Abstract
The outer cores of the lipooligosaccharides (LOS) of many strains of Campylobacter jejuni mimic human gangliosides in structure. A population of cells of C. jejuni strain 81-176 produced a mixture of LOS cores which consisted primarily of structures mimicking GM2 and GM3 gangliosides, with minor amounts of structures mimicking GD1b and GD2. Genetic analyses of genes involved in the biosynthesis of the outer core of C. jejuni 81-176 revealed the presence of a homopolymeric tract of G residues within a gene encoding CgtA, an N-acetylgalactosaminyltransferase. Variation in the number of G residues within cgtA affected the length of the open reading frame, and these changes in cgtA corresponded to a change in LOS structure from GM2 to GM3 ganglioside mimicry. Site-specific mutation of cgtA in 81-176 resulted in a major LOS core structure that lacked GalNAc and resembled GM3 ganglioside. Compared to wild-type 81-176, the cgtA mutant showed a significant increase in invasion of INT407 cells. In comparison, a site-specific mutation of the neuC1 gene resulted in the loss of sialic acid in the LOS core and reduced resistance to normal human serum but had no affect on invasion of INT407 cells.
Campylobacter jejuni is one of the most common causes of bacterial diarrhea worldwide (10, 25) and has been shown to be frequently associated with the development of Guillain-Barré syndrome (GBS), a postinfectious polyneuropathy (23). The association of C. jejuni and the development of GBS is thought to result from the molecular mimicry between outer core structures of bacterial lipooligosaccharides (LOS) and human gangliosides (18, 19). Thus, for example, the cores of different isolates of C. jejuni have been shown to mimic GM1, GM2, GD3, GD1a, GT1a, and GQ1b (1-3, 18-24, 29, 30, 42, 43). However, similar ganglioside mimicry can be found in strains associated with both uncomplicated enteritis and GBS (18). While considerable research efforts have focused on the role of LOS in the development of GBS, little attention has been placed on the function of these sialylated LOS structures in the pathogenesis of gastrointestinal disease. A locus involved in LOS biosynthesis in C. jejuni MSC57360, the type strain of the HS:1 serogroup, has been described previously (12). It has also been shown that the loss of sialic acid (NeuNAc) from the LOS core results in the increased immunogenicity of the core and an increased sensitivity to normal human serum (12). However, strain MSC57360 is noninvasive in vitro (P. Guerry and C. P. Ewing , unpublished data), and little is known about its virulence potential. In this report, we have characterized the LOS core of C. jejuni 81-176 (serogroups HS:23 and HS:36), which is one of the best-characterized strains of C. jejuni (5-7, 14, 15, 26, 28, 41) and one which has been shown to induce diarrhea in human volunteers in two separate studies (7; D. Tribble et al., unpublished data). These results indicate that the LOS core of C. jejuni 81-176 is composed of structures that mimic several gangliosides, although the predominant structure is a GM2 ganglioside mimic. Slip strand mismatch recombination of the cgtA gene, which encodes an N-acetylgalactosaminyl (GalNAc) transferase, results in changes in the major core structure from GM2 to GM3 ganglioside mimicry. Insertional inactivation of the cgtA gene results in a mutant lacking GM2 mimicry, and this mutant shows enhanced invasion of intestinal epithelial cells in vitro.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Characteristics of C. jejuni 81-176 have been described previously (7, 15). The strain was grown at 37°C under microaerophilic conditions on Mueller-Hinton (MH) agar (Difco) supplemented with chloramphenicol (CHL; 20 μg/ml) or kanamycin (KAN; 25 μg/ml) when appropriate. Strains for invasion assays were grown in MH biphasic cultures supplemented with antibiotics when appropriate. Escherichia coli XL1-Blue was the host for λ ZAP Express (Stratagene, La Jolla, Calif.), and DH5α was the host for subcloning experiments.
DNA cloning and sequence analyses.
C. jejuni genes were cloned from a partial Sau3A library constructed in λ ZAP Express. The library was probed with a 515-bp PCR product specific for the neuC1 gene of C. jejuni MSC57360 (12), which had been purified by agarose gel electrophoresis and labeled by random priming with [32P]dCTP (New England Nuclear, Boston, Mass.). The PCR primers have been described previously (12). Positive clones were plaque purified and rehybridized, and pure phage populations were excised to the phagemid pBK-CMV according to the instructions of the manufacturer. Three overlapping clones, pCJ8-6, pCJ8-8, and pSG886, were subjected to DNA sequence analyses with a Perkin-Elmer Applied Biosystems model 373A automated DNA sequencer. Custom sequencing primers were synthesized on a Perkin-Elmer Applied Biosystems model 292 DNA synthesizer. Additional DNA sequences were generated by sequencing with primers within transposon insertions into the plasmid (see below).
Generation of cgtA and neuC1 mutants.
cgtA and neuC1 mutants were constructed using a Tn5-based in vitro transposition system (Epicentre, Madison, Wis.) in which the Cmr cassette from pRY109 (40) was cloned into pEZ::TN pMOD, as described previously (12). The transposon was PCR amplified with primers as described by Epicentre and used in an in vitro reaction with pCJ8-6 as the target. The reaction product was transformed into E. coli DH5α by standard methods, and plasmid DNAs from individual transformants were sequenced using primers which read out from within the Cmr cassette (12) to determine the insertion point and the orientation within the gene. Insertions were selected in which the Cmr cassette had been inserted in the same orientation as the target gene had been transcribed to minimize polarity on downstream genes. Plasmids were used to transform C. jejuni 81-176, with selection on MH agar supplemented with 15 μg of CHL/ml (37). Transformants were analyzed by PCR with primers bracketing the Cmr insertion point to confirm that the DNA had been inserted by a double crossover.
Complementation in trans.
The DNA cloned in pSG886 was transferred from pBK-CMV into the E. coli-C. jejuni shuttle plasmid pRY107 (40) as an EcoRI-PstI-ended fragment. This clone, pRY886, was transformed into E. coli DH5α containing RK212.2 (9) and then conjugally transferred into C. jejuni 81-176 cgtA and into C. jejuni MSC57360 cgtA (12), with selection on MH agar supplemented with 10 μg of trimethoprim/ml, 25 μg of KAN/ml, and 15 μg of CHL/ml.
LOS isolation.
For electrophoretic analyses of LOS, whole-cell lysates were digested with proteinase K as described by Hitchcock and Brown (13). For analyses by fast atom bombardment-mass spectrometry (FAB-MS), the biomass was subjected to hot phenol-water extraction (39) and the crude LOS were purified by enzymatic treatments with RNase A, DNase II, and proteinase K and by ultracentrifugation, as described previously (21).
Electrophoresis.
Proteinase K-digested whole-cell lysates containing LOS were separated on 16% Tricine gels (Invitrogen, Carlsbad, Calif.) and then silver stained (Bio-Rad, Hercules, Calif.).
Characterization of LOS and cgtA sequence from individual colonies.
Individual colonies of C. jejuni 81-176 were washed in phosphate-buffered saline and then digested with proteinase K as described by Hitchcock and Brown (13). Fractions of the same individual colonies of 81-176 were boiled, the cell debris was centrifuged, and the supernatants were used as templates for PCRs. The primers cgtA-F1 (5"-CGATGTGGATCATTACTACGATGC-3") and cgtA-B1 (5"-CGTTTCGGCGGTATTTTAAGGC-3") were used. PCR conditions were 30 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 30 s. The 354-bp PCR product was purified with QIAquick PCR purification kits (Qiagen, Chatsworth, Calif.) and sequenced with the cgtA-B1 oligonucleotide as the primer.
INT407 cell adherence and invasion assays.
Adherence and invasion assays were done by previously described methods (5, 6, 14, 26, 41) with modifications. Approximately 1.5 × 106 bacteria were added to a layer of approximately 4 × 105 INT407 cells (multiplicity of infection, ∼4) and incubated at 37°C for 2 h. For determination of adherence, the cells were washed four times in Hanks' balanced salt solution with strong agitation for 2 min, the monolayer was lysed with 0.01% Triton X-100 for 30 min at room temperature on an orbital shaker, and the total numbers of bacteria were enumerated by plate count. For determination of invasion, the monolayer was incubated with 100 μg of gentamicin/ml in prewarmed minimal essential medium (Gibco, Gaithersburg, Md.) for an additional 2 h prior to lysis of the monolayer with Triton X-100. Internalized bacteria were enumerated by plate count. The data were calculated as the percentage of bacteria in the inoculum which adhered or invaded.
Serum sensitivity.
Sensitivity tests were performed as described previously (12).
FAB-MS.
Individual colonies were examined for LOS type as described above and restreaked onto one MH agar plate each. Growth from a colony shown to produce 3.6-kDa LOS and from another colony shown to produce 3.8-kDa LOS was expanded to several plates. The next day, the biomass was collected into phosphate-buffered saline and frozen prior to analysis by FAB-MS. In addition, a small amount of the biomass was restreaked for individual colonies, which were subjected to proteinase K digestion and analyzed on Tricine gels to determine the LOS type. Among the population which had produced the 3.8-kDa LOS, 2 of 27 (7.4%) colonies had shifted to expression of the 3.6-kDa LOS; of the colonies which had originally produced the 3.6-kDa LOS, 0 of 18 colonies had shifted to expression of the 3.8-kDa LOS. FAB-MS analyses were conducted on permethylated core oligosaccharides as described previously (4, 12).
Nucleotide sequence accession number.
The sequences described in this paper have been submitted to GenBank under accession number AF305571.
RESULTS
Analysis of natural LOS variants of C. jejuni 81-176.
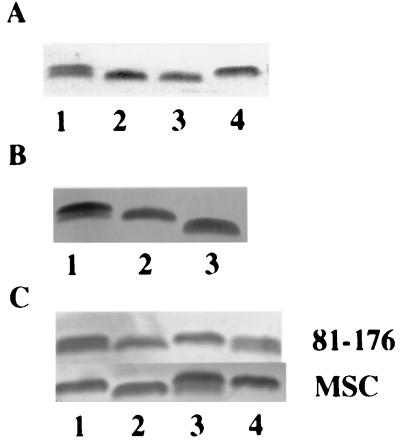
LOS preparations from a population of 81-176 cells were visualized following electrophoresis as a doublet, as shown in Fig. 1A, lane 1. The upper and lower bands ran at apparent molecular masses of 3.8 and 3.6 kDa, respectively. However, when LOS preparations from individual colonies were analyzed, the LOS appeared as a single band of either 3.8 or 3.6 kDa (Fig. 1A, lanes 2 through 4). An individual colony expressing the 3.8-kDa LOS core and another colony expressing the 3.6-kDa LOS core were restreaked, and, after a 48-h incubation at 37°C, proteinase K-treated whole-cell preparations of individual colonies were examined electrophoretically again. Among the progeny of the colony which had been expressing the 3.8-kDa LOS, 12 of 52 colonies (23%) had shifted to expression of the 3.6-kDa form. Among the progeny of the cell which originally expressed the 3.6-kDa form, only 1 of 52 colonies (2%) produced the 3.8-kDa LOS core.
