Abstract
Chlamydia pneumoniae, a human pathogen causing respiratory infections and probably contributing to the development of atherosclerosis and heart disease, is an obligate intracellular parasite which for replication needs to productively interact with and enter human cells. Because of the intrinsic difficulty in working with C. pneumoniae and in the absence of reliable tools for its genetic manipulation, the molecular definition of the chlamydial cell surface is still limited, thus leaving the mechanisms of chlamydial entry largely unknown. In an effort to define the surface protein organization of C. pneumoniae, we have adopted a combined genomic-proteomic approach based on (i) in silico prediction from the available genome sequences of peripherally located proteins, (ii) heterologous expression and purification of selected proteins, (iii) production of mouse immune sera against the recombinant proteins to be used in Western blotting and fluorescence-activated cell sorter (FACS) analyses for the identification of surface antigens, and (iv) mass spectrometry analysis of two-dimensional electrophoresis (2DE) maps of chlamydial protein extracts to confirm the presence of the FACS-positive antigens in the chlamydial cell. Of the 53 FACS-positive sera, 41 recognized a protein species with the expected size on Western blots, and 28 of the 53 antigens shown to be surface-exposed by FACS were identified on 2DE maps of elementary-body extracts. This work represents the first systematic attempt to define surface protein organization in C. pneumoniae.
Chlamydia pneumoniae is an obligate intracellular bacterium and a common human pathogen (48). It is a significant cause of pneumonia in both hospital and outpatient settings, accounting for approximately 7 to 10% of cases of community-acquired pneumonia among adults. C. pneumoniae has also been associated with atherosclerotic and cardiovascular disease, as suggested by results of seroepidemiologic studies, detection of the organism in atherosclerotic plaque specimens, experimental in vitro cell culture studies, animal model studies, and two small secondary prevention antibiotic treatment trials (12, 13, 15, 19, 20, 28, 45).
Like all obligate intracellular parasites, for its survival and propagation C. pneumoniae must accomplish several essential tasks which include adhering to and entering host cells, creating an intracellular niche for replication, exiting host cells for subsequent invasion of neighboring cells, and also avoiding host defense mechanisms. To carry out all these functions, C. pneumoniae has developed a unique biphasic life cycle involving two developmental forms, a spore-like infectious form (elementary bodies [EBs]) and an intracelluar replicative form (reticulate bodies [RBs]). Adhesion, host cell colonization capabilities, and the ability to cope with host defense mechanisms when outside the cell presumably rely in large part on EB surface organization.
Because of the intrinsic difficulty in working with C. pneumoniae and the lack of adequate methods for its genetic manipulation, there is still a paucity of information regarding protein components of the EB cell surface. Apart from surface localization of a number of proteins inferred by computer analysis (50), at present only a few proteins have been characterized and shown to be surface associated. These include OmpA, the homolog of the major outer membrane protein (MOMP) of Chlamydia trachomatis (41, 57), PorB (an OmpA analog shown to be surface-exposed in C. trachomatis by Kubo and Stephens [29]), OmcB (a protein thought to anchor heparin-like molecules to the chlamydial surface) (49), and a 76-kDa protein shown to induce neutralizing antibodies in vitro (39). Furthermore, evidence of expression and possible surface localization of 11 out of the 21 members of the Pmp family of polymorphic outer membrane proteins has been reported (6, 16, 27). Surface localization of all these proteins has been assessed by using a variety of immunological assays including Western blot analysis, dot blot on whole chlamydial cells, microimmunofluorescence (MIF), and immunoelectron microscopy. In general, to reduce the risk of false positive results due to antibody cross-reactivity with antigens sharing similar epitopes, antigens were annotated as surface-exposed when converging evidence from more than one immunological assay was collected. More recently, transcriptional activities of surface candidate genes have also been investigated and taken as a relevant piece of information to further support immunological data (16).
Here we describe a new genomic-proteomic approach to identify EB surface proteins based on high-throughput expression and purification of C. pneumoniae antigens, Western blotting, flow cytometry (fluorescence-activated cell sorter [FACS]) analysis, two-dimensional electrophoresis (2DE), and mass spectrometry analysis.
This work represents the first systematic analysis of chlamydial surface proteins and is intended to open the way to further studies on the mechanisms underlying C. pneumoniae entry into and infection of eukaryotic host cells. Furthermore, the data presented will help to identify new candidates for the development of diagnostics and vaccines against this important human pathogen.
MATERIALS AND METHODS
Preparation of C. pneumoniae EBs and chromosomal DNA.
C. pneumoniae FB/96, a clinical isolate from a patient with pneumonia at the Sant’Orsola Polyclinic, Bologna, Italy, was grown in LLC-MK2 cells. EBs were harvested 72 h after cell culture infection and purified by gradient centrifugation as described previously (47). Purified chlamydiae were resuspended in sucrose-phosphate transport buffer and stored at −80°C until use. When required, prior to storage EB infectivity was heat inactivated by 2 h of incubation at 56°C. Chromosomal DNA was prepared from gradient-purified EBs by lysing the cells overnight at 37°C with 10 mM Tris-HCl, 150 mM NaCl, 2 mM EDTA, 0.6% sodium dodecyl sulfate (SDS), 100 μg of proteinase K/ml, and sequential extraction with phenol, phenol-chloroform, and chloroform.
In silico analyses.
C. pneumoniae genomic sequences were obtained from the following web sites: the Berkeley Genome Project (http://chlamydia-www.berkeley.edu:4231/), The Institute for Genomic Research (http://www.tigr.org), the STD sequence database of Los Alamos National Laboratory Bioscience Division (http://www.stdgen.lanl.gov/), and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/micr.html). In silico analyses of genomes and open reading frame (ORF) selection were carried out essentially as already described (42) using several computer programs, including the Genetics Computer Group package of the University of Wisconsin, FASTA, ClustalW, BLAST, ProDom, and PROTEAN (DNAstar, Inc. Madison, Wis.). Theoretical molecular weights and isoelectric points were calculated using the Compute pI/Mw tool (3) available at http://www.expasy.ch/tools/pi_tool.html.
Cloning and expression of recombinant proteins.
Selected ORFs from the C. pneumoniae CWL029 genome (26) were cloned into plasmid expression vectors so as to obtain three kinds of recombinant proteins: (i) proteins with a six-histidine tag at the C terminus (cpn-His); (ii) proteins fused with glutathione S-transferase (GST) at their N terminus (Gst-cpn); and (iii) proteins having a C-terminal six-histidine tag and an N-terminal GST domain (Gst-cpn-His). Cpn-His proteins were obtained by cloning in the pET21b+ vector (Novagen), and Gst-cpn proteins were obtained by using pGEX-KG vectors (18). Escherichia coli BL21(DE3) (Novagen) was the recipient of pET21b+-derived recombinant plasmids, whereas E. coli BL21 (Novagen) was used for pGEX-NN- and pGEX-NNH-derived plasmids. PCR primers were designed so as to amplify genes without the signal peptide coding sequence. When a signal peptide or processing site was not clearly predictable, the ORF sequence was cloned as annotated by Kalman and coworkers (26), starting from the first predicted codon. PCRs were performed on 50 ng of genomic DNA, using 2 U of Taq DNA polymerase (Platinum Taq; Gibco-BRL) in a final volume of 100 μl and the Gene Amp PCR System 9600 (Perkin Elmer). PCR products were purified from agarose gels and ligated to the appropriate vector. Recombinant clones were grown in Luria-Bertani medium (500 ml) containing 100 μg of ampicillin/ml and grown at 37°C until an optical density at 600 nm (OD600) of 0.5 was reached. Expression of recombinant proteins was then induced by adding 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG). Three hours after IPTG induction, cells were collected by centrifugation. Before protein purification, aliquots of the cell pellets (corresponding to an OD600 of 0.1) were resuspended in sample loading buffer (60 mM Tris-HCl [pH 6.8], 5% [wt/vol] SDS, 10% [vol/vol] glycerol, 0.1% [wt/vol] bromophenol blue, 100 mM dithiothreitol [DTT]), boiled for 5 min, and analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
Purification of recombinant proteins.
Recombinant E. coli cells were resuspended in 50 mM Tris-HCl (pH 8.0) and broken in a French press apparatus (SLM Aminco, Rochester, N.Y.). After centrifugation at 30,000 × g, the supernatants of the pET-transformed E. coli extracts were loaded onto nickel-activated chelating Sepharose columns (Amersham Pharmacia Biotech), and after column washing with 20 mM imidazole recombinant proteins were eluted with 250 mM imidazole. The supernatants carrying pGEX derivatives were loaded onto glutathione-Sepharose 4B columns (Amersham Pharmacia Biotech), and proteins were eluted with 50 mM Tris-HCl, 10 mM reduced glutathione, pH 8.0. After purification, the recombinant proteins were stored at −20°C after addition of glycerol (30% vol/vol). Water-insoluble His-tagged proteins were purified from the inclusion bodies. After bacterial cell disruption (using a French press) and centrifugation, the pellets were solubilized with 50 mM Tris-HCl, 1 mM tris(2-carboxyethyl)phosphine (TCEP) (Pierce), 6 M guanidine-HCl, pH 8.5. After centrifugation at 30,000 × g for 15 min the supernatants were loaded on nickel-activated chelating Sepharose (Pharmacia) affinity columns. The columns were washed with 50 mM Tris-HCl, 6 M urea, 1 mM TCEP (pH 8.5) containing 20 mM imidazole and eluted with the same buffer containing 250 mM imidazole (pH 8.5). Finally, 400 μl of each protein (1 mg/ml) was diluted with 800 μl of 50 mM Tris-HCl, 1.5 M arginine, 10 mM DTT, pH 8.5, and stored at −20°C after addition of 800 μl of glycerol. Protein concentrations were determined using the Bradford method.
Immunological assays.
Groups of four 6- to 7-week-old CD1 female mice (Charles River, Como, Italy) were immunized intraperitoneally at days 1, 15, and 29 with 20 μg of purified recombinant protein in Freund’s adjuvant. Immune sera were prepared from blood samples collected on day 43 and pooled before use. In order to reduce the amount of antibodies possibly elicited by contaminating E. coli antigens, the immune sera were incubated overnight at 4°C with nitrocellulose strips adsorbed with an E. coli BL21 total protein extract.
For Western blot analysis (53), total proteins from purified C. pneumoniae FB/96 EBs (2 μg per lane) were separated by SDS-PAGE (30) and electroblotted onto nitrocellulose membranes. The membranes were incubated with preimmune sera or with sera from immunized mice (diluted 1:100) and then with a peroxidase-conjugated anti-mouse antibody (Sigma; diluted 1:3,000). After washing with phosphate-buffered saline (PBS), 0.1% Tween 20, blots were developed using an Opti-4CN Substrate Kit (Bio-Rad).
FACS analysis was performed by a newly developed assay (unpublished data). Essentially, 2 × 105 gradient-purified FB/96 EBs in PBS, 0.1% bovine serum albumin (BSA) were incubated for 30 min at 4°C with mouse antisera (standard dilution, 1:400). After centrifugation and washing with 200 μl of PBS, samples were incubated for 30 min at 4°C with goat anti-mouse immunoglobulin G, F(ab′)2-specific, conjugated with R-phycoerythrin (dilution 1:100) (Jackson ImmunoResearch Laboratories, Inc.). The samples were then washed with PBS-BSA, resuspended in 150 μl of PBS-BSA, and analyzed by FACS analysis using a FACScalibur apparatus (Becton Dickinson, Mountain View, Calif.). When the assay was performed with infectious samples, chlamydiae-antibody complexes were fixed with 0.1% formaldehyde (55) before processing through the FACS apparatus. Control samples were similarly prepared. Positive control antibodies were a commercial anti-MOMP C. pneumoniae-specific monoclonal antibody (catalogue no. 11-215; Argene Biosoft, Varilhes, France) and an anti-chlamydial lipopolysaccharide (LPS) antibody (monoclonal anti-Chlamydia, fluorescein isothiocyanate conjugated; catalogue no. 5000111 from Meridian Diagnostics, Inc., Cincinnati, Ohio). An immune mouse serum prepared against purified C. pneumoniae whole cells was also used. Background control sera were obtained from mice immunized with either GST (GST fusion control) or with the protein fraction eluted from Ni columns loaded with a BL21(pET21b+) protein extract (His-tag fusion controls). FACS data were analyzed using the Cell Quest Software (Becton Dickinson). The shift between the histograms was taken as a measure of antibody binding to the EB cell surface. The Kolmorov-Smirnov (K-S) two-sample test (58) was performed on the two overlaid histograms. The D/s(n) values (an index of dissimilarity between the two curves) are reported as K-S score in Table 1.
TABLE 1.
The FACS-positive set of proteinsa
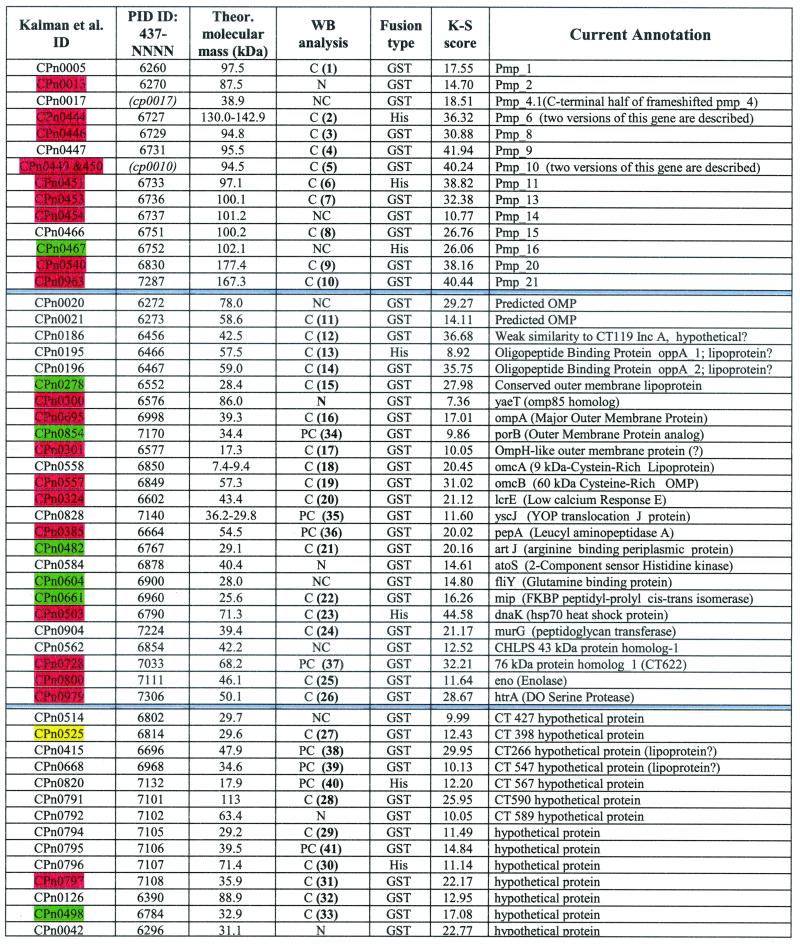
Gene identification (ID) in the CWL029 genome (26) and the last four digits of PID protein database accession numbers (427-NNNN) are reported.Two ORF products not included in the PID database are identified by arbitrary codes (shown in parentheses and italics). Theoretical molecular masses (in kilodaltons) were calculated for putative mature forms. The WB analysis column summarizes the results obtained by probing total EB proteins with antisera against the recombinant proteins; numbers in parentheses refer to the blot images shown in Fig. 2. WB results are classified as follows: C, consistent (the predominant band is consistent with the expected molecular weight; additional minor bands may also be present); PC, partially consistent (a band of the expected molecular weight is present together with additional bands of higher molecular weight or greater intensity); NC, nonconsistent (the detected bands do not correspond to predictable molecular weight values); N, negative (no band pattern obtained). FACS results are reported as K-S scores. The 28 proteins which have been detected in C. pneumoniae extracts by 2DE mapping and MALDI-TOF analysis are color highlighted as follows: red, proteins detected both in this work and by Vandahl et al. (54); yellow, a protein reported only in reference 54; green, proteins detected only in this work.
Mass spectrometry analysis of 2DE protein maps.
