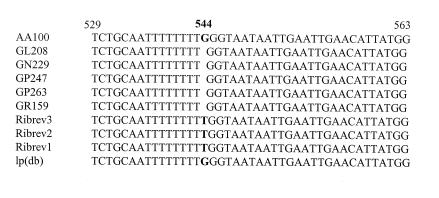Abstract
The final step of the intracellular life cycle of Legionella pneumophila and other intracellular pathogens is their egress from the host cell after termination of intracellular replication. We have previously isolated five spontaneous mutants of L. pneumophila that replicate intracellularly similar to the wild-type strain but are defective in pore formation-mediated cytolysis and egress from mammalian and protozoan cells, and the mutants have been designated rib (release of intracellular bacteria). Here, we show that the rib mutants are not defective in the activity of enzymes secreted through the type II secretion system, including phospholipase A, lysophospholipase A, and monoacylglycerol lipase, although they are potential candidates for factors that lyse host cell membranes. In addition, the pilD and lspG mutants, which are defective in the type II secretion system, are not defective in the pore-forming toxin. We show that all five rib mutants have an identical point mutation (deletion) following a stretch of poly(T) in the icmT gene. Spontaneous revertants of the rib mutants, due to an insertion of a nucleotide following the poly(T) stretch in icmT, have been isolated and shown to have regained the wild-type phenotype. We constructed an icmT insertion mutant (AA100kmT) in the chromosome of the wild-type strain by allelic exchange. The AA100kmT mutant was as defective as the rib mutant in pore formation-mediated cytolysis and egress from mammalian and protozoan cells. Both the rib mutant and the AA100kmT mutant were complemented by the icmT gene for their phenotypic defect. rtxA, a gene that is thought to have a minor role in pore formation, was not involved in pore formation-mediated cytolysis and egress from mammalian and protozoan cells. We conclude that the icmT gene is essential for pore formation-mediated lysis of mammalian and protozoan cells and the subsequent bacterial egress.
Legionella pneumophila, the etiologic agent of Legionnaires’ disease, is a gram-negative bacterium, ubiquitous in the aquatic environment, where it survives as an intracellular parasite of amoebae and ciliated protozoa (21, 31). In humans, L. pneumophila replicates within alveolar macrophages and possibly epithelial cells (1, 19) and utilizes similar mechanisms to parasitize the protozoan host (18, 36). After phagocytosis by mammalian and protozoan cells, the bacteria modulate the biogenesis of their vacuole into a niche that permits it to escape the classical endosomal-lysosomal degradation pathways (9, 23). This vacuole is subsequently surrounded by mitochondria and rough endoplasmic reticulum (2, 22). A group of 23 genes designated dot (defect in organelle trafficking)/icm (intracellular multiplication) are implicated in modulating the formation of the replicative vacuole and subsequent intracellular replication (33, 38). The dot/icm complex is thought to constitute a type IV-like secretion system capable of transferring effector molecules into the host cell to evade the endocytic pathways.
A fundamental step in the pathogenic life cycle of intracellular pathogens is their ability to lyse and egress from the host cell after termination of intracellular replication, which subsequently leads to infection of uninfected neighboring cells or to transmission to a new host. The mechanisms by which intracellular pathogens egress from the host cell are not well understood. It has been presumed that the physical and metabolic burden on the host cell by a large number of intracellular bacteria is sufficient to rupture the host cell by nonspecific means. Recent data about L. pneumophila have shed some light on a specific and pathogen-regulated mechanism for bacterial egress from the host cell. After termination of intracellular replication within mammalian and protozoan cells, L. pneumophila induces cytolysis of the host cell, which is mediated by a pore-forming activity (6, 17). This pore formation-mediated cytolysis is triggered in vitro and in vivo upon growth transition into the postexponential phase (6, 10). Five mutants of L. pneumophila defective in the pore-forming toxin are not defective in modulating biogenesis of their replicative vacuole and replicate intracellularly similar to the wild-type strain. However, these mutants are defective in egress from mammalian and protozoan cells upon termination of intracellular replication, and the mutants have been designated rib (release of intracellular bacteria) (6). However, the mutants are released at a later time, most likely due to apoptotic changes in the infected cell (6). We propose to designate the pore forming-toxin Rib toxin. The rib mutants are also defective in acute cytotoxic lethality to mice (6). We have shown that the miniTn10::Kan insertions used to generate these five mutants are located in different chromosomal regions distinct from the two regions of the dot/icm genes (6). However, cosmid clones harboring the chromosomal regions defective in the mutants failed in complementation studies for the defect in the pore-forming toxin. In addition, reconstruction of the original five mutants by allelic exchange of the Kan-inserted loci into the wild-type chromosome showed that the newly constructed mutants are indistinguishable from the wild-type strain in intracellular replication, cytotoxicity, and pore formation-mediated egress from mammalian and protozoan cells. Therefore, the mutations in the five mutants are spontaneous (6).
In this report, we show that the rib mutants are not defective in any of the enzymes secreted through the type II secretion system, which are potential factors that disrupt host cell membranes. This was further confirmed by the pilD and lspG mutants, both of which are defective in the type II secretion system and both of which are not defective in the pore-forming toxin. We show that the icmT gene is the defective locus in the rib mutants and is essential for egress of intracellular bacteria from both U937 macrophages and Acanthamoeba polyphaga upon termination of intracellular replication. The defect in icmT is a point deletion after a stretch of poly(T) in the carboxy terminus of the gene. In addition, we show that the rtxA gene that has been thought to be required for pore formation (12) is not required for the pore formation-mediated egress from macrophages and protozoa.
MATERIALS AND METHODS
Bacterial strains and media.
L. pneumophila serogroup I strain AA100 was the parental strain used for all experiments in this study. L. pneumophila strains were grown on buffered charcoal-yeast extract (BCYE) plates or in buffered yeast extract broth. All the strains and the plasmids used in this study are described in Table 1. For L. pneumophila, antibiotics were added to the media at the following concentrations: kanamycin, 50 μg/ml; chloramphenicol, 2.5 and 5 μg/ml. Escherichia coli strains were cultured on Luria-Bertani (LB) agar plates or in LB broth. Antibiotics were added to the media at the following concentrations: kanamycin, 50 μg/ml; chloramphenicol, 20 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or sequence | Reference or source |
|---|---|---|
| Strains | ||
| L. pneumophila | ||
| AA100 | 3 | |
| GN229 | Kanr | 6 |
| GR159 | Kanr | 6 |
| GL208 | Kanr | 6 |
| GP247 | Kanr | 6 |
| GP263 | Kanr | 6 |
| AA100kmT | AA100; icmT:: Kanr | This study |
| AA100kmS | AA100; icmS:: Kanr | This study |
| φp24 | ΔRtxA | 13, 14 |
| NU259 | 130b; LpsG::Kanr | 30 |
| NU243 | 130b; PilD::Kanr | 28 |
| ribrev1 | This study | |
| ribrev2 | This study | |
| ribrev3 | This study | |
| E. coli DH5α | supE44 ΔlacU169 (ψ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | BRL |
| Plasmids | ||
| pBC SK+ | ColE1 origin, lacZα, Cmr | Stratagene |
| pMW100 | pMMB207αβ; Cmr; icmGCDJB, tphA, and icmF region | 29 |
| pGS-LC-32 | pMMB207αβ; Cmr; icmTSRQPO | 34 |
| pGS-LC-47 | pMMB207αβ; Cmr; icmNMLKEG | 33 |
| pTSRQ | pBC SK+; Cmr; icmTSRQ | This study |
| pTS | pBC SK+; Cmr; icmTS | This study |
| pT | pBC SK+; Cmr; icmT gene | This study |
| pkanT | pBC SK+; Cmr; ; icmT:: Kanr | This study |
| pS | pBC SK+; Cmr; icmS gene | This study |
| pkanS | pBC SK+; Cmr; ; icmS:: Kanr | This study |
| pR | pBC SK+; Cmr; icmR gene | This study |
| pkanR | pBC SK+; Cmr; icmR:: Kanr | This study |
| pQ | pBC SK+; Cmr; icmQ gene | This study |
| pkanQ | pBC SK+; Cmr; icmQ:: Kanr | This study |
Cell culture.
U937 cells were cultured in RPMI 1640 containing 10% fetal bovine serum (Gibco). U937 cells were differentiated with phorbol 12-myristate 13-acetate (Sigma) for 48 h before use as previously described (18). A. polyphaga cells were maintained in peptone yeast glucose medium. Glucose was omitted from the medium to prevent growth of amoebae during infection with Legionella.
DNA manipulations.
Transfections, restriction enzyme digestions, and DNA manipulation were performed as previously described (5). Restriction enzymes and T4 DNA ligase were purchased from Promega (Madison, Wis.). L. pneumophila chromosomal DNA was prepared by using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, Minn.). Plasmid and cosmid DNA preparations were performed with the Bio-Rad (Hercules, Calif.) Quantum miniprep kit and by the polyethylene glycol DNA extraction procedure as described before (32). Electroporations were performed with a Bio-Rad Gene Pulser, as recommended by the manufacturer. Purification of DNA fragments from agarose gels for subcloning was carried out with a QIAquick gel purification kit (Qiagen Inc., Chatsworth, Calif.). Oligonucleotide synthesis for PCR and sequencing were performed by Integrated DNA Technologies, Inc. (Coralville, Calif.). Sequencing was carried out by ACGT Inc. (Northbrook, Ill.). Sequence analysis and comparisons were carried out with DNAMAN software and NCBI molecular biology software (www.ncbi.nlm.nih.gov).
Enrichment of rib transconjugants in A. polyphaga.
A cosmid library of L. pneumophila in E. coli was mobilized into the rib mutants GN229 and GL208 by conjugation. The selection was carried out on BCYE containing kanamycin (50 μg/ml) and chloramphenicol (5 μg/ml). The 2resulting transconjugants were pooled and frozen at −80°C until use. The pools were used for infection of A. polyphaga cells to enrich for clones that were able to egress from A. polyphaga after intracellular replication. Briefly, A. polyphaga was infected with the pool of transconjugants of the rib mutant GN229 at a multiplicity of infection (MOI) of 10 in six-well tissue culture dishes containing 106 A. polyphaga per well, and the dishes were incubated for 1 h at 37°C. At the end of the infection period, the monolayers were treated with gentamicin (50 μg/ml). The bacteria released into the culture medium of amoebae after 48 h incubation were used to infect freshly grown A. polyphaga as described above, and the supernatant of the infection was obtained after 24 h of incubation. Three rounds of enrichment were carried out. Representatives of the transconjugant clones that egress from amoebae were used in a contact-dependent hemolysis assay and cytotoxicity assay for U937 cells.
Genetic characterization of the rib mutants.
Three plasmids (pMW100, pGS-Lc-47, and pGS-Lc-32) containing different regions of dot/icm genes (kind gifts of H. Shuman) (Table 1) were used to transform the rib mutants by electroporation. The icmTSRQ locus was amplified from AA100 genomic DNA, and SalI and NotI sites were introduced in the forward (P1) and reverse (P2) primers (Table 2), respectively, by PCR using long-template Taq (Boehringer Mannheim). The annealing temperature used was 50°C, with the extension temperature of 68°C for 25 cycles. The PCR product was purified from the agarose gel and ligated to the SalI- and NotI-digested pBC sk+ plasmid. The resulting ligation mixture was used to transform DH5α. icmR and icmQ genes were cloned in a similar fashion as described above to create pR and pQ, respectively. AvaII digestion was used to delete the icmRQ 2region from the plasmid pTSRQ, hence resulting in pTS. Plasmid pT contains an in-frame deletion of the icmS gene created by inverse PCR of the plasmid pTS using the primers P3 and P4 (Table 2) with the NheI restriction enzyme sequence linked to the 5′ end of the primers. The same strategy was utilized in creating in-frame deletion of the icmT gene with the resulting plasmid pS, using primers P5 and P6 (Table 2). The following primer combinations were used for amplification of different icm genes: P7 and P8 for the icmR gene and P9 and P2 for the icmQ gene.
TABLE 2.
Primers used in this study
| Primer | Sequencea | Restriction enzyme |
|---|---|---|
| P1 (salIIcmTS) | ACGCGTCGACCACAGTTAAAACTTCAAGCTGAACC | SalI |
| P2 (notIRQ2) | ATAAGAATGCGGCCGCTGCTCAGAGCTATTTTT | NotI |
| P3 | CTAGCTAGCACACTTGCTAATATCTCGCT | NheI |
| P4 | CTAGCTAGCCGTTTGTTATGTATGACGG | NheI |
| P5 | CTAGCTAGCCCAGTGGCTCGTTGCT | NheI |
| P6 | CTAGCTAGCGGTTCAGTTAGATCTGCTAG | NheI |
| P7 | ACGCGTCGACATGAGCTTATTCAAGGGGGC | SalI |
| P8 | ATAAGAATGCGGCCGCGGGAACCAAGAATTAGGGG | NotI |
| P9 | ACGCGTCGACAATGTTTGGTGCTGCTGAATC | SalI |
Underlined letters are the sequences of restriction enzymes used for cloning.
In vitro transposon mutagenesis was carried out on the plasmid of choice using an EZ::TN<KAN-2> insertion kit as described by the manufacturer (Epicenter Technologies, Madison, Wis.). A transposon library of the plasmid was used to transform E. coli DH5α. Transformants were selected on LB agar plates containing kanamycin-chloramphenicol and screened by PCR for location of insertion. The resulting plasmid extracted from E. coli DH5α was used to transform the rib mutant. Some of the plasmid constructs containing the transposon kanamycin insertion were used for allelic exchange to L. pneumophila AA100 chromosome by natural transformation (37).
Complementation of the defects was carried out for the resulting transformants by contact-dependent hemolysis assay, cytolysis of A. polyphaga and U937 cells following intracellular replication, and cytotoxicity assay as described below.
Contact-dependent pore formation assay.
Contact-dependent pore formation in plasma membrane was determined by examining hemolysis of sheep red blood cells (sRBCs) by L. pneumophila at an MOI of 25 following 2 h of bacterium-sRBC contact, as previously described (6, 26).
Cytotoxicity of L. pneumophila to U937 macrophages and A. polyphaga.
L. pneumophila strains were grown on BCYE plates for 3 days prior to infection of U937 macrophages or amoebae. Infection was performed, in triplicate, in 96-well plates containing 105 cells/well at an MOI of 1 for 1 h for U937, and in 24-well plates with coverslips containing 5 × 105 cells/well at an MOI of 10 for A. polyphaga. The infection period was followed by gentamicin (50 μg/ml) treatment for 1 h, which was followed by three washes to remove extracellular bacteria. At several time points, the monolayers of macrophages were treated for 4 h with 10% Alamar Blue dye (Alamar Bioscience Inc., Sacramento, Calif.) as previously described (4). Viability of the monolayers of macrophages was determined by measurement of optical density (OD) of Alamar Blue-treated monolayers by using a VMAX Kinetic Microplate reader (Molecular Devices, Menlo Park, Calif.) and expressed as percent cell death compared to uninfected cells by using the formula [1 − (mean OD of treated cells/mean OD of untreated cells)] × 100%.
In A. polyphaga infection experiments, the cytolysis of amoebae was documented by images of the infected monolayers at different time intervals to provide a qualitative assessment of the cytolysis. Cells were observed with an Axiophot Photomicroscope (Carl Zeiss, Inc., Oberkochen, Germany).
Growth kinetics of released L. pneumophila strains from U937 macrophages.
Infections of U937 macrophages by L. pneumophila strains were performed, in triplicate, in 96-well plates containing 105 cells/well at an MOI of 1 for U937 macrophages. At the end of the infection period, the monolayers were treated with gentamicin (50 μg/ml) for 1 h as described above. The number of released bacteria in the monolayers at several time intervals after washing of the gentamicin was determined.
Lipolytic activities in supernatants and cell lysates.
The bacteria were grown in buffered yeast extract broth to an OD at 600 nm (OD660) of 1.85 (late-logarithmic phase). The bacteria were pelleted and lysed with Triton X-100-lysozyme, and the volume of the cell lysate was adjusted to the volume of the supernatant. Due to their ability to release high levels of fatty acids from dipalmitoylphosphatidylglycerol (DPPG) and monopalmitoyllysophosphatidylcholine, dilutions of 1:2 or 1:10 of the lysates were made, prior to the enzymatic assays.
Phospholipase A (PLA), lysophospholipase A (LPLA), and monoacylglycerol lipase activities were determined as we described previously (7, 16). Briefly, PLA activity was determined by hydrolysis of the substrates DPPG and dipalmitoylphosphatidylcholine (final concentrations, 5 mg/ml). The LPLA activity was determined by hydrolysis of the substrate monopalmitoylphosphatidylcholine (1-MPG) (final concentration, 3.4 mg/ml). Mono-acylglycerol lipase activity was determined by hydrolysis of the 1-monopalmitoylglycerol substrate (final concentration, 2.2 mg/ml).
RESULTS
Enrichment for cosmid clones to complement the rib mutants for their defect in egress from A. polyphaga.
Since the rib mutants are completely defective in egress from A. polyphaga, we exploited this phenotype to isolate cosmid clones that complement the mutants for egress from amoebae. The cosmid library of L. pneumophila was mobilized into the rib mutants GN229 and GL208, and the two pools of transconjugants were used, separately, to infect A. polyphaga. The bacteria released into the culture medium after 48 h incubation were used to infect freshly grown A. polyphaga to enrich for transconjugants that are capable of egress. The two mutants (without the cosmid library) were used as controls in these infections to monitor the increase in the number of GN229 and GL208 transconjugants that are able to egress compared to the mutants. The defect in egress for the two rib mutants was completely complemented to the wild-type level after three enrichments for transconjugant clones that were able to egress from A. polyphaga. In contrast, the defect in egress of the two mutant controls was stable during the three enrichments (data not shown). Surprisingly, several trials have failed to isolate cosmids from 30 random transconjugants of GL208 and 15 random transconjugants of GN229, which regained the wild-type phenotype for egress from A. polyphaga. We concluded that these transconjugants were spontaneous revertants that have been able to repair the genetic lesion responsible for the rib phenotype.
Lipolytic activities of enzymes secreted through the type II secretion system by the rib mutants.
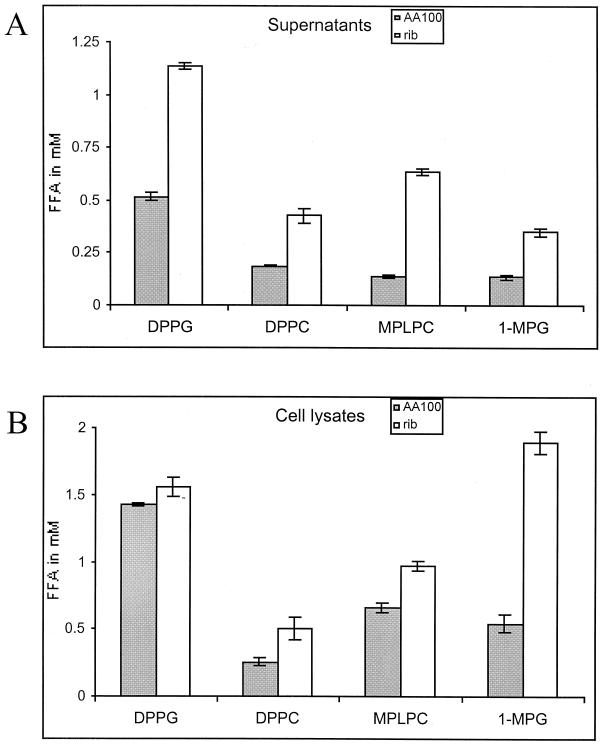
Many lipolytic enzymes secreted by the type II secretion system of L. pneumophila (lsp, for Legionella secretion pathway) are potential candidates for enzymes that may lyse the host cell membrane by the hydrolysis of lipid constituents or generation of the hemolytic agent lysophosphatidylcholine. Therefore, we tested the rib mutants for these activities (7, 16). Our data show that none of the previously described enzyme activities of PLA, LPLA, and monoacylglycerol lipase were reduced in the rib mutant GN229 (Fig. 1A and 1B). On the contrary, all activities were slightly higher in the rib mutant than in the wild-type strain. The acid phosphatase enzyme, which is also secreted by the type II secretion system (30), was also elevated in culture supernatants of the rib mutants (data not shown). It is not clear why the activities are elevated in the rib mutant.
FIG. 1.
The rib mutants are not defective in lipolytic enzymes secreted through the type II secretion system. Lipid hydrolysis by supernatants (A) and cell lysates (B) of AA100 and the rib mutant was tested for the ability to release free fatty acids (FFA) from dipalmitoylphosphatidylcholine (DPPC), DPPG, monopalmitoyllysophosphatidylcholine (MPLPC), and 1-MPG. These results are representative of two additional experiments, with the exception of a variation observed between experiments for the cell lysates activity of the wild-type strain and the mutant on 1-MPG (data not shown). Error bars represent standard deviations.
The data were further confirmed by examination of the pore-forming activity, using contact-dependent hemolysis of sRBCs, of the two mutants defective in the type II secretion system. These mutants are NU259, which carries a Kan insertion in lspG, which encodes an inner membrane structural protein, and the NU243 mutant, which is disrupted in pilD, the prepilin peptidase gene. The result showed that these two mutants were indistinguishable from the wild-type strain in pore formation activity (data not shown). Thus, the type II secretion system is not required for the pore-forming activity, and the rib mutants are not defective in lipolytic enzymes secreted by the type II secretion system.
The icmTSRQPO locus complements the phenotype of the five rib mutants.
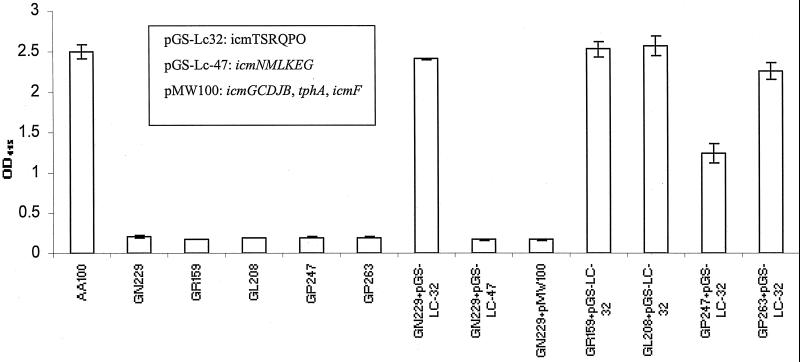
Since the dot/icm genes have been shown to be involved in the pore-forming activity of L. pneumophila (26), we decided to introduce different regions of the 23 dot/icm genes to the rib mutants for possible complementation. The icmTSRQPO region; the icmGCDJB, tphA, and icmF regions; and the icmNMLKEG region were tested for the ability to restore the pore-forming toxin activity to the rib mutant GN229, using contact-dependent hemolysis of sRBCs. The data showed that only the plasmid harboring icmTSRQPO complemented the GN229 phenotype (Fig. 2). Interestingly, the other four rib mutants were also complemented by the same icmTSRQPO locus. The data showed that the rib mutants’ genetic lesion was within icmTSRQPO.
FIG. 2.
The icmTSRQPO-containing plasmid (pGS-Lc32) complements the rib mutants. Contact-dependent hemolysis of sRBC was performed at MOI of 25 for 2 h at 37°C and measured by OD415 for the release of hemoglobin. The genes harbored by each plasmid are indicated in the box insert. These data are representative of three different experiments, each done in triplicate, and error bars represent standard deviations.
The icmT gene is essential for the pore-forming activity.
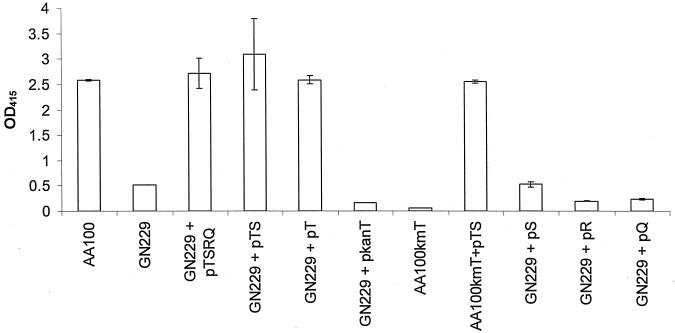
In order to determine which gene(s) was defective in the rib mutants, two approaches were adopted. The first approach was to clone various regions of the icmTSRQPO and test them for complementation of the rib mutants (Fig. 3). As icmPO has homologies to proteins involved in the DNA transfer by conjugation and essential for intracellular replication (33), the icmTSRQ region was cloned first from AA100 and tested for complementation of the GN229 mutant. The contact-dependent hemolysis assay (Fig. 4) showed that pTSRQ complemented the rib mutant GN229. The plasmid pTS, obtained by deletion of icmRQ from the plasmid pTSRQ, and the plasmids pT and pS, obtained by in-frame deletions in icmS and icmT, respectively, were tested for complementing the defect in the GN229 mutant. In addition, the plasmids pR and pQ, which contain icmR and icmQ, respectively, were also tested for complementation. The data showed that only icmTS and icmT complemented the defect of GN229 in the pore-forming toxin (Fig. 4). Plasmids harboring icmS, icmR, or icmQ, failed to complement GN229.
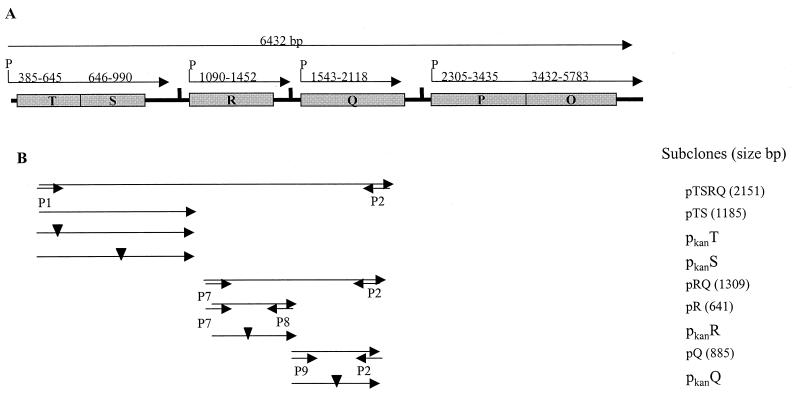
FIG. 3.
Genetic organization of the icmTSRQPO locus (EMBL accession no. Y12705). (A) Size and predicted transcriptional units of the icmTSRQPO locus are indicated by the arrows, while the numbers above the arrows indicate the nucleotide range of the sequence. (B) The genetic regions contained in several subclones and the primers used for their PCR amplification are shown. Inverted filled triangles indicate Kan insertions, and the small arrows indicate the primers used for PCR.
FIG. 4.
The icmT gene is essential for pore-forming activity. Contact-dependent hemolysis of sRBCs was performed using the strain GN229 or AA100kmT transformed by plasmids carrying different subclones of icmTSRQ or by plasmids harboring Kan insertions in icmT. In each experiment, AA100 and GN229 were used as positive and negative controls, respectively. These data are representative of three different experiments, each done in triplicate, and error bars represent standard deviations.
The second approach was to disrupt the individual genes within pTSRQ by transposon mutagenesis to create new plasmids for complementation and also to facilitate construction of L. pneumophila AA100 isogenic mutants within this region (Table 1). The rib mutant GN229 was transformed with the pTSRQ-derived plasmids, pkanT, pkanS, pkanR, and pkanQ (Kan indicates insertion within the gene), and the hemolysis assay was performed to test for complementation for the pore-forming toxin (Fig. 4). The mutation in GN229 was complemented by pkanS, pkanR, and pkanQ but was not complemented by pkanT.
To further confirm the role of icmT, we performed allelic exchange of pKanT into the chromosome of the wild-type strain AA100 to create an isogenic insertion mutant (AA100kmT). The AA100kmT mutant was defective for the pore-forming toxin and was complemented with icmTS and icmT, similar to GN229 (Fig. 4). Taken together, the data clearly showed that icmT was defective in GN229 and was essential for the pore-forming toxin.
Genetic lesion within icmT of the rib mutants.
To ascertain the fact that the rib mutant phenotype was the result of a mutation in icmT, the icmTS region of the wild-type strain AA100 and of the five rib mutants was sequenced. The results showed that the only difference in the sequence between the rib mutants and the wild-type strain was located in the icmT gene (Fig. 5). All five rib mutants had an identical deletion of a single nucleotide (G) at position 544. This deletion in icmT occurred after a stretch of poly(T) regions and is predicted to result in the expression of a truncated protein of 54 amino acids instead of a native protein of 86 amino acids. Since all five rib mutants were complemented for the defect in the pore-forming toxin by icmT alone, this point deletion in icmT of the rib mutants is responsible for their defect in the pore-forming toxin-mediated lysis and egress from the host cells.
FIG. 5.
The mutation of the five rib mutants and their revertants is located in icmT. The sequences of icmT of the five rib mutants and three of the rib revertants are aligned. L. pneumophila (strain AA100) from our laboratory and that from the sequence deposited by Segal and Shuman (34), designated here lp(db), were also included.
Genetic bases of reversion of the rib mutants phenotype.
The first section of our results showed that our trial to isolate cosmid clones for complementation of two of the rib mutants (GN229 and GL208) for the defective egress from A. polyphaga resulted in spontaneous revertants that had no cosmids but regained the wild-type phenotype. This prompted us to analyze the icmTS region in the revertants. Sequencing of the icmTS region in three GN229 revertants (Fig. 5) showed an insertion of a T at nucleotide 544 in icmT, which is the exact location of the point deletion in the rib mutants. Although the insertion was a T in the revertants and not a G as in AA100, this point insertion resulted in a codon for the same amino acid as the wild-type strain (GGG for wild type and TGG for revertants; both encode glycine).
The icmT gene is essential for cytolysis of macrophages and A. polyphaga by L. pneumophila and subsequent bacterial egress.
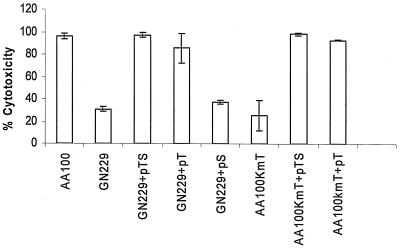
Cytotoxicity assays were carried out in U937 macrophages and A. polyphaga to examine the rib mutants and the complemented strains for cytotoxicity to the host cells. The Alamar Blue assays, carried out after 72 h of infection at an MOI of 1 (Fig. 6) showed that the rib mutant GN229 or AA100kmT had minimal cytotoxicity compared to the wild-type strain. The microscopic observation of the infected monolayers showed that the bacteria were trapped in the cells and failed to lyse and egress from the host cells (data not shown), consistent with our previous observations (6). The GN229 and AA100kmT complemented by icmTS or icmT showed a phenotype similar to that of the wild-type strain in cytolysis and egress from the host cell.
FIG. 6.
The icmT gene of L. pneumophila is essential for cytotoxicity to U937 macrophages. Cytotoxicity to U937 macrophages infected at an MOI of 1 by L. pneumophila strains was measured at 72 h postinfection for the reduction of the Alamar Blue dye, and results were compared to those for noninfected cells. These data are representative of three different experiments, each done in triplicate, and error bars represent standard deviations.
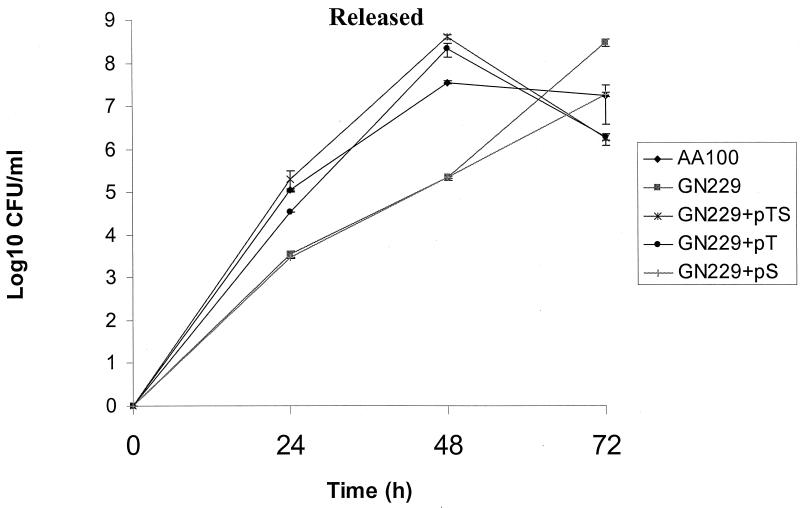
We have previously shown that the rib mutants are defective in egress from the host cell after termination of intracellular replication but are subsequently released 24 to 48 h later, which is most likely due to apoptosis of the infected cell (6). The kinetics of bacterial egress from U937 macrophages was performed at an MOI of 1 to test the ability of the mutants and the complemented strains to egress from the host cell after intracellular growth. As expected, there was an ∼100-fold decrease in the egress of GN229 (105 CFU/ml) compared to that of AA100 (107 CFU/ml) at 24 h. At 48 h postinfection, there was a 500-fold reduction in egress of GN229 compared to the parental strain (Fig. 7). Importantly, GN229 complemented by icmTS or icmT egressed from U937 macrophages similar to AA100, while GN229 complemented with icmS was similar to the GN229 mutant (Fig. 7). These data showed that icmT was essential for egress from macrophages upon termination of intracellular replication.
FIG. 7.
The icmT gene is essential for bacterial egress from macrophages. U937 macrophages were infected by strains of L. pneumophila at an MOI of 1 for 1 h followed by gentamicin treatment for 1 h to kill extracellular bacteria. The bacteria released from the supernatants were plated for colony enumeration. These data are representative of three different experiments, each done in triplicate, and error bars represent standard deviations.
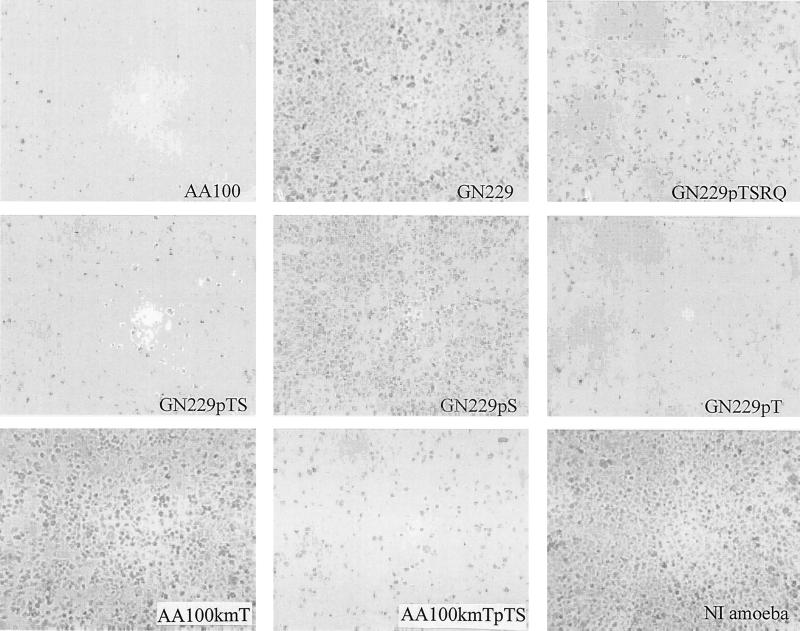
The infection of A. polyphaga by the wild-type strain, the mutants, and the complemented strains (Fig. 8) showed that GN229 and AA100kmT were defective in cytolysis of the host cell, in sharp contrast to the wild-type and the complemented strains. As with macrophages, microscopic observation of cells infected by the rib mutant GN229 and AA100KmT showed that the mutants failed to lyse the protozoan cells (Fig. 8). The defect in egress of the mutants was complemented by plasmids carrying icmTSRQPO (data not shown), icmTSRQ, icmTS, and icmT. We concluded that the icmT gene was essential for cytotoxicity and lysis of both human and protozoan cells by L. pneumophila.
FIG. 8.
The icmT gene of L. pneumophila is essential for cytolysis of A. polyphaga. Shown are representative phase-contrast images of A. polyphaga infected by L. pneumophila AA100, GN229, and GN229 harboring different plasmids at an MOI of 10 and examined at 96 h postinfection. NI, noninfected A. polyphaga.
The rtxA gene is not involved in pore-forming toxin-mediated egress from macrophages and A. polyphaga.
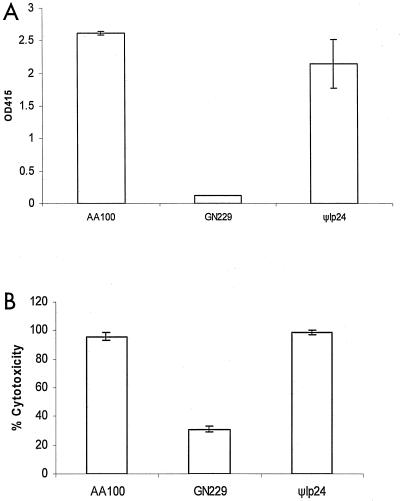
The rtxA gene has been shown to be involved in entry into macrophages and is thought to have a minor role in pore formation when tested for enhanced permeability of the plasma membrane to ethidium bromide (13, 14). Since it was proposed that RtxA might play a role in pore formation, we examined its role in pore-forming toxin-mediated egress from macrophages and protozoa. The role of this gene in egress, contact-dependent hemolysis, and cytotoxicity to macrophages and amoebae was examined using an isogenic mutant obtained by an in-frame deletion in rtxA (Φlp24) (13, 14) and compared to the wild-type strain AA100. Amplification by PCR of the rtxA gene confirmed the deletion of rtxA within Φlp24 (data not shown). In all assays (Fig. 9 and data not shown), the ψlp24 mutant had an indistinguishable phenotype from that of AA100. Thus, rtxA was not required for cytolysis or for pore formation-mediated egress of Legionella from mammalian and protozoan cells.
FIG. 9.
The rtxA gene is not required for pore formation-mediated hemolysis of sRBCs and for cytotoxicity to U937 macrophages. (A) Contact-dependent hemolysis assays of sRBCs; (B) cytotoxicity assays to U937 macrophages. Strains tested are the wild-type strain AA100, GN229, and ψlp24 (in-frame rtxA deletion mutant). These data are representative of three different experiments, each done in triplicate, and error bars represent standard deviations.
DISCUSSION
We have shown recently that the five rib mutants replicate intracellularly similar to the parental strain but are defective in pore formation-mediated cytolysis and egress from the host cell upon termination of intracellular replication (6, 17). The data show that icmTS and icmT all complement the rib mutants for the defect in pore formation-mediated cytolysis and egress from host cells upon termination of intracellular replication. Since icmS is required for intracellular growth of L. pneumophila in macrophages (15) and does not complement the rib mutants, our studies have been focused on icmT.
Although the defective phenotype of the rib mutants is due to a spontaneous point deletion in icmT, an insertion mutation in icmT has a similar phenotype to the rib mutants, in the defect in pore formation-mediated egress upon termination of intracellular replication. Importantly, since this insertion mutant is also complemented by icmT, the kanamycin insertion does not have a polar effect on the expression of the downstream icmS. This is rather surprising, since the termination codon of icmT is followed immediately downstream with the initiation codon of IcmS. Our observations suggest that icmS may be regulated separately by its own promoter. However, promoter and transcriptional studies would be required to determine this speculation.
Complementation of the pore-forming activity and its role in cytolysis and egress from both mammalian and protozoan cells by icmTSRQPO are only partial (34), in contrast to the complete complementation by icmTSRQ, icmTS, and icmT, all of which are cloned in the pBC plasmid. The pGS-Lc-32 plasmid harboring icmTSRQPO contains the mobilization region, mob, which is essential for conjugation. It has been previously shown that the presence of the mobilization genes on a complementing plasmid is also responsible for the low level of complementation of other dot/icm mutants (35). It is thought that the Mob proteins compete with export of bacterial effectors required for cytotoxicity and intracellular multiplication (35). Thus, the partial complementation by the pGS-Lc-32 plasmid is most likely due to the interference of the Mob proteins in export of the pore-forming toxin.
The rib mutants are spontaneous (6), and our trials to complement them for egress from A. polyphaga with the cosmid library have also resulted in isolation of spontaneous revertants that regained the wild-type phenotype but do not contain cosmids. Interestingly, two of the mutants (without being transformed by the cosmid library) that have been used as controls in the amoebal enrichment do not revert to the wild-type phenotype. The spontaneous revertants do not have the wild-type sequence of icmT, suggesting that there has been no recombination with the cosmid harboring the icmT region. However, it is not clear how harboring the cosmid library would enhance the spontaneous reversion process, which occurred in the two tested mutants. Sequencing of the icmT gene of the five rib mutant strains and three of the rib revertants has shown that a single nucleotide deletion or insertion, respectively, is responsible for the phase variation in the pore-forming activity. This deletion or insertion in the rib mutants and their revertants, respectively, occurs after a poly(T) stretch and is most likely due to slipped-strand mispairing during DNA replication. This mode of phase variation has been described for virulence genes of many pathogens such as the pilC of Neisseria gonorrhoeae, in which a slipped-strand mispairing after a stretch of G nucleotides results in a frameshift mutation (24). It would be interesting to determine the rate of phase variation of icmT in vitro and intracellularly. It would be also interesting to examine phase variation at this locus and its role in the defect in the pore-forming toxin of other Legionella species, such as L. micdadei (8), that have been shown to have the dot/icm loci (25).
In addition to its essential role in pore formation-mediated lysis of the host cell, IcmT is also involved in the DNA conjugation (35). The IcmT protein sequence is similar to the TraK protein encoded by the transfer region of Incl plasmid (27). Although our data clearly show an essential role for IcmT in pore formation-mediated egress from the host cell upon termination of intracellular replication, it is not known how IcmT is involved in this process at the molecular and biochemical level. It is unlikely that IcmT is a common regulator of several virulence traits that are induced upon termination of intracellular replication (10), including the pore-forming toxin (6), evasion of phagolysosomal fusion, and motility, since the rib mutant evades phagolysosomal fusion and is motile (6). It is possible that IcmT is the structural toxin or is a cofactor that is specifically required for export of the pore-forming toxin, as a cofactor or a chaperon. Future studies are designed to determine whether IcmT is the pore-forming toxin or is a cofactor required for export and/or activity of the pore-forming toxin.
Whether the Lsp type II and the Dot/Icm type IV secretion systems of L. pneumophila interact in export of effector molecules through the cytoplasmic and outer membrane, respectively, is not known. The pertussis toxin of Bordetella pertussis is exported through the type II secretion systems into the periplasm, where the holotoxin is assembled and is subsequently exported by the type IV secretion system through the outer membrane (11). We have shown that mutants of L. pneumophila defective in the type II secretion system are not defective in the pore-forming toxin. Therefore, in contrast to the pertussis toxin, export of the pore-forming toxin is independent of the type II secretion system. We have also recently shown that export of the effector molecule responsible for the induction of apoptosis occurs through the Dot/Icm system and its export is also independent of the type II secretion system (Zink et al., submitted for publication). Since the type II secretion system is not required for intracellular replication of L. pneumophila within mammalian cells (20), it is most likely that it is also not required for export of effector molecules responsible for modulation of the phagosome to evade maturation along the endosomal-lysosomal degradation pathway. Therefore, export of many virulence factors through the Dot/Icm type IV secretion system is independent of the type II secretion system.
The RtxA-like protein of L. pneumophila (repeats in structural toxin) has been shown to be involved in entry into macrophages and to play a minor role in pore formation when examined by the increase in permeability to ethidium (13). We have shown that RtxA plays no role in pore formation when detected by hemolysis of RBCs. We propose that there are at least two pores of different sizes generated by L. pneumophila in host cell membranes. One pore can be detected by the increase in permeability of the host cell membrane to ethidium bromide and is mediated by RtxA upon bacterial entry. The second and larger pore is generated by the pore-forming toxin that is required for bacterial egress and is detected by hemolysis of RBCs. This hypothesis is supported by our data, which show that rtxA is not required for pore formation-mediated bacterial egress and that the rtxA mutant is not impaired for pore formation-mediated hemolysis of RBCs. However, the rtxA mutant is partially defective in causing an increased permeability of the host cell membrane to ethidium bromide (13). Kirby et al. have shown that polyethylene glycol (PEG) 3350 (average molecular weight, 3350) prevents blebbing of macrophages infected by L. pneumophila, whereas PEG 1000 does not. Interestingly, PEG 3350 does not protect infected cells from becoming permeable to ethidium bromide (26). It is likely that the pore used for bacterial entry is the pore detected by increased permeability of the host cell to ethidium bromide, and RtxA may a partial play a role in this process. This smaller pore is likely to be distinct from the larger pore utilized for cytolysis and bacterial egress from the host cell and is detected by hemolysis of RBCs.
In summary, we have shown that the rib mutants, which replicate intracellularly but are defective in pore formation-mediated egress from the host cell, are defective in icmT due to a point deletion after a stretch of poly(T) in the sequence encoding the C terminus of the protein. We have also shown that the type II secretion system of L. pneumophila is not involved in export of the pore-forming toxin.
Acknowledgments
We thank H. Shuman and R. Isberg for their kind gifts of the dot/icm clones. We thank Jeff Cirillo for the kind gift of the rtxA in-frame deletion mutant.
N.P.C. is supported by Public Health Service Award RO1AI43987, and Y.A.K. is supported by Public Health Service Award RO1AI43965 and R29AI38410.
Editor: J. T. Barbieri
REFERENCES
- 1.Abu Kwaik, Y. 1998. Fatal attraction of mammalian cells to Legionella pneumophila. Mol. Microbiol. 30: 689–696. [DOI] [PubMed] [Google Scholar]
- 2.Abu Kwaik, Y. 1996. The phagosome containing Legionella pneumophila within the protozoan Hartmanella vermiformis is surrounded by the rough endoplasmic reticulum. Appl. Environ. Microbiol. 62: 2022–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abu Kwaik, Y., B. I. Eisenstein, and N. C. Engleberg. 1993. Phenotypic modulation by Legionella pneumophila upon infection of macrophages. Infect. Immun. 61: 1320–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abu Kwaik, Y., L.-Y. Gao, O. S. Harb, and B. J. Stone. 1997. Transcriptional regulation of the macrophage-induced gene (gspA) of Legionella pneumophila and phenotypic characterization of a null mutant. Mol. Microbiol. 24: 629–642. [DOI] [PubMed] [Google Scholar]
- 5.Abu Kwaik, Y., and L. L. Pederson. 1996. The use of differential display-PCR to isolate and characterize a Legionella pneumophila locus induced during the intracellular infection of macrophages. Mol. Microbiol. 21: 543–556. [DOI] [PubMed] [Google Scholar]
- 6.Alli, O. A. T., L.-Y. Gao, L. L. Pedersen, S. Zink, M. Radulic, M. Doric, and Y. Abu Kwaik. 2000. Temporal pore formation-mediated egress from macrophages and alveolar epithelial cells by Legionella pneumophila. Infect. Immun. 68: 6431–6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aragon, V., S. Kurtz, A. Flieger, B. Neumeister, and N. P. Cianciotto. 2000. Secreted enzymatic activities of wild-type and pilD-deficient Legionella pneumophila. Infect. Immun. 68: 1855–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bezanson, G., S. Burbridge, D. Haldane, C. Yoell, and T. Marrie. 1992. Diverse populations of Legionella pneumophila present in the water of geographically clustered institutions served by the same water reservoir. J. Clin. Microbiol. 30: 570–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bozue, J. A., and W. Johnson. 1996. Interaction of Legionella pneumophila with Acanthamoeba castellanii: uptake by coiling phagocytosis and inhibition of phagosome-lysosome fusion. Infect. Immun. 64: 668–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrne, B., and M. S. Swanson. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66: 3029–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8: 354–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cirillo, J. D., S. L. Cirillo, L. Yan, L. E. Bermudez, S. Falkow, and L. S. Tompkins. 1999. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect. Immun. 67: 4427–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cirillo, S. L., L. E. Bermudez, S. H. El-Etr, G. E. Duhamel, and J. D. Cirillo. 2001. Legionella pneumophila entry gene rtxA is involved in virulence. Infect. Immun. 69: 508–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cirillo, S. L., J. Lum, and J. D. Cirillo. 2000. Identification of novel loci involved in entry by Legionella pneumophila. Microbiology 146: 1345–1359. [DOI] [PubMed] [Google Scholar]
- 15.Coers, J., J. C. Kagan, M. Matthews, H. Nagai, D. M. Zuckman, and C. R. Roy. 2000. Identification of icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol. 38: 719–736. [DOI] [PubMed] [Google Scholar]
- 16.Flieger, A., S. Gong, M. Faigle, S. Stevanovic, N. P. Cianciotto, and B. Neumeister. 2001. Novel lysophospholipase A secreted by Legionella pneumophila. J. Bacteriol. 183: 2121–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, L.-Y., and Y. Abu Kwaik. 2000. The mechanism of killing and exiting the protozoan host Acanthamoeba polyphaga by Legionella pneumophila. Environ. Microbiol. 2: 79–90. [DOI] [PubMed] [Google Scholar]
- 18.Gao, L.-Y., O. S. Harb, and Y. Abu Kwaik. 1997. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant hosts, mammalian and protozoan cells. Infect. Immun. 65: 4738–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao, L.-Y., B. J. Stone, J. K. Brieland, and Y. Abu Kwaik. 1998. Different fates of Legionella pneumophila pmi and mil mutants within human-derived macrophages and alveolar epithelial cells. Microb. Pathog. 25: 291–306. [DOI] [PubMed] [Google Scholar]
- 20.Hales, L. M., and H. A. Shuman. 1999. Legionella pneumophila contains a type II general secretion pathway required for growth in amoebae as well as for secretion of the Msp protease. Infect. Immun. 67: 3662–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harb, O. S., L.-Y. Gao, and Y. Abu Kwaik. 2000. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ. Microbiol. 2: 251–265. [DOI] [PubMed] [Google Scholar]
- 22.Horwitz, M. A. 1983. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 158: 1319–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horwitz, M. A. 1983. The Legionnaires’ disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 158: 2108–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jonsson, A. B., G. Nyberg, and S. Normark. 1991. Phase variation of gonococcal pili by frameshift mutation in pilC, a novel gene for pilus assembly. EMBO J. 10: 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joshi, A. D., and M. S. Swanson. 1999. Comparative analysis of Legionella pneumophila and Legionella micdadei virulence traits. Infect. Immun. 67: 4134–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirby, J. E., J. P. Vogel, H. L. Andrews, and R. R. Isberg. 1998. Evidence for pore-forming ability by Legionella pneumophila. Mol. Microbiol. 27: 323–336. [DOI] [PubMed] [Google Scholar]
- 27.Komano, T., T. Yoshida, K. Narahara, and N. Furuya. 2000. The transfer region of IncI1 plasmid R64: similarities between R64 tra and Legionella icm/dot genes. Mol. Microbiol. 35: 1348–1359. [DOI] [PubMed] [Google Scholar]
- 28.Liles, M. R., P. H. Edelstein, and N. P. Cianciotto. 1999. The prepilin peptidase is required for protein secretion by and the virulence of the intracellular pathogen Legionella pneumophila. Mol. Microbiol. 31: 959–970. [DOI] [PubMed] [Google Scholar]
- 29.Purcell, M., and H. A. Shuman. 1998. The Legionella pneumophila icmGCDJBF genes are required for killing of human macrophages. Infect. Immun. 66: 2245–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossier, O., and N. P. Cianciotto. 2001. Type II protein secretion is a subset of the PilD-dependent processes that facilitate intracellular infection by Legionella pneumophila. Infect. Immun. 69: 2092–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33: 1179–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila chromosome. Proc. Natl. Acad. Sci. USA 95: 1669–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segal, G., and H. A. Shuman. 1997. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect. Immun. 65: 5057–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segal, G., and H. A. Shuman. 1998. Intracellular multiplication and human macrophage killing by Legionella pneumophila are inhibited by conjugal components of IncQ plasmid RSF1010. Mol. Microbiol. 30: 197–208. [DOI] [PubMed] [Google Scholar]
- 36.Segal, G., and H. A. Shuman. 1999. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 67: 2117–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stone, B. J., and Y. Abu Kwaik. 1999. Natural competency for DNA uptake by Legionella pneumophila and its association with expression of type IV pili. J. Bacteriol. 181: 1395–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279: 873–876. [DOI] [PubMed] [Google Scholar]