FIG. 1.
Silver-stained LOS cores. Proteinase K-digested whole-cell lysates were separated on 16% Tricine gels for silver staining. (A) A population of 81-176 cells (lane 1) was compared to individual colonies of 81-176 (lanes 2 through 4). (B) Lane 1, 81-176; lane 2, 81-176 cgtA; lane 3, 81-176 neuC1. (C) Complementation in trans in either 81-176 or MSC57360 (12) backgrounds. Lane 1, wild type; lane 2, cgtA mutant; lane 3, cgtA mutant complemented with pRY886; lane 4, wild type.
FAB-MS characterization of LOS.
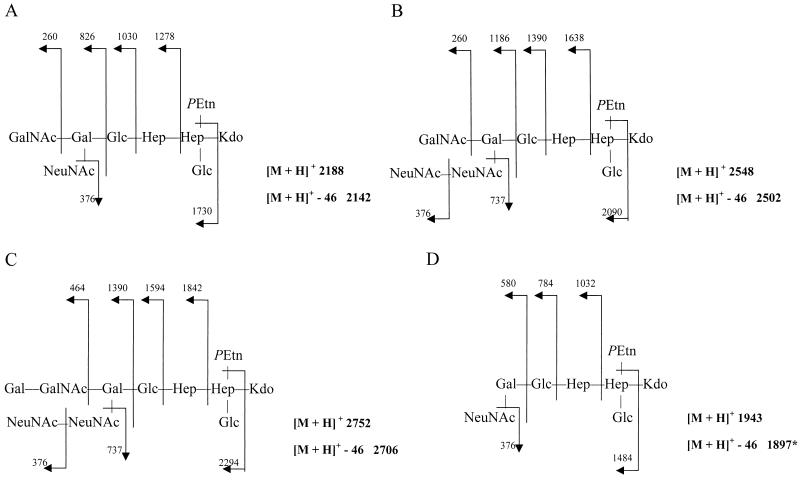
LOS preparations that had been enriched for the 3.8- and 3.6-kDa forms were subjected to FAB-MS, and four distinct structures were observed, as shown in Fig. 2. The major structure found in the preparation enriched for the 3.8-kDa LOS core was structure A, which, like the cores of the type strains of serogroups HS:23, HS:36, and HS:1, mimics GM2 ganglioside (Fig. 2A) (3). Structures B and C, which mimic GD2 and GD1b gangliosides, respectively, were also observed at lower concentrations in the 3.8-kDa LOS core preparation (Fig. 2B and C). Structure D, which mimics GM3 ganglioside, was the major component of the 3.6-kDa LOS form; a minor amount of structure A (GM2-like) was also observed (Fig. 2A).
FIG. 2.
Natural ganglioside mimics. FAB-MS of LOS isolated from 81-176 phase variants. (A) Structure A. Predominant component of 3.8-kDa LOS cores and minor component of 3.6-kDa LOS cores (GM2 like). (B) Structure B. Minor component of 3.8-kDa LOS core (GD2 like). (C) Structure C. Minor component of 3.8-kDa LOS core (GD1b like). (D) Structure D. Predominant component of 3.6-kDa LOS core (GM3 like).
Cloning and sequence analysis of the LOS genes of C. jejuni 81-176.
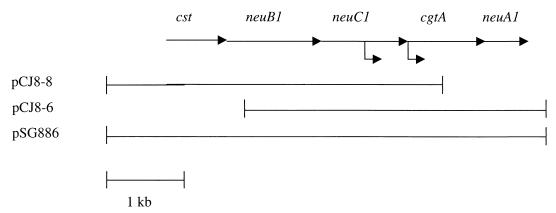
The DNA encoding the enzymes involved in biosynthesis of the outer core of C. jejuni 81-176 was cloned as described in Materials and Methods. DNA sequence analysis revealed the presence of five open reading frames (ORFs), as shown in Fig. 3, in an organization similar to that previously described for other strains (11, 12, 16, 17, 27). Table 1 lists the ORFs and their sequence similarities to those reported for other C. jejuni strains. The cluster includes genes encoding proteins with significant sequence similarity to enzymes involved in NeuNAc biosynthesis, which have been designated neuB1, neuC1, and neuA1 by Parkhill et al. (27). These genes encode a putative NeuNAc synthase; an N-acetylglucosamine(GlcNAc)-6-phosphate 2-epimerase/GlcNAc-6-phosphatase, which is involved in the synthesis of mannosamine; and aCMP-NeuNAc synthetase, respectively (17). In addition,ORF1 encodes a predicted protein which is 95 and 92% identical to bifunctional sialyltransferases (CstII) described for the type strain of serogroup HS:19, ATCC 43446, and a GBS isolate, OH4384, respectively (11). These enzymes are capable of transferring NeuNAc to the O-3 of galactose and to the O-8 of another NeuNAc (11). In all cases, the predicted proteins from strain 81-176 were more similar to the proteins from serogroup HS:19 than to those from NCTC 11168.
FIG. 3.
Schematic representation of the genes encoding the outer core of C. jejuni 81-176. The lengths of each ORF were as follows: cst, 876 bp; neuB1, 1,041 bp; neuC1, 1,065 bp; cgtA, 948 bp; and neuA1, 666 bp. The positions of insertion of a Cmr cassette are indicated by the arrows below the line. The insertion into neuC1 was 820 bp into the coding region of the gene, and the insertion into cgtA was 4 bp into the ORF. In both cases, the Cmr cassette was inserted in the same orientation in which the genes were transcribed. The regions of DNA cloned in pCJ8-8, pCJ8-6, and pSG886 are shown.
TABLE 1.
Homology of predicted proteins of strain 81-176 to other C. jejuni proteins
| Strain 81-176 | Strain (protein)b | % Identity, % similarityc | Predicted protein | ||
|---|---|---|---|---|---|
| ORF | Predicted molecular mass (kDa)a | Gene | |||
| 1 | 34.6 (291) | cstlI | 43446 | 95, 95 (291) | Sialyltransferase |
| OH4384 | 92, 93 (291) | ||||
| 11168 (Cj1140) | 52, 66 (289) | ||||
| 2 | 38.1 (346) | neuB1 | OH4384 | 95, 95 (346) | Sialic acid synthase |
| 43446 | 94, 94 (346) | ||||
| 11168 (Cj1141) | 74, 85 (343) | ||||
| 3 | 40.5 (354) | neuC1 | 43446 | 98, 98 (352) | Mannosamine synthesis |
| OH4384 | 98, 98 (352) | ||||
| 11168 (Cj1142) | 69, 84 (352) | ||||
| 4 | 37.4 (315) | cgtA | 11168 (Cj1143) | 65, 80 (308) | N-Acetylgalactosaminyltransferase |
| OH4384 | 47, 64 (292) | ||||
| 5 | 25.0 (221) | neuA | OH4384 | 84, 85 (221) | CMP-NeuNAc synthetase |
| 11168 (Cj1143) | 61, 72 (218) | ||||
Numbers in parentheses indicate the predicted numbers of amino acid residues.
Strains 43446 and OH4384 were described by Gilbert et al. (11), and strain 11168 has been described by Parkhill et al. (27). Gene product Cj1143 of NCTC 11168 was identified as a CMP-NeuNAc synthetase (27), but the ORF appears to encode a fusion protein in that strain and in MSC57360 (12). Mutational analysis of this gene in MSC57360 showed that the predicted protein functioned as a GalNAc transferase (12). In 81-176, there are two distinct ORFs that correspond to the amino (ORF4) and carboxy (ORF5) ends of the Cj1143 protein.
Numbers in parentheses indicate actual total numbers of amino acid residues in each protein.
ORF4 was sequenced from two independent clones, pCJ8-6 and pSG886. As cloned in pSG886, ORF4 encoded a protein of 315 amino acids that showed 47% identity and 64% similarity to a GalNAc transferase (CgtA) described for OH4384 (11). The predicted protein also showed higher homology (65% identity and 80% similarity) to the N-terminal 315 amino acids of protein Cj1143 from C. jejuni NCTC 11168, identified as a CMP-NeuNAc synthetase by Parkhill et al. (27), and to the corresponding gene product in C. jejuni MSC57360 (12), as shown in Table 1. However, Cj1143 in NCTC 11168 and its ortholog in MSC57360 (12) appear to be fusions of the products of cgtA and neuA1, but this apparent fusion protein has been shown to function as a GalNAc transferase in MSC57360 (12). Interestingly, as sequenced from pCJ8-6, the strain 81-176 cgtA gene encoded a truncated protein of 208 amino acids. The truncation occurred 23 bp after a run of 11 G nucleotides; the corresponding sequence from the pSG886 clone contained 10 G nucleotides.
Generation and characterization of neuC1 and cgtA mutants.
Site-specific insertional mutants in the cgtA and neuC1 genes were constructed as described in Materials and Methods. The electrophoretic mobilities of the LOS cores of the mutants and wild-type strain 81-176 were compared by analysis of proteinase K-digested whole cells on 16% Tricine gels, and the results are shown in Fig. 1B. The LOS core of a population of wild-type C. jejuni 81-176 cells again resolved in a silver-stained gel as a doublet with approximate molecular masses of 3.8 and 3.6 kDa (Fig. 1B, lane 1). The core of the cgtA mutant migrated with the lower, 3.6-kDa, band of the strain 81-176 doublet (Fig. 1B, lane 2), whereas the core of the neuC1 mutant migrated below both of the wild-type 81-176 bands (Fig. 1B, lane 3).
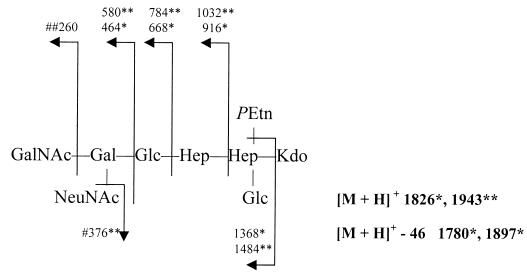
FAB-MS analyses of the cores of the neuC1 and cgtA mutants confirmed the loss of NeuNAc and GalNAc, respectively (12), as shown in Fig. 4. Thus, the major core structure of the cgtA mutant appears identical to that of the major component of the naturally occurring 3.6-kDa LOS band and displayed GM3 mimicry.
FIG. 4.
FAB-MS analyses of LOS cores of C. jejuni 81-176 mutants. Numbers refer to m/z values for ions. HexNAc, N-acetylhexosamine; Hep, heptose; Kdo, 3-deoxy-d-manno-2-octulosonic acid. The pseudomolecular ion and daughter ions observed for the oligosaccharide of the mutant in neuC1 are indicated by ∗ (derivatization results in partial degradation of the Kdo with loss of 46 mass units), whereas those in cgtA are indicated by ∗∗. The ion for NeuNAc, indicated by #, was absent from the neuC1 mutant, and that for HexNAc, indicated by ##, was absent in the cgtA mutant.
Phase variation of cgtA expression.
Based on the observations that the core of the wild-type strain 81-176 appeared to have a component with the same mobility as that of a cgtA mutant and the occurrence of a variable, homopolymeric G tract within the gene, the ability of cgtA to undergo slip strand mismatch repair was examined. The electrophoretic mobilities of the cores of 51 individual colonies of wild-type 81-176 cells were analyzed on Tricine gels, and a portion of the cgtA gene from each colony was amplified by PCR and subjected to DNA sequence analysis, as described in Materials and Methods. The results are summarized in Table 2. In 31 of 51 colonies (61%), the LOS core migrated at approximately 3.8 kDa, similar to that seen in Fig. 1A, lane 4. In all 31 colonies producing the 3.8-kDa core, the PCR-amplified cgtA gene contained a tract of 10 G's, corresponding to the full-length ORF. The LOS cores of 20 of 51 colonies (39%) ran at approximately 3.6 kDa, and the PCR-amplified cgtA genes of all 20 were truncated, although the number of G's varied. Thus, 9 of 20 colonies (45%) had 9 G's, 10 of 20 colonies (50%) had 11 G's, and 1 of 20 colonies (5%) had 12 G's.
TABLE 2.
Characterization of LOS and the cgtA gene of 51 individual colonies of strain 81-176
| No. of colonies | Apparent LOS core mass (kDa)a | No. of G nucleotides in cgtA geneb | Predicted length of cgtA ORF (nucleotides) |
|---|---|---|---|
| 31 | 3.8 | 10 | 316 |
| 10 | 3.6 | 11 | 208 |
| 9 | 3.6 | 9 | 206 |
| 1 | 3.6 | 12 | 207 |
Determined by electrophoresis on 16% Tricine gels.
Determined by DNA sequence analysis as described in Materials and Methods.
The DNA cloned in pSG886 was transferred into the Kmr shuttle vector pRY108 (40) to generate plasmid pRY886, which was conjugally transferred into the 81-176 cgtA mutant. The presence of the plasmid in trans complemented the defect in the mutant, as determined by electrophoretic analysis of the LOS core (Fig. 1C, lane 3, panel labeled 81-176). However, upon subculture of 81-176 cgtA(pRY886), variation in core masses was observed among individual colonies, indicating that slip strand mismatch recombination was also occurring on the plasmid copy of cgtA (data not shown).
C. jejuni strain MSC57360 also has a LOS core which mimics GM2 ganglioside (3, 12). Although the LOS biosynthesis genes of MSC57360 show high homology to those of 81-176 in general, the cgtA gene of MSC57360 lacked the homopolymeric G tract responsible for the phase variation found in the 81-176 cgtA gene (12). The LOS core of wild-type MSC57360 and a cgtA mutant (12) are shown in the panel marked “MSC” in Fig. 1C, lanes 1 and 2, respectively. When pRY886 was transferred into the MSC57360 cgtA mutant, the core appeared as a doublet (Fig. 1C, lane 3), similar to that seen in wild-type 81-176 but unlike that seen in wild-type MSC57360 (Fig. 1C, lane 1). Again, examination of individual colonies of MSC57360 cgtA (pRY886) revealed differences in core mass (data not shown).
Biological effects of mutation of neuC1 and cgtA.
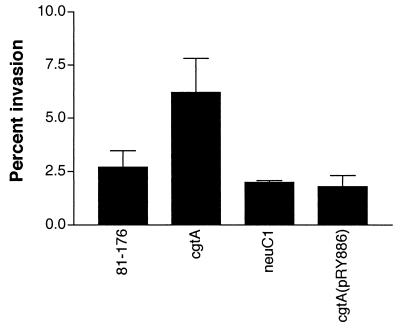
The abilities of wild-type strain 81-176 and the neuC1 and cgtA mutants to adhere to and invade INT407 cells were compared. There was no statistically significant difference among the three strains in their abilities to adhere to INT407 cells (data not shown). The invasion level of wild-type 81-176 into the inoculum was 2.7% ± 0.76% (mean ± standard deviation), and the neuC1 mutant invasion level was comparable (2.0% ± 0.8%) (Fig. 5) . However, the cgtA mutant displayed a statistically significant increase in invasion (6.2% ± 1.6%; P = 0.017) compared to that of the wild type. When the cgtA mutant was complemented in trans with pRY886, the level of invasion was reduced to levels similar to those of the wild type (1.8% ± 0.51%).
FIG. 5.
Invasion of INT407 cells. Invasion levels are shown as the percentages of the input inoculum internalized. Negative controls of E. coli HB101 invaded at 0.04% ± 0.007%. The results represent the means of three to eight different invasion assays.
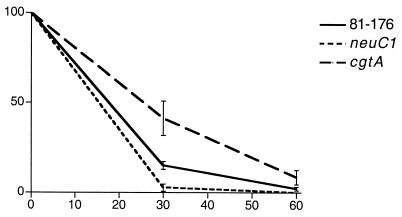
A neuC1 mutant of C. jejuni MSC57360 has been shown to decrease resistance to normal human serum, and a cgtA mutant has shown a slight increase in serum resistance (12). The neuC1 mutant of 81-176 also showed decreased resistance to human serum (P < 0.05), and the cgtA mutant showed an increased resistance after both 30- and 60-min incubations (P < 0.05), as shown in Fig. 6. Controls in which complement was inactivated by heat showed no killing (data not shown).
FIG. 6.
Comparison of serum resistance levels of wild-type 81-176 and mutants. Strains were incubated with pooled normal human serum for 0, 30, and 60 min. Results represent the means ± standard errors for three experiments.
DISCUSSION
Studies on the biosynthesis of LOS of other mucosal pathogens, such as Neisseria spp., have indicated that there is an inherent heterogeneity in core length and carbohydrate content because of differing additions of glycose groups at the nonreducing termini such that a single bacterial strain can produce a repertoire of LOS molecules (34). Thus, for example, a population of Neisseria gonorrhoeae cells from a single strain consists of up to six phenotypically different variants (31, 32). Similarly, the LOS core of C. jejuni 81-176 cells resolved electrophoretically as two distinct bands, and FAB-MS analyses confirmed the presence of two major structural variants, as well as minor structures not resolved on Tricine gels. Conversion between the two major LOS structures, which resemble GM2 and GM3 gangliosides, appears to occur at a relatively high frequency via a slip strand mismatch recombination which affects expression of the cgtA gene. High-frequency slip-stranded mismatch recombination has been observed in genomic sequence analyses of C. jejuni (27), and slip strand mismatch recombination of aβ-1,3-galactosyltransferase gene (wlaN) has been shown to affect LOS structure in C. jejuni NCTC 11168 (16). DNA sequence analyses have indicated that the structural differences between two GBS isolates, C. jejuni OH4384, which displayed GT1a mimicry, and C. jejuni OH4382, which exhibited GD3 mimicry, result from truncation of cgtA in strain OH4382 following a homopolymeric tract of eight A residues (11). However, the ability of either C. jejuni OH4384 or OH4382 to reversibly express cgtA has not been reported. The slip strand mismatch recombination observed in the present study with strain 81-176 occurred at a site in the cgtA gene different from that in which the mutation described by Gilbert et al. (11) in these two HS:19 isolates occurred. Moreover, the absence of similar homopolymeric tracts in the cgtA gene of either C. jejuni NCTC 11168 or MSC57360 would suggest that the ability of this gene to undergo phase variation is strain dependent. Thus, introduction of the 81-176 cgtA gene into the MSC57360 background resulted in the expression of LOS cores of variable mass, similar to those observed in wild-type 81-176.
Loss of NeuNAc had no apparent effect on the ability of 81-176 neuC1 to be internalized into INT407 cells. In contrast, loss of GalNAc from the core in the cgtA mutant enhanced internalization into INT407 cells. Although ganglioside mimicry of C. jejuni LOS has been considered only in terms of pathogenesis of GBS, LOS structure may directly affect the ability of C. jejuni to cause gastrointestinal disease. Moreover, the ability to alter the LOS structure may afford selective advantages to C. jejuni, as described for several other pathogens (33-36, 38). The major LOS structure produced by the cgtA mutant is presumably the GM3 mimic, since the inability to add GalNAc to the core precludes synthesis of the three other observed core structures (Fig. 2). Thus, the presence of GM3 mimicry in LOS appears to enhance invasion into epithelial cells, since the cgtA insertional mutant invaded at levels higher than those of the wild-type strain containing a mixture of at least four core structures. However, electrophoretic examination of the LOS cores of wild-type 81-176 colonies isolated following invasion into INT407 cells indicated that they also contained a mixture of LOS types (data not shown). Expression of other ganglioside mimics may be beneficial during other steps of pathogenesis, but the high frequency of the switch between LOS types precluded biological studies with the natural variants. Interestingly, we have also observed that 81-176 undergoes a high-frequency phase variation in the expression of a kpsM-dependent, high-molecular-weight glycan (6). A mutant that did not express the high-molecular-weight glycan showed reduced invasion levels in vitro and was attenuated in a ferret model of diarrheal disease (6), a model that appears to correlate with invasion in vitro (5, 6, 41). However, the relative insensitivity of the existing ferret model, which simply measures the presence or absence of diarrhea, is not likely to discriminate strains of slightly increased virulence such as the cgtA mutant.
Current data on C. jejuni 81-176 and that of Linton et al. on strain NCTC 11168 (16) would suggest that the LOS core structures of C. jejuni are dynamic and that the capacity to undergo variations can be strain dependent. Moreover, there are increasing indications that there may be differences in the mechanisms by which C. jejuni strains cause diarrheal disease. Thus, we have recently described a plasmid in C. jejuni 81-176 which encodes a putative type IV secretion system involved in internalization into intestinal epithelial cells (5). Interestingly, a virulence gene contained in this plasmid was found in only a limited number of fresh isolates in documented cases of human disease (5). Although many C. jejuni strains have the ability to synthesize ganglioside mimics, the trigger for GBS may also require a particular mechanism of presentation of that mimic to the host as well as a certain genetic background of the host (21). We are further exploring the role of LOS and other carbohydrate antigens in the virulence of C. jejuni 81-176.
Acknowledgments
This work was supported by NIAID grant 1 ROI AI43559 (to P.G.), Naval Medical Research and Development Command work no. 61102A3M161102BS13AK.11 (to P.G.), and a grant from the Irish Health Research Board (to A.P.M). M.M.P. is a recipient of an Irish Health Research Board Postdoctoral Fellowship.
We thank Gary Majam and Isabelle Walker for technical assistance.
Editor: V. J. DiRita
REFERENCES
- 1.Aspinall, G. O., A. Mainkar, M. M. Prendergast, B. Jacobs, and A. P. Moran. 1998. Lipopolysaccharides from a neuropathy-associated infection from Campylobacter jejuni serotype O:23, p. 93-96. In A. J. Lastovica, D. G. Newell, and E. E. Lastovica (ed.), Campylobacter, Helicobacter and related organisms. Institute of Child Health, Cape Town, South Africa.
- 2.Aspinall, G. O., A. G. McDonald, H. Pang, L. A. Kurjanczyk, and J. L. Penner. 1994. Lipopolysaccharides of Campylobacter jejuni serotype O:19: structures of core oligosaccharide regions from the serostrain and two bacterial isolates from patients with the Guillain-Barré syndrome. Biochemistry 33:241-249. [DOI] [PubMed] [Google Scholar]
- 3.Aspinall, G. O., A. G. McDonald., T. S. Raju, H. Pang, A. P. Moran, and J. L. Penner. 1993. Chemical structures of the core regions of Campylobacter jejuni serotypes O:1, O:4, O:23, and O:36 lipopolysaccharides. Eur. J. Biochem. 213:1017-1027. [DOI] [PubMed] [Google Scholar]
- 4.Aspinall, G. O., A. G. McDonald, T. S. Raju, H. Pang, J. L. Kurjanczyk, J. L. Penner, and A. P. Moran. 1993. Chemical structures of the core region of Campylobacter jejuni serotype O:2 lipopolysaccharide. Eur. J. Biochem. 213:1029-1037. [DOI] [PubMed] [Google Scholar]
- 5.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bacon, D. J., C. M. Szymanski, D. H. Burr, R. P. Silver, R. A. Alm, and P. Guerry. 2001. A phase variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol. Microbiol. 40:769-778. [DOI] [PubMed] [Google Scholar]
- 7.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472-479. [DOI] [PubMed] [Google Scholar]
- 8.Blaser, M. J., P. F. Smith, and P. F. Kohler. 1985. Susceptibility of Campylobacter isolates to the bactericidal activity of human serum. J. Infect. Dis. 151:227-235. [DOI] [PubMed] [Google Scholar]
- 9.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman, C. R., J. Neimann, H. C. Wegener, and R. V. Tauxe. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 121-138. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 11.Gilbert, M., J. R. Brisson, M. F. Karwaski, J. Michniewicz, A. M. Cunningham, Y. Wu, N. M. Young, and W. W. Wakarchuk. 2000. Biosynthesis of ganglioside mimics in Campylobacter jejuni OH4384. J. Biol. Chem. 275:3896-3906. [DOI] [PubMed] [Google Scholar]
- 12.Guerry, P., C. P. Ewing, T. E. Hickey, M. M. Prendergast, and A. P. Moran. 2000. Sialylation of lipooligosaccharide cores affects immunogenicity and serum resistance of Campylobacter jejuni. Infect. Immun. 68:6656-6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu, L., and D. J. Kopecko. 1999. Campylobacter jejuni 81-176 associates with microtubules and dynein during invasion of human intestinal cells. Infect. Immun. 67:4171-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korlath, J. A., M. T. Osterholm, L. A. Judy, J. C. Forfang, and R. A. Robinson. 1985. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152:592-596. [DOI] [PubMed] [Google Scholar]
- 16.Linton, D., M. Gilbert, P. G. Hitchen, A. Dell, H. R. Morris, W. Wakarchuk, N. A. Gregson, and B. W. Wren. 2000. Phase variation of a β-1,3 galactosyltransferase involved in generation of the ganglioside GM1-like lipo-oligosaccharide of Campylobacter jejuni. Mol. Microbiol. 37:501-514. [DOI] [PubMed] [Google Scholar]
- 17.Linton, D., A. V. Karlyshev, P. G. Hitchen, H. R. Morris, A. Dell, N. A. Gregson, and B. W. Wren. 2000. Multiple N-acetyl neuraminic acid synthetase (neuB) genes in Campylobacter jejuni: identification and characterization of the gene involved in sialylation of lipo-oligosaccharide. Mol. Microbiol. 35:1120-1134. [DOI] [PubMed] [Google Scholar]
- 18.Moran, A. P., B. J. Appelmelk, and G. O. Aspinall. 1996. Molecular mimicry of host structures by lipopolysaccharides of Campylobacter and Helicobacter spp.: implications in pathogenesis. J. Endotoxin Res. 3:521-531. [Google Scholar]
- 19.Moran, A. P., and D. T. O'Malley. 1995. Potential role of lipopolysaccharides of Campylobacter jejuni in the development of Guillain-Barré syndrome. J. Endotoxin Res. 2:233-235. [Google Scholar]
- 20.Moran, A. P., J. L. Penner, and G. O. Aspinall. 2000. Campylobacter lipopolysaccharides, p. 241-257. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 21.Moran, A. P., and M. M. Prendergast. 1998. Molecular mimicry in Campylobacter jejuni lipopolysaccharides and the development of Guillain-Barré syndrome. J. Infect. Dis. 178:1549-1550. [DOI] [PubMed] [Google Scholar]
- 22.Moran, A. P., E. T. Rietschel, T. U. Kosunen, and U. Zähringer. 1991. Chemical characterization of Campylobacter jejuni lipopolysaccharides containing N-acetylneuraminic acid and 2,3-diamino-2,3-dideoxy-d-glucose. J. Bacteriol. 173:618-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nachamkin, I., B. M. Allos, and T. W. Ho. 2000. Campylobacter jejuni infection and the association with Guillain-Barré syndrome, p. 155-175. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 24.Nam Shin, J. E., S. Ackloo, A. S. Mainkar, M. A. Monteiro, H. Pang, J. L. Penner, and G. O. Aspinall. 1998. Lipo-oligosaccharides of Campylobacter jejuni serotype O:10. Structures of core oligosaccharide regions from a bacterial isolate from a patient with the Miller-Fisher syndrome and from the serotype reference strain. Carbohydr. Res. 305:223-232. [DOI] [PubMed] [Google Scholar]
- 25.Oberhelman, R. A., and D. N. Taylor. 2000. Campylobacter infections in developing countries, p. 139-153. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 26.Oelschlaeger, T. A., P. Guerry, and D. J. Kopecko. 1993. Unusual microtubule-dependent endocytosis mechanisms triggered by Campylobacter jejuni and Citrobacter freundii. Proc. Natl. Acad. Sci. USA 90:6884-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. M. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 28.Pei, Z., C. Burucoa, B. Grignon, S. Baqar, X.-Z. Huang, D. J. Kopecko, A. L. Bourgeois, J.-L. Fauchere, and M. J. Blaser. 1998. Mutation in the peb1A locus of Campylobacter jejuni reduces interactions with epithelial cells and intestinal colonization of mice. Infect. Immun. 66:938-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prendergast, M. M., A. J. Lastovica, and A. P. Moran. 1998. Lipopolysaccharides from Campylobacter jejuni O:41 strains associated with Guillain-Barré syndrome exhibit mimicry of GM1 ganglioside. Infect. Immun. 66:3649-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salloway, S., L. A. Mermel, M. Seamans, G. O. Aspinall, J. E. Nam Shin, L. A. Kurjanczyk, and J. L. Penner. 1996. Miller-Fisher syndrome associated with Campylobacter jejuni bearing lipopolysaccharide molecules that mimic human ganglioside GD3. Infect. Immun. 64:2945-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider, H., T. L. Hale, W. D. Zollinger, R. C. Seid, Jr., C. A. Hammack, and J. M. Griffiss. 1984. Heterogeneity of molecular size and antigenic expression within lipooligosaccharides of individual strains of Neisseria gonorrhoeae and Neisseria meningitidis. Infect. Immun. 45:544-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider, H., C. A. Hammack, M. A. Apicella, and J. M. Griffiss. 1988. Instability of expression of lipooligosaccharides and their epitopes in Neisseria gonorrhoeae. Infect. Immun. 56:942-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Putten, J. P. 1993. Phase variation of lipopolysaccharide directs interconversion of invasive and immuno-resistant phenotypes of Neisseria gonorrhoeae. EMBO J. 12:4043-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Putten, J. P. M., and B. D. Robertson. 1995. Molecular mechanisms and implications for infection of lipopolysaccharide variation in Neisseria. Mol. Microbiol. 16:847-853. [DOI] [PubMed] [Google Scholar]
- 35.Vogel, U., H. Claus, G. Heinze, and M. Frosch. 1997. Functional characterization of an isogenic meningococcal alpha-2,3-sialyltransferase mutant: the role of lipooligosaccharide sialylation for serum resistance in serogroup B meningococci. Med. Microbiol. Immunol. 186:159-166. [DOI] [PubMed] [Google Scholar]
- 36.Vogel, U., A. Weinberger, R. Frank, A. Müller, J. Köhl, J. P. Atkinson, and M. Frosch. 1997. Complement factor C3 deposition and serum resistance in isogenic capsule and lipooligosaccharide sialic acid mutants of serogroup B Neisseria meningitidis. Infect. Immun. 65:4022-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, Y., and D. E. Taylor. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiser, J. N., and N. Pan. 1998. Adaptation of Haemophilus influenzae to acquired and innate humoral immunity based on phase variation of lipopolysaccharide. Mol. Microbiol. 30:767-775. [DOI] [PubMed] [Google Scholar]
- 39.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharides: extraction with phenol water and further applications of the technique. Methods Carbohydr. Chem. 5:83-92. [Google Scholar]
- 40.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127-130. [DOI] [PubMed] [Google Scholar]
- 41.Yao, R., D. H. Burr, P. Doig, T. J. Trust, H. Niu, and P. Guerry. 1994. Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: role of motility in adherence and invasion of eukaryotic cells. Mol. Microbiol. 14:883-893. [DOI] [PubMed] [Google Scholar]
- 42.Yuki, N., T. Taki, F. Inagaki, T. Kasama, M. Takahashi, K. Saito, S. Handa, and T. Miyatake. 1993. A bacterium lipopolysaccharide that elicits Guillain-Barré syndrome has a GM1 ganglioside-like structure. J. Exp. Med. 178:1771-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuki, N., T. Taki, M. Takahashi, K. Saito, H. Yoshino, T. Tai, S. Handa, and T. Miyatake. 1994. Molecular mimicry between GQ1b ganglioside and lipopolysaccharides of Campylobacter jejuni isolated from patients with Fisher syndrome. Ann. Neurol. 36:791-793. [DOI] [PubMed] [Google Scholar]