Gradient-purified FB/96 EBs were solubilized at a final concentration of 5.5 mg/ml with Immobiline rehydration buffer {7 M urea, 2 M thiourea, 2% (wt/vol) 3-[3-cholamidopropyl)-demethylammonio]-1-propanesulfonate, 2% (wt/vol) ASB 14 (5), 2% (vol/vol) C.A 3-10NL (Amersham Pharmacia Biotech, Piscataway, N.J.), 2 mM tributyl phosphine, 65 mM DTT}. Samples (250 μg of protein) were adsorbed overnight on Immobiline DryStrips (7 cm; pH 3 to 10; nonlinear). Electrofocusing was performed in an IPGphor Isoelectric Focusing Unit (Amersham Pharmacia Biotech). Before PAGE separation, the focused strips were incubated in 4 M urea, 2 M thiourea, 30% (vol/vol) glycerol, 2% (wt/vol) SDS, 5 mM tributyl phosphine, 2.5% (wt/vol) acrylamide, 50 mM Tris-HCl (pH 8.8), as described previously (21, 22). SDS-PAGE was performed on linear 9-to-16% acrylamide gradients. Gels were stained with colloidal Coomassie (Novex, San Diego, Calif.) (8). Stained gels were scanned with the Personal Densitometer SI (Molecular Dynamics) at 8 bits and 50 μm per pixel. Map images were annotated with the software Image Master 2D Elite (version 3.10; Amersham Pharmacia Biotech). Protein spots were excised from the gel using an Ettan Spot Picker (Amersham Pharmacia Biotech) and dried in a vacuum centrifuge. In-gel digestion of samples for mass spectrometry and extraction of peptides were performed as described by Wilms and coworkers (56). Samples were desalted with a Zip Tip (Millipore), eluted with a saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile, 0.1% trifluoroacetic acid and directly loaded onto a SCOUT 381 multiprobe plate (Bruker, Bremen, Germany). Spectra were acquired on a Bruker Biflex II matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) apparatus. Spectra were calibrated using a combination of known standard peptides, located in spots adjacent to the samples. Resulting values for monoisotopic peaks were used for database searches using the computer program Mascot (40), which is available at the website http://www.matrixscience.com/. All searches were performed using an error of 200 to 500 ppm as a constraint.
RESULTS
In silico selection.
Antigens likely to be exposed on the surface of the chlamydial EBs were selected by a multistep computational analysis of the entire genome of C. pneumoniae strain CWL029. The general strategy was to maximize the chance of identifying bacterial surface components by selecting not only proteins predicted by protein localization algorithms to be outer membrane components in gram-negative bacteria but also those predicted as periplasmic or inner membrane proteins. In addition, we selected proteins on the basis of their homology to proteins described as surface exposed in other bacteria, independently from the in silico prediction. In practice, we started by following the published annotation of Kalman et al. (26), which is available at http://chlamydia-www.berkeley.edu:4231/. In this database, out of a total of 1,073 genes, 636 currently have an assigned biological function. From this group, we selected for subsequent cloning and expression 72 ORFs encoding proteins predicted to be peripherally located in the chlamydial cell. Due to the lack of striking sequence homologies to other well-characterized bacterial proteins, the other 437 ORFs are generically annotated as hypothetical in public databases. We submitted this group of sequences to primary sequence analysis, searching for the presence of potential signal peptides and/or transmembrane regions. However, proteins with more than two predicted transmembrane segments were excluded, since previous experience (42) showed that these proteins are not successfully obtainable in high-throughput heterologous expression programs. In this way, 61 additional hypothetical proteins were selected as having potentially surface-exposed domains.
Furthermore, after sequence homology searches in the nonredundant databases available at the website of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/micr.html), we selected a group of C. pneumoniae proteins with remote similarities to surface proteins of other bacteria. This group included a small number of proteins which, lacking an identifiable N-terminal signal peptide for secretion, are predicted as cytoplasmic by algorithms like PSORT. A further selection added a number of proteins which have been described as immunogenic in patients with C. trachomatis infection (46). In total, 141 ORFs were selected in silico for further experimental screening and subjected to high-throughput cloning and expression in E. coli.
Antigen cloning and expression.
The 141 ORFs were amplified by PCRs and cloned in two different E. coli expression vectors so as to obtain each antigen as both GST and His-tag fusion proteins. When the presence of an N-terminal signal peptide for secretion could be clearly predicted, the corresponding nucleotide sequence was excluded from the expression construct in order to avoid possible targeting of the recombinant protein toward the E. coli cytoplasmic membrane. When ORF expression was analyzed, we found that 86.5 and 71.6% of the genes could be expressed as GST and His-tag fusions, respectively. Eight genes for which expression as a His-tag fusion was not successful and GST-fusion products appeared to undergo marked degradation were cloned in a third vector designed to express a recombinant protein flanked by two affinity handles, the GST moiety at the N terminus and the six-His tail at the C terminus. Seven out of these eight genes could be expressed using this vector.
The recombinant fusion proteins were obtained either in a water-soluble form or precipitated as inclusion bodies. Since correct folding is known to be important for eliciting antibodies able to recognize native antigens, the water-soluble form was used when available. When no soluble protein could be obtained, antigen solubilization from the inclusion bodies of the His-tag fusions and subsequent purification was undertaken. In total, 173 recombinant C. pneumoniae fusion proteins deriving from 124 different genes were selected for preparing immune sera in mice.
Identification of surface proteins by flow cytometry.
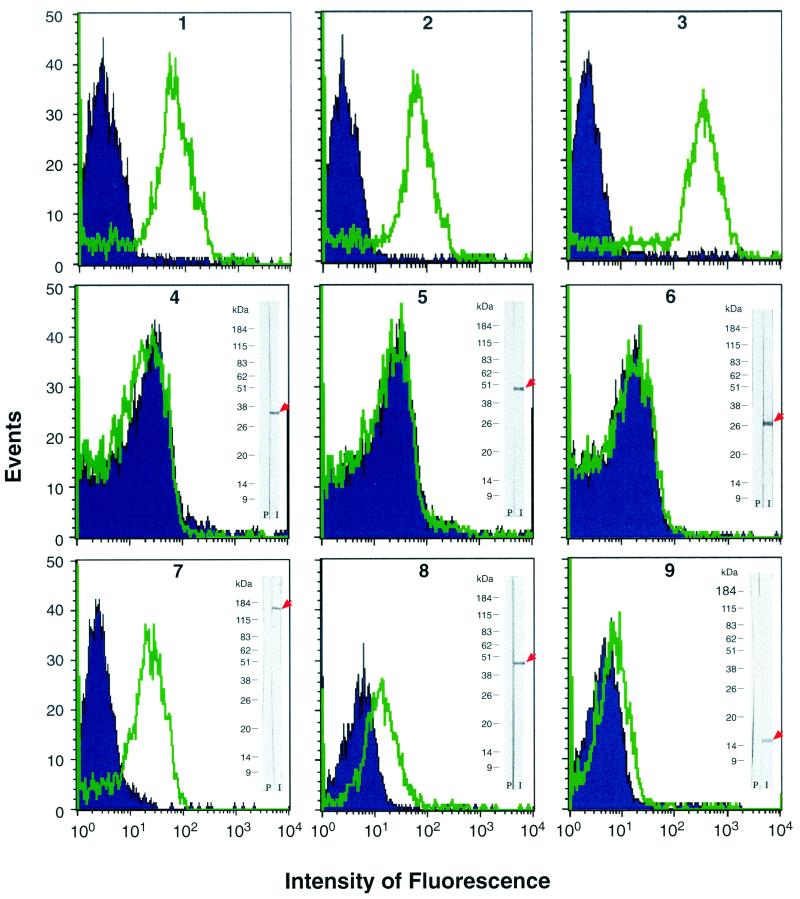
We first verified whether FACS analysis could be used to detect the binding to the chlamydial cell surface of specific antibodies against well-known surface antigens. In fact, while flow cytometry has been used in studies with C. trachomatis (52), its application on C. pneumoniae has not been reported so far. As shown in Fig. 1 (panel 2), a commercial anti-MOMP C. pneumoniae-specific monoclonal antibody could significantly shift the distribution curve of a control chlamydial cell population (blue peaks in Fig. 1). Anti-MOMP antibodies were used as a positive control since a surface location of this antigen has recently been shown in C. pneumoniae (41, 57). Similar results were obtained with a second commercial anti-chlamydial LPS antibody (Meridian Diagnostics) and also with a hyperimmune mouse serum which we prepared against purified whole cells of C. pneumoniae (Fig. 1, panels 1 and 3).
FIG. 1.
FACS analysis of antibody binding to whole C. pneumoniae EBs. Blue histograms (event counts versus fluorescence channels) are the FACS output for EBs stained with background control antibodies. Green histograms are the FACS output of EBs stained with antigen-specific antibodies. Positive controls (panels 1 to 3) were as follows: panel 1, anti-C. pneumoniae LPS monoclonal antibody, with an irrelevant monoclonal antibody (SEAM-3) specific for the type B meningococcal capsule as background control; panel 2, C. pneumoniae-specific anti-MOMP monoclonal antibody (Argene Biosoft), with an irrelevant fluorescein isothiocyanate-conjugated anti-immunoglobulin G2a monoclonal antibody as background control; panel 3, mouse hyperimmune serum against whole EBs, with the corresponding preimmune mouse serum as background control. Negative controls (panels 4 to 6), with mouse anti-GST serum as background control, were as follows: panel 4, mouse serum against 6482-GST fusion protein (PID accession no. 4376582; predicted as a cytoplasmic 36.8-kDa protein); panel 5, mouse serum against 6732-GST fusion protein (PID accession no. 4376732; predicted as a cytoplasmic 43.5-kDa protein); panel 6, mouse serum against 6881-GST fusion protein (PID accession no. 4376881; predicted as a cytoplasmic 26.0-kDa protein). Examples of FACS-positive sera (panels 7 to 9), with mouse anti-GST serum as background control, were as follows: panel 7, antiserum to 7287-GST antigen (Pmp-21); panel 8, antiserum to 6602-GST antigen (LrcE); panel 9, antiserum to 6577-GST antigen (annotated as an OmpH-like OMP). Western blotting data obtained from total EB proteins stained with the same antiserum used for the FACS assays are also shown.
Having demonstrated that flow cytometry can indeed be used to follow antibody binding to the surface of C. pneumoniae EBs, we set up a series of negative controls to exclude the possibility that FACS analysis could reveal proteins not exposed on the bacterial surface. To this aim we produced mouse sera against three C. pneumoniae antigens, all predicted to be cytoplasmic proteins by the PSORT algorithm. When used in the FACS assay, none of the three polyclonal antibodies could bind to C. pneumoniae EBs (Fig. 1, panels 4, 5, and 6). On the contrary, the same sera specifically recognized protein species with the expected molecular weight when C. pneumoniae EB total extracts were used in Western blotting experiments (Fig. 1, panels 4, 5, and 6). Altogether, these data indicate that FACS analysis can be used to follow antibody interaction to C. pneumoniae EBs and that only interactions with surface-exposed proteins are revealed.
We then analyzed all sera against each recombinant C. pneumoniae antigen for the ability to bind to the surface of chlamydial cells. Of 157 sera tested, 59 yielded positive results in the FACS binding assay, leading to the identification of 53 putative surface-exposed proteins. In general, sera derived from the same antigen expressed as GST and His fusions gave similar results in terms of EB recognition (data not shown). The list of the 53 surface protein candidates is given in Table 1, in which the FACS assay data (extent of peak shift and dissimilarity of curve shape) are elaborated by calculating the K-S statistics score (see Materials and Methods). Typical FACS data are shown in Fig. 1, panels 7, 8 and 9, where examples of high (K-S score = 40.44), intermediate (K-S score = 21.12), and low (K-S score = 10.05) positivity are presented. It has to be pointed out that the K-S score cannot be used to correlate the amount of antigen on the EB surface. In fact, many factors contribute to the K-S value, including the concentration of the antigen on the cell surface, its accessibility, the quality (purity and folding) of the recombinant antigen used in immunization, and the concentrations and affinities of the antibodies elicited by each recombinant antigen.
The protein list in Table 1 is divided into three sections: (i) proteins belonging to the well-described family of chlamydial polymorphic OMPs (top section); (ii) 14 proteins so far described only as hypothetical proteins, 7 of which are encoded by genes with orthologs in C. trachomatis and 7 of which are encoded by C. pneumoniae-specific genes (bottom section); and (iii) a mixed group of 25 proteins which include proteins previously shown to be outer membrane components in Chlamydia spp. or predicted as outer membrane components in gram-negative bacteria, and proteins which in silico predictions assign to the periplasmic, inner membrane, or cytoplasmic compartments (middle section). In some cases, this apparent discrepancy can be simply explained by the inadequacy of the in silico predictions (see Discussion below), whereas other cases are open to further investigation.
Analysis of FACS-positive antigens by Western blotting and mass spectrometry.
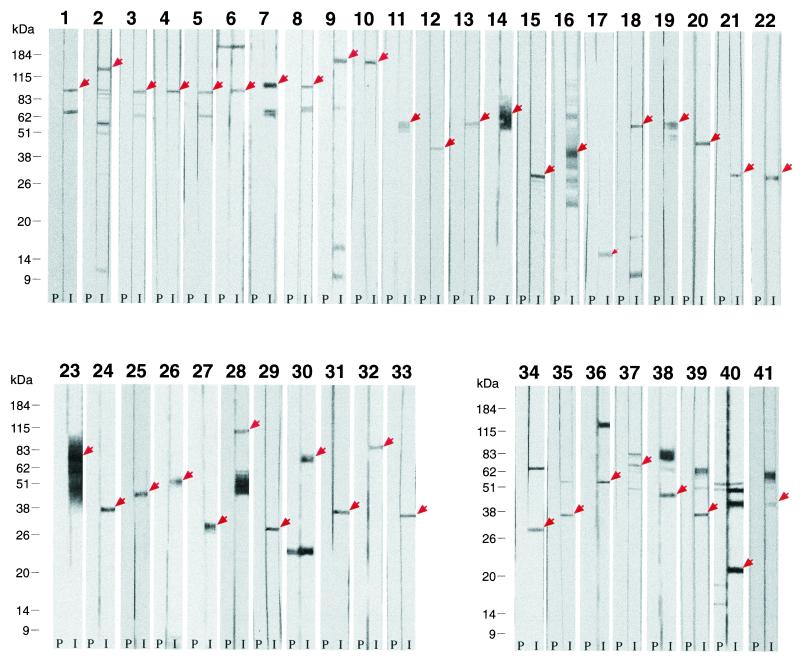
The 53 FACS-positive sera were also screened by Western blot analysis on whole-protein extracts of EB chlamydial preparations. The results of this analysis are given in Table 1 and Fig. 2. In total, 33 sera were specific, in that they recognized a protein species having the expected molecular mass (Fig. 2, panels 1 to 33). Eight additional sera did react with proteins having a molecular mass consistent with the theoretical calculation, but they also recognized a few protein bands having different molecular masses (panels 34 to 41). Finally, 12 sera were scored negative, in that they were either Western blot negative (5 sera) or they reacted with EB antigens of unexpected size (not shown). FACS positivity of the 5 sera which failed to recognize any protein species in chlamydial total extracts was unexpected. These apparently contradictory results can be reconciled assuming either a different sensitivity between the two assays or that sufficient conformation needs to be preserved for these proteins to be recognized. These possible explanations are supported by our 2DE map analysis of total EB proteins (see below), which allowed the identification of two out of five Western blot-negative, FACS-positive proteins.
FIG. 2.
Western blot analysis of total protein extracts from C. pneumoniae EBs, performed using mouse immune sera against recombinant antigens. For antigen identification, refer to Table 1. The panel identification numbers correspond to the numbers reported in the WB analysis column of Table 1. In each panel, the strip on the right shows the results obtained with the antigen-specific immune serum (I), and the strip on the left shows the results obtained with the corresponding preimmune serum (P). Panels 1 to 33 are data that were scored as “consistent” in Table 1, and panels 34 to 41 show results that were scored as “partially consistent” in Table 1.
The control experiments showing that antibodies against three cytoplasmic proteins were FACS negative strongly support the conclusion that the 53 FACS-positive sera indeed recognize proteins exposed on the EB surface. Furthermore, the Western blotting data show that at least 41 of the same 53 sera recognize protein species with the expected molecular mass from EB total protein extracts. However, the possibility that antibody cross-reactivity could generate some false positive results cannot be ruled out, especially in the case of Western blot-negative sera. In an attempt to further support our FACS data, we carried out a proteomic analysis of EB total proteins by using 2DE combined with spot identification by mass spectrometry. The rationale behind this approach is based on the assumption that identification of FACS-positive antigens on 2DE maps of C. pneumoniae would reduce the risk of improper annotation in that it would indicate that a given antigen not only is recognized by a specific antiserum but also that it is unequivocally present in the cell extract.
A typical 2DE protein map of C. pneumoniae EB proteins in the 3 to 10 pI range is shown in Fig. 3. Most of the spots visible in the figure were identified by MALDI-TOF analysis (only relevant data are shown in the figure). Out of a total of 130 identified gene products, 27 belonged to the group of 53 FACS-positive antigens shown in Table 1. Details of protein identification by mass spectrometry are reported in Table 2. While the writing of this paper was in progress, Vandahl et al. (54) published a valuable and extensive annotation of the 2DE protein map of C. pneumoniae. Among the annotated proteins, they found 21 of our FACS-positive antigens (highlighted in red in Table 1), one of which (CPn0525, highlighted in yellow in Table 1) was not detectable in our 2DE maps. Combining their results with our data, overall 28 out of 53 FACS-positive antigens can be identified on C. pneumoniae 2DE maps.
FIG. 3.
2DE map of proteins from purified EBs of C. pneumoniae FB/96. The protein spots marked with the gene ID (see Table 2) correspond to FACS-positive antigens identified by MALDI-TOF analysis.
TABLE 2.
Proteins identified by MALDI-TOFd
| Current annotation | Kalman ID | PID ID 437-NNNN | No. of matching peptides | Coveragea (%) | Theor. pI | Theor. MW | GRAVY valuec | m/z and deduced sequence for peptide selected for PSD analysisc |
|---|---|---|---|---|---|---|---|---|
| Pmp_2 | CPn0013 | 6270 | 20 | 30 | 5.84 | 87.5 | −0.220 | |
| Conserved outer membrane lipoprotein protein | CPn0278 | 6552 | 20 | 66 | 6.49 | 28.4 | −0.162 | |
| VaeT omp85 homolog | CPn0300 | 6576 | 18 | 29 | 7.69 | 86.0 | −0.355 | |
| OmpH-like OMP | CPn0301 | 6577 | 10 | 58 | 4.75 | 17.3 | −0.778 | 1607.01 156TTEIIAILNESYFKK169 |
| LcrE (low calcium response E) | CPn0324 | 6602 | 13 | 39 | 4.98 | 43.4 | −0.468 | |
| PepA (leucyl aminopeptidase A) | CPn0385 | 6664 | 14 | 28 | 5.95 | 54.5 | +0.012 | |
| Pmp_6 | CPn0444 | 6727 | 8 | 8b | 5.31 | 142.9 | −0.187 | |
| Pmp_7 | CPn0445 | 6728 | 19 | 22 | 5.73 | 100.0 | −0.113 | |
| Pmp_8 | CPn0446 | 6729 | 13 | 21 | 5.16 | 94.8 | −0.170 | |
| Pmp_10 | CPn0449 and -50 | 26 | 32 | 5.22 | 94.5 | −0.143 | ||
| Pmp_11 | CPn0451 | 6733 | 15 | 21 | 5.91 | 97.1 | −0.203 | |
| Pmp_13 | CPn0453 | 6736 | 17 | 24 | 6.55 | 100.1 | −0.116 | |
| Pmp_14 | CPn0454 | 6737 | 18 | 23 | 6.76 | 101.2 | −0.036 | |
| Pmp_16 | CPn0467 | 6752 | 11 | 13 | 6.04 | 102.1 | −0.273 | |
| Art J (arginine binding periplasmic protein) | CPn0482 | 6767 | 9 | 38 | 5.45 | 29.1 | +0.050 | 1471.91 192VVLKDFPNLVATR204 |
| Hypothetical protein | CPn0498 | 6784 | 15 | 39 | 5.55 | 32.9 | −0.244 | |
| DnaK (heat shock protein) | CPn0503 | 6790 | 12 | 16 | 4.99 | 71.3 | −0.394 | |
| Pmp_20 | CPn0540 | 6830 | 18 | 12 | 5.36 | 177.4 | −0.191 | |
| OmcB (60-kDa cysteine-rich OMP) | CPn0557 | 6849 | 23 | 47 | 5.62 | 57.3 | −0.116 | |
| FliY (glutamine binding protein) | CPn0604 | 6900 | 8 | 24 | 7.66 | 28.0 | +0.098 | 1296.78 201IISKPLNADGLR212 |
| 5.7 | 25.3 | 0.056 | ||||||
| Mip (FKBP-type peptidyl-prolyl cis-trans isomerase) | CPn0661 | 6960 | 3 | 13 | 5.22 | 25.6 | −0.488 | 914.78 58TFGHLLAR65 |
| OmpA (MOMP) | CPn0695 | 6998 | 13 | 48 | 6.13 | 39.3 | −0.037 | |
| CHLPN 76-kDa protein homolog_1 (CT622) | CPn0728 | 7033 | 18 | 36 | 4.85 | 68.2 | −0.351 | |
| Hypothetical protein | CPn0797 | 7108 | 6 | 21 | 5.16 | 35.9 | −0.181 | 1398.77 128EGYTHAFVFDGR139 |
| Eno (enolase) | CPn0800 | 7111 | 14 | 32 | 4.66 | 46.1 | −0.079 | |
| PorB (OMP analog) | CPn0854 | 7170 | 10 | 23b | 5.10 | 34.4 | −0.048 | |
| Pmp_21 | CPn0963 | 7287 | 13 | 7b | 4.84 | 167.3 | −0.175 | |
| HtrA (DO serine protease) | CPn0979 | 7306 | 12 | 22 | 6.39 | 50.1 | −0.063 |
Calculated from the whole protein.
Peptides from the C-terminal part of the protein.
GRAVY, grand average of hydropathicity (available at http://www.expasy.ch/tools/protparam.html; PSD, postsource decay.
The results were obtained from protein spots in the 2DE map shown in Fig. 3 and other maps obtained by isoelectrofocusing, at different pH ranges.
DISCUSSION
The recent availability of extensively annotated genomic sequences has opened the way to new experimental approaches in both basic and applied research. DNA microarrays, proteomic technologies, and whole-genome expression cloning programs allow us to address scientific issues from a completely different perspective which is expected to accelerate the path to new discoveries (14). For example, by using in silico genome analysis coupled to high-throughput cloning and expression, we have recently identified vaccine candidates against meningococcus type B (MenB) (42). The fact that no satisfactory vaccines have been discovered against this human pathogen, in spite of an intensive research activity in the last three decades, pinpoints a potentiality of the new genomic technologies.
In this work we adopted an approach similar to the one designed for MenB vaccine identification to address a more basic issue, namely, the elucidation of surface protein organization in C. pneumoniae. Until now this bacterial compartment, which includes components crucial for initiating the chlamydial replicative cycle and possibly also for survival against host immune responses, had been poorly characterized, in large part because of the technical difficulty of working with this pathogen. In fact, C. pneumoniae requires eukaryotic cells for growth and proliferation; also, procedures for C. pneumoniae transformation and genetic manipulation are still unavailable. Our approach overcomes these limitations, thus representing an effective way to rapidly and systematically tackle this important issue. The approach is based on six main experimental steps: (i) in silico analysis of the C. pneumoniae genome sequence to identify genes potentially encoding proteins destined for the periphery of the bacterial cell (including outer and inner membrane and periplasmic proteins); (ii) cloning, expression, and purification of selected candidates; (iii) use of purified antigens to generate mouse immune sera; (iv) analysis of sera specificity by Western blotting of total EB extracts; (v) assessment of antigen localization by FACS analysis on whole EBs; and (vi) identification of FACS-positive antigens on 2DE maps of C. pneumoniae EB proteins.
An intrinsic limit of this experimental approach is that, like the other approaches so far utilized to define C. pneumoniae surface antigens, the assessment of protein localization and accessibility ultimately relies on the specificity of antigen-antibody recognition. Therefore, the occurrence of false positive results due to antibody cross-reactivity cannot be excluded. However, the fact that in most cases FACS positivity is accompanied by either a specific response in Western blot analysis or the identification of the corresponding antigen on 2DE maps, suggests that cross reactions do not represent a dominant feature in this study. In fact, inspection of the data reported in Table 1 leads to the conclusion that for 24 antigens, FACS data are supported by both Western blot analysis and 2DE-mass spectrometry, whereas for 21 additional sera FACS analysis is strengthened by either Western blotting (17 antigens) or 2DE-mass spectrometry (4 antigens). Therefore, for 8 antigens only, surface localization is inferred only by FACS data.
An important observation that, in our opinion, further validates the experimental approach proposed here is that, as mentioned before, a few antigens have been reported in the literature to be surface exposed in C. pneumoniae. All of them have also been classified as surface exposed by our analysis. A few comments on the 53 surface antigens identified in this study are given below.
Known or expected surface components.
A few of the 53 proteins in Table 1 have already been identified by others as surface components in either C. pneumoniae or in other chlamydial species. As already pointed out, they include OmpA (MOMP homolog) (41, 57), PorB (29), the 76-kDa protein homolog (39), and the 11 members of the Pmp family (separately discussed below) which was recently characterized by Grimwood and coworkers (16). This group also includes the cysteine-rich 60-kDa protein encoded by omcB, which was recently shown to be a chlamydial surface component to which heparin-like molecules can anchor (49), and the smaller associated protein OmcA, currently annotated as a 9-kDa cysteine-rich outer membrane complex lipoprotein generally considered to be associated with differentiation of the intracellular RBs into extracellular infectious EBs and necessary for structural integrity of the EB outer envelope.
