Abstract
Leptospiral protein antigens are of interest as potential virulence factors and as candidate serodiagnostic and immunoprotective reagents. We identified leptospiral protein antigens by screening a genomic expression library with serum from a rabbit hyperimmunized with formalin-killed, virulent Leptospira kirschneri serovar grippotyphosa. Genes expressing known outer membrane lipoproteins LipL32 and LipL41, the heat shock protein GroEL, and the α, β, and β′ subunits of RNA polymerase were isolated from the library. In addition, a new leptospiral gene that in Escherichia coli expressed a 45-kDa antigen with an amino-terminal signal peptide followed by the spirochetal lipobox Val−4-Phe−3-Asn−2-Ala−1↓Cys+1 was isolated. We designated this putative lipoprotein LipL45. Immunoblot analysis of a panel of Leptospira strains probed with LipL45 antiserum demonstrated that many low-passage strains expressed LipL45. In contrast, LipL45 was not detected in high-passage, culture-attenuated strains, suggesting that LipL45 is a virulence-associated protein. In addition, all leptospiral strains tested, irrespective of culture passage, expressed a 31-kDa antigen that was recognized by LipL45 antiserum. Southern blot and peptide mapping studies indicated that this 31-kDa antigen was derived from the carboxy terminus of LipL45; therefore, it was designated P31LipL45. Membrane fractionation studies demonstrated that P31LipL45 is a peripheral membrane protein. Finally, we found that P31LipL45 levels increased as Leptospira entered the stationary phase, indicating that P31LipL45 levels were regulated. Hamsters infected with L. kirschneri formed an antibody response to LipL45, indicating that LipL45 was expressed during infection. Furthermore, the immunohistochemistry of kidneys from infected hamsters indicated that LipL45 was expressed by L. kirschneri that colonized the renal tubule. These observations suggest that expression of LipL45 responds to environmental cues, including those encountered during infection of a mammalian host.
Spirochetes of the genus Leptospira are the causative agents of leptospirosis, an emerging zoonosis encountered worldwide. Many animals serve as reservoir hosts and transmit the pathogen directly or indirectly to humans. Leptospires persist in the proximal renal tubules of kidneys in reservoir hosts and are shed in the urine into the surrounding environment. Leptospires can survive for weeks in soil or water before they are acquired by a new host via abrasions or by penetration of mucous membranes. Human victims of leptospirosis present with symptoms that vary in severity from mild flu-like symptoms to lethal pulmonary hemorrhage or hepatorenal failure (36).
At least 268 serovars and 17 species of Leptospira and Leptonema have been described to date (9). The antigenic basis for serovar specificity resides in the carbohydrate side chain structure of lipopolysaccharides (LPS) (12). Antibodies targeting leptospiral LPS provide protective immunity (32, 33). Because of the variety of LPS forms presented to the host immune system by different serovars during leptospiral infection, the currently available whole-cell vaccines do not provide cross-protection against infection by heterologous serovars. Moreover, in some cases, these vaccines may not even protect against homologous challenge (6). New vaccine strategies are necessary to provide effective protection against the diversity of serovars that a susceptible host may encounter.
A recent study showed that an LPS-depleted outer membrane protein extract provided hamsters with complete protection against homologous leptospiral challenge and partial protection against heterologous lethal challenge (49). The protection appeared to be protein mediated since LPS from the same organism did not protect hamsters from lethal infection by a heterologous serovar (49). These results suggest that an effective subunit vaccine can be developed by using one or more leptospiral proteins. Proteins located in the leptospiral outer membrane are of the greatest interest in this regard because outer membrane proteins are potentially exposed to the host immune system on the leptospiral surface. Antigenic conservation of leptospiral proteins was demonstrated in immunoblot studies that showed that sera from rabbits hyperimmunized with one strain recognized numerous proteins of a wide variety of serovars (11, 39). Several of these protein antigens have been identified, including the outer membrane lipoproteins LipL32 (24) and LipL41 (45) and the porin OmpL1 (23), as well as the heat shock proteins DnaK (2) and GroEL (3). In an initial study to examine the feasibility of using outer membrane proteins as subunit vaccines, recombinant OmpL1 and LipL41 expressed as Escherichia coli membrane proteins synergistically protected Golden Syrian hamsters from homologous challenge (26). Although the hamsters were not challenged with heterologous serovars of Leptospira in this study, the antigenic conservation of LipL41 and OmpL1 among all pathogenic serovars of Leptospira suggests that these outer membrane proteins could provide at least partial protection against heterologous challenge (23, 45).
The complexity of the leptospiral protein antigen profile indicates that additional protein antigens remain to be identified (39). To isolate leptospiral genes that express such antigens, we screened a leptospiral genomic expression library with serum from a rabbit hyperimmunized with Leptospira kirschneri serovar grippotyphosa. In this report, we describe initial characterization of one of these proteins, a novel putative lipoprotein that we designated LipL45.
MATERIALS AND METHODS
Bacterial strains and media.
L. kirschneri serovar grippotyphosa strain RM52 was isolated during an outbreak of porcine abortion in 1983 (53). Low- and high-passage L. kirschneri serovar grippotyphosa strain RM52 and other leptospiral strains were obtained from the National Leptospirosis Reference Center (National Animal Disease Center, Agricultural Research Service, U.S. Department of Agriculture, Ames, Iowa) and were cultivated at 30°C in Johnson-Harris bovine serum albumin-Tween 80 medium (Bovuminar PLM-5 microbiological medium; Intergen) (31). Low-passage samples of the RM52 isolate were either stored in liquid nitrogen or passaged in liquid media at least 200 times to generate a high-passage form. The high-passage strain was unable to produce a lethal infection in hamsters at any dose and was able to infect hamsters only at a dose of ≥107 cells per animal by intraperitoneal inoculation. Low-passage forms of L. kirschneri serovar grippotyphosa strain ISU 82, Leptospira interrogans serovar canicola/portlandvere strains Mex 1 and CDC Nic 1808, and L. interrogans serovar pomona type kennewicki strain P10637-46 were generated by challenging 3-week-old Golden Syrian hamsters intraperitoneally with 5 × 104 cells of each organism. When clinical signs of illness developed or at least 14 days postinoculation, the hamsters were euthanized, and blood and kidney tissue were collected at necropsy and cultured in a semisolid medium which consisted of Bovuminar PLM-5 medium supplemented with 0.2% agar and 100 μg of 5-fluorouracil (Sigma) per ml. Experimental protocols involving rabbits and hamsters were approved by the VA Greater Los Angeles Healthcare System Animal Research Committee.
E. coli XL1-Blue (endA1 gyrA96 hsdR17 lac− recA1 relA1 supE44 thi-1 [F′ lacIqZΔM15]), BM25.8 [supE44 thi Δlac-proAB (F′ traD36 proAB+ lacIqZΔM15) λimm434(kanr) P1(camr) hsdR(rK12− mK12−)], and BLR(DE3)/pLysS [F− ompT hsdSB(rB− mB−) gal dcm Δ(srl-recA)306::Tn10(DE3) pLysS] were grown in Luria broth supplemented with 100 μg of ampicillin per ml, 100 μg of carbenicillin per ml, or 25 μg of chloramphenicol per ml when appropriate. The antibiotics were purchased from Sigma.
Isolation of genes encoding leptospiral protein antigens.
DNA restriction and modifying enzymes were purchased from New England Biolabs. Genomic DNA was prepared from virulent L. kirschneri RM52 by the method of Yelton and Charon (55). The DNA was partially digested with AluI, purified with a QIAquick PCR purification kit (Qiagen), modified with EcoRI methylase by using the instructions of the manufacturer (New England Biolabs), and purified again with a QIAquick PCR purification kit. The methylated DNA was then ligated to an EcoRI linker (pCGGATTCCG), purified with a QIAquick PCR purification kit, digested with EcoRI, and separated by electrophoresis in a 0.8% preparative agarose gel. DNA that was 1 to 5 kb long was excised from the gel, purified with a QIAquick gel extraction kit (Qiagen), ligated to λTriplEx arms by following the instructions provided by the supplier (Clontech), and packaged into λ heads with Gigapack III Gold packaging extract (Stratagene). The phage titer of the library was determined by infecting E. coli XL1-Blue.
To screen the library, approximately 103 PFU was plated by using E. coli XL1-Blue, transferred to nitrocellulose membranes (Schleicher & Schuell) soaked with isopropyl-β-D-thiogalactopyranoside (IPTG), and processed as recommended by the manufacturer (Schleicher & Schuell). The membranes were treated for 30 min with blocking solution, which consisted of 5% nonfat dry milk in phosphate-buffered saline (PBS) containing 0.1% Tween 20 (PBS-T); then they were incubated for 2 h with rabbit antiserum to virulent L. kirschneri RM52 (27) diluted 1:1,000 in blocking solution, washed three times in PBS-T, incubated for 30 min with donkey anti-rabbit immunoglobulin-horseradish peroxidase conjugate (Amersham) diluted 1:4,000 in blocking buffer, and finally washed three times in PBS-T. Lifts were developed with the ECL Western blot detection reagents (Amersham) and exposed to Hyperfilm (Amersham) to identify plaques that reacted with the rabbit antiserum. Each reactive plaque was stored at 4°C in 1 ml of SM (0.1 M NaCl, 8 mM MgSO4, 50 mM Tris-HCl [pH 7.5], 0.01% [wt/vol] gelatin) with 1 to 2 drops of chloroform. λ clones that reacted with the serum were purified further by two rounds of plaque purification. Finally, the plasmid embedded in the phage DNA was excised by infecting E. coli BM25.8 with the λ clones as described by the manufacturer of the expression vector (Clontech).
