrupt the binding between Xenopus Wnt8 and Fz2 CRD (Hsieh et al, 1999b) were engineered into a homologous position in the Fz CRD (gray triangles). FzΔCRD1 removes the entire region shown, leaving the flanking N- and C-terminal sequences unmodified. The breakpoints of FzΔCRD2 and Fz2ΔCRD are indicated with open triangles. FzΔCRD2 includes eight amino acids not present in FzΔCRD1. These eight amino acids are deleted in FzΔCRD3 with the remainder of the construct identical to FzΔCRD2.
Expression of fz transgenes in S2 cells
To characterize initially our constructs, we expressed them in Drosophila S2 cells. S2 cells are well suited for studying fz-mediated Arm signaling since they lack endogenous fz and wg expression, providing a clean background (Bhanot et al, 1996; Sato et al, 1999). We performed a Western blot of whole-cell extracts made from cells transfected with fz variants to determine whether the transgenes produce the expected proteins. Probing the samples with an antibody directed against an epitope in the extracellular ‘hinge' region present in all variants (except for fz2ΔCRD) showed that all proteins are synthesized (Figure 2A). When we compared abundance for the three fzΔCRD variants, we observed there was less FzΔCRD2 protein synthesized compared to either FzΔCRD1 or FzΔCRD3. The same difference in abundance between FzΔCRD2 and FzΔCRD3 was observed when extracts were probed with an anti-Flu antibody, which additionally shows that fz2ΔCRD is abundantly produced (Figure 2A).
Figure 2.
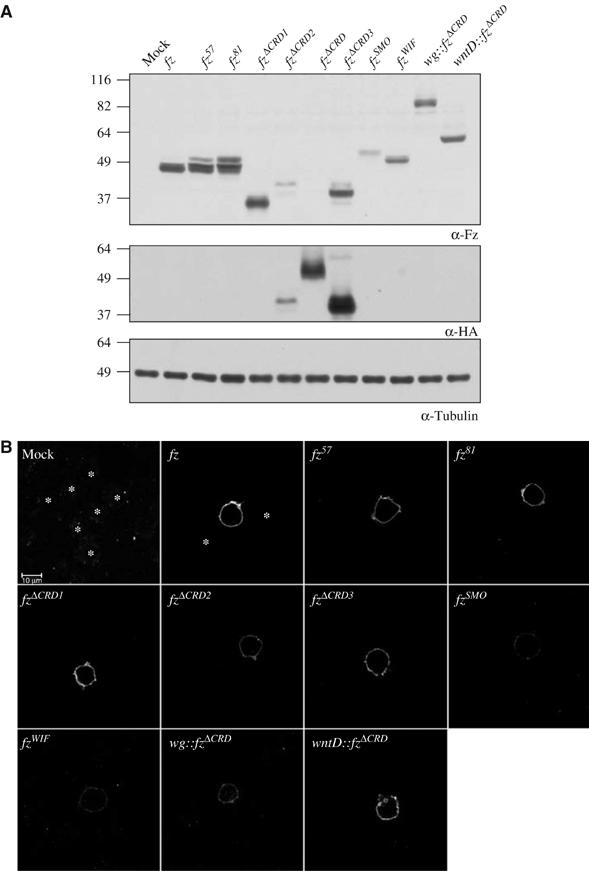
Fz protein abundance and surface localization in S2 cells. (A) Whole-cell protein extracts of S2 cells transfected with fz transgenes probed with a polyclonal antibody directed against the ‘hinge' region of Fz (α-Fz, top panel), an epitope, which Fz2 lacks. When the same samples are probed with an HA antibody, we observed that Fz2ΔCRD is expressed comparably to FzΔCRD3, which are both expressed at higher levels than FzΔCRD2. The molecular mass in kDa of a known protein standard is indicated. (B) Cell surface localization of Fz protein was assayed in S2 cells transfected with fz transgenes and stained using an antibody directed against the ‘hinge' region of Fz. Fz2 was not tested as it lacks this epitope. The same settings were used for all samples aside from mock-transfected cells where the gain was intentionally set higher to emphasize lack of signal. Staining is specific, as untransfected cells (*) are not detectable. The scale bar in upper left panel is 10 μm.
To determine whether the fz transgenes we constructed produce proteins that are localized to the plasma membrane, we transfected cells and immunostained them with our extracellular fz antibody (Figure 2B). There is no specific signal observed in untransfected or mock-transfected cells (Figure 2B). Like wild-type Fz, all Fz variants are targeted to the cell surface. The abundance of each Fz protein on the membrane correlates with the protein level observed in whole-cell extracts. Therefore, similar to wild type, all Fz variants are synthesized and targeted to the plasma membrane.
fz transgenes with CRD mutations are compromised for Arm signaling in cell culture
As a measure of signaling, we tested transcriptional activation of Arm signaling by a reporter assay (Figure 3A). S2 cells were transiently transfected with a TCF/LEF-dependent luciferase reporter (Veeman et al, 2003b) and an fz transgene alone or together with a wg transgene. None of the fz transgenes activated the reporter in the absence of wg showing, as reported previously, that the Fz protein does not activate signaling in the absence of a Wnt (Bhanot et al, 1996). Expression of wg alone did not substantially activate the reporter (1.7±0.3-fold). Cells transfected with both wild-type fz and wg potently activated the reporter (80.6±0.3-fold). In comparison, cells expressing wg and either fz57 or fz81 show a dramatic reduction in their response, with fz81 being more compromised than fz57 (29.8±3.1- and 43.9±3.4-fold, respectively). Cells expressing fzΔCRD1 or fzSMO only modestly elevated the reporter in the presence of wg (2.6±0.3- and 3.9±0.3-fold). The fz2ΔCRD moderately activated the reporter when cotransfected with wg (15.4±2.1-fold). Interestingly, fzΔCRD2 displayed a marked increase in wg-dependent reporter activation compared to fzΔCRD1 (7.6±1.0- versus 2.6±0.3-fold). Either the wg signal sequence or Flu tags are responsible for the enhanced wg responsiveness since fzΔCRD3 shows a similar elevation (8.0±1.7-fold) compared to fzΔCRD1. Taken together, these data show that the CRD of fz is required for robust signaling.
Figure 3.
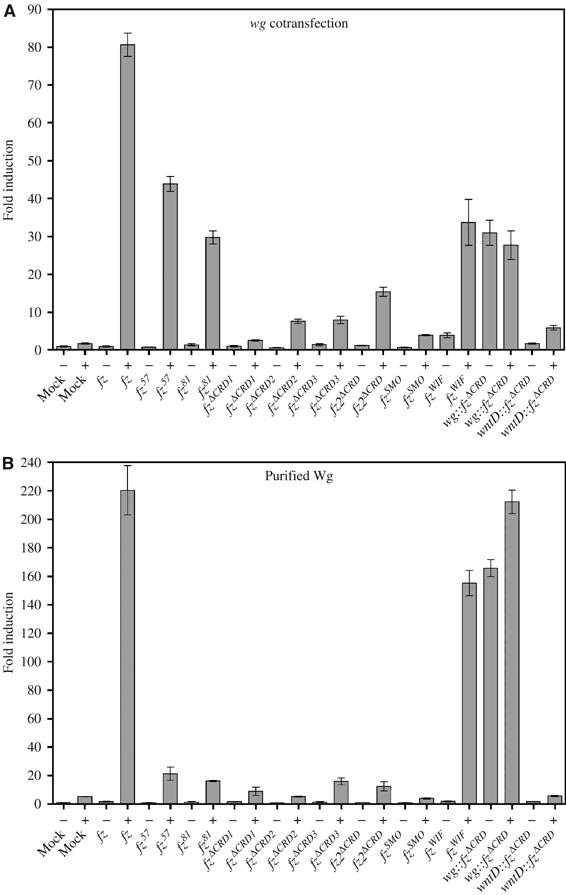
Arm signaling of fz CRD variants in S2 cells. (A) To measure Arm signaling, TCF/LEF-dependent luciferase reporter assays were performed on cells cotransfected with pTub (Mock) or a pTub-fz variant and empty pTub (−) or pTub-wg (+). (B) TCF/LEF-dependent luciferase reporter assays were performed on cells cotransfected with pTub (Mock) or a pTub-fz variant and challenged with plain medium (−) or medium containing purified Wg (+).
Importantly, expression of fzWIF, which adds back to the fzΔCRD1 receptor a Wnt-binding domain, partially restored wg responsiveness (33.7±10.5-fold). Expression of the wg∷fzΔCRD fusion transgene alone led to the activation of the reporter (31.0±5.8-fold) that was not enhanced by cotransfection of wg (27.7±6.6-fold). Activation is specific to the wg∷fzΔCRD since another wnt-fz fusion, wntD∷fzΔCRD, did not activate the reporter on its own (1.7±0.2-fold) and showed only minimal activation in the presence of wg (5.9±1.0-fold). To determine whether the fz CRD variants can interact with Wg protein, we transfected the receptor transgenes with a TCF/LEF-dependent reporter, and after allowing time for protein expression, we challenged the cells with Wg protein purified from conditioned medium of S2 cells constitutively expressing wg (Figure 3B) (Willert et al, 2003). We observed a similar trend with cotransfection. However, whereas the cells transfected with wild-type fz more robustly activated Arm signaling in this assay (220.5±24.4-fold), cells transfected with fz57 and fz81 showed a diminished capacity to activate the reporter (21.4±6.5- and 16.2±0.4-fold, respectively). Importantly, all recombinant Fz proteins localize to the cell surface (Figure 2B, fz2ΔCRD was not tested), which suggests that in the cotransfection assay, the Fz57 and Fz81 proteins might in part interact with Wg protein in an intracellular compartment during synthesis, which might augment an otherwise weak interaction. The CRD deletion variants and fzSMO behave similarly in this assay and the cotransfection assay. Strikingly, cells expressing fzWIF are significantly more responsive to purified soluble Wg protein than cotransfected wg (155±12.7 and 33.7±10.5, respectively). wg∷fzΔCRD-expressing cells showed a modest increase in reporter activation in the presence of soluble Wg protein (165.9±8.4 versus 212.3±11.7). Cells expressing wntD∷fzΔCRD failed to activate the reporter in the absence or presence of Wg protein.
wg∷fzΔCRD requires arr to activate Arm signaling
Wg is thought to activate Arm signaling by bridging the two membrane coreceptors Fz and Arr and bringing together their associated intracellular signaling components (Tamai et al, 2000; Tolwinski et al, 2003; Cong et al, 2004). It has been reported that lowering the amount of Arr protein by RNA interference (RNAi) interferes with Arm signaling in cell culture (Schweizer and Varmus, 2003). As shown above, expression of wg∷fzΔCRD constitutively activates Arm signaling. We therefore asked whether the Wg∷FzΔCRD protein requires Arr to elicit signaling. We performed TCF/LEF reporter assays on S2 cells expressing either wg∷fzΔCRD or wntD∷fzΔCRD and lowered the amount of Arr protein by RNAi (Figure 4). This results in a 3.5-fold reduction in reporter activation in cells expressing wg∷fzΔCRD. Thus, the Wg∷FzΔCRD protein requires the Arr coreceptor to constitutively activate Arm signaling.
Figure 4.
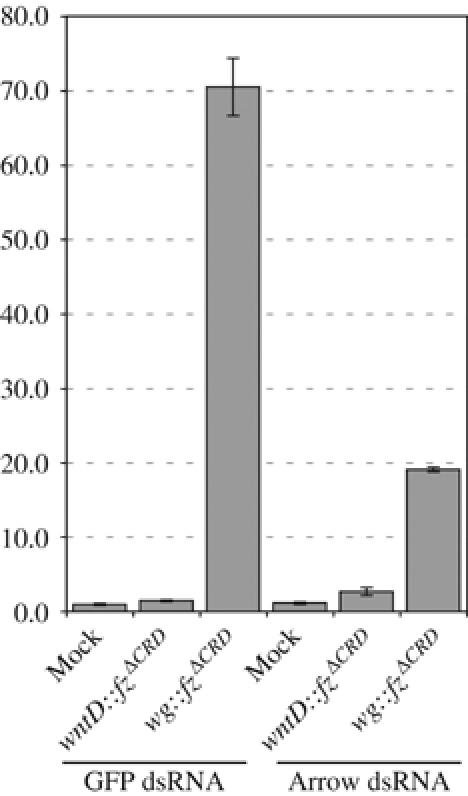
wg∷fzΔCRD requires arr to activate Arm signaling in S2 cells. TCF/LEF reporter assays on two sets of cells transfected with empty plasmid (Mock), wntD∷fzΔCRD or wg∷fzΔCRD transgenes. One set was cotransfected with control double-stranded RNA (GFP dsRNA) and the second set was cotransfected with arr dsRNA (Arrow dsRNA). There is a 3.7-fold reduction in signaling when cells are cotransfected with arr dsRNA compared to the control dsRNA.
fz transgenes with altered CRDs are compromised for Arm signaling in vivo
To assess whether CRD mutations interfere with Arm signaling mediated by Wg in vivo, we tested a subset of the transgenes to determine whether they could rescue fz,fz2 mutant embryos. We removed fz and fz2 by making germline clones (Perrimon et al, 1996). These mutant embryos exhibit defects in the specification of naked cuticle, indicating loss of Arm signaling (compare Figure 5A and B). When ubiquitously expressed throughout the embryo, wild-type fz is fully capable of restoring normal patterning to the mutant embryos (Figure 5C). In fz57, fz81 and fzΔCRD rescue crosses, denticles are present in regions that should be entirely naked. We also found fusions of denticle belts (Figure 5D–F). Consistent with the TCF/LEF reporter assays for fz function (Figure 3), the ability of fz57 to rescue the mutant embryos was stronger than the two other transgenes, but rescue was not complete (Figure 5D). fz81 and fzΔCRD1 gave incomplete rescue, although in each case the resulting cuticle phenotype was less severe than the fz,fz2 mutant.
Figure 5.
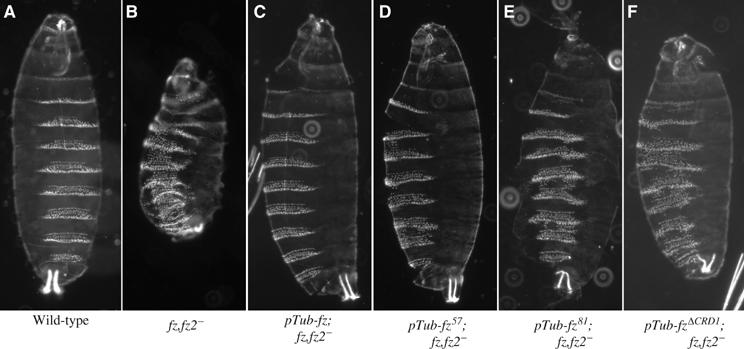
fz CRD mutations show Arm signaling defects in embryos. (A) Cuticle of a wild-type Drosophila embryo. (B) fz,fz2 germline clone zygotically mutant for fz and fz2. The following panels are the same genotype as in (B), zygotically expressing via the tubulin-1α promoter (pTub) one copy of (C) wild-type fz, (D) fz57, (E) fz81 and (F) fzΔCRD1. For each of the transgenes, one insert line was tested.
We also tested whether the transgenes could rescue lethality caused by the absence of endogenous fz-mediated Arm signaling. Only a wild-type fz transgene is capable of rescuing fz,fz2 germline clones to adulthood (Figure 5C, Table I). In contrast, the CRD mutant transgenes were incapable of restoring viability to fz,fz2 mutants (Table I).
Table 1.
Rescue of viability in fz,fz2− germline clones
| Rescue construct | n | % |
|---|---|---|
| pTub-fz | 71 | 11 |
| pTub-fz57 | 43 | 0 |
| pTub-fz81 | 42 | 0 |
| pTub-fzΔCRD1 | 60 | 0 |
In addition, we tested whether fz transgenes carrying CRD mutations can rescue Arm signaling in the Drosophila wing. Activation of Arm signaling by wg and fz is required for the formation of bristles that normally pattern the margin of the wing (Chen and Struhl, 1999). In addition, fz has another, wg-independent function that is unrelated to Arm signaling. It is responsible for the planar cell polarization (PCP) of Drosophila tissues including the hairs and bristles of the wing and clones of cells mutant for fz also nonautonomously affect the polarization of surrounding wild-type cells (Vinson and Adler, 1987). Wing margin bristles made by fz mutant clones have an abnormal elevation, rotated away from the margin (Figure 6, compare panel C with A and B). Homozygous clones for fz and fz2 are identifiable by the recessive pigmentation marker, yellow (y) (open arrows), and develop unpigmented (yellow) margin bristles where heterozygous tissue produces darker pigmented bristles (closed arrows). As in the embryo, fz and fz2 act redundantly as the receptor for Wg. Similar to control clones (Figure 6A), clones of cells mutant for either fz or fz2 alone (Figure 6B and C) are still capable of Wg-mediated activation of Arm signaling as evidenced by numerous yellow bristles. Clones of cells lacking both fz and fz2 fail to generate yellow bristles and often lack bristles altogether (Figure 6D) (Chen and Struhl, 1999). Loss of Arm signaling, such as what occurs in fz,fz2 mutant clones, results in the ectopic expression of wg (Rulifson et al, 1996), which can ultimately result in the formation of an ectopic bristle due to the inappropriate activation of Arm signaling in heterozygous cells neighboring the clone (arrowheads) (Chen and Struhl, 1999). Uniform expression of wild-type fz in the wing completely restores the patterning to fz fz2 mutant cells present at the margin (Figure 6E). Importantly, in contrast to wild-type fz, expression of the CRD mutants fz57, fz81 and fzΔCRD1 does not fully restore patterning to fz,fz2 mutant clones (Figure 6F–H). In addition to polarization defects, the Arm signaling defects manifest as abnormal bristle spacing and ectopic bristles near the margin. However, the order of the Arm signaling strength of the CRD mutants deviated from that observed in the embryo and cell culture. In the latter assays, rescue of Arm signaling by fz81 was less efficient than either fz57 or fzΔCRD1, which rescue Arm signaling to a similar degree. As was the case in the embryo, all of the CRD mutants were able to significantly, but not fully rescue Arm signaling.
Figure 6.
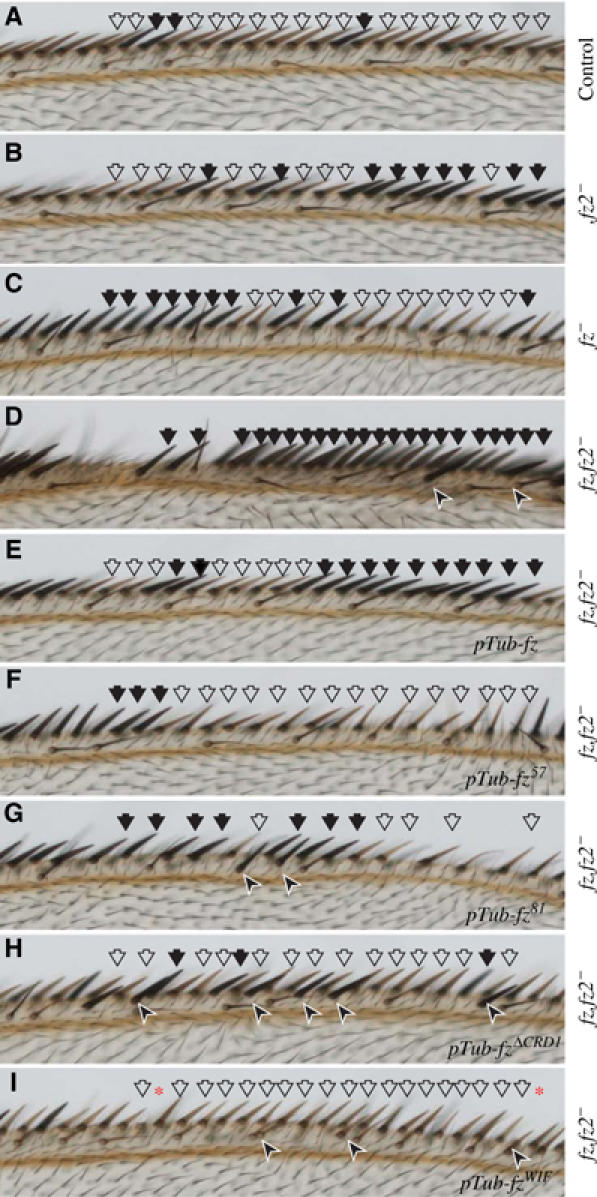
fz CRD mutations show Arm signaling defects in wings. Shown here are the wing margins of flies possessing different clones as indicated by the vertical label to the right of each panel. Clones that cross the margin elaborate yellow bristles due to the loss of the y+ marker and are indicated with open arrows. Darker bristles that are elaborated by heterozygous cells are indicated with closed arrows. Arrowheads indicate the presence of a bristle formed inappropriately away from the margin. (A) Control clones that do not carry any mutations produce numerous yellow bristles, similar to clones of either fz2− (B) or fz− (C). (D) fz,fz2− clones do not generate yellow margin bristles and have large patches along the margin lacking bristles. The bristles generated by surrounding heterozygous tissue show tighter spacing and occasional ectopic bristles that form away from the margin (arrowheads). (E) The margin gaps and ectopic bristle phenotypes of fz,fz2− clones are fully rescued by the ubiquitous expression of one copy of wild-type fz. These wings generate many yellow margin bristles. (F) fz,fz2− clones rescued by fz57 have subtle defects in the spacing of the margin bristles. (G) fz,fz2− clones rescued by fz81 have fewer yellow margin bristles and more severe bristle spacing problems. (H) fz,fz2− clones rescued by fzΔCRD1 have ectopic margin bristles, which are almost invariably dark, indicating they are derived from nearby heterozygous cells. (I) fz,fz2− clones rescued by fzWIF produce numerous yellow bristles. There are places along the margin where a bristle appears to be missing. However, unlike the defects in bristle spacing seen with fz57 and fz81 rescued clones, in fzWIF rescued clones a structure resembling a bristle socket forms in the place of the apparently missing bristle (*). For each of the transgenes, two separate insert lines were tested.
fz transgenes with CRD mutations have Arm signaling defects. Does adding back a Wnt-binding domain restore full function? Expression of fzWIF rescues fz,fz2 clones, although rescue is incomplete, with observable defects in the patterning of the margin (Figure 6I). In some cases, a bristle within a clone of mutant tissue is missing from the margin, appearing to leave a gap. However, these instances differ qualitatively from gaps seen with the expression of the CRD mutants since in clones rescued by fzWIF the socket for the absent bristle is formed (*). Therefore, fzWIF is capable of restoring Arm signaling to cells mutant for fz and fz2 and rescues more efficiently than the fz mutants, but it is not as efficient as wild-type fz.
Overexpresion of fzWIF and wg∷fzΔCRD activate Arm signaling in vivo
In addition to rescue of fz,fz2 mutant clones in the wing, we overexpressed fzWIF in the wing using the Gal4/UAS system (Brand and Perrimon, 1993) to assess whether it is capable of signaling in vivo. Interestingly, overexpression of fzWIF throughout the wing blade results in a global disruption of PCP and the formation of ectopic bristles near the wing margin, a gain-of-function Arm signaling phenotype (Figure 7A and B). Bristle formation near the source of Wg suggests that fzWIF requires a Wnt ligand to activate Arm signaling in vivo, as it does in vitro (Figure 3). Interestingly, the phenotypes observed following fzWIF overexpression are the composite of the individual overexpression phenotypes of wild-type fz and fz2, which lead exclusively to disruption of PCP and generation of ectopic bristles, respectively (Figure 7C and D) (Zhang and Carthew, 1998). The presence of ectopic bristles shows that fzWIF is capable of activating Arm signaling via Wg in vivo as it does in cell culture.
Figure 7.
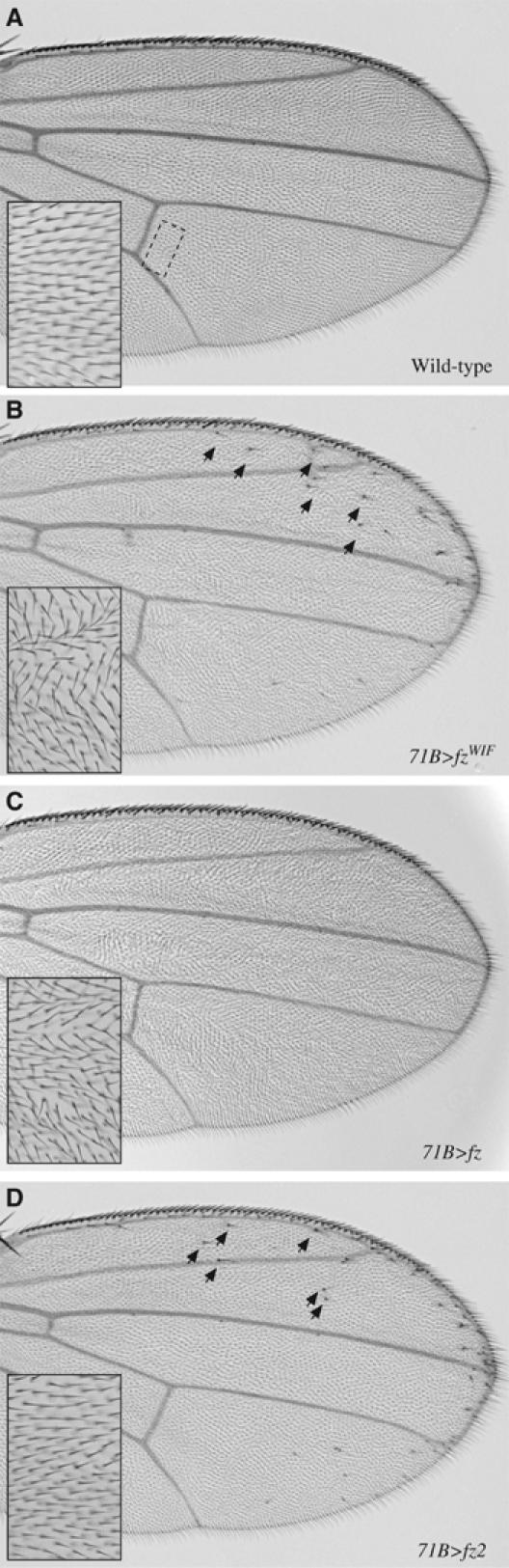
fzWIF activates Arm signaling in vivo. (A) Picture of a wild-type Drosophila wing. The boxed region distal to the posterior crossvein indicates the location of the inset in all panels. Note all wing hairs uniformly point towards the distal portion of the wing. (B) Overexpression of fzWIF generates both Arm-dependent ectopic bristles near the wing margin (arrows) and Arm-independent PCP defects throughout the wing. (C) Overexpression of fz alone only leads to a PCP disruption throughout the wing. (D) Overexpression of fz2 only produces ectopic bristles near the wing margin (arrows).
Overactivation of Arm signaling throughout the entire wing, as caused by overexpression of wg and wg∷fzΔCRD using the same driver as above, leads to an overall disruption of normal development preventing the assay of ectopic bristle formation (data not shown). Therefore, we returned to Drosophila embryos to ask whether overexpression of wg∷fzΔCRD activates Arm signaling in vivo. wg∷fzΔCRD and wntD∷fzΔCRD were expressed in the domain of the gene hairy (h), which is expressed in alternating segments. Importantly, similar to overexpression of wild-type wg, overexpression of wg∷fzΔCRD leads to the activation of Arm signaling resulting in the formation of ectopic naked cuticle (Figure 8B and C). As a control, expression of wntD or wntD∷fzΔCRD did not promote naked cuticle formation (Figure 8D and E). Thus, both fzWIF and wg∷fzΔCRD can activate Arm signaling when overexpressed in vivo. However, whereas fzWIF requires the endogenous Wg protein (in cell culture and in the wing tissue, where ectopic bristles form close to th Wg source at the margin), wg∷fzΔCRD constitutively activates Arm signaling.
Figure 8.
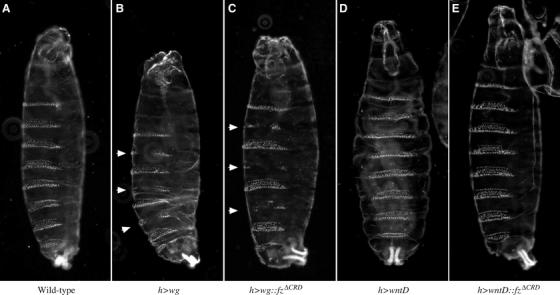
wg∷fzΔCRD activates Arm signaling in vivo. (A) Wild-type cuticle pattern displayed by an embryo with one copy of the hairy-gal4 (h) driver but lacking a UAS transgene. (B) Expression of wg in alternating segments results in the formation of ectopic naked cuticle (arrows). (C) Expression of wg∷fzΔCRD results in the formation of ectopic naked cuticle (arrows). Either expression of wntD (D) or wntD∷fzΔCRD (E) does not result in naked cuticle formation.
Discussion
What is the function of the Fz CRD in Arm signaling?
Our principle finding is that the Fz CRD is required for efficient Arm signaling. Fz transgenes carrying CRD mutations have compromised Arm signaling function in cell culture and cannot fully restore Arm signaling to fz,fz2 mutants in vivo. In addition, we found that adding a heterologous Wnt-binding domain (WIF) to a CRD-deleted fz restores its ability to activate Arm signaling via Wg in cell culture. Based on the manipulations and results, we hypothesized that the function of the CRD is to bring Wg in close proximity with the membrane portion of the receptor, a function that can be taken over by other Wnt-binding domains. We tested this idea by creating a transgene fusing Wg to Fz, eliminating the CRD in the process, which results in a constitutively active receptor.
While both in vivo and in vitro tests reveal that mutants with a defective Wnt interaction domain are compromised for Arm signaling, the requirement for the CRD is most evident in cell culture where all of the mutants show a reduced activity, particularly the one where the entire CRD is lacking. In the cell culture experiments, where we can measure the Wg signaling in a quantitative manner, we find a range of responses to the CRD mutants (Figure 3) corresponding to the differences in Wnt-binding strength (Hsieh et al, 1999b). In vivo, by examining cuticles and the abilities of the CRD mutants to restore signaling, we also notice a range of phenotypes (Figure 5). While these rescue data are more difficult to measure, the phenotypes correspond in strength to the in vitro signaling levels. We infer from this relationship that signaling operates through the same mechanism in vivo as in cell culture. As an extension of this argument, we suggest that the CRD plays a similar role in cell culture as in the embryo. However, signaling in vivo is less stringently dependent on the presence of the CRD, suggesting that its absence is being compensated for by other factors. If the function of the CRD (or other Wnt-binding domains such as the WIF) is, as we propose, to bring Wg in close proximity to the membrane domain of Fz, it is possible this function is taken over by other molecules acting in trans and that these factors are not present in vitro. Candidates for such molecules are members of the CRD containing ROR family and the RYK receptor tyrosine kinase, which has a WIF domain (Yoshikawa et al, 2003; Forrester et al, 2004; Inoue et al, 2004; Lu et al, 2004). It is also possible that extracellular matrix molecules provide such an accessory function, by presenting or concentrating Wg close to the Fz signaling domain.
Is the only function of the CRD (or another Wg-binding domain, such as WIF) to capture Wg and to present it to the coreceptor Arrow? In that view, there would be no need for the seven-transmembrane domain of the Fz receptors; Fz would solely act to promote Wg interacting with Arrow. We find this unlikely; there are several studies that point to a requirement of specific residues in the Fz membrane domain in signaling (Umbhauer et al, 2000; Cong et al, 2004; Toomes et al, 2004; Povelones et al, 2005). Mutations in those residues, either engineered or present in natural alleles, disrupt signaling. In addition, it has been recently proposed that in Drosophila, fz activates PCP and Arm signaling through heterotrimeric G proteins (Katanaev et al, 2005). Finally, expressing the CRD on the cells surface as a GPI-linked membrane molecule does not promote signaling, but instead acts as a dominant negative (Cadigan et al, 1998). Taken together, these data suggest that the transmembrane portion of fz is a dynamic signal activating molecule and not merely a Wg presentation module.
Planar cell polarity phenotypes
As shown in Figure 7, overexpression of fzWIF in the Drosophila wing leads to both gain-of-function PCP and Arm signaling phenotypes. This is the composite of the consequences of fz and fz2 overexpression, which individually activate PCP and Arm signaling, respectively (Zhang and Carthew, 1998). There is much interest in determining how each receptor couples to a particular pathway (Adams et al, 2000; Boutros et al, 2000; Rulifson et al, 2000; Wu et al, 2004). Although there is some disagreement in these studies, it is generally concluded that the transmembrane portion of fz, including the cytoplasmic tail, couples it to PCP signaling. Since fzWIF contains this portion of fz, it is not surprising that it too affects PCP signaling. What structural feature of fz2 is responsible for coupling it exclusively to Arm signaling? We found that specifically replacing the fz CRD with the WIF domain results in a receptor that, like fz2, can activate Arm signaling. This finding is consistent with a study of fz/fz2 chimeras where the ability to activate Arm signaling was shown to be a property of the fz2 CRD (Rulifson et al, 2000). It was proposed that the feature conferring Arm coupling was the 10-fold higher affinity of the fz2 CRD for the Wg protein. By analogy, the WIF domain, like the fz2 CRD, may have a higher affinity for Wg than the fz CRD.
Materials and methods
Construction of pTub-fz transgenes
Wild-type Drosophila fz was PCR amplified using the following primers containing restriction sites for cloning:
5′-fz Spe I: GGACTAGTATGTGGCGTCAAATCCTG 3′-fz Xba I: GCTCTAGACTAGACGTACGCCTGCGC
The fz57, fz81 and fzΔCRD transgenes were constructed using Splicing by Overlapping Extension PCR (SOE–PCR). Two overlapping fragments (frag 1 and frag 2) with the specific lesion were generated in separate reactions using wild-type fz as a template. Frag 1 was generated using 5′-fz Spe I and 3′-fz frag 1 primers (below). Frag 2 was generated using 5′-fz frag 2 and 3′-fz Xba I primers. The two fragments were combined as a template in a third reaction with the primers 5′-fz Spe I and 3′-fz Xba I:
3′-fz57 frag 1 CTTGCAGATCGATATTCCGGATCCGGTGATGGGTT CACAGCG
5′-fz57 frag 2 CGCTGTGAACCCATCACCGGATCCGGAATATCGAT CTGCAAG
3′-fz81 frag 1 CCTCCAGACCCGCCTCTCCGGATCCCTCCTGCTTG GTATGGCC
5′-fz81 frag 2 GGCCATACCAAGCAGGAGGGATCCGGAGAGGCGG GTCTGGAGG
3′-fzΔCRD1 frag 1 GGTATTCTCCGCGCGATTGTGATGTGGCAGTCC
5′-fzΔCRD1 frag 2 CATCACAATCGCGCGGAGAATACCACATCATCG
We obtained the previously described pTub-fzΔCRD, pTub-fz2ΔCRD (Chen et al, 2004). The fzΔCRD transgene constructed by Chen et al (2004) (in this work called fzΔCRD2) differs from ours in three ways: it utilizes the signal sequence of wg, contains three Flu epitopes and includes eight more residues, including the 10th cysteine residue of the CRD. We modified the fzΔCRD2 transgene to remove the eight fz residues (157–164) and generate a molecule with the identical C-terminal portion as fzΔCRD1 (fzΔCRD3). The fzΔCRD3 transgene was constructed by SOE–PCR using fzΔCRD2 as a template. Since this construct uses the wg signal sequence, frag 1 was produced with 5′-wg Spe I and the 3′-frag 1 primer (below). Frag 2 was amplified using the 5′ primers listed below and 3′-fz Xba I. The full-length product was amplified with 5′-wg Spe I and 3′-fz Xba I:
3′-fzΔCRD3 frag1 GGTATTCTCCGCGAGGGGCAAGCTAGCGTAATC 5′-fzΔCRD3 frag2 AGCTTGCCCCTCGCGGAGAATACCACATCATCG
The wg∷fzΔCRD and wntD∷fzΔCRD transgenes were constructed by SOE–PCR using wg and wntD as a template for frag 1 and unique cloning primers containing a SpeI site. fz was the template for frag 2 using the primers below and the same 3′-fz Xba I primer as above:
5′-wg Spe I: GGACTAGTATGGATATCAGCTATATC 3′-wg∷fzΔCRD frag1: GGTATTCTCCGCCAGACACGTGTAGATGACC 5′-wg∷fzΔCRD frag2: TACACGTGTCTGGCGGAGAATACCACATCA TCG
5′-wntD Spe I: GGACTAGTATGATTTTTGCCATCACATTC 3′-wntD∷fzΔCRD frag1: GGTATTCTCCGCGTAGCAGGAGT ACTGCCTTTC 5′-wntD∷fzΔCRD frag2: TACTCCTGCTACGCGGAGAATAC CACATCATCG
The fzWIF and fzSMO transgenes were constructed by SOE–PCR using three overlapping fragments. The 5′-fz Spe I and 3′-fz Xba I primers were used to generate frag 1 and 3, respectively. These same primers were used to amplify the full-length product:
3′-fzWIF frag1: GGCCTCCGCCCGGCGATTGTGATGTGGCAGTCC 5′-fzWIF frag2: CATCACAATCGCCGGGCGGAGGCCGGGCCGCCG 3′-fzWIF frag2: GGTATTCTCCGCACATGTTTTAAAGAAGATAGC 5′-fzWIF frag3: CTTTAAAACATGTGCGGAGAATACCACATCATCG
3′-fzSMO frag1: GGTGGGGTAGCAGCGATTGTGATGTGGCAGTCC 5′-fzSMO frag2: CATCACAATCGCTGCTACCCCACCTCGAACGCAACC 3′-fzSMO frag2: GGTATTCTCCGCACACTGGCCAGTTCCGTTG 5′-fzSMO frag3: ACTGGCCAGTGTGCGGAGAATACCACATCATCG
All final PCR products were cut with SpeI and XbaI and inserted into XbaI cut P-element vector pTub for constitutive expression in Drosophila via the tubulin-α1 promoter (Chen et al, 2004).
Arm signaling reporter assays
Drosophila S2 cells were transfected with the Arm signaling reporter superTOPFLASH (Veeman et al, 2003b), a loading control pIB/His/lacz (Invitrogen) and a fz transgene in combination with a wg transgene or empty vector. wg and fz variant transgenes were expressed via the constitutive tubulin-α1 promoter (Chen et al, 2004). Transfection complexes were formed using 1.5 μl of Fugene6 (Roche) in 50 μl of DES serum-free medium+2 mM GlutaMAX (Invitrogen) and 250 ng superTOPFLASH, 125 ng pIB/lacZ, 385 ng each of fz and wg or empty vector. Complexes were divided into three wells of a 96-well plate containing 2 × 105 cells/well. Cells were disrupted in 20 μl of lysis buffer and luciferase activity was measured in 5 μl of lysate 40 h post-transfection using the Dual-Light Assay System (Applied Biosystems) and a LB960 Luminometer (Berthold). β-Galactosidase activity was used to normalize the values. In experiments using purified Wg, 40 h post-transfection with receptor, superTOPFLASH and pIB/lacZ, 2 × 105 cells were placed into wells of a 96-well plate in 50 μl of medium. A measure of 50 μl of culture medium containing purified Wg diluted at 1:125 (a final concentration of 120 ng/ml) or vehicle (PBS+1% CHAPS+250 mM NaCl). Each sample was done in duplicate. Cells were incubated for an additional 20 h and reporter assays were done as above.
arr and GFP RNAi
Double-stranded arr and GFP RNA was prepared from a PCR product using previously described primers (Schweizer and Varmus, 2003) that incorporate a binding site for the T7 polymerase (GGATTAATACGACTCACTATAGGGAGA). RNA was synthesized from PCR products using the MEGAScript RNAi kit (Ambion). S2 cells were transfected with 250 ng of pTub, pTub-wg∷fzΔCRD or pTub-wntD∷fzΔCRD, 250 ng superTOPFLASH, 125 ng pIB/lacZ and 500 ng of double-stranded RNA of arr or GFP. Complexes were formed using 3 μl of Fugene6 and 100 μl of serum-free medium and were divided into three wells of a 24-well plate containing 1 × 106 cells/well. Reporter assays were performed as described above:
dsRNA arr (700 bp product),
5′-GGATTAATACGACTCACTATAGGGAGACAGATCGA TGTGATCGTTAGG 3′-GGATTAATACGACTCACTATAGGGAGACCATGCTC TGACAGAGTTCG
dsRNA GFP (700 bp),
5′-GGATTAATACGACTCACTATAGGGAGAATGAGTAA AGGAGAAGAACTTTTC 3′-GGATTAATACGACTCACTATAGGGAGATTATTTGT ATAGTTCATCCATGCC
Purification of Wg
Wg protein was purified according to the Wnt purification method previously described (Willert et al, 2003) from 6 l of conditioned medium taken from S2 cells stably expressing wg. The concentration of the purified protein is approximately 15 ng/μl.
Western blot and surface staining of fz variants
For Western blots, cells transfected with fz transgenes alone, identical to reporter assays with purified Wg and were lysed with TNT buffer (150 mM NaCl, 50 mM Tris (pH 7.5) and 1% Triton X-100). Protein was run on a 10% SDS–PAGE gel and probed with an N-terminal rabbit anti-Fz antibody (NFz) that recognizes the ‘hinge' region of Fz, present on all constructs except fz2ΔCRD. Identical blots were probed with affinity-purified 16B12 mouse anti-HA and 3A5 mouse anti-Tubulin antibodies. For surface staining, cells from the same transfection were allowed to adhere to glass coverslips for 10 min. They were washed once with PBS and fixed for 10 min at room temperature in PBS+4% formaldehyde. The cells were blocked in PBS+10% normal donkey serum and incubated with NFz antibody in block buffer overnight at 4°C. Cells were visualized by incubation with Alexa488-labeled donkey anti-rabbit secondary antibody (Molecular Probes).
Rescue of Arm signaling in embryos
Germline clones of the genotype fzGL31th st fz2e2FRT2A were induced using the FLP/ovoD/FRT system (Perrimon et al, 1996). Adler et al (Jones et al, 1996) described the fzGL31 allele. The fz2e2 allele is a nonsense mutation (W285*) in the extracellular ‘hinge' region. Clones were made by heat-shocking yw hs-FLP/w; ovoDw+FRT2A/fzGL31th st fz2e2FRT2A females every 12 h following a 24 h egg lay. Females with germline clones were crossed to males of the genotype w; pTub-fzrescue; fzGL31th st fz2e2FRT2A/TM6B. The ability to fully rescue Arm signaling was measured by the determining the percentage of non-TM6B flies to eclose. The genotype of these flies was confirmed by scoring th, a recessive marker mapping between fz and fz2. There was also a residual PCP phenotype in a small compartment previously shown to be refractory to rescue using a different fz transgenes (Krasnow and Adler, 1994). We interpret this as incomplete rescue by pTub-fz of the PCP phenotype in the of fzGL31 background:
w; fzGL31th st fz2e2FRT2A/TM6B w; pTub-fzWT.7; fzGL31th st fz2e2FRT2A/TM6B w; pTub-fz57.8; fzGL31th st fz2e2FRT2A/TM6B w; pTub-fz81.3; fzGL31th st fz2e2FRT2A/TM6B w; pTub-fzΔCRD; fzGL31th st fz2e2FRT2A/TM6B
Rescue of Arm signaling in the wing
Males from following stocks were crossed to y;vgQ1206-Gal4 UAS-flp; hsCD2,y+ri FRT2A/TM2 and raised at 25°C to generate marked clones in adult wings as previously described (Strapps and Tomlinson, 2001). Mutants clones located at the margin are positively marked by the loss of y+, which makes the bristles yellow. Heterozygous tissue produces dark bristles. For each of the transgenes, two separate insert lines were tested:
yw hsflp; nGFP FRT2A w; th st fz2e3FRT2A/TM3 w; fzGL31FRT2A/TM6B w; fzGL31th st fz2e2FRT2A/TM3 w; pTub-fzWT.3/CyOZ; fzGL31th st fz2e2FRT2A/TM3Z w; pTub-fzWT.7; fzGL31th st fz2e2FRT2A/TM6B w; pTub-fz57.7; fzGL31th st fz2e2FRT2A/TM3Z w; pTub-fz57.8; fzGL31th st fz2e2FRT2A/TM6B w; pTub-fz81.2; fzGL31th st fz2e2FRT2A/TM3Z w; pTub-fz81.3; fzGL31th st fz2e2FRT2A/TM6B w; pTub-fzΔCRD; fzGL31th st fz2e2FRT2A/TM6B w; pTub-fzWIF.5/CyO; fzGL31th st fz2e2FRT2A/TM6B
Overexpression in the wing and in embryos
fz variants were cloned into a UAS expression vector and transformed into Drosophila by standard techniques. UAS lines were crossed to the 71B-Gal4 and h-Gal4 (Flybase IDs: FBst0001747 and FBst0001734) drivers for expression in the wing and embryo, respectively. All crosses were raised at 25°C.
Acknowledgments
We would like to acknowledge Catriona Logan and Michael Gordon for insightful discussions and comments during the preparation of this manuscript and Matt Fish for help in making transgenic lines. The Wingless protein was kindly provided by Kim Harnish in our lab. This work was supported by the Howard Hughes Medical Institute and by a grant from the National Institutes of Health (R01 GM60388).
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Siden-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC (2000) The genome sequence of Drosophila melanogaster. Science 287: 2185–2195 [DOI] [PubMed] [Google Scholar]
- Alcedo J, Ayzenzon M, Von Ohlen T, Noll M, Hooper JE (1996) The Drosophila smoothened gene encodes a seven-pass membrane protein, a putative receptor for the hedgehog signal. Cell 86: 221–232 [DOI] [PubMed] [Google Scholar]
- Bejsovec A (2005) Wnt pathway activation: new relations and locations. Cell 120: 11–14 [DOI] [PubMed] [Google Scholar]
- Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R (1996) A new member of the Frizzled family from Drosophila functions as a Wingless receptor. Nature 382: 225–230 [DOI] [PubMed] [Google Scholar]
- Bhanot P, Fish M, Jemison JA, Nusse R, Nathans J, Cadigan KM (1999) Frizzled and Dfrizzled-2 function as redundant receptors for Wingless during Drosophila embryonic development. Development 126: 4175–4186 [DOI] [PubMed] [Google Scholar]
- Boutros M, Mihaly J, Bouwmeester T, Mlodzik M (2000) Signaling specificity by Frizzled receptors in Drosophila. Science 288: 1825–1828 [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Fish MP, Rulifson EJ, Nusse R (1998) Wingless repression of Drosophila Frizzled 2 expression shapes the Wingless morphogen gradient in the wing. Cell 93: 767–777 [DOI] [PubMed] [Google Scholar]
- Chen CM, Strapps W, Tomlinson A, Struhl G (2004) Evidence that the cysteine-rich domain of Drosophila Frizzled family receptors is dispensable for transducing Wingless. Proc Natl Acad Sci USA 101: 15961–15966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CM, Struhl G (1999) Wingless transduction by the Frizzled and Frizzled2 proteins of Drosophila. Development 126: 5441–5452 [DOI] [PubMed] [Google Scholar]
- Cong F, Schweizer L, Varmus H (2004) Wnt signals across the plasma membrane to activate the {beta}-catenin pathway by forming oligomers containing its receptors, Frizzled and LRP. Development 131: 5103–5115 [DOI] [PubMed] [Google Scholar]
- Dann CE, Hsieh JC, Rattner A, Sharma D, Nathans J, Leahy DJ (2001) Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains. Nature 412: 86–90 [DOI] [PubMed] [Google Scholar]
- Forrester WC, Kim C, Garriga G (2004) The C. elegans Ror RTK CAM-1 collaborates with EGL-20/Wnt to regulate cell migration. Genetics 204: 1951–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A, Jiang J, Ip Y T (2005) Drosophila WntD is a target and an inhibitor of the Dorsal/Twist/Snail network in the gastrulating embryo. Development 132: 3419–3429 [DOI] [PubMed] [Google Scholar]
- Gordon MD, Dionne MS, Schneider DS, Nusse R (2005) WntD is a feedback inhibitor of Dorsal/NF-kB in Drosophila development and immunity. Nature [E-pub ahead of print: doi:10.1038/Nature04073] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JC, Kodjabachian L, Rebbert ML, Rattner A, Smallwood PM, Samos CH, Nusse R, Dawid IB, Nathans J (1999a) A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature 398: 431–436 [DOI] [PubMed] [Google Scholar]
- Hsieh JC, Rattner A, Smallwood PM, Nathans J (1999b) Biochemical characterization of Wnt-Frizzled interactions using a soluble, biologically active vertebrate Wnt protein. Proc Natl Acad Sci USA 96: 3546–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Oz HS, Wiland D, Gharib S, Deshpande R, Hill RJ, Katz WS, Sternberg PW (2004) C. elegans LIN-18 is a Ryk ortholog and functions in parallel to LIN-17/Frizzled in Wnt signaling. Cell 118: 795–806 [DOI] [PubMed] [Google Scholar]
- Jones KH, Liu J, Adler PN (1996) Molecular analysis of EMS-induced Frizzled mutations in Drosophila melanogaster. Genetics 142: 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katanaev VL, Ponzielli R, Semeriva M, Tomlinson A (2005) Trimeric G protein-dependent Frizzled signaling in Drosophila. Cell 120: 111–122 [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW (1998) Use of dsRNA-mediated genetic interference to demonstrate that Frizzled and Frizzled 2 act in the wingless pathway. Cell 95: 1017–1026 [DOI] [PubMed] [Google Scholar]
- Krasnow RE, Adler PN (1994) A single Frizzled protein has a dual function in tissue polarity. Development 120: 1883–1893 [DOI] [PubMed] [Google Scholar]
- Logan CY, Nusse R (2004) The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20: 781–810 [DOI] [PubMed] [Google Scholar]
- Lu W, Yamamoto V, Ortega B, Baltimore D (2004) Mammalian Ryk is a Wnt coreceptor required for stimulation of neurite outgrowth. Cell 119: 97–108 [DOI] [PubMed] [Google Scholar]
- Oishi I, Suzuki H, Onishi N, Takada R, Kani S, Ohkawara B, Koshida I, Suzuki K, Yamada G, Schwabe GC, Mundlos S, Shibuya H, Takada S, Minami Y (2003) The receptor tyrosine kinase Ror2 is involved in non-canonical Wnt5a/JNK signalling pathway. Genes Cells 8: 645–654 [DOI] [PubMed] [Google Scholar]
- Perrimon N, Lanjuin A, Arnold C, Noll E (1996) Zygotic lethal mutations with maternal effect phenotypes in Drosophila melanogaster. II. Loci on the second and third chromosomes identified by P-element-induced mutations. Genetics 144: 1681–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povelones M, Howes R, Fish M, Nusse R (2005) Genetic evidence that Drosophila Frizzled controls planar cell polarity and Armadillo signaling by a common mechanism. Genetics [E-pub ahead of print: doi:10.1534/genetics105.045245] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson EJ, Micchelli CA, Axelrod JD, Perrimon N, Blair SS (1996) wingless refines its own expression domain on the Drosophila wing margin. Nature 384: 72–74 [DOI] [PubMed] [Google Scholar]
- Rulifson EJ, Wu CH, Nusse R (2000) Pathway specificity by the bifunctional receptor Frizzled is determined by affinity for wingless. Mol Cell 6: 117–126 [PubMed] [Google Scholar]
- Sato A, Kojima T, Ui-Tei K, Miyata Y, Saigo K (1999) Dfrizzled-3, a new Drosophila Wnt receptor, acting as an attenuator of Wingless signaling in wingless hypomorphic mutants. Development 126: 4421–4430 [DOI] [PubMed] [Google Scholar]
- Schweizer L, Varmus H (2003) Wnt/Wingless signaling through beta-catenin requires the function of both LRP/Arrow and Frizzled classes of receptors. BMC Cell Biol 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strapps WR, Tomlinson A (2001) Transducing properties of Drosophila Frizzled proteins. Development 128: 4829–4835 [DOI] [PubMed] [Google Scholar]
- Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, Hess F, Saint-Jeannet JP, He X (2000) LDL-receptor-related proteins in Wnt signal transduction. Nature 407: 530–535 [DOI] [PubMed] [Google Scholar]
- Tolwinski NS, Wehrli M, Rives A, Erdeniz N, DiNardo S, Wieschaus E (2003) Wg/Wnt signal can be transmitted through arrow/LRP5,6 and Axin independently of Zw3/Gsk3beta activity. Dev Cell 4: 407–418 [DOI] [PubMed] [Google Scholar]
- Tolwinski NS, Wieschaus E (2004) Rethinking WNT signaling. Trends Genet 20: 177–181 [DOI] [PubMed] [Google Scholar]
- Toomes C, Bottomley HM, Jackson RM, Towns KV, Scott S, Mackey DA, Craig JE, Jiang L, Yang Z, Trembath R, Woodruff G, Gregory-Evans CY, Gregory-Evans K, Parker MJ, Black GC, Downey LM, Zhang K, Inglehearn CF (2004) Mutations in LRP5 or FZD4 underlie the common familial exudative vitreoretinopathy locus on chromosome 11q. Am J Hum Genet 74: 721–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbhauer M, Djiane A, Goisset C, Penzo-Mendez A, Riou JF, Boucaut JC, Shi DL (2000) The C-terminal cytoplasmic Lys-thr-X-X-X-Trp motif in Frizzled receptors mediates Wnt/beta-catenin signalling. EMBO J 19: 4944–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel M, Ingham PW (1996) Smoothened encodes a receptor-like serpentine protein required for hedgehog signalling. Nature 382: 547–551 [DOI] [PubMed] [Google Scholar]
- Veeman MT, Axelrod JD, Moon RT (2003a) A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev Cell 5: 367–377 [DOI] [PubMed] [Google Scholar]
- Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT (2003b) Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol 13: 680–685 [DOI] [PubMed] [Google Scholar]
- Vinson CR, Adler PN (1987) Directional non-cell autonomy and the transmission of polarity information by the Frizzled gene of Drosophila. Nature 329: 549–551 [DOI] [PubMed] [Google Scholar]
- Wehrli M, Dougan ST, Caldwell K, O'Keefe L, Schwartz S, Vaizel-Ohayon D, Schejter E, Tomlinson A, DiNardo S (2000) arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature 407: 527–530 [DOI] [PubMed] [Google Scholar]
- Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, Yates JR III, Nusse R (2003) Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423: 448–452 [DOI] [PubMed] [Google Scholar]
- Wu CH, Nusse R (2002) Ligand receptor interactions in the WNT signaling pathway in Drosophila. J Biol Chem 277: 41762–41769 [DOI] [PubMed] [Google Scholar]
- Wu J, Klein TJ, Mlodzik M (2004) Subcellular localization of Frizzled receptors, mediated by their cytoplasmic tails, regulates signaling pathway specificity. PLoS Biol 2: E158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa S, McKinnon RD, Kokel M, Thomas JB (2003) Wnt-mediated axon guidance via the Drosophila Derailed receptor. Nature 422: 583–588 [DOI] [PubMed] [Google Scholar]
- Zhang J, Carthew RW (1998) Interactions between Wingless and DFz2 during Drosophila wing development. Development 125: 3075–3085 [DOI] [PubMed] [Google Scholar]