Also reported in the literature for C. trachomatis are data on the surface exposure of DnaK and Mip-like proteins, both immunogenic antigens in human chlamydial genital infections (46). DnaK (a member of the hsp70 family predictable in silico as an inner membrane protein with a PSORT probability score of 0.151) has been described as being associated with the outer membrane of C. trachomatis (43) and exposed on the chlamydial surface, as suggested by the fact that it can elicit neutralizing antibodies in vitro (7). Also, a recent study (25) has proposed that the hsp70 ortholog protein of Helicobacter pylori is a stress-induced surface adhesin. A similar outer membrane association of DnaK has been reported for the intracellular pathogen Coxiella burnetii by Macellaro et al. (34). In this last study, the dual localization of DnaK was confirmed by immuno-electronmicroscopy. As DnaK is usually described as a chaperonin normally expected to be located in the cytoplasm or the inner side of the cytoplasmic membrane, a possible explanation for the surface localization is that DnaK may artificially bind to the outer membrane due to its peptide binding properties, an event which may occur in vivo following release of the protein from bacteria undergoing autolysis. The fact remains that DnaK is found on the surface of bacteria and is often a dominant immunogen in human infections.
The chlamydial Mip protein, predicted by PSORT as periplasmic with a probability score of 0.930, is homologous to the surface-exposed macrophage infectivity potentiator (Mip) protein of Legionella pneumophila, has peptidyl-prolyl cis/trans isomerase activity that is inhibited by FK506 and rapamycin, and is implicated in initiation of chlamydial infection (33). In C. trachomatis, it was not possible to demonstrate surface-exposed Mip epitopes on infectious EB or RB forms either by immunofluorescence or immuno-gold electron microscopy. However, when antibodies to the N-terminal segment of Mip were used, a complement-dependent inhibition of up to 91% of infectivity for cell cultures was observed, suggesting that antibody-accessible Mip epitopes are in fact present on infectious EBs (32). Our results therefore support the finding that at least a portion of Mip emerges on the chlamydial surface and is immunoaccessible.
By analogy with other bacterial species, one could also expect to find in the surface candidate list the Omp85-like protein, shown to be on the surface of Neisseria gonorrhoeae and Treponema pallidum, and the protein encoded by the ORF CPn301 and currently annotated by homology with a C. trachomatis gene as the OmpH-like OMP, although the C. pneumoniae ORF has in fact lost significant similarity to OmpH.
The PMP family.
The list of candidate surface proteins shown in Table 1 includes 14 members of the polymorphic membrane protein (PMP) family, a large superfamily with 9 members in C. trachomatis and 21 members in C. pneumoniae, all having the common feature of two conserved amino acid motifs, GGAI and FXXN, repeated in variable copy numbers in the N-terminal portion (17, 44, 47). The function of this intriguing group of proteins and the reason for such an expansion of this set of paralogous genes in C. pneumoniae are not understood. A first issue currently being addressed is whether they are simultaneously expressed or their expression is regulated during the C. pneumoniae life cycle. Our data show that at least 15 of the pmp gene set are actually expressed components of EB cells, thus confirming and expanding the data recently published by Grimwood and coworkers (16) for strains of C. pneumoniae which are different from the one we used in this study. The expression of 11 Pmps, 10 of which were FACS positive, was confirmed by mass spectrometry, whereas 4 Pmps (Pmp1, Pmp4, Pmp9, and Pmp15) not identified on 2DE maps were positive in both Western blotting and FACS analyses.
Although the role of Pmps remains to be elucidated, involvement in adhesion, molecular transport, signaling, and other cell wall-associated functions have been proposed (44).
Hypothetical proteins.
An interesting finding is represented by the presence in Table 1 of a group of proteins which so far have only been annotated as hypothetical. Seven of these proteins are encoded by genes with corresponding orthologs in C. trachomatis, while the remaining seven are specific protein components of C. pneumoniae. Considering the high similarity between the C. trachomatis and C. pneumoniae genomes, it is reasonable to assume that C. pneumoniae-specific genes play important roles in C. pneumoniae biology. In particular, these surface-exposed hypothetical proteins may be implicated in C. pneumoniae-specific tissue tropism.
On the basis of in silico analysis, four of the hypothetical C. pneumoniae-specific antigens (the products of the CPn0794, -0795, -0796, and -0797 genes), together with the CPn0798 and CPn0799 genes, may be grouped in a new family of related outer membrane-associated proteins. These proteins have a repeat structure in common, which is somehow suggestive of the analogous situation in the Pmp set. Whether these proteins are also immunogenic in human infections and may represent possible vaccine candidates remain to be explored in future studies.
Unexpected findings.
A number of entries in Table 1 are proteins which, according to the current annotation and by in silico localization prediction algorithms such as PSORT, could be expected to be periplasmic, inner membrane, or even cytoplasmic components. The reason why the antibodies against some of these proteins bind to the chlamydial surface is somewhat surprising. However, in silico analyses (which are usually based either on the recognition of known protein sequence motifs signaling cellular localization or on sequence homologies of variable reliability) are still far from being infallible. Examples are the already-discussed OmcB, OmcA, DnaK, and Mip-like proteins. Similarly, the 76 kDa homolog 1 CPn0728 protein, which gave a strong positive signal in our FACS assay, is predicted by PSORT to be cytoplasmic (K-S score = 0.272), but its surface localization is supported by the fact that it was reported to induce antibodies which neutralize the infectivity of C. pneumoniae (39). Another example of misleading in silico predictions is represented by enolase (see below).
In addition to the failure of computer programs in predicting protein compartmentalization, one should always keep in mind that chlamydiae are notably very atypical bacteria, and some proteins may have a different role and/or localization with respect to other bacteria. If one considers that chlamydiae could have developed some specific localization or secretion signals which are not recognized by currently available prediction algorithms, then for chlamydiae the discrepancies between in silico prediction and experimental results may well be a frequent occurrence. For instance, one of the unexpected results is the positivity of MurG. The murG product is similar to a protein that is involved in other bacteria in the synthesis of peptidoglycan (PG) and that is active at the inner membrane level (44). However, in Chlamydia spp., PG synthesis is clearly anomalous (44). Until recently, the chlamydial cell was thought to be totally deprived of PG, and after the surprising finding of the PG synthesis genes in the genome sequence, chlamydial PG is thought to be synthesized not as a cell wall structure but for a different and still undefined purpose (1).
A particularly interesting case in which in silico predictions are contradicted by experimental data is represented by the product of the eno gene. This protein belongs to the family of well-known glycolytic enzymes and should not be expected to be FACS positive, since in silico analysis predicts it to be associated to the cytoplasmic inner membrane compartment. However, the localization of an enolase ortholog on the bacterial surface has been recently described for gram-positive, group A streptococci (37, 38). So, in this case, the positive FACS assay results for C. pneumoniae enolase are in fact supported by literature data, and to our knowledge this is the first report of the presence of this enzyme on the surface of a gram-negative bacterium. It is important to note that in streptococci the surface enolase is a multifunction protein with plasmin(ogen) binding properties, and Fontan and coworkers (10) suggested that surface enolase could facilitate host tissue invasion by preventing the generation of fibrin clots. Furthermore, this report (10) shows that streptococcal enolase can induce antibodies cross-reacting with the human enolase expressed on the surface of hematopoietic cells. It is tempting to speculate that also in chlamydial infections the induction of antienolase antibodies could be the cause of autoimmune inflammatory reactions.
It is worth noting the presence in Table 1 of two putative surface proteases, PepA and HtrA, which are part of a group of proteins selected for experimental screening essentially because their C. trachomatis orthologs were reported to be immunogenic in patients with C. trachomatis infection (46). Interestingly, the Haemophilus influenzae HtrA homolog has been shown to be a potential vaccine candidate since it can induce a protective immunity in animal models (31).
Finally to be noted is the identification on the C. pneumoniae surface of the products of the lcrE and yscJ genes, both predicted to be part of a type III secretion (TTS) system on the basis of homologies with other well-described secretion systems in other bacteria, like those in Yersinia and Salmonella. The TTS systems of gram-negative pathogens appear to be derived from flagellar structures and are involved in contact-dependent secretion of virulence factors (24). The possible existence of such a secretion system in Chlamydia was first described by Hsia and colleagues (23) for Chlamydia psittaci, then confirmed by the subsequent genome sequencing projects, and more recently has been described in functional studies (9, 51). The current hypothesis envisages this system as being active during the intracellular phase of the chlamydial replicative cycle for the insertion of chlamydial proteins (like the Inc set) into the inclusion membrane that separates the growing chlamydial microcolony from the host cell cytoplasm (1, 44), and also for secreting into the host cell cytoplasm proteins which modulate the cell response to ongoing chlamydial replication. The protein encoded by lcrE is homologous to Yersinia YopN, a surface protein (11) thought to be a TTS response regulator, sensing either a host cell contact in vivo or Ca2+ concentration in vitro and located at the most outward position in the TTS structure (4, 44). The finding that LcrE is actually exposed in the spore-like EB form suggests that the TTS apparatus may also be fully assembled in extracellular chlamydiae, possibly to be used in early events of cell infection, e.g., in order to assist the entry of chlamydiae into the host cell and a successful establishment of the early chlamydial inclusion vacuole. This would be in agreement with the proposed hypothesis (2) that the needle-like projections first observed by Matsumoto on the EB surface in the early fundamental electron microscopic studies on C. trachomatis (35), and later confirmed in C. pneumoniae (36), are in fact TTS structures.
Another putative component of a chlamydial TTS that induced FACS-positive antibodies is YscJ (Table 1). According to the Yersinia model of TTS structure, this protein would be expected to be located in the periplasmic space. It is, however, possible that part of this protein protrudes through the outer membrane or perhaps, in the chlamydial TTS structure, this protein plays a different role. In fact, it has been noted (44) that when the set of genes thought to form the chlamydial TTS structure is compared with its homologous set of genes in other bacteria, several components appear to be missing in the chlamydial genome. The fact that LcrE appears to be present and accessible to antibodies on the surface of the infectious EB form makes this protein a possible vaccine candidate, since an efficient block of the TTS organelle may in turn inhibit the infection process by “freezing” the LcrE negative regulator.
Concluding remarks.
This work represents the first successful attempt of a systematic analysis of proteins located on the outer surface of C. pneumoniae. The data presented here, besides providing important clues for further investigation on the molecular mechanisms of chlamydial pathogenicity, may be relevant for developing new and more specific diagnostic assays for the assessment of C. pneumoniae infections in humans. We are currently exploiting C. pneumoniae protein chips with the aim of analyzing antibody response profiles in sera from infected patients and healthy carriers. Such an analysis might lead to the identification of specific protein recognition patterns that may eventually be correlated with diverse types of infection, and they eventually might help elucidate the involvement of C. pneumoniae in the development of atherosclerosis and heart disease. Furthermore, since a prophylactic immunization against C. pneumoniae has to rely at least in part on immune responses against proteins exposed on the surface of infectious chlamydiae, our data open the way to a rational selection of new vaccine candidates.
Acknowledgments
We thank Rino Rappuoli, Isaac Smith, and Dave Dubnau for their critical reading of the manuscript, G. Corsi for the artwork, and A. Maiorino for her expert secretarial assistance.
This work was partially supported by the Italian Ministry of University, Scientific and Technological Research (MURST), and by a grant from the European Community.
Editor: D. L. Burns
REFERENCES
- 1.Bavoil, P. M., R. C. Hsia, and D. Ojcius. 2000. Closing in on Chlamydia and its intracellular bag of tricks. Microbiology 146: 2723–2731. [DOI] [PubMed] [Google Scholar]
- 2.Bavoil, P. M., and R. C Hsia. 1998. Type III secretion in Chlamydia: a case of deja vu? Mol. Microbiol. 28: 860–862. [DOI] [PubMed] [Google Scholar]
- 3.Bjellqvist, B., G. J. Hughes, C. Pasquali, N. Paquet, F. Ravier, J. C. Sanchez, S. Frutiger, and D. Hochstrasser. 1993. The focusing positions of poly-peptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 14: 1023–1031. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, L. W., and O. Schneewind. 2000. Yersinia enterocolitica type A, an intracellular regulator of the type III machinery, is required for specific targeting of YopE, YopH, YopM, and YopN into the cytosol of eukaryotic cells. J. Bacteriol. 182: 3183–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chevallet, M., V. Santoni, A. Poinas, D. Rouquie, A. Fuchs, S. Kieffer, M. Rossignol, J. Lunardi, J. Garin, and T. Rabilloud. 1998. New zwitterionic detergents improve the analysis of membrane proteins by two-dimensional electrophoresis. Electrophoresis 19: 1901–1909. [DOI] [PubMed] [Google Scholar]
- 6.Christiansen, G., A.-S. Pedersen, K. Hjernø, B. Vandahl, and S. Birkelund. 2000. Potential relevance of Chlamydia penumoniae surface proteins to an effective vaccine. J. Infect. Dis. 181: 528–537. [DOI] [PubMed] [Google Scholar]
- 7.Danilition, S., I. W. Maclean, R. Peeling, S. Winston, and R. C. Brunham. 1990. The 75-kilodalton protein of Chlamydia trachomatis: a member of the heat shock protein 70 family? Infect. Immun. 58: 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doherty, N. S., B. H. Littman, K. Reilly, A. C. Swindell, J. M. Buss, and N. L. Anderson. 1998. Analysis of changes in acute-phase plasma proteins in an acute inflammatory response and in rheumatoid arthritis using two-dimensional gel electrophoresis. Electrophoresis 19: 355–363. [DOI] [PubMed] [Google Scholar]
- 9.Fields, K. A., and T. Hackstadt. 2000. Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol. Microbiol. 38: 1048–1060. [DOI] [PubMed] [Google Scholar]
- 10.Fontan, P. A., V. Pancholi, M. M. Nociari, and V. A. Fischetti. 2000. Antibodies to streptococcal surface enolase react with human alpha-enolase: implications in poststreptococcal sequelae. J. Infect. Dis. 182: 1712–1721. [DOI] [PubMed] [Google Scholar]
- 11.Forsberg, A., A. M. Viitanen, M. Skurnik, and H. Wolf-Watz. 1991. The surface-located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis. Mol. Microbiol. 5: 977–986. [DOI] [PubMed] [Google Scholar]
- 12.Gaydos, C. A., J. T. Summersgill, N. N. Sahney, J. A. Ramirez, and T. C. Quinn. 1996. Replication of Chlamydia penumoniae in vitro in human macrophages, endothelial cells, and aortic smooth muscle cells. Infect. Immun. 64: 1614–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goth, S. R., and R. S. Stephens. 2001. Rapid, transient phosphatidylserine externalization induced in host cells by infection with Chlamydia spp. Infect. Immun. 69: 1109–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grandi, G. 2001. Antibacterial vaccine design using genomics and proteomics. Trends Biotechnol. 19: 181–188. [DOI] [PubMed] [Google Scholar]
- 15.Grayston, J. T. 2000. Background and current knowledge of Chlamydia penumoniae and atherosclerosis. J. Infect. Dis. 181: S402–S410. [DOI] [PubMed] [Google Scholar]
- 16.Grimwood, J., L. Olinger, and R. S. Stephens. 2001. Expression of Chlamydia penumoniae polymorphic membrane protein family genes. Infect. Immun 69: 2383–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimwood, J., and R. S. Stephens. 1999. Computational analysis of the polymorphic membrane protein superfamily of Chlamydia trachomatis and Chlamydia pneumoniae. Microb. Comp. Genomics 4: 187–201. [DOI] [PubMed] [Google Scholar]
- 18.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192: 262–267. [DOI] [PubMed] [Google Scholar]
- 19.Gupta, S., E. W. Leatham, D. Carrington, M. A. Mendall, J. C. Kaski, and A. J. Camm. 1997. Elevated Chlamydia pneumoniae antibodies, cardiovascular events, and azithromycin in male survivors of myocardial infarction. Circulation 96: 404–407. [DOI] [PubMed] [Google Scholar]
- 20.Gurfinkel, E., G. Bozovich, A. Daroca, E. Beck, and B. Mautner. 1997. Randomised trial of roxithromycin in non-Q-wave coronary syndromes: ROXIS pilot study. Lancet 350: 404–407. [DOI] [PubMed] [Google Scholar]
- 21.Gygi, S. P., G. L. Corthals, Y. Zhang, Y. Rochon, and R. Aebersold. 2000. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc. Natl. Acad. Sci. USA 97: 9390–9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herbert, B. R., M. P. Molloy, A. A. Gooley, B. J. Walsh, W. G. Bryson, and K. L. Williams. 1998. Improved protein solubility in two-dimensional electrophoresis using tributyl phosphine as reducing agent. Electrophoresis 19: 845–851. [DOI] [PubMed] [Google Scholar]
- 23.Hsia, R. C., Y. Pannekoek, E. Ingerowski, and P. M. Bavoil. 1997. Type III secretion genes identify a putative virulence locus of Chlamydia. Mol. Microbiol. 25: 351–359. [DOI] [PubMed] [Google Scholar]
- 24.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62: 379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huesca, M., A. Goodwin, A. Bhagwansingh, P. Hoffman, and C. A. Lingwood. 1998. Characterization of an acidic-pH-inducible stress protein (hsp70), a putative sulfatide binding adhesin, from Helicobacter pylori. Infect. Immun. 66: 4061–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21: 385–389. [DOI] [PubMed] [Google Scholar]
- 27.Knudsen, K., A. S. Madsen, P. Mygind, G. Christiansen, and S. Birkelund. 1999. Identification of two novel genes encoding 97- to 99-kilodalton outer membrane proteins of Chlamydia penumoniae. Infect. Immun. 67: 375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krull, M., A. C. Klucken, F. N. Wuppermann, O. Fuhrmann, C. Magerl, J. Seybold, S. Hippenstiel, J. H. Hegemann, C. A. Jantos, and N. Suttorp. 1999. Signal transduction pathways activated in endothelial cells following infection with Chlamydia pneumoniae. J. Immunol. 162: 4834–4841. [PubMed] [Google Scholar]
- 29.Kubo, A., and R. S. Stephens. 2000. Characterization and functional analysis of porB, a Chlamydia porin and neutralizing agent. Mol. Microbiol. 38: 772–780. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 31.Loosmore, S. M., Y. P. Yang, R. Oomen, J. M. Shortreed, D. C. Coleman, and M. H. Klein. 1998. The Haemophilus influenzae HtrA protein is a protective antigen. Infect. Immun. 66: 899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lundemose, A. G., D. A. Rouch, S. Birkelund, G. Christiansen, and J. H. Pearce. 1992. Chlamydia trachomatis Mip-like protein. Mol. Microbiol. 6: 2539–2544. [DOI] [PubMed] [Google Scholar]
- 33.Lundemose, A. G., J. E. Kay, and J. H. Pearce. 1993. Chlamydia trachomatis Mip-like protein has peptidyl-prolyl cis/trans isomerase activity that is inhibited by FK506 and rapamycin and is implicated in initiation of chlamydial infection. Mol. Microbiol. 7: 777–783. [DOI] [PubMed] [Google Scholar]
- 34.Macellaro, A., E. Tujulin, K. Hjalmarsson, and L. Norlander. 1998. Identification of a 71-kilodalton surface-associated Hsp70 homologue in Coxiella burnetii. Infect. Immun. 66: 5882–5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsumoto, A. 1981. Electron microscope observations of surface projections and related intracellular structures of Chlamydia organisms. J. Electron Microsc. 30: 315–320. [PubMed] [Google Scholar]
- 36.Miyashita, N., Y. Kanamoto, and A. Matsumoto. 1993. The morphology of Chlamydia pneumoniae. J. Med. Microbiol. 38: 418–425. [DOI] [PubMed] [Google Scholar]
- 37.Pancholi, V., and V. A. Fischetti. 1997. A novel plasminogen/plasmin binding protein on the surface of group A streptococci. Adv. Exp. Med. Biol. 418: 597–599. [DOI] [PubMed] [Google Scholar]
- 38.Pancholi, V., and V. A. Fischetti. 1998. Alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273: 14503–14515. [DOI] [PubMed] [Google Scholar]
- 39.Perez Melgosa, M., C. C. Kuo, and L. A. Campbell. 1994. Isolation and characterization of a gene encoding a Chlamydia pneumoniae 76-kilodalton protein containing a species-specific epitope. Infect. Immun. 62: 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567. [DOI] [PubMed] [Google Scholar]
- 41.Peterson, E. M., X. Cheng, Z. Qu, and L. M. De La Maza. 1996. Characterization of the murine antibody response to peptides representing the variable domains of the major outer membrane protein of Chlamydia pneumoniae. Infect. Immun. 64: 3354–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pizza, M., V. Scarlato, V. Masignani, M. M. Giuliani, B. Arico, M. Comanducci, G. T. Jennings, L. Baldi, E. Bartolini, B. Capecchi, C. L. Galeotti, E. Luzzi, R. Manetti, E. Marchetti, M. Mora, S. Nuti, G. Ratti, L. Santini, S. Savino, M. Scarselli, E. Storni, P. Zuo, M. Broeker, E. Hundt, B. Knapp, E. Blair, T. Mason, H. Tettelin, D. W. Hood, A. C. Jeffries, N. J. Saunders, D. M. Granoff, J. C. Venter, E. R. Moxon, G. Grandi, and R. Rappuoli. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287: 1816–1820. [DOI] [PubMed] [Google Scholar]
- 43.Raulston, J. E., C. H. Davis, D. H. Schmiel, M. W. Morgan, and P. B. Wyrick. 1993. Molecular characterization and outer membrane association of a Chlamydia trachomatis protein related to the hsp70 family of proteins. J. Biol. Chem. 268: 23139–23147. [PubMed] [Google Scholar]
- 44.Rockey, D. D., J. Lenart, and R. S. Stephens. 2000. Genome sequencing and our understanding of chlamydiae. Infect. Immun. 68: 5473–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saikku, P., M. Leinonen, K. Mattila, M. R. Ekman, M. S. Nieminen, P. H. Makela, J. K. Huttunen, and V. Valtonen. 1988. Serological evidence of an association of a novel Chlamydia, TWAR, with chronic heart disease and acute myocardial infarction. Lancet ii: 983–986. [DOI] [PubMed] [Google Scholar]
- 46.Sanchez-Campillo, M., L. Bini, M. Comanducci, R. Raggiaschi, B. Marzocchi, V. Pallini, and G. Ratti. 1999. Identification of immunoreactive proteins of Chlamydia trachomatis by Western blot analysis of a two-dimensional electrophoresis map with patient sera. Electrophoresis 20: 2269–2279. [DOI] [PubMed] [Google Scholar]
- 47.Schachter, J., and P. B. Wyrick. 1994. Culture and isolation of Chlamydia trachomatis. Methods Enzymol. 236: 377–390. [DOI] [PubMed] [Google Scholar]
- 48.Stephens, R. S. (ed.). 1999. Chlamydia: intracellular biology, pathogenesis, and immunity. American Society for Microbiology, Washington, D.C.
- 49.Stephens, R. S., K. Koshiyama, E. Lewis, and A. Kubo. 2001. Heparin-binding outer membrane protein of chlamydiae. Mol. Microbiol. 40: 691–699. [DOI] [PubMed] [Google Scholar]
- 50.Stephens, R. S., and C. Lammel. 2001. Chlamydia outer membrane protein discovery using genomics. Curr. Opin. Microbiol. 4: 16–20. [DOI] [PubMed] [Google Scholar]
- 51.Subtil, A., C. Parsot, and A. Dautry-Varsat. 2001. Secretion of predicted Inc proteins of Chlamydia pneumoniae by a heterologous type III machinery. Mol. Microbiol. 39: 792–800. [DOI] [PubMed] [Google Scholar]
- 52.Taraktchoglou, M., A. A. Pacey, J. E. Turnbull, and A. Eley. 2001. Infectivity of Chlamydia trachomatis serovar LGV but not E is dependent on host cell heparan sulfate. Infect. Immun. 69: 968–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76: 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vandahl, B. B., S. Birkelund, H. Demol, B. Hoorelbeke, G. Christiansen, J. Vandekerckhove, and K. Gevaert. 2001. Proteome analysis of the Chlamydia pneumoniae elementary body. Electrophoresis 22: 1204–1223. [DOI] [PubMed] [Google Scholar]
- 55.Wang, S. P., C. C. Kuo, and J. T. Grayston. 1979. Formalinized Chlamydia trachomatis organisms as antigen in the micro-immunofluorescence test. J. Clin. Microbiol. 10: 259–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilm, M., A. Shevchenko, T. Houthaeve, S. Breit, L. Schweigerer, T. Fotsis, and M. Mann. 1996. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379: 466–469. [DOI] [PubMed] [Google Scholar]
- 57.Wolf, K., E. Fischer, D. Mead, G. Zhong, R. Peeling, B. Whitmire, and H. D. Caldwell. 2001. Chlamydia pneumoniae major outer membrane protein is a surface-exposed antigen that elicits antibodies primarily directed against conformation-dependent determinants. Infect. Immun. 69: 3082–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Young, I. T. 1977. Proof without prejudice: use of the Kolmogorov-Smirnov test for the analysis of histograms from flow systems and other sources. J. Histochem. Cytochem. 25: 935–941. [DOI] [PubMed] [Google Scholar]