Gel electrophoresis and immunoblotting.
Samples were solubilized in final sample buffer consisting of 50 mM Tris-HCl (pH 6.8), 100 mM dithiothreitol, 2% sodium dodecyl sulfate (SDS), and 0.1% bromophenol blue in 20% glycerol. Proteins were separated in 12% PAGEr Gold precast Tris-glycine gels (BioWhittaker Molecular Applications) or SDS-12 or 15% polyacrylamide gel electrophoresis (PAGE) gels by using a discontinuous buffer system essentially as described previously (35). Following electrophoresis, each gel was stained with 0.2% Coomassie brilliant blue R-250 (in 10% acetic acid–45% methanol) or transferred electrophoretically to a polyvinylidene difluoride membrane (Millipore). The membranes were incubated with rabbit serum in blocking solution for 30 min (see below for the serum titers used), washed three times with PBS-T, incubated with donkey anti-rabbit immunoglobulin–horseradish peroxidase conjugate (Amersham) at a dilution of 1:5,000 in blocking solution for 30 min, and again washed three times with PBS-T. The membranes were developed with the ECL Western blot detection system (Amersham), and bands were visualized with Hyperfilm (Amersham).
DNA sequencing.
The nucleotide sequences of the clones were assembled from individual sequences obtained by using a combination of primer walking and sequencing of nested deletions. Deletions were generated from plasmid clones by removing restriction fragments extending from inside an insert into the multicloning sites flanking the insert. Oligonucleotides were synthesized by GIBCO BRL. The UCLA Core Sequencing Facility and the Yale/Keck Core DNA Sequencing Facility performed the sequencing reactions.
Expression and purification of recombinant LipL45.
The portion of 5orf2 encoding the mature part of LipL45 was amplified with Pfu Turbo DNA polymerase (Stratagene) by using the upstream primer 5orf2(Bm)-1up (5′-TTGGATCCTGTAAGAAACCTACCGAAAGTT-3′) and the downstream primer 5orf2(Ps)-2dn (5′-TCCCTGCAGATCTATCGATTGGCTCTGGAACC-3′); the primers contained a BamHI site and a PstI site, respectively, near their 5′ ends (underlined). After an initial 1 min of denaturation at 95°C, the reaction mixture was subjected to 15 cycles of denaturation at 95°C for 30 s, annealing at 52°C for 30 s, and extension at 72°C for 1.25 min. The final cycle was followed by an additional extension step consisting of 72°C for 2 min. The PCR product was purified with a Qiagen QIAquick PCR purification kit, digested with BamHI and PstI, and cloned into the His6 expression vector pRSETA (Invitrogen). Similarly, the DNA corresponding to the last 88 amino acid residues encoded by 5orf2 was amplified with the upstream primer 5orf2(Bm)-4up (5′-AAGGATCCGTAGCGGCTCGTCAAGAAGAA-3′) and the downstream primer 5orf2(Ps)-2dn; DNA corresponding to the first 96 amino acid residues encoded by 5orf2 was amplified with 5orf2(Bm)-1up and 5orf2(Ec)-3dn (5′-GGGAATTCATTAGAGTCTTGTATCGGAATTGCCTA-3′). The amplification conditions were identical to those described above, except that the extension time during the 15 cycles was reduced to 30 s. The PCR products were digested with BamHI and either PstI or EcoRI (at the sites underlined) and cloned into pRSETA (Invitrogen).
To express the His6 fusion proteins, plasmids were transformed into E. coli BLR(DE3)/pLysS, which was grown in 500 ml of Luria broth containing carbenicillin (100 μg/ml) to the mid-logarithmic phase with vigorous shaking (optical density at 600 nm, 0.5). IPTG was then added to the culture to a final concentration of 0.5 mM, and incubation was continued for an additional 3 h to induce expression of the fusion protein. The bacteria were lysed by sonication, and both the His6-LipL45 and His6-5ORF2C fusion proteins were purified from the soluble phase by using Ni2+-nitrilotriacetic acid (NTA) affinity columns according to the instructions of the manufacturer (Qiagen). The His6-5ORF2N fusion protein formed inclusion bodies, which were solubilized in 8 M urea-10 mM Tris-HCl-100 mM NaH2PO4 (pH 8.0) and subsequently purified by using an Ni2+-NTA affinity column.
Rabbit antisera.
Purified His6 fusion proteins were loaded onto a preparative SDS-PAGE gel containing 12% polyacrylamide (for His6-LipL45) or 15% polyacrylamide (for His6-5ORF2N and His6-5ORF2C) and allowed to migrate into the separating gel during electrophoresis. A band containing 250 to 500 μg of fusion protein was excised from the gel, desiccated, ground to a powder, dissolved in 1 ml of water, mixed with 1 ml of complete Freund’s adjuvant (Sigma), and inoculated subcutaneously and intramuscularly into New Zealand White rabbits (Harlan Sprague Dawley) that were free of leptospiral antibodies. Additional immunizations with similar amounts of fusion protein in powdered acrylamide gel mixed with incomplete Freund’s adjuvant (Sigma) were administered 4 and 8 weeks after the primary immunization. Blood was collected from the rabbits 10 weeks after the primary immunization and processed to obtain serum (28).
Antibody to GroEL from L. interrogans serovar copenhageni was a generous gift from B. Adler (Monash University, Clayton, Victoria, Australia) (1).
Southern blottiing.
Genomic DNA was extracted from L. kirschneri with a Wizard genomic DNA purification kit (Promega). A 2.5-μg portion of DNA was digested with 5 to 20 U of EcoRI or ClaI overnight in a 20-μl (final volume) mixture. The digested DNA was separated in a 0.8% agarose gel. The gel was then incubated twice for 30 min in denaturing buffer (3 M NaCl, 0.4 M NaOH) and once for 15 min in transfer buffer (3 M NaCl, 8 mM NaOH). The DNA was transferred from the gel to a nylon membrane (Bio-Rad GT genomic tested membrane) by using the rapid downward transfer method (14). Following transfer, the membrane was soaked for 5 min in neutralizing buffer (0.2 M sodium phosphate, pH 6.8) and baked at 80°C until it was dry.
The probe was synthesized with the upstream primer 5orf2-5up (5′-CCAATCTTAAGGAAAATGGGTATGA-3′) and the downstream primer 5orf2-6dn (5′-ATTACTTCTTGAACATCTGCTTGAT-3′), which was labeled at its 5′ end with biotin. The reaction mixture (final volume, 50 μl) was assembled according to the instructions of the supplier of the Taq polymerase (Qiagen). The PCR mixture was subjected to an initial denaturation step consisting of 95°C for 1 min and then 20 cycles of denaturation at 95°C for 30 s, annealing at 54°C for 30 s, and extension at 72°C for 2 min and a final extension step consisting of 4 min at 72°C.
The membrane was subjected to prehybridization for 3 h at 42°C in hybridization solution containing 40% formaldehide, 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 10 mM Tris-HCl (pH 7.4), 10% dextran sulfate, 1× Denhardt’s solution, and 50 μg of salmon sperm DNA per ml in a hybridization tube in a rotisserie oven (Hybaid). Prior to hybridization, the biotin-labeled PCR probe was boiled for 10 min and snap cooled on ice. The denatured probe was mixed with hybridization buffer and incubated overnight with the membrane at 42°C. Following hybridization, the blot was washed twice (15 min each) in 2× SSC-0.1% SDS at room temperature (low stringency). A Phototope-Star chemiluminescent detection kit from New England Biolabs was used to visualize bands with Hyperfilm (Amersham).
Peptide mapping of the C termini of LipL45 and P31LipL45.
To purify P31LipL45 by immunoprecipitation, 4 × 1010 leptospire cells were centrifuged, washed once with 10 ml of PBS-5 mM MgCl2, resuspended in 5 ml of a solution containing 1% protein grade Triton X-100 (Calbiochem), 20 mM Tris-HCl (pH 8), 150 mM NaCl, 2 mM EDTA, and 0.1 mM phenylmethylsulfonyl fluoride, and mixed with a Nutator for 30 min at 4°C to solubilize the outer membrane. The Triton X-100-insoluble protoplasmic cylinder was pelleted by centrifugation for 15 min at 10,000 rpm in a Sorvall SM24 rotor. The supernatant fluid was mixed with 25 μl of LipL45 antiserum and 62.5 μl of a suspension containing 0.2 g of protein A-Sepharose (Sigma) per ml and mixed on a Nutator for 30 min at 4°C. The mixture was centrifuged for 10 min at 3,500 rpm in the SM24 rotor to pellet the P31-antibody-Sepharose complex. The pellet was washed twice with 10 mM Tris-HCl (pH 8)-0.01% Triton X-100 and resuspended in 50 μl of final sample buffer containing 0.25 mM phenylmethylsulfonyl fluoride. The proteins in the mixture were separated in an SDS-12% PAGE gel and stained with Coomassie brilliant blue. Affinity-purified His6-LipL45 fusion protein was electrophoresed similarly in an SDS-PAGE gel and stained.
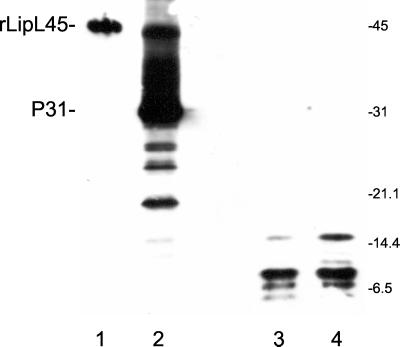
Both the 31-kDa band and the 46-kDa band, representing native P31LipL45 and recombinant His6-LipL45, respectively (see Fig. 6, lanes 1 and 2), were excised from the gel and soaked in 0.5 ml of water for 15 min at 4°C. Each gel slice was then cut into small pieces, which were soaked in 50 μl of V8 sample buffer (0.1 M Tris-HCl [pH 6.8], 0.2% SDS, 20% glycerol, 0.2% bromphenol blue) for 15 min at 4°C, heated for 10 min at 50°C, and placed in a well of an SDS-15% PAGE gel in a Hoefer SE 600 unit. A 25-μl portion of a solution containing 10 μg of V8 protease per ml in V8 sample buffer was added to the same well, and electrophoresis was immediately started. Electrophoresis was interrupted when the stack was 1 cm above the stacking gel-resolving gel interface to permit digestion of P31LipL45 by V8 protease. Electrophoresis was resumed 30 min later to separate the peptides in the resolving gel.
FIG. 6.
Peptide mapping of recombinant LipL45 and P31LipL45. Recombinant His6-LipL45 (rLipL45) and native P31LipL45 (P31) were gel purified, digested with V8 protease, and subjected to immunoblot analysis. The membrane was probed with serum generated against the C-terminal 88 amino acid residues of LipL45 (1:4,000). Lane 1, purified recombinant His6-LipL45; lane 2, high-passage L. kirschneri RM52; lane 3, gel-purified P31 treated with V8 protease; lane 4, gel-purified His6-LipL45 treated with V8 protease. The positions of molecular mass standards (in kilodaltons) are indicated on the right. The His6-LipL45 protein migrated as a 46-kDa band (lane 1). Note that LipL45 was detected when a large number of high-passage leptospires were loaded onto the gel (lane 2).
Cellular fractionation of Leptospira.
The outer membrane of L. kirschneri was solubilized with Triton X-114 as described previously (24), except that 0.5% protease inhibitor cocktail (catalog no. P8849; Sigma) was included in the lysis buffer. The membrane was isolated and washed as described previously (44), with some minor modifications. Cells were suspended in lysis buffer (20 mM Tris-HCl [pH 8], 50 mM NaCl, 2 mM EDTA, 0.5% protease inhibitor cocktail). Then 2 mg of lysozyme was added, and the cell suspension was incubated on ice for 5 min. The cell suspension was frozen at −80°C and thawed three times with vigorous vortexing. The membrane was pelleted by centrifugation at 16,000 × g for 30 min and resuspended in 600 μl of lysis buffer. Samples (100 μl) of the membrane suspension were mixed separately with 100 μl of 400 mM NaCl, 100 μl of 3.6 M urea, 100 μl of 1.2 M Na2CO3, and 100 μl of 1% Triton X-100 with the Nutator for 15 min. The membrane was recovered by centrifugation in a microcentrifuge for 15 min and resuspended in 166 μl of final sample buffer. Protein released into the supernatant fluid was precipitated with acetone and resuspended in 166 μl of final sample buffer. Each membrane pellet was resuspended in 166 μl of final sample buffer.
Immunohistochemistry.
The methods used to perform an immunohistochemistry analysis and to obtain L. kirschneri-infected hamster kidney tissue have been described previously (4). Briefly, groups of three 5-week-old Golden Syrian hamsters (Harlan Sprague Dawley) were inoculated intraperitoneally with 105 virulent L. kirschneri RM52 cells. Hamsters that were still alive 28 days after this challenge were euthanized, and liver and kidney tissues were removed, fixed in formalin, and embedded in paraffin. Tissue sections were stained with hematoxylin and eosin stain or silver stain by the technique of Steiner and Steiner (50).
Serial 5-μm sections of kidneys taken 28 days after infection with L. kirschneri were cut and placed on Pro-bond Plus slides. Paraffin was removed from the sections with xylene and ethanol by standard procedures. The tissue sections were pretreated with 0.1% trypsin in 0.1 M Tris-HCl (pH 7.6)-0.1% CaCl2 for 5 min at 37°C. Nonspecific staining of tissue sections was blocked by using 10% normal goat serum and incubation at room temperature for 20 min prior to overnight incubation at 4°C with primary antibody. LipL45 antiserum was used at a 1:6,000 dilution. The controls included kidney sections from uninfected hamsters treated with hyperimmune serum and from infected hamsters treated with normal rabbit serum or no primary antibody. Unbound primary antibody was removed, and the tissues were incubated at room temperature for 30 min with biotinylated goat anti-rabbit immunoglobulin (Vector). After washing, sections were incubated for 20 min at room temperature with supersensitive streptavidin-alkaline phosphatase (Biogenex). Enzyme reactions were developed with new Fuchsin (Biogenex). All slides were counterstained with hematoxylin before dehydration in alcohols and Propar (a xylene substitute), and coverslips were mounted.
Nucleotide sequence accession number.
The sequence of the 2,264-nucleotide insertion of clone 5, which encodes the lipL45 gene from L. kirschneri serovar grippotyphosa strain RM52, has been deposited in the GenBank database under accession number AF379683.
RESULTS
Isolation of the gene encoding LipL45.
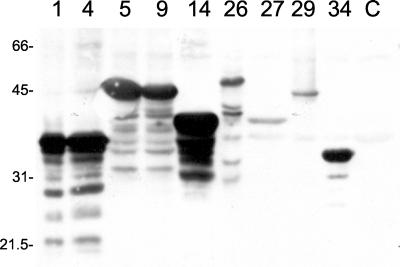
We constructed a leptospiral expression library by using genomic DNA from virulent L. kirschneri serovar grippotyphosa strain RM52. The library was screened with serum from a rabbit hyperimmunized with formalin-killed, virulent L. kirschneri, which resulted in isolation of 104 reactive λ bacteriophage clones. The plasmid DNA embedded in the λ bacteriophage sequence was excised from these clones and introduced into E. coli for immunoblot analysis. Figure 1 shows the reactivity of the serum used to screen the library with E. coli transformed with nine representative clones. The serum reacted with antigens ranging in size from 20 to 150 kDa and did not react with any protein expressed from plasmids excised from four nonreactive λ clones. Antigens ranging in size from 33 to 50 kDa are shown in the immunoblot in Fig. 1. Immunoblotting did not detect a signal with 28 clones, apparently because of the instability of the foreign protein expressed in E. coli or the small size of the antigen (data not shown).
FIG. 1.
Expression of leptospiral antigens in E. coli. Several clones obtained from the expression library were transformed as plasmid DNA into E. coli XL1-Blue. Overnight cultures of the transformants were resuspended in final sample buffer and analyzed in an immunoblot with serum (1:5,000) from a rabbit hyperimmunized with formalin-killed, virulent L. kirschneri. The clone number is indicated above each lane (lanes 1 and 4, LipL32; lanes 5, 9, and 29, LipL45; lane 14, unknown; lane 26, GroEL; lane 27, unknown; lane 34, LipL41; lane C, negative control that does not express an antigen). The positions of molecular mass standards (in kilodaltons) are indicated on the left.
To determine the identities of the clones, the 76 plasmid clones expressing an antigen that was detected in the immunoblot screening analysis were examined either by immunoblot analysis with sera specific for known leptospiral antigens (LipL32, LipL41, OmpL1, and GroEL) or by partial sequence analysis. Sequences were analyzed with BLASTX (http://www.ncbi.nlm.nih.gov/BLAST/). For clones harboring multiple open reading frames, a deletion analysis was performed to identify the gene expressing the antigen. Nine clones encoded one of the outer membrane lipoproteins (LipL32 or LipL41) (24, 45). Fifteen clones encoded the α, β, or β′ subunit of RNA polymerase. Of the 13 clones that encoded the β or β′ subunit (43), 11 expressed an antigen that was larger than 80 kDa. Based on this observation, many of the 14 unsequenced clones that expressed a high-molecular-mass antigen (>80 kDa) probably encoded the β or β′ subunit. In addition, three clones encoded dihydrolipoamide succinyltransferase, an enzyme involved in intermediary metabolism, and four clones encoded the heat shock protein GroEL (3). The sequences of four antigens expressed by nine clones did not significantly match any sequence in the databases (GenBank, EMBL). Two of the four novel antigens were predicted to have a cleavable signal sequence.
The remaining 22 of the 76 clones encoded one of two putative lipoproteins. LipL32, LipL41, OmpL1, and GroEL antisera did not react with any of these clones (data not shown). One of the 22 clones, clone 5, was selected for further analysis. The sequence of clone 5 revealed that there are two open reading frames harbored in a 2,264-bp insert (Fig. 2) (accession number AF379683). 5orf1 consists of the last 756 nucleotides of an open reading frame fused to vector sequences upstream of the insert, and 100 nucleotides downstream of 5orf1 is 5orf2, which is a 1,164-nucleotide open reading frame that is predicted to express a protein which is 388 amino acid residues long (Fig. 2). 5orf2 has two potential ATG start codons separated by three nucleotides. Because the upstream ATG is separated from the probable Shine-Dalgarno sequence by only two nucleotides (Fig. 2), translation is more likely to start from the downstream ATG. 5orf2 is followed by two consecutive stop codons. A potential E. coli ϕ70 promoter consisting of the sequences aTGAtt and aAcAAT in the −35 and −10 regions (with matches to consensus indicated by uppercase) separated by 16 nucleotides is upstream of 5orf2. Immediately downstream of 5orf2 is a sequence resembling a ρ-independent transcription terminator, an inverted repeat followed by several thymines. No sequence resembling a ρ-independent terminator was found between 5orf1 and 5orf2, raising the possibility that the genes are cotranscribed.
FIG. 2.
Sequence of the leptospiral gene encoding LipL45. The nucleotide sequence of clone 5 (accession number AF379683) from position 760 through position 2109 is shown. Probable −35 and −10 promoter sequences are indicated by large letters. The Shine-Dalgarno sequence is underlined. The spirochetal lipobox is indicated by boldface type, and the proposed signal peptidase II processing site is indicated by an arrow. The sequences of the PCR primers used to generate the probe for the Southern blot are indicated by multiple arrowheads. The putative ρ-independent transcriptional terminator is indicated by dashed arrows. The amino acid sequence used to generate serum to the C terminus of LipL45 (5ORF2C) is shaded.
Deletion analysis of the insert harbored by clone 5 indicated that 5orf2 expresses the 45-kDa protein in E. coli (data not shown). The amino terminus of the protein encoded by 5orf2 appears to consist of a 21-amino-acid signal peptide sequence with a spirochetal lipobox containing the lipoprotein signal peptidase cleavage site Val−4-Phe−3-Asn−2-Ala−1↓Cys+1 (22). The processed protein is 367 amino acid residues long, and its calculated molecular weight is 39,813. A BLAST search revealed 98% amino acid sequence identity with the uncharacterized putative lipoprotein qlp42 from L. interrogans (accession number AF320329). We designated the protein LipL45 to conform to the nomenclature used for other leptospiral lipoproteins (22).
LipL45 levels are associated with culture passage.
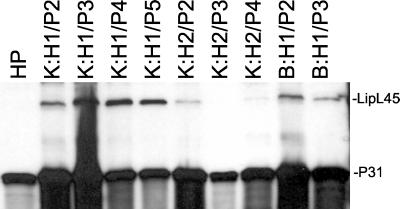
To generate antibody against LipL45, the protein without its signal sequence was expressed with an amino-terminal His6 tag in E. coli and purified by Ni2+-NTA affinity chromatography. A rabbit was immunized with the fusion protein to generate antibody against LipL45. An immunoblot of various isolates of L. kirschneri RM52 cultured from the blood or kidneys of two hamsters infected in the laboratory was probed with the LipL45 antiserum (Fig. 3). These isolates were passaged serially in vitro less than five times. The LipL45 antiserum detected a 45-kDa antigen in all of the low-passage isolates (Fig. 3, lanes 2 to 10). Hamster 1 isolates expressed more LipL45 than hamster 2 isolates, possibly because of outgrowth of different clonal populations that adapted to culture conditions at different rates. Interestingly, the 45-kDa band was not detected in the culture-attenuated form of L. kirschneri RM52 (Fig. 3, lane 1). The LipL45 antiserum also reacted with a 31-kDa antigen (P31) expressed by both low- and high-passage strains.
FIG. 3.
LipL45 levels in low- and high-passage L. kirschneri isolates. L. kirschneri was cultured from kidney (K) or blood (B) samples, and isolates are designated by the hamster (H1 or H2) from which they were isolated and the number of passages in culture (P2 to P5). High-passage L. kirschneri (HP) was obtained by multiple passages of the organism in vitro. The immunoblot was probed with LipL45 antiserum (1:5,000).
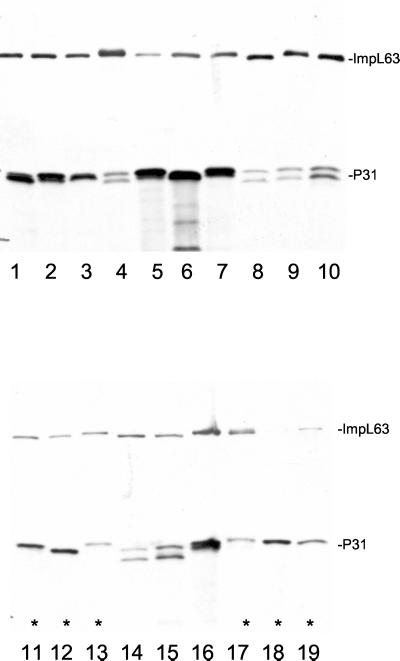
The relationship of LipL45 detection with culture passage was examined in other low- and high-passage leptospiral strains. Nine virulent leptospiral strains representing L. kirschneri serovar grippotyphosa, L. interrogans serovar pomona, and L. interrogans serovar canicola were inoculated into hamsters for the purpose of culturing low-passage hamster isolates. Immunoblots of these low-passage strains demonstrated that four of the nine hamster isolates expressed full-length LipL45 (Fig. 4, lanes 1 to 5; data not shown). Expression of LipL45 was also examined in a variety of saprophytic and high-passage pathogenic strains of Leptospira and Leptonema representing most species and serogroups (Fig. 5). To control for loading, the blot was also probed with serum to ImpL63, an inner membrane protein expressed by all pathogenic and saprophytic serovars of Leptospira examined to date. LipL45 was not detected in any of the strains, but all of the strains expressed the P31 antigen (Fig. 5). Interestingly, pathogenic serovars produced a P31 doublet, whereas saprophytic serovars produced a single band, a potentially useful phenotypic marker for pathogenicity. The results of a short exposure that revealed the doublets are shown in Fig. 5. Longer exposures did not reveal full-length LipL45 (data not shown). These observations suggest that LipL45 levels depend in part on culture passage. Because the 45-kDa band was observed only in low-passage isolates and because low-passage Leptospira isolates are expected to be virulent, LipL45 is a virulence-associated protein.
FIG. 4.
LipL45 expression in low-passage isolates of various serovars of Leptospira. Leptospira isolates were cultured from the kidneys of infected hamsters and passaged serially in Bovuminar PLM-5 medium up to five times (P2 to P5). Leptospires were resuspended in final sample buffer prior to immunoblot analysis with LipL45 antiserum (1:4,000). Lane 1, L. kirschneri serovar grippotyphosa strain RM52; lane 2, L. interrogans serovar pomona strain P10637-46; lane 3, L. interrogans serovar canicola/portlandvere strain Mex 1; lane 4, L. interrogans serovar canicola/portlandvere strain CDC Nic 1808; lane 5, L. kirschneri serovar grippotyphosa strain ISU 82; lane 6, L. kirschneri serovar grippotyphosa strain RM52, high passage.
FIG. 5.
LipL45 expression in high-passage pathogenic and saprophytic isolates of various serovars of Leptospira. A panel of leptospiral serovar isolates was examined by immunoblot analysis with LipL45 (1:5,000) and ImpL63 (1:4,000) antisera. Lane 1, L. kirschneri serovar grippotyphosa strain Moskva V; lane 2, L. interrogans serovar pomona strain PO-01; lane 3, L. interrogans serovar bratislava strain AS-05; lane 4, Leptospira noguchii serovar proechymis strain LT 796; lane 5, L. noguchii serovar fort bragg strain Fort Bragg; lane 6, L. kirschneri serovar mozdok strain 5621; lane 7, L. kirschneri serovar grippotyphosa strain RM52; lane 8, Leptospira borgpetersenii serovar hardjo strain HB-15B/93U; lane 9, Leptospira santarosai serovar canalzonae strain CZ 188; lane 10, L. santarosai serovar bakeri strain 79; lane 11, Leptospira wolbachii serovar biflexa strain codice; lane 12, Leptonema illini 3055; lane 13, Leptospira inadai serovar lyme strain 10; lane 14, L. borgpetersenii serovar javanica strain Veldrat Batavia 46; lane 15, L. borgpetersenii serovar tarassovi strain Perepelicin; lane 16, L. interrogans serovar pomona strain RZ11; lane 17, Leptospira weilii serovar celledoni strain Celledoni; lane 18, Leptospira biflexa serovar patoc strain Patoc I; lane 19, Leptospira meyeri serovar semaranga strain Veldrat Semarang 173. The lanes containing the saprophytic species L. wolbachii, L. illini, L. inadai, L. weilii, L. biflexa, and L. meyeri are indicated by asterisks (lanes 11 to 13 and 17 to 19).
LipL45 is converted into a 31-kDa antigen.
To determine the relationship of P31 to LipL45, we first created His6 fusion proteins fused to the amino-terminal 96 amino acid residues of LipL45 (5ORF2N) and to the carboxy-terminal 88 amino acid residues of LipL45 (5ORF2C) (Fig. 2) and raised antiserum against each fusion protein. Immunoblot analysis showed that 5ORF2C antiserum reacted with both LipL45 and P31 (data not shown), demonstrating that P31 is antigenically related to the C-terminal 88 amino acid residues of LipL45. On the basis of these results, 5ORF2N antiserum would be expected to react with LipL45 and not with P31. However, we were not able to raise 5ORF2N antiserum; sera from two rabbits immunized with the N terminus of LipL45 did not react with either protein in immunoblots of low- and high-passage L. kirschneri isolates. Because the 5ORF2N fusion protein used for immunization had been solubilized in urea prior to purification, the leptospires were subjected to equivalent denaturing conditions by boiling the bacteria in final sample buffer containing urea prior to electrophoresis. Despite these precautions, the 5ORF2N antisera did not react with LipL45 or P31 (data not shown).
We wished to determine whether P31 was a fragment of LipL45 or a cross-reactive antigen expressed from a gene other than lipL45. While eight independent clones expressing LipL45 were isolated from our expression library, no clone expressing a 31-kDa or smaller protein with homology to LipL45 was isolated. This observation is consistent with P31 being a processing product of LipL45 rather than a 31-kDa protein expressed from a separate gene. Nevertheless, it is possible that antibodies that react with the 31-kDa protein were not present in the serum used to screen the expression library. Therefore, we performed two experiments to examine this question more carefully.
First, the C termini of LipL45 and P31 were compared by proteolytic digestion. If the C terminus of LipL45 encompasses the 31-kDa protein, then serum raised against the C terminus of LipL45 should react with identical peptides generated by V8 protease digestion of LipL45 and P31. Gel-purified His6-tagged LipL45 fusion protein and native P31 (see Materials and Methods) were used as substrates for proteolysis. The digestion products were subjected to immunoblot analysis with 5ORF2C antiserum (Fig. 6). Figure 6 shows that the digestion patterns for LipL45 (lane 4) and P31 (lane 3) were identical, indicating that P31 is derived from the C terminus of LipL45.
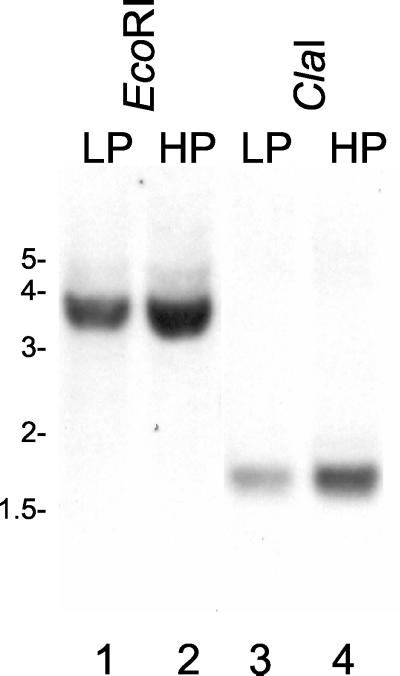
Second, a low-stringency Southern blot analysis in which low- and high-passage L. kirschneri genomic DNA were used was performed (Fig. 7). Each DNA was cut with EcoRI (Fig. 7, lanes 1 and 2) or ClaI (lanes 3 and 4); restriction sites for these enzymes are not found in the 5orf2 probe, which spans most of the reading frame (Fig. 2). As expected for a single-copy lipL45 gene, both types of digestion resulted in a single band. The probe hybridized with a 1.6-kb ClaI fragment and a 3.5-kb EcoRI fragment. Examination of the sequence of clone 5 revealed two ClaI restriction sites 1.6 kb apart (Fig. 2). This observation suggests that other genes containing sequences related to the lipL45 gene are not present in the L. kirschneri genome. To indicate the relationship between LipL45 and P31, we designated the 31-kDa protein P31LipL45.
FIG. 7.
Low-stringency Southern blot analysis of the gene encoding LipL45. Genomic DNA isolated from low-passage (LP) (lanes 1 and 3) or high-passage (HP) (lanes 2 and 4) L. kirschneri isolates were digested with EcoRI (lanes 1 and 2) or ClaI (lanes 3 and 4) and subjected to a low-stringency Southern blot analysis with a 5orf2 probe (Fig. 2). The positions of molecular weight standards (in kilobases) are indicated on the left.
P31LipL45 is a peripheral membrane protein.
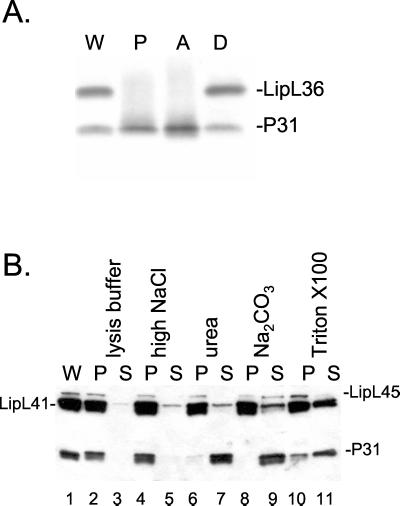
The locations of LipL45 and P31LipL45 in L. kirschneri were determined by exploiting the unique properties of the nonionic detergent Triton X-114 (7). Previous studies have shown that Triton X-114 solubilizes the outer membrane of L. kirschneri, leaving behind the protoplasmic cylinder consisting of the cytoplasm surrounded by the inner membrane (27, 56). As expected, these studies localized the penicillin-binding proteins to the protoplasmic cylinder of Leptospira (10, 27). Furthermore, when the temperature of the solubilized material is raised above the clouding point of the detergent to 37°C, LPS and outer membrane proteins LipL32, LipL36, LipL41, and OmpL1 partition into the detergent phase and periplasmic proteins partition into the aqueous phase (24, 25, 45). When Triton X-114 fractions were probed with LipL45 antiserum, P31LipL45 was found to be distributed among the protoplasmic cylinder, the detergent phase, and the aqueous phase (Fig. 8A). One interpretation of these results is that P31LipL45 is loosely bound to the inner membrane and/or the outer membrane. In contrast, outer membrane protein LipL36 was located entirely in the detergent phase, as described in a previous study (Fig. 8A) (25).
FIG. 8.
Localization of LipL45 and P31LipL45. (A) High-passage L. kirschneri was fractionated with Triton X-114, and each fraction was subjected to immunoblot analysis with LipL45 (1:5,000) and LipL36 (1:4,000) antisera. Lane W, whole cells; lane P, insoluble pellet; lane A, aqueous fraction; lane D, detergent fraction. (B) Membrane fractions of virulent L. kirschneri were washed with buffer (lanes 2 and 3), NaCl (lanes 4 and 5), urea (lanes 6 and 7), Na2CO3 (lanes 8 and 9), or Triton X-100 (lanes 10 and 11) for 15 min. After this, the membrane was pelleted by centrifugation, and the pellets (P) and supernatant fluids (S) were subjected to immunoblot analysis with LipL45 and LipL41 antisera. Lane 1 contained unfractionated L. kirschneri (W).
To investigate the relationship of LipL45 and P31LipL45 with the lipid bilayer further, L. kirschneri was lysed with lysozyme, followed by several rounds of freezing and thawing. The membrane was pelleted by centrifugation and washed with various reagents (Fig. 8B) (44). Peripheral membrane proteins are solubilized by high salt concentrations, high urea concentrations, or high pH values, whereas integral membrane proteins are retained in the lipid bilayer following any of these treatments (20, 44). Both LipL45 and P31LipL45 were found in the membrane pellet (Fig. 8B, lane 1). Neither protein could be removed with a high-salt wash (lanes 3 and 4). On the other hand, P31LipL45 was washed off the membrane by urea, whereas LipL45 was resistant to urea treatment (lanes 5 to 8). Integral membrane protein OmpL1 could not be washed off the membrane by any treatment (data not shown) (44). Lipoproteins LipL41 and LipL45 were released from the membrane by sodium carbonate treatment. Extremes of pH are known to hydrolyze the covalently attached lipid moieties that anchor lipoproteins to membranes (8). The results of the urea experiment suggest that P31LipL45 is a peripheral membrane protein loosely associated with the outer or inner membrane and can be dislodged under conditions that do not affect integral membrane proteins. LipL45 appears to be an integral membrane protein, presumably due to integration of its N-terminal lipid moiety. Apparently, Triton X-100 did not completely solubilize the membrane; LipL41 and P31LipL45 were partially removed from the membrane, and all LipL45 remained in the membrane pellet (Fig. 8B, lanes 10 and 11).
P31LipL45 levels increase during the stationary phase.
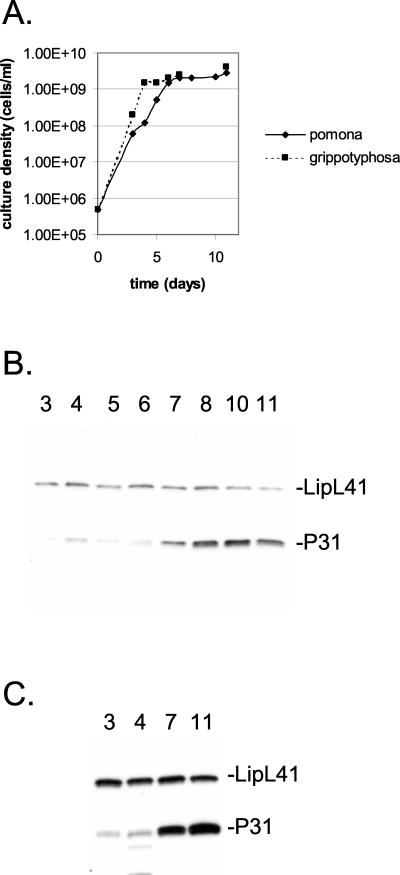
In our studies of LipL45, we noticed variation in the levels of P31LipL45 after different cultures of Leptospira were harvested. We suspected that this variation resulted from changes in P31LipL45 expression with growth phase. To test this hypothesis, a small inoculum of L. interrogans serovar pomona was introduced into culture medium and allowed to grow to the stationary phase (Fig. 9A). Bacteria were harvested at daily intervals, and the level of P31LipL45 in each sample was determined by immunoblot analysis with the LipL45 antiserum (Fig. 9B). The immunoblot was simultaneously probed with the LipL41 antiserum; LipL41 levels are not affected by the growth phase of Leptospira (25). Equal loading was also confirmed by staining the total protein on the immunoblot with amido blue stain (data not shown). The amount of P31LipL45 increased up to fourfold as the bacteria entered the stationary phase (Fig. 9A and B, day 7). The level of P31LipL45 remained high for at least several days in the stationary phase (Fig. 9B, days 8 to 11). Similar results were obtained with L. kirschneri (Fig. 9A and C).
FIG. 9.
Variation of P31LipL45 levels with growth phase. (A) L. interrogans serovar pomona and L. kirschneri serovar grippotyphosa were inoculated into Bovuminar PLM-5 medium at a density of 5 × 105 cells/ml, and the cultures were incubated at 30°C. The number of cells in each culture was determined by dark-field microscopy at different times. (B and C) Samples (1 × 108 cells) obtained from the cultures at different times were examined by immunoblot analysis with LipL45 (1:4,000) and LipL41 (1:8,000) antisera. The strains used were L. interrogans serovar pomona strain P10637-46 (B) and L. kirschneri serovar grippotyphosa strain RM52 (C).
LipL45 is expressed by leptospires residing in the kidney.
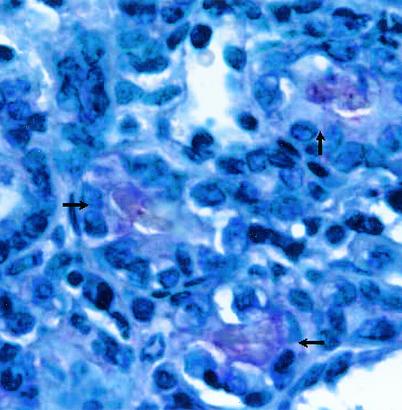
To examine LipL45 expression during infection, kidney sections of hamsters infected with virulent L. kirschneri were examined by immunohistochemical staining. We have used this technique to examine LipL32, LipL36, LipL41, and OmpL1 expression in infected hamsters in previous studies (4, 24). Kidney sections treated with preimmune rabbit antiserum showed no detectable reactivity (data not shown). In contrast, the LipL45 antiserum moderately stained the lumina of some kidney tubules, presumably only those containing leptospires (Fig. 10). This result indicates that LipL45 is expressed in the kidney during infection. Consistent with this interpretation, serum from an infected hamster reacted with LipL45 expressed by E. coli (data not shown).
FIG. 10.
Immunohistochemistry of the kidney of an infected hamster. A kidney from an infected Golden Syrian hamster obtained 28 days after infection with virulent L. kirschneri was examined by performing an immunohistochemistry analysis with LipL45 antibody. The arrows indicate the reactive lumina of three kidney tubules. The lumen of one tubule (left) and one venule (top, containing a red blood cell) did not react with the LipL45 antibody.
DISCUSSION
We isolated a leptospiral gene encoding a novel protein designated LipL45. This protein is predicted to be a lipoprotein based on the sequence Val−4-Phe−3-Asn−2-Ala−1↓Cys+1 at the putative signal peptidase cleavage site found at the end of the 21-amino-acid signal peptide. The phenylalanine at the −3 position and the asparagine at the −2 position do not conform with the definition of the consensus lipobox sequence (Leu, Ala, Val)−4-Leu−3-(Ala, Ser)−2-(Gly, Ala)−1↓Cys+1, which is based on gram-negative lipoprotein signal peptidase cleavage sites (29). However, a recent alignment of spirochetal lipoproteins revealed a more relaxed consensus sequence (22). For example, several lipoproteins, including OspC from Borrelia burgdorferi and TpN29-35 from Treponema pallidum, have a phenylalanine residue at the −3 position. In addition, all members of the 2.9 lipoprotein family of B. burgdorferi have an asparagine residue at the −2 position (42). Although we have not determined whether LipL45 in virulent Leptospira isolates can be labeled by radiolabeled palmitate, the pH-sensitive association of LipL45 with the membrane and the inability of a high salt or urea concentration to remove LipL45 from the membrane are consistent with LipL45 being a lipoprotein (Fig. 8B) (8).
LipL45 is predicted to be a 39.8-kDa protein after removal of its signal peptide. The discrepancy between the predicted molecular mass (39.8 kDa) and the apparent molecular mass (45 kDa) (Fig. 1) is similar to discrepancies observed with other spirochetal lipoproteins (24, 45, 52). Nevertheless, we cannot rule out the possibility that LipL45 is posttranslationally modified. Such modifications, if present, must be conserved in L. kirschneri and E. coli since LipL45 migrated as a 45-kDa protein when it was expressed in either organism. It is unlikely that the lipL45 gene that we have described is a product of ligation of two unrelated fragments of leptospiral DNA because the entire lipL45 gene was found in six independent clones.
Our initial characterization of LipL45 revealed several properties that are unique among known leptospiral proteins. LipL45 is a putative 45-kDa lipoprotein that is converted into a 31-kDa species that we designated P31LipL45. Our data indicate that P31LipL45 is derived from the C terminus of LipL45. First, serum raised against the C-terminal 88 amino acid residues of LipL45 reacted with P31LipL45 (data not shown). Second, peptide mapping demonstrated that the sizes of V8 proteolytic fragments of the C termini of LipL45 and P31LipL45 are identical (Fig. 6). Because LipL45 appears to lack an internal start codon at a position expected for a 31-kDa protein (Fig. 2), we favor the possibility that P31LipL45 originates from LipL45 by proteolytic processing. The processing event may or may not depend on removal of the signal peptide, but the two processing steps may be tightly coupled. At least one export strategy that utilizes two processing reactions during secretion has been characterized. Numerous proteins, including the immunoglobulin A1 proteases of Haemophilus influenzae and Neisseria species, use the autotransporter (type V) secretion mechanism, which involves cleavage of the N-terminal signal peptide during transit through the inner membrane, followed by autoproteolysis during export through the outer membrane (30). The second cleavage step is thought to involve formation of a transmembrane pore by the C terminus of the protein in the outer membrane, through which the passenger N-terminal domain passes prior to autoproteolytic cleavage. However, LipL45 does not resemble an autotransporter. First, P31LipL45 does not form an integral membrane protein characteristic of the transmembrane segment of the autotransporter (Fig. 8). Second, we have not detected the N-terminal fragment of LipL45 in culture medium or in the bacteria (data not shown). Regardless of the actual mechanism of P31LipL45 export, it is the first nonlipidated bacterial protein of which we are aware that exploits the lipoprotein secretion pathway to target itself to its final destination.
P31LipL45 appeared as a doublet in all pathogenic Leptospira species and as a single band in saprophytic Leptospira species (Fig. 5). This observation indicates that P31LipL45 is a marker for pathogenic Leptospira isolates. We do not understand the molecular basis for this observation, but it may involve posttranslational modification or cleavage at multiple sites of LipL45 in the pathogens. The rate at which these events occur may be influenced by the growth rate, with the modification or cleavage event completed in the faster-growing saprophytic species.
P31LipL45 is the first leptospiral peripheral membrane protein to be identified. Unlike integral membrane proteins, P31LipL45 was washed off with urea, indicating that it is a peripheral membrane protein (Fig. 8B). P31LipL45 was not washed off by high salt concentrations, indicating that electrostatic charge is not the primary mode of association of P31LipL45 with the membrane. Our hypothesis is that following removal of the signal peptide and lipidation of the N-terminal cysteine, full-length LipL45 associates via its lipid moieties with the cytoplasmic membrane, with the protein extending into the periplasm. The protein is subsequently processed by an unknown protease, releasing P31LipL45, which seems to associate with both the inner and outer membranes of Leptospira (Fig. 8). The only other spirochetal peripheral membrane proteins described to date are the GlpQ and TpLRR proteins of T. pallidum and the P22-A antigen of B. burgdorferi (46–48).
Another novel feature of P31LipL45 is that its level changes with the growth phase of the organism. The P31LipL45 level increases as a Leptospira culture enters the stationary phase (Fig. 9). This effect may result either from increased transcriptional and/or translational synthesis of LipL45 or from a diminished rate of P31LipL45 breakdown as Leptospira enters the stationary phase. During infection of a mammalian host, virulent Leptospira cells multiply rapidly in the blood before they are cleared by the reticuloendothelial system (17). Some of the leptospires that survive migrate to the proximal convoluted tubule of the kidney, where they are able to multiply to a high density (4, 16, 37). LipL45 was detected in infected hamster kidneys by immunohistochemical staining (Fig. 10). The microenvironment in the kidney tubules may simulate the stationary phase, causing leptospires to upregulate expression of certain proteins, including the P31LipL45 form of LipL45. Expression of other leptospiral proteins may be downregulated while Leptospira resides in the kidney. Indeed, LipL36 levels decline as Leptospira enters the stationary phase and are not detected in leptospires colonizing the kidney (4, 25). We do not know whether the inducing signal for P31LipL45 upregulation is created by nutrient depletion, cell density, or some other factor.
We found that detection of LipL45 is correlated with virulence (Fig. 3 to 5). We therefore identified LipL45 as a virulence-associated protein to distinguish it from true virulence factors. Construction of a virulent L. kirschneri strain with a disrupted lipL45 gene will be necessary to determine whether LipL45 is a virulence factor (41). The lipL45 gene is the first leptospiral gene that expresses a virulence-associated protein to be isolated. A virulence-associated fibronectin-binding protein was recently identified as a band on a blot, but the observed molecular mass of this protein, 35 kDa, is too low for it to be LipL45 (38). LipL45 is a highly specific marker for virulence because it is not detected in culture-attenuated (high-passage) strains (Fig. 5), but it is a somewhat insensitive marker for virulence because of our failure to detect it in many low-passage (virulent) strains (Fig. 4; data not shown). The inability to detect LipL45 after culture passage of Leptospira is probably due to increased expression or activity of the protease that cleaves LipL45 into P31LipL45. However, LipL45 is not completely suppressed in high-passage organisms; LipL45 was detected when large numbers of leptospires were probed in an immunoblot (Fig. 6, lane 2).
We have not eliminated the possibility that P31LipL45 is a decay product rather than a processed product of LipL45. If it is a decay product, it is plausible that expression or activity of the protease or proteases that target LipL45 is suppressed in an infected host. A similar phenomenon has been described for the SpeB cysteine protease in group A streptococcus. Examination of different clinical isolates of group A streptococcus demonstrated that SpeB expression is inversely correlated with the severity of disease (34). A potential substrate of the SpeB protease is the surface-exposed M protein, a virulence factor essential for group A streptococcus to evade complement-mediated lysis (34). Failure to downregulate SpeB expression or activity may result in inappropriate cleavage and inactivation of M protein during infection. Other bacterial pathogens are known to regulate protease expression in response to environmental cues (21).
The antigenicity of LipL45 allowed us to isolate the lipL45 gene from a leptospiral genomic expression library. A total of 40% of the clones isolated from the expression library harbored genes encoding lipoproteins, including LipL32 and LipL41. Previous studies with B. burgdorferi and T. pallidum have shown that lipoproteins are highly immunogenic when animals are immunized with whole bacteria (8, 13). Lipid moieties dramatically increase the antibody response to the peptide or protein to which they are attached (5, 15). Undoubtedly, other lipoprotein genes remain to be isolated from the leptospiral genome. At least eight proteins are labeled when Leptospira is grown in the presence of radiolabeled palmitate (25). In addition, other leptospiral lipoproteins may be expressed only during infection, as demonstrated for several B. burgdorferi lipoproteins (18, 51, 54). Surprisingly, up to 38% of the clones harbored genes for RNA polymerase subunits. It is possible that like GroEL, which represented 5% of the clones isolated from the expression library, the RNA polymerase subunits contain sequences that are highly stimulatory to the immune system (40).
Although the serum used to screen the library contained OmpL1 antibody (data not shown), we failed to clone the OmpL1 gene from the library, possibly because of its extreme lethality when it is expressed in E. coli, even at low levels (26). Similarly, we were not able to isolate the gene encoding DnaK from the library; the immune system of the rabbit may not have recognized this ubiquitous protein because it could be a self-antigen. It is also possible that some genes encoding protein antigens were not isolated due to chance. In addition to the RNA polymerase subunits, we isolated two other genes that are found in a wide range of bacteria and encode two enzyme subunits involved in metabolism. The isolation of genes encoding cytoplasmic proteins indicates that whole-cell immunization results in production of antibody against antigens that may not be accessible to the host immune system during infection by live Leptospira. Indeed, serum from infected hamsters did not react with some antigens recognized by the rabbit serum used to screen the library (data not shown).
In conclusion, we isolated a gene encoding a virulence-associated lipoprotein, LipL45. Our results suggest that LipL45 is an inner membrane protein whose C terminus is released as a 31-kDa peripheral membrane protein. P31LipL45 is associated with both the inner and outer membranes. The amount of P31LipL45 in leptospires increases dramatically during the stationary phase, indicating that expression is environmentally regulated. Immunohistochemical and serologic studies with the hamster model of leptospirosis indicated that LipL45 or P31LipL45 or both are expressed during infection. The level of expression appears to be relatively low, since the immunohistochemical reaction is less intense that the reactions seen with other leptospiral antigens (Fig. 10) and few sera from human leptospirosis patients react with recombinant LipL45, as determined by enzyme-linked immunosorbent assays (4, 19, 24). An important survival strategy of pathogenic bacteria is to restrict expression of key immunoprotective antigens during infection of their mammalian hosts. Further studies are under way to determine whether LipL45 or its processed form, P31LipL45, is a target of a protective immune response.
Acknowledgments
We thank Ann Bauernfiend (University of Southern Indiana) and LaDonna Elpers (BAS, Inc.) for their technical assistance with the immunohistochemical experiments and Mary Mazel (Veterans Affairs Greater Los Angeles Healthcare System) for her assistance with the hamster challenge procedure. We also thank Ben Adler for his generous gift of GroEL antiserum.
This work was supported by VA Medical Research Funds (J.M.) and by Public Health Service grant AI-34431 to D.A.H.
Editor: D. L. Burns
REFERENCES
- 1.Ballard, S. A., S. Faine, and B. Adler. 1990. Purification and characterization of a protein antigen from Leptospira interrogans serovar hardjo, common to a wide range of bacteria. J. Gen. Microbiol. 136: 1849–1857. [DOI] [PubMed] [Google Scholar]
- 2.Ballard, S. A., M. Go, R. P. Segers, and B. Adler. 1998. Molecular analysis of the dnaK locus of Leptospira interrogans serovar Copenhageni. Gene 216: 21–29. [DOI] [PubMed] [Google Scholar]
- 3.Ballard, S. A., R. P. A. M. Segers, N. Bleumink-Pluym, J. Fyfe, S. Faine, and B. Adler. 1993. Molecular analysis of the hsp (groE) operon of Leptospira interrogans serovar copenhageni. Mol. Microbiol. 8: 739–751. [DOI] [PubMed] [Google Scholar]
- 4.Barnett, J. K., D. Barnett, C. A. Bolin, T. A. Summers, E. A. Wagar, N. F. Cheville, R. A. Hartskeerl, and D. A. Haake. 1999. Expression and distribution of leptospiral outer membrane components during renal infection of hamsters. Infect. Immun. 67: 853–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bessler, W. G., M. Cox, A. Lex, B. Suhr, K. H. Wiesmuller, and G. Jung. 1985. Synthetic lipopeptide analogs of bacterial lipoprotein are potent polyclonal activators for murine B lymphocytes. J. Immunol. 135: 1900–1905. [PubMed] [Google Scholar]
- 6.Bolin, C. A., R. L. Zuerner, and G. Trueba. 1989. Effect of vaccination with a pentavalent leptospiral vaccine containing Leptospira interrogans serovar hardjo type hardjo-bovis on type hardjo-bovis infection of cattle. Am. J. Vet. Res. 50: 2004–2008. [PubMed] [Google Scholar]
- 7.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256: 1604–1607. [PubMed] [Google Scholar]
- 8.Brandt, M. E., B. S. Riley, J. D. Radolf, and M. V. Norgard. 1990. Immunogenic integral membrane proteins of Borrelia burgdorferi are lipoproteins. Infect. Immun. 58: 983–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brenner, D. J., A. F. Kaufmann, K. R. Sulzer, A. G. Steigerwalt, F. C. Rogers, and R. S. Weyant. 1999. Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int. J. Syst. Bacteriol. 49: 839–858. [DOI] [PubMed] [Google Scholar]
- 10.Brenot, A., D. Trott, I. Saint Girons, and R. Zuerner. 2001. Penicillin-binding proteins in Leptospira interrogans. Antimicrob. Agents Chemother. 45: 870–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, J. A., R. B. LeFebvre, and M. J. Pan. 1991. Protein and antigen profiles of prevalent serovars of Leptospira interrogans. Infect. Immun. 59: 1772–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bulach, D. M., T. Kalambaheti, A. de la Peña-Moctezuma, and B. Adler. 2000. Lipopolysaccharide biosynthesis in Leptospira. J. Mol. Microbiol. Biotechnol. 2: 375–380. [PubMed] [Google Scholar]
- 13.Chamberlain, N. R., M. E. Brandt, A. L. Erwin, J. D. Radolf, and M. V. Norgard. 1989. Major integral membrane protein immunogens of Treponema pallidum are proteolipids. Infect. Immun. 57: 2872–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chomczynski, P. 1992. One-hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal. Biochem. 201: 134–139. [DOI] [PubMed] [Google Scholar]
- 15.Erdile, L. F., M. A. Brandt, D. J. Warakomski, G. J. Westrack, A. Sadziene, A. G. Barbour, and J. P. Mays. 1993. Role of attached lipid in immunogenicity of Borrelia burgdorferi OspA. Infect. Immun. 61: 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faine, S. 1962. The growth of Leptospira australis B in the kidneys of mice in the incipient experimental carrier state. J. Hyg. 60: 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faine, S. 1964. Reticuloendothelial phagocytosis of virulent leptospires. Am. J. Vet. Res. 25: 830–835. [PubMed] [Google Scholar]
- 18.Fikrig, E., S. W. Barthold, W. Sun, W. Feng, S. R. Telford III, and R. A. Flavell. 1997. Borrelia burgdorferi P35 and P37 proteins, expressed in vivo, elicit protective immunity. Immunity 6: 531–539. [DOI] [PubMed] [Google Scholar]
- 19.Flannery, B., D. Costa, F. P. Carvalho, H. Guerreiro, J. Matsunaga, E. D. Da Silva, A. G. Ferreira, L. W. Riley, M. G. Reis, D. A. Haake, and A. I. Ko. 2001. Evaluation of recombinant leptospira antigen-based enzyme-linked immunosorbent assays for the serodiagnosis of leptospirosis. J. Clin. Microbiol. 39: 3303–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujiki, Y., A. L. Hubbard, S. Fowler, and P. B. Lazarow. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93: 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guina, T., E. C. Yi, H. Wang, M. Hackett, and S. I. Miller. 2000. A PhoP-regulated outer membrane protease of Salmonella enterica serovar Typhimurium promotes resistance to alpha-helical antimicrobial peptides. J. Bacteriol. 182: 4077–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haake, D. A. 2000. Spirochetal lipoproteins and pathogenesis. Microbiology 146: 1491–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haake, D. A., C. I. Champion, C. Martinich, E. S. Shang, D. R. Blanco, J. N. Miller, and M. A. Lovett. 1993. Molecular cloning and sequence analysis of the gene encoding OmpL1, a transmembrane outer membrane protein of pathogenic Leptospira spp. J. Bacteriol. 175: 4225–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haake, D. A., G. Chao, R. L. Zuerner, J. K. Barnett, D. Barnett, M. Mazel, J. Matsunaga, P. N. Levett, and C. A. Bolin. 2000. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect. Immun. 68: 2276–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haake, D. A., C. Martinich, T. A. Summers, E. S. Shang, J. D. Pruetz, A. M. McCoy, M. K. Mazel, and C. A. Bolin. 1998. Characterization of leptospiral outer membrane lipoprotein LipL36: downregulation associated with late-log-phase growth and mammalian infection. Infect. Immun. 66: 1579–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haake, D. A., M. K. Mazel, A. M. McCoy, F. Milward, G. Chao, J. Matsunaga, and E. A. Wagar. 1999. Leptospiral outer membrane proteins OmpL1 and LipL41 exhibit synergistic immunoprotection. Infect. Immun. 67: 6572–6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haake, D. A., E. M. Walker, D. R. Blanco, C. A. Bolin, M. N. Miller, and M. A. Lovett. 1991. Changes in the surface of Leptospira interrogans serovar grippotyphosa during in vitro cultivation. Infect. Immun. 59: 1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Hayashi, S., and H. C. Wu. 1990. Lipoproteins in bacteria. J. Bioenerg. Biomembr. 22: 451–471. [DOI] [PubMed] [Google Scholar]
- 30.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69: 1231–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, R. C., and V. G. Harris. 1967. Differentiation of pathogenic and saprophytic leptospires. I. Growth at low temperatures. J. Bacteriol. 94: 27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jost, B. H., B. Adler, and S. Faine. 1989. Experimental immunisation of hamsters with lipopolysaccharide antigens of Leptospira interrogans. J. Med. Microbiol. 29: 115–120. [DOI] [PubMed] [Google Scholar]
- 33.Jost, B. H., B. Adler, T. Vinh, and S. Faine. 1986. A monoclonal antibody reacting with a determinant on leptospiral lipopolysaccharide protects guinea pigs against leptospirosis. J. Med. Microbiol. 22: 269–275. [DOI] [PubMed] [Google Scholar]
- 34.Kansal, R. G., A. McGeer, D. E. Low, A. Norrby-Teglund, and M. Kotb. 2000. Inverse relation between disease severity and expression of the streptococcal cysteine protease, SpeB, among clonal M1T1 isolates recovered from invasive group A streptococcal infection cases. Infect. Immun. 68: 6362–6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 36.Levett, P. N. 2001. Leptospirosis. Clin. Microbiol. Rev. 14: 296–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marshall, R. B. 1976. The route of entry of leptospires into the kidney tubule. J. Med. Microbiol. 9: 149–152. [DOI] [PubMed] [Google Scholar]
- 38.Merien, F., J. Truccolo, G. Baranton, and P. Perolat. 2000. Identification of a 36-kDa fibronectin-binding protein expressed by a virulent variant of Leptospira interrogans serovar icterohaemorrhagiae. FEMS Microbiol. Lett. 185: 17–22. [DOI] [PubMed] [Google Scholar]
- 39.Nunes-Edwards, P. L., A. B. Thiermann, P. J. Bassford, Jr., and L. V. Stamm. 1985. Identification and characterization of the protein antigens of Leptospira interrogans serovar hardjo. Infect. Immun. 48: 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, S. H., B. Y. Ahn, and M. J. Kim. 1999. Expression and immunologic characterization of recombinant heat shock protein 58 of Leptospira species: a major target antigen of the humoral immune response. DNA Cell Biol 18: 903–910. [DOI] [PubMed] [Google Scholar]
- 41.Picardeau, M., A. Brenot, and I. Saint Girons. 2001. First evidence for gene replacement in Leptospira spp. Inactivation of L. biflexa flaB results in non-motile mutants deficient in endoflagella. Mol. Microbiol. 40: 189–199. [DOI] [PubMed] [Google Scholar]
- 42.Porcella, S. F., T. G. Popova, D. R. Akins, M. Li, J. D. Radolf, and M. V. Norgard. 1996. Borrelia burgdorferi supercoiled plasmids encode multicopy tandem open reading frames and a lipoprotein gene family. J. Bacteriol. 178: 3293–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Renesto, P., K. Lorvellec-Guillon, M. Drancourt, and D. Raoult. 2000. rpoB gene analysis as a novel strategy for identification of spirochetes from the genera Borrelia, Treponema, and Leptospira. J. Clin. Microbiol. 38: 2200–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shang, E. S., M. M. Exner, T. A. Summers, C. Martinich, C. I. Champion, R. E. W. Hancock, and D. A. Haake. 1995. The rare outer membrane protein, OmpL1, of pathogenic Leptospira species is a heat-modifiable porin. Infect. Immun. 63: 3174–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shang, E. S., T. A. Summers, and D. A. Haake. 1996. Molecular cloning and sequence analysis of the gene encoding LipL41, a surface-exposed lipoprotein of pathogenic Leptospira species. Infect. Immun. 64: 2322–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shevchenko, D. V., D. R. Akins, E. Robinson, M. Li, T. G. Popova, D. L. Cox, and J. D. Radolf. 1997. Molecular characterization and cellular localization of TpLRR, a processed leucine-rich repeat protein of Treponema pallidum, the syphilis spirochete. J. Bacteriol. 179: 3188–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shevchenko, D. V., T. J. Sellati, D. L. Cox, O. V. Shevchenko, E. J. Robinson, and J. D. Radolf. 1999. Membrane topology and cellular location of the Treponema pallidum glycerophosphodiester phosphodiesterase (GlpQ) ortholog. Infect. Immun. 67: 2266–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simpson, W. J., M. E. Schrumpf, S. F. Hayes, and T. G. Schwan. 1991. Molecular and immunological analysis of a polymorphic periplasmic protein of Borrelia burgdorferi. J. Clin. Microbiol. 29: 1940–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sonrier, C., C. Branger, V. Michel, N. Ruvoen-Clouet, J. P. Ganiere, and G. Andre-Fontaine. 2000. Evidence of cross-protection within Leptospira interrogans in an experimental model. Vaccine 19: 86–94. [DOI] [PubMed] [Google Scholar]
- 50.Steiner, G., and G. Steiner. 1944. New simple silver stain for demonstration of bacteria, spirochetes, and fungi in sections of paraffin embedded tissue blocks. J. Lab. Clin. Med. 29: 868–871. [Google Scholar]
- 51.Suk, K., S. Das, W. Sun, B. Jwang, S. W. Barthold, R. A. Flavell, and E. Fikrig. 1995. Borrelia burgdorferi genes selectively expressed in the infected host. Proc. Natl. Acad. Sci. USA 92: 4269–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swancutt, M. A., B. S. Riley, J. D. Radolf, and M. V. Norgard. 1989. Molecular characterization of the pathogen-specific, 34-kilodalton membrane immunogen of Treponema pallidum. Infect. Immun. 57: 3314–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thiermann, A. B., R. D. McClellan, and H. T. Hill. 1984. Improved techniques for the isolation of leptospires from swine abortion cases. Ann. Proc. Am. Assoc. Vet. Lab. Diagn. 27: 233–244. [Google Scholar]
- 54.Wallich, R., C. Brenner, M. D. Kramer, and M. M. Simon. 1995. Molecular cloning and immunological characterization of a novel linear-plasmid-encoded gene, pG, of Borrelia burgdorferi expressed only in vivo. Infect. Immun. 63: 3327–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yelton, D. B., and N. W. Charon. 1984. Cloning of a gene required for tryptophan biosynthesis from Leptospira biflexa serovar patoc into Escherichia coli. Gene 28: 147–152. [DOI] [PubMed] [Google Scholar]
- 56.Zuerner, R. L., W. Knudtson, C. A. Bolin, and G. Trueba. 1991. Characterization of outer membrane and secreted proteins of Leptospira interrogans serovar pomona. Microb. Pathog. 10: 311–322. [DOI] [PubMed] [Google Scholar]