Abstract
5S rRNA extends from the central protuberance of the large ribosomal subunit, through the A-site finger, and down to the GTPase-associated center. Here, we present a structure-function analysis of seven 5S rRNA alleles which are sufficient for viability in the yeast Saccharomyces cerevisiae when expressed in the absence of wild-type 5S rRNAs, and extend this analysis using a large bank of mutant alleles that show semidominant phenotypes in the presence of wild-type 5S rRNA. This analysis supports the hypothesis that 5S rRNA serves to link together several different functional centers of the ribosome. Data are also presented which suggest that in eukaryotic genomes selection has favored the maintenance of multiple alleles of 5S rRNA, and that these may provide cells with a mechanism to post-transcriptionally regulate gene expression.
Keywords: 5S rRNA, Ribosome, Virus, Frameshifting, Drugs
Introduction
The availability of detailed structural information, biochemical assays, and molecular genetic test systems make the ribosome a robust model for studying how the structures of complex molecules dictate function at the molecular level. The ribosome is a megadalton complex, composed of multiple proteins and RNAs, in which the activities of numerous functional centers are coordinated to synthesize proteins with great accuracy. In simple terms, the ribosome orchestrates the various stages in peptide synthesis by coordinating the activities of least nine functional centers through at least seven discrete unidirectional steps (reviewed in Noller et al. 2001). Analyses of static X-ray crystal structures have been useful in localizing the functional centers at the atomic level revealing, for example, that the catalytic activity of the ribosome is mediated by RNA, and identifying the binding sites for antibiotics (reviewed in Yonath et al. 1998; Noller et al. 2001; Steitz and Moore 2003; Wilson and Nierhaus 2003). Cryo-EM studies have supplied complementary information, providing dynamic views of intra-ribosomal movements during many of the different phases of the translation cycle (reviewed in Frank 2003).
5S rRNA is the smallest RNA component of the ribosome, and its secondary structure has been elucidated in many living organisms (Szymanski et al. 2002). Although the tertiary structure of 5S rRNA has been determined for the free molecule (Funari et al. 2000; Lorenz et al. 2000) and its isolated domains (Betzel et al. 1994; Xiong and Sundaralingam 2000; Huber et al. 2001), and as a part of a ribosomal complex (Ban et al. 2000; Harms et al. 2001; Spahn et al. 2001; Yusupov et al. 2001), its precise function in protein synthesis is not fully understood. Biochemical studies with ribosomes from Escherichia coli led to the hypothesis that 5S rRNA acts to facilitate communication between the different functional centers, helping to coordinate the multiple events catalyzed by the ribosome (Bogdanov et al. 1995; Dokudovskaya et al. 1996), and this view was further supported by a later study of yeast ribosomes (Smith et al. 2001). There are 100–200 rDNA genes in the haploid yeast genome, all of which are located in a single tandem array on chromosome XII called the RDN1 locus (Petes 1979a, b). The genes encoding 5S, 5.8S, 18S and 25S rRNA are named RDN5, RDN5.8, RDN18 and RDN25, respectively. Using a yeast strain as host in which all chromosomally encoded 5S rDNA genes were deleted (Wai et al. 2000), we found that all but seven of 246 mutant 5S rRNA alleles tested were incompatible with viability when expressed as the sole form of the molecule. Genetic analyses suggested that these alleles might affect the binding of peptidyl-tRNA to ribosomes, and molecular analyses using ribosomes isolated from these strains revealed changes in chemical protection patterns in both 5S and 25S rRNA. Given the limited number of viable alleles and their relatively weak phenotypes, a more detailed phenotypic analysis was conducted by expressing each member of the entire library of alleles in a strain background containing only four wild-type chromosomal 5S rDNA genes (Oakes et al. 1998; Wai et al. 2000). These studies revealed the possible involvement of different regions of 5S rRNA with different functional centers of the ribosome. A bioinformatic analysis revealed the apparent conservation of multiple 5S rRNA alleles in the genomes of all eukaryotes examined. The observation of allele-specific semi-dominant phenotypes in our mutant analysis led us to ask whether naturally occurring variants of this molecule might also affect translational fidelity. Overexpression of naturally occurring 5S rDNA variants in a wild-type RDN1 background indeed resulted in allele-specific changes in −1 and +1 ribosomal frameshifting, suggesting that changes in the expression patterns of endogenous allelic 5S rRNA variants may be used to alter gene expression.
Materials and methods
Strains and genetic methods
Escherichia coli strain DH5a was used to amplify plasmids, and transformations of E. coli were performed using the standard calcium chloride method as described previously (Sambrook et al. 1989). Yeast cells were transformed using the alkali cation method (Ito et al. 1983). YPAD, YPG, SD, synthetic complete medium (H-) and 4.7 MB plates used for testing the killer phenotype were prepared and used as described previously (Wickner and Leibowitz 1976). Plasmid shuffling techniques using 5-flouroorotic acid (5-FOA) were performed as previously described (Rose et al. 1990). For dilution spot assays, growing yeast cells in mid-logarithmic phase were initially diluted to 2 × 107 colony forming units (CFU)/ml. Subsequently, 104 CFU (5 μl) aliquots, and tenfold dilutions from the same cultures thereof were spotted either onto rich medium and incubated at 15, 30 and 37°C, or incubated at 30°C on rich medium containing anisomycin (10 μg/ml) or sparsomycin (30 μg/ml). Assays for programmed ribosomal frameshifting followed previously described protocols (Smith et al. 2001; Harger and Dinman 2003). These involve the use of 0-frame control and −1 or +1 ribosomal frameshift test vectors in which the production of a reporter enzyme (either firefly luciferase of β-galactosidase) is dependent upon a programmed ribosomal frameshift event. Percentage frameshifting is calculated by dividing the enzymatic activities measured from cells expressing the frameshift test plasmids by those from cells expressing the 0-frame controls, and multiplying the resulting values by 100%. All assays were performed with enough replicates (generally approximately 30) to achieve confidence levels of >95%, and standard errors were calculated as previously described (Jacobs and Dinman 2004). Cytoduction of the L–A and M1 killer virus from strain JD759 into rho-o strains was carried out as previously described (Dinman and Wickner 1992). Ty1 retrotransposition assays were performed using pJEF 1105 as described by Boeke et al. (1988). Briefly, transcription of a neor-tagged Ty1 cDNA clone was induced by incubation in 2% galactose at 20°C for 3 days, after which cells that had lost pJEF1105 were identified by screening for growth on dextrose-containing medium in the presence of 5-FOA. The cells were then grown in liquid culture overnight, and tenfold dilutions of mid-log cells were spotted onto a medium containing 100 μg/ml Gentcin (Life Technologies, Gaithersburg, Md.) to select for cells in which retrotransposition had occurred, or onto rich medium as a control.
Plasmids and yeast strains
The pJD 180 series of plasmids contain one complete RDN1 repeat cloned into the pRS400 vector series (Christianson et al. 1992), the pJD106 and pJD209 series consist of high-copy-number vectors containing variants of the 5S rDNA gene, and members of the pJD211 series harbor a single copy of the 35S rDNA operon (Oakes et al. 1998). The pJD373 series of plasmids is based on the pJD211 series, and carry the C1495U mutation in helix 44 of 18S rDNA. This allele was previously shown to confer a recessive hygromycin-resistant phenotype on yeast cells (Velichutina et al. 2001). Standard site-directed mutagenesis was used to generate the naturally occurring allelic variants of 5S rRNA found in yeast (RDN5-2 to RDN5-7) and the RDN5-Ooc and RDN5-Som hybrids of yeast and Xenopus 5S rRNAs, using the RDN5-1 allele cloned into pRS424, a high copy-number 2 m TRP1 vector (Christianson et al. 1992) as the template.
Generation of the rdn1ΔΔ yeast strain JD1111 (MATa ade2-1 ura3-1 leu2-3 his3-11 trp1 can1-100 rdn1δ ::HIIS3 pJD106.URA pJD211. LEU [LA-HN M1]), and of strains harboring mutant alleles of 5S rDNA on a 2μ-TRP1 vector was described previously (Smith et al. 2001). The starting strain was based on NOY891 (Oakes et al. 1998), which was subsequently found to contain four telomere-proximal 5S rDNA repeats on chromosome XII (Wai et al. 2000). These repeats are deleted in NOY1049 [MATa ade2-1 ura3-1 trp1-1 his3-11 leu2-3, 112 can1-100ΔrDNA::his3::hisG + pNOY353 (GAL7-35S rDNA, 5S rDNA, TRP1, 2μ, Ampr)] (Wai et al. 2000). Strain JD932 (MATa ade 2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 can1-100) has a complete, wild-type RDN1 locus.
To construct a new series of strains lacking the chromosomal 5S rDNA repeats, NOY1049 was transformed with pJD180 Ura. Transformants were incubated in liquid standard defined medium lacking uracil (-ura) for 4 days, streaked for single colonies on - ura solid medium, and subsequently replica-plated onto medium lacking tryptophan (-trp). Trp− auxotrophs were picked and designated JD1248. The observed rate of loss of tryptophan prototrophy was approximately 10%. JD1253 was created by introducing the killer virus into JD1248 cells by cytoduction, and these cells were used for subsequent transformation with pJD373.Leu and pJD106.Trp (wild-type or mutant 5S alleles). Transformants were selected on medium lacking tryptophan and leucine. Colonies were allowed to grow to diameters of approximately 2 mm, and cells were subsequently streaked for single colonies on medium lacking tryptophan and leucine, and containing 5-FOA (Rose et al. 1990) and hygromycin (300 μg/ml).
Purification and analysis of mutant yeast ribosomes
Yeast cells were grown with constant shaking at 30°C in H–Leu–Trp to an OD595 of 0.5–0.6. Cells were harvested by centrifugation, washed thrice with water, and resuspended (1 ml/g wet weight) in lysis buffer A [20 mM HEPES-KOH pH 7.4, 5 mM magnesium acetate, 50 mM KCl, 10% (v/v) glycerol, 1 mM DTT, 1 × RNASecure (Ambion) and 1 tablet/50 ml Complete proteinase inhibitor cocktail (Roche)]. Cells were lysed using a Bead-beater, lysates were cleared of cellular debris by centrifugation at 15,000× g for 30 min at 4°C with subsequent adjustment of KCl concentration to 500 mM. Lysates were layered onto an equal volume of buffer B [20 mM HEPES-KOH pH 7.4, 5 mM magnesium acetate, 500 mM KCl, 25% glycerol, 1 mM DTT, 1 × RNA Secure (Ambion), 1 tablet/50 ml Complete proteinase inhibitor cocktail (Roche)], and centrifuged at 43,000 rpm in a Beckman Type 70TI rotor for 10 h. Ribosome pellets were resuspended in dilution buffer C (50 mM HEPES-KOH pH7.4, 5 mM magnesium acetate, 50 mM NH4Cl, 10% glycerol, 1 mM DTT), and aliquots were flash-frozen in liquid nitrogen and stored at −70°C.
Wild-type to mutant ratios of 5S rRNA in ribosome preparations were estimated by primer extension (Sigmund et al. 1988). Oligonucleotides complementary to 5S rRNA were labeled with γ[32P]ATP using T4 polynucleotide kinase, and annealed with 5S rRNA isolated from purified ribosomes. Primer extension reactions were performed using AMV reverse transcriptase (Roche) in a buffer containing 50 mM Tris–HCl pH 8.3, 60 mM NaCl, 6 mM magnesium acetate, 10 mM DTT, and chain terminating mixes containing 0.25 mM dNTPs (three of the four) plus 0.25 mM ddNTP (the fourth). Reactions were incubated at 45°C for 30 min, terminated by the addition of 2 × formamide sequencing dye, heated to 90°C, quenched on ice and fractionated by electrophoresis thorough 12% polyacrylamide urea denaturing gels. Labeled bands were visualized by autoradiography.
Chemical protection analyses
Chemical probing with dimethylsulphate (DMS), kethoxal, and carbodiimide metho- p-toluenesulfonate (CMCT), followed by RT primer extension analysis of modified RNAs were performed as described by Stern et al. (1988). The following primers (named according to first transcribed base) were used: 2,957 (5′-AACCTGTCTCACGACGG-3′), 3,057 (5′-CCTGATCAGACAGCCGC-3′), 1,231 (5′-GACTTCC ATGGCCACCG-3′), 1,343 (5′-GGGCATCATATCAA CCC-3′) and 1,112 (5′-CTTACCAAAAATGGCCC-3′) for S. cerevisiae 25S rRNA, and 99 (5′-AGATTGCAGCACCTGAGTTTCG- 3′) for 5S rRNA.
Bioinformatic methods
GenBank was first queried for 5S rRNA sequences according to species. The resulting sequences were then used in BLAST searches (Altschul et al. 1990) to identify homologous sequences in the database. Sequences were hand curated to identify those that (1) were represented by at least three independent GenBank accessions, and (2) did not contain overly large deletions of sequence (i.e., did not appear to be truncation products or pseudogenes). The resulting sequences were then aligned with one another using Clustal W (Thompson et al. 1994).
Results
Generation and characterization of 5S rRNA mutants in the absence of wild-type 5S rRNA
In a previous study, we described the generation of a near saturation library of 5S rRNA variants; specifically, 246 out of the 363 possible alleles of the yeast molecule were expressed from a high-copy-number 2μ vector (Smith et al. 2001). With the aim of examining the effects of the 5S rRNA mutants in the absence of the wild-type molecule, we obtained the strain NOY1049, in which the chromosomal 5S rDNA repeats were completely deleted (Wai et al. 2000). This strain was subjected to two additional rounds of genetic manipulation. The final strain, JD1253, contained one complete copy of all of the rRNA genes (i.e., one entire RDN1 repeat) on a 2μ-URA3 plasmid. In addition, the yeast-killer virus was introduced into this strain. The killer phenotype is caused by infection of yeast cells by the endogenous L–A and M1 dsRNA viruses (reviewed in Wickner 1996). The M1 virus, and hence the killer phenotype, is highly susceptible to defects in the translational apparatus, e.g., to defects in 60S ribosomal subunit biogenesis (Ohtake and Wickner 1995b), to the presence of translational inhibitors (Sommer and Wickner 1982; Carroll and Wickner 1995), to mutations in ribosomal proteins (Wickner et al. 1982; Carroll and Wickner 1995; Ohtake and Wickner 1995a; Meskauskas and Dinman 2001), and to changes in the efficiency of programmed −1 ribosomal frameshifting (−1 PRF; Dinman and Wickner 1992). In order to minimize recombination and select cells containing only mutant forms of 5S rRNA, a rigorous selection scheme involving negative selection for both the wild-type RDN1 gene and the plasmid on which it was encoded was devised (see Materials and methods). Having established the new strain and selection protocol, we examined the effects of all 246 5S rRNA alleles on cell growth and viability. Surprisingly, only seven of the alleles were viable: all the others were incompatible with viability when expressed as the sole form of 5S rRNA. The seven viable alleles were A20C, C69U, A76U, A79G, U81C, A84U and C93U. Direct sequence analyses of 5S rRNAs extracted from ribosomes demonstrated that these alleles could be stably maintained as the only forms of the 5S molecule in yeast cells (Fig. 1). Interestingly, base-pair interactions are conserved in all of these mutants, with the exception of the A20C allele (see below).
Fig. 1.
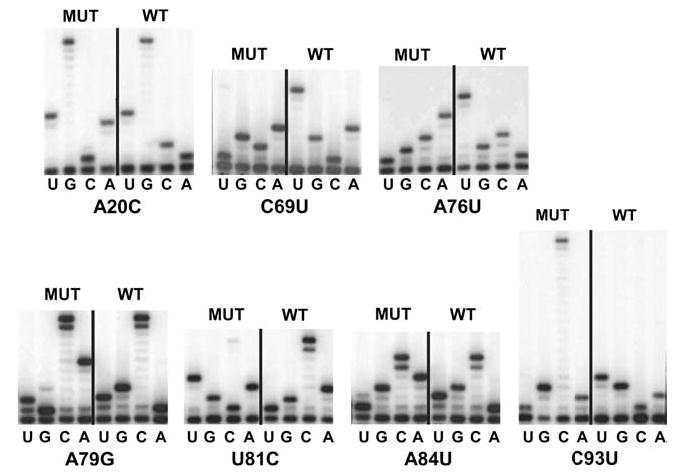
Direct rRNA sequence analyses of “pure” 5S rRNA mutants. rRNAs were extracted from ribosomes purified from JD1253 cells expressing wild-type or mutant forms of 5S rRNA. Oligonucleotides complementary to 5S rRNA were labeled with γ[32P]ATP using T4 polynucleotide kinase, and annealed with 5S rRNA isolated from purified ribosomes. Primer-extension reactions were performed using AMV reverse transcriptase, fractionated by electrophoresis through denaturing 12% polyacrylamide-urea gels, and labeled bands were visualized by autoradiography
The new mutant strains were first characterized with regard to their growth rates. In general, expression of each of the seven functional 5S rRNA mutant alleles was associated with significantly reduced rates of growth at 30°C as compared to wild-type (Fig. 2, top left). Although some of the mutants may have been slightly temperature-sensitive (Fig. 2, center panel), they were surprisingly resistant to cold (Fig. 2, top right).
Fig. 2.
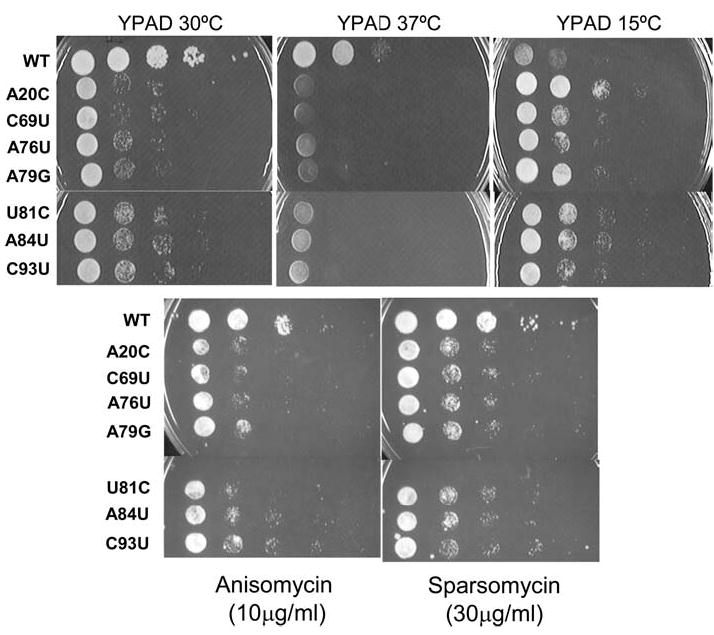
Growth phenotypes of “pure” 5S rRNA mutants. Mid-logarithmically growing JD1253 cells were diluted to 2 × 107 colony forming units (CFU)/ml. Subsequently, aliquots (5 μl) containing 104 CFU, and tenfold dilutions from the same cultures thereof were spotted either onto rich medium (YPAD) and incubated at 15, 30, and 37°C, or onto YPAD containing anisomycin (10 μg/ml) or sparsomycin (30 μg/ml) and incubated at 30°C
Translational inhibitors provide expedient probes for changes in ribosome function (reviewed in Ogle and Ramakrishnan 2005). Anisomycin has been shown to decrease ribosomal affinity for aminoacyl-tRNA (aa-tRNA) at the ribosomal A-site, and sparsomycin increases binding of peptidyl-tRNA at the P-site (reviewed in Pestka 1977). Though the results with anisomycin were generally equivocal (Fig. 2, lower left), sparsomycin actually improved the growth of all of the mutants (Fig. 2, lower right). Both the cold- and sparsomycin-resistant phenotypes could be explained by decreased affinities of ribosomes harboring the mutant 5S rRNAs for peptidyl-tRNAs. Specifically, sparsomycin would directly correct such a defect, while environmental conditions of decreased entropy would indirectly help to stabilize the ribosome/peptidyl-tRNA interaction.
Programmed ribosomal frameshifting (PRF), in which specific cis-acting signals in mRNAs induce translating ribosomes to change the translational reading frame, provides a powerful tool with which to monitor translational fldelity (reviewed in Harger et al. 2002). There are two types of PRF: −1 PRF directs a translational shift by one base in the 5′ direction, while +1 PRF results in a net slip by one base in the 3′ direction (reviewed in Farabaugh 1996). The PRF signals from two endogenous yeast viruses, L–A (which utilizes a −1 PRF mechanism) and the Ty1 retrotransposable element (which uses +1 PRF) were employed to monitor the effects of the 5S alleles on these processes using dual-luciferase reporter plasmids (Harger and Dinman 2003). All seven of the viable mutants promoted modest but statistically significant changes in −1 PRF. The greatest effect was observed with A84U (Fig. 3a), and similar results were observed with regard to +1 PRF (Fig. 3b). Both −1 and +1 PRF require slippage by the peptidyl-tRNA (reviewed in Harger et al. 2002). Thus, decreases in the affinities of ribosomes containing the 5S rRNA mutants for peptidyl-tRNA could result in increased slipperiness of this tRNA species at both the −1 and +1 PRF signals
Fig. 3.
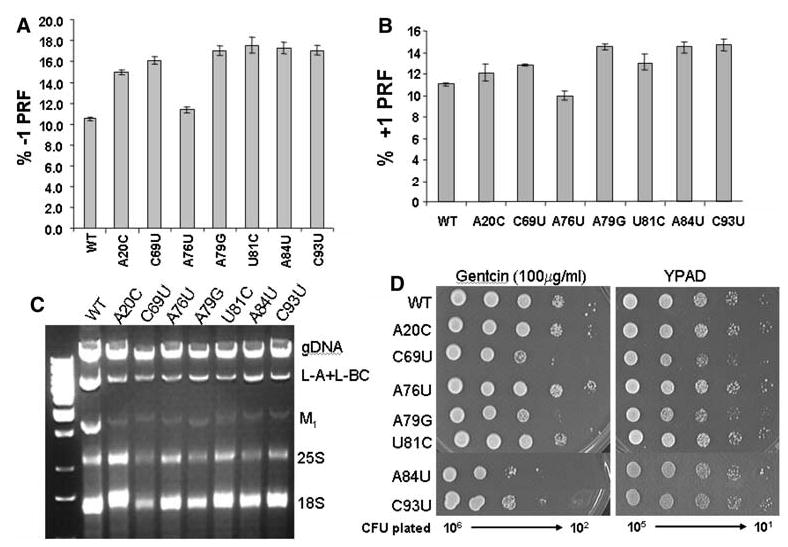
a–d Effects of pure 5S rRNA mutants on programmed ribosomal frameshifting and virus propagation. JD1253 cells expressing the indicated “pure” 5S rRNA were assayed with respect to the following phenotypes. a Cells were transformed with 0-frame control and L–A derived −1 PRF test dual luciferase reporter plasmids, and −1 PRF efficiencies were determined as described by Harger and Dinman (2003). All assays were replicated at least 30 times, and standard errors (represented by the error bars) were calculated as described by Jacobs and Dinman (2004). b Programmed +1 ribosomal frameshifting was analyzed using a Ty1 derived +1 PRF signal cloned into the dual luciferase reporter plasmids as described above. c Total nucleic acids were extracted from JD1253 cells expressing the indicated 5S rRNA alleles, fractionated by electrophoresis on a 1.5% native agarose gel, and stained with ethidium bromide. Genomic DNA (gDNA), viral dsRNAs, and rRNAs are indicated. L-BC is a dsRNA virus unrelated to L–A and M1. d Cells were transformed with pJEF1105, a galactose-inducible Ty1 cDNA clone containing the neor selectable reporter, incubated at 20°C for 4 days on medium containing 2% galactose, replica plated onto medium containing 100 μg/ml 5-FOA, and incubated at 30°C to select for cells that had lost pJDF1105. Viable colonies were grown on YPAD medium overnight, and 10-fold dilutions of cells ranging from 106 to 102 CFU were spotted onto YPAD medium containing 100 μg/ml of Genticin. In parallel, 105–101 CFU were spotted onto YPAD medium alone
The efficiency of PRF determines the stoichiometric ratios of structural to enzymatic proteins available for virus particle assembly, and therefore changes in PRF efficiencies can have profound impacts on killer virus propagation, as well as on frequencies of Ty1 retrotransposition (reviewed in Dinman 1995). The changes in −1 PRF that were promoted by the 5S rRNA mutants were qualitatively reflected in assays for yeast “killer” activity (reviewed in Wickner 1986), where A84U had the killer− phenotype, while the zones of killer activity for the other mutants were significantly decreased in size relative to wild-type (data not shown). Analysis of total nucleic acids shows that copy numbers of both the L–A and M1 dsRNA viral genomes were significantly decreased relative to wild-type in cells expressing the mutant 5S rRNAs (Fig. 3c), demonstrating that changes in −1 PRF were affecting virus maintenance. Similarly, Ty1 retrotransposition frequencies were most strongly reduced by mutants expressing 5S rDNA alleles that promoted the largest increases in +1 PRF (Fig. 3d).
Structural characterization of ribosomes containing mutant forms of 5S rRNA
Isolated ribosomes containing only mutant forms of 5S rRNA were probed for structural changes in 25S and 5S rRNAs using three base-specific reagents: dimethylsulfate (DMS), kethoxal and carbodiimide metho-p-toluenesulfonate (CMCT). Specifically, DMS preferentially donates a methyl group to hydrogen bond accepting ring nitrogens of C, G and A residues; kethoxal reacts with the N1 and N2 amine of solvent exposed G residues, and CMCT reacts with non-interacting N1 and N3 nitrogens of G and U residues, respectively. Base modification by these reagents causes n−1 reverse transcriptase stops, allowing for the identification of base-specific changes in rRNA structure. A set of primers for reverse transcription analysis was employed to monitor structural changes in domains II, V and VI of 25S rRNA, and throughout the entire length of 5S rRNA.
The results of these studies revealed that the mutants showed structural changes in both 5S and 25S rRNAs. Figure 4a shows representative autoradiograms from these studies, while Fig. 4b depicts the bases in question in relation to two-dimensional representations of 5S rRNA and helix 95 of the 25S rRNA. The reactivities of three bases in 5S rRNA were specifically altered by the mutations. In particular, in all of the mutants (with the exception of C69U), G85, which is normally unpaired, was protected from chemical modification. Allele-specific effects were also observed with regard to G91 and A92, which lie in the loop D region of the molecule: all of the mutants except A84G promoted increased protection at position G91, while only U81C altered the protection pattern of A92. Two of the 5S rRNA mutants also had effects on 25S rRNA. Specifically, the A20C and U81C alleles conferred protection on G3027, and weaker but consistent reactivity at position U3013. Interestingly, G3027 is located in the sarcin/ricin loop (SRL) which is involved in elongation factor binding. U3013 is near the base of the helix 95, close to where helices 94–97 coordinate to interact with ribosomal protein L3 (see Fig. 7).
Fig. 4.
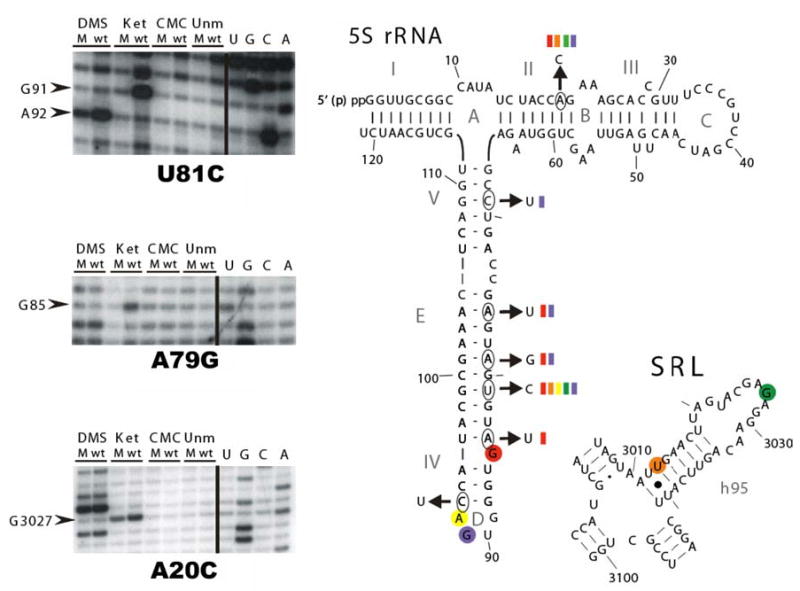
Chemical protection analyses. Ribosomes purified from JD1253 cells expressing either wild-type or mutant 5S rRNAs were chemically probed with DMS, kethoxal and CMCT. The products of reverse transcriptase primer extension reactions were subjected to PAGE analysis on denaturing PA-urea gels, and visualized by autoradiography. Left panel Representative autoradiographs showing altered reactivities of bases in 5S rRNA (top and middle), and 25S rRNA (bottom). Right panel Summary of the effects of pure mutants mapped onto 5S rRNA and the helix 95-sarcin/ricin loop (SRL) region of 25S rRNA. The open circles indicate positions of wild-type bases, and the arrows indicate the relevant mutant alleles. The color-coded bars indicate which mutant 5S rRNAs alter the chemical reactivities of other similarly coded, circled, bases in 5S and 25S rRNAs
Fig. 7.

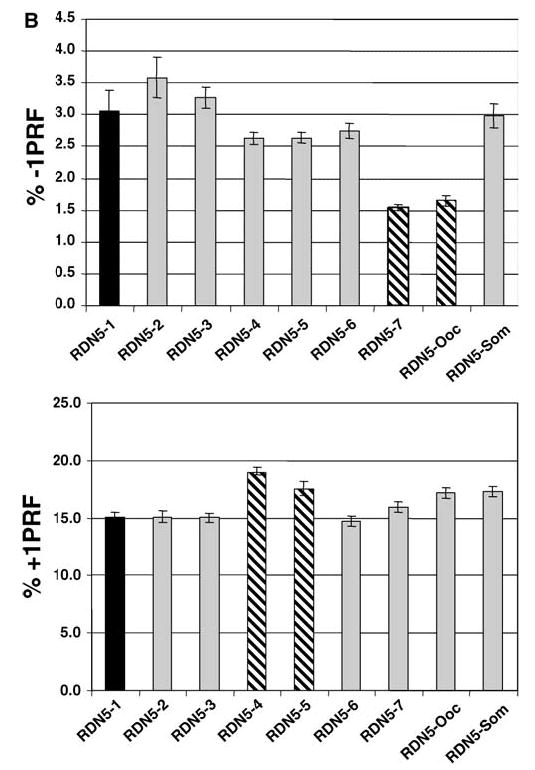
a, b Allelic variants of 5S rRNAs in eukaryotic genomes. a Conservation of multiple 5S rDNA alleles in eukaryotic genomes. GenBank was first queried for 5S rRNA sequences according to species, and the resulting sequences were used in BLAST searches (Altschul et al. 1990) to identify homologous sequences in the database. Sequences were hand curated to ensure their validity, and then aligned with one another as described in Materials and methods. RDN5-som and RDN5-ooc show the sequences of the hybrid yeast/Xenopus clones. Color coding is used to denote base substitutions. b Semi-dominant effects of naturally occurring yeast and hybrid yeast/Xenopus 5S rRNA alleles on L–A directed −1, and Ty1 promoted +1 PRF. Using the yeast RDN5-1 allele cloned into a high-copy-number 2μ vector, oligonucleotide-primed site-directed mutagenesis was used to create the other six naturally occurring yeast RDN5 alleles (RDN5-2 to RDN5-7), and the hybrid yeast/Xenopus RDN5-ooc and RDN5-som alleles (see Fig. 6a). These alleles were episomally expressed in JD932 cells, a wild-type yeast strain containing a full complement of chromosomal rDNA genes. Programmed −1 and +1 ribosomal frameshifting efficiencies were monitored as described in Fig. 6
Phenotypic analyses of 5S alleles in a mixed mutant/wild-type context: correlation between mutational clusters, changes in programmed ribosomal frameshifting and virus propagation
In previous studies, we demonstrated that expression of mutant 5S rRNA alleles from high-copy-number plasmids could induce semi-dominant PRF phenotypes in otherwise wild-type strain backgrounds (Dinman and Wickner 1995). In light of the limited number of mutants that were viable in the absence of wild-type 5S rRNA, we exploited this property to analyze the other members of the library of mutant 5S rRNA alleles. In an effort to enhance the probability of observing semidominant effects of the mutations, we also employed a yeast strain containing only four chromosomal copies of the RDN5 gene (JD1111) instead of a true wild-type, which typically harbors >100 copies (Oakes et al. 1998; Wai et al. 2000; Smith et al. 2001). Reverse transcriptase primer extension sequence analyses of 5S rRNA obtained from ribosomes purified from JD1111 cells harboring the mutant 5S rDNA plasmids confirmed the presence of mixed populations of mutant and wild-type 5S rRNA, with ratios of mutant to wild-type ranging from approximately 1:10 to 1:1 depending on the strain (Fig. 5). These results directly demonstrate that the cells express ribosomes containing mixed populations of wild-type and mutant 5S rRNAs. These cells were then transformed with −1 PRF reporter, and 0-frame control vectors, and the efficiency of −1 PRF was determined for all of the mutants that had previously been shown to affect the killer phenotype (Smith et al. 2001). The results show a broad correlation between changes in frameshifting efficiencies and defects in virus maintenance (Fig. 6a). The effects of the mutants on −1 PRF are allele specific: there was no apparent correlation between increased or decreased −1 frameshifting and physical location of the mutation along 5S rRNA. We note that these experiments were performed with monocistronic lacZ reporters (Dinman et al. 1991), which tend to enhance the absolute differences in −1PRF (Harger and Dinman 2004). As previously reported, since these 5S rDNA alleles promoted loss of the killer virus and did not promote nonsense-mediated mRNA decay (NMD) defects (Smith et al. 2001), changes in observed −1 PRF efficiencies reflect true changes in frameshifting (Harger and Dinman 2004). Based on their physical locations along the molecule and their abilities to suppress the expression of nonsense-containing reporter genes (Smith et al. 2001), 22 of the 5S rRNA mutants were also selected for characterization of +1 PRF efficiency and Ty1 retrotransposition frequencies. Expression of the majority of these alleles in JD1111 cells resulted in moderate to severe effects on Ty1 retrotransposition, and strong correlations were observed between this inhibition of retrotransposition and changes in +1 PRF efficiency (Fig. 6b). In addition, +1 PRF efficiencies tended to be elevated by mutations in the segment extending from position 7 to 57, whereas +1 PRF was generally inhibited in mutants spanning positions 79–100. As discussed in greater detail below, this observation suggests that different regions of the molecule are linked to distinct ribosomal functions.
Fig. 5.
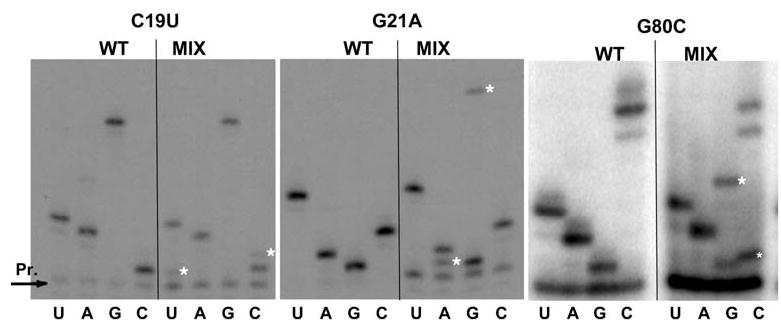
Direct rRNA sequence analyses of “mixed” 5S rRNA mutants. rRNAs were extracted from ribosomes purified from JD1111 cells expressing wild-type or mixtures of mutant and wild-type forms of 5S rRNA. Oligonucleotides complementary to 5S rRNA were labeled with γ[32P]ATP using T4 polynucleotide kinase, and annealed with 5S rRNA isolated from purified ribosomes. Primer-extension reactions were performed using AMV reverse transcriptase, fractionated by electrophoresis through denaturing 12% polyacrylamide-urea gels, and visualized by autoradiography. The asterisks indicate the 5S rRNA mutations in the mixed populations
Fig. 6.
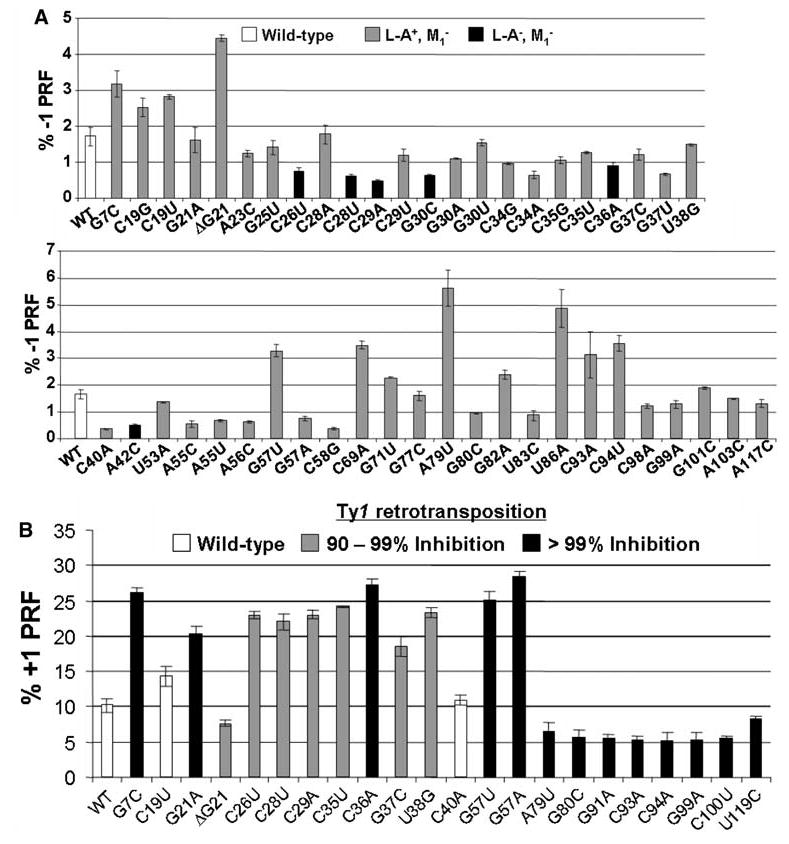
a, b Semi dominant effects of 5S rRNA alleles on programmed ribosomal frameshifting and virus propagation. a Effects on −1 PRF and killer virus propagation. b Effects on +1 PRF and Ty1 retrotransposition. JD1111 cells expressing wild-type or mixtures of wild-type and mutant 5S rRNA alleles were transformed with monocistronic lacZ-based 0-frame, −1 or +1 frameshift reporter plasmids and L–A virus promoted −1, or Ty1 promoted +1 PRF efficiencies were determined as previously described (Dinman et al. 1991; Dinman and Wickner 1992). Alleles of 5S rRNA are shown on the X-axis and PRF efficiencies are indicated on the Y-axis. The error bars correspond to standard error. In panel a, the different shadings indicate inhibitory effects of 5S rRNA alleles on maintenance of either the M1 satellite virus alone or both L– A and M1, as previously described (Smith et al. 2001). In panel b, the shadings correspond to inhibitory effects of 5S rRNA alleles on Ty1 retrotransposition frequencies assessed using pJEF1105
A naturally occurring allelic variant of 5S rRNA specifically inhibits programmed −1 ribosomal frameshifting
The observation that mutant forms of 5S rRNA can exert semi-dominant effects on translation is interesting, but could be entirely artifactual, without any true biological relevance. However, eukaryotic genomes contain >100 copies of rDNA genes. Though it is generally assumed either that they are all identical, or that minor differences are inconsequential, it is possible that allelic 5S rDNA variants may have been functionally selected for. To examine this issue, GenBank was searched for species-specific 5S rDNA alleles that fulfilled the following criteria (1) each variant had to be represented by at least three independent entries, and (2) obvious pseudogenes (e.g., those containing large deletions or insertions) were rejected. These searches revealed the presence of multiple 5S rDNA alleles in every genome examined. Specifically, the S. cerevisiae genome contains at least seven different allelic variants, Homo sapiens has ≥13, Xenopus laevis includes ≥6, Mus musculus contains ≥16, and Drosophila melanogaster has ≥11 (Fig. 7a). To test the hypothesis that different forms of 5S rRNA may differentially affect translational fidelity, site-directed mutagenesis was used to create the seven different naturally occurring yeast 5S rRNA allelic variants, and these were expressed from high-copy-number 2μ vectors. All but the “wild-type” (RDN5-1) variants of the molecule were inviable, either as the sole form of 5S rRNA (in JD1253 cells), or in the presence of four chromosomal copies (in JD1111 cells) (data not shown). Intriguingly, expression of the RDN5-7 variant in a wild-type RDN1 strain background (JD932) resulted in significant inhibition of −1 PRF, while the RDN5-4 and RDN5-5 alleles promoted a slight increase in +1 PRF (Fig. 7b).
Effects of hybrid yeast/Xenopus 5S rRNAs on programmed ribosomal frameshifting
Early studies on 5S rRNA in X. laevis revealed that the ribosomes of oocytes and somatic cells contained different forms 5S rRNA (Ford and Southern 1973). In light of the observation that 5S rRNA allelic variants can have differential effects on translational fidelity, we hypothesize that Xenopus may use the two different forms of 5S rRNAs to post-transcriptionally regulate gene expression during the developmental program. To examine this question, yeast 5S rRNA clones were mutagenized to mimic the Xenopus oocyte and somatic 5S rRNA variants. Specifically, bases in yeast 5S rRNA at positions 30, 48, 54, 56 and 57 were altered to the corresponding residues in either Xenopus somatic (RDN5-som), or oocyte (RDN5-ooc) 5S rRNAs (see Fig. 6a). Although both forms were inviable in JD1253 and JD1111 strain backgrounds, overexpression of RDN5-ooc promoted significant and specific inhibition of −1 PRF in the context of the wild-type RDN1 locus (JD932 cells; Fig. 6b). The potential significance of these findings is discussed below.
Discussion
This study presents a number of important additions to our understanding of 5S rRNA. The observation that the overwhelming majority of the mutants tested were inviable when expressed as the sole forms of 5S rRNA implies that 5S rRNA plays a critical role in ensuring the proper functioning of the ribosome. The finding of seven mutants that were viable as the sole forms of 5S rRNA has made possible the first real structure/function analysis of this molecule in a eukaryotic system. Functionally, the ability of these mutants to antagonize the effects of sparsomycin, which increases binding of peptidyl-tRNA to the P-site (reviewed in Pestka 1977), suggests that they may promote decreased binding of peptidyl-tRNAs; similar observations were made with mutant forms of ribosomal protein L5 (Meskauskas and Dinman 2001). Though A20 and C69 are located near the L5 binding site (see Fig. 8a), the other mutants are not located near this region. Thus, the general effects observed here are probably indirect. The observation of enhanced growth at lower temperature is consistent with this hypothesis, in which conditions of decreased entropy would help to stabilize the interactions between peptidyl-tRNA and the ribosome. The observation that these alleles also tend simultaneously to promote increased +1 PRF and −1 PRF, which both require slippage of the peptidyl-tRNA, is also consistent with this hypothesis, as previously observed in cells expressing mutant derivatives of ribosomal protein L5 (Meskauskas and Dinman 2001).
Fig. 8.
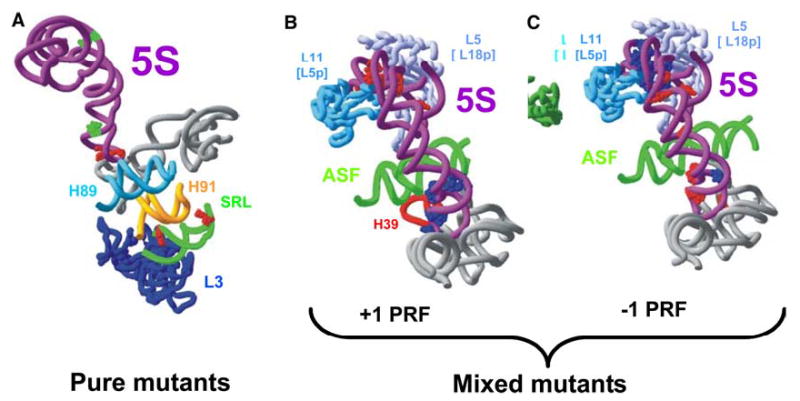
a–c Correlation of 5S rRNA structure with function. Mutations in 5S rRNA that produced phenotypic and physical changes are mapped onto the high-resolution structure of the yeast ribosome described by Spahn et al. (2004). a Mapping of sites affected in “pure” mutants onto 5S and 25S rRNA structures. Positions marked in green in 5S rRNA show locations of A20, A79, and U81. Positions marked in red show the locations of bases whose chemical reactivities were altered as a consequence of the 5S rRNA mutations. b, c “Mixed” 5S rRNA alleles that affected +1 (b) or −1 PRF (c). Mutations at bases shown in red stimulated PRF, and those shown in blue inhibited PRF
These alleles also provide unique insights into 5S rRNA structure. For example, although the mutants were scattered throughout different regions of 5S rRNA (open circles in Fig. 4b and green spheres in Fig. 8a), five of them specifically influenced the chemical reactivities of nucleotides G85 and G91 (see Fig. 4b and red spheres in Fig. 8a). The most prominent effects were observed for A20C and U81C mutants. By replacing an A–U basepair with a C–U mismatch, A20C should destabilize helix II. In contrast, substitution of a G–U pair with the more stable G–C (U81C) should stabilize helix IV. The decreased reactivity of G85 and G91 towards kethoxal in these mutants indicates a tighter association of the Helix IV/Loop D region of the molecule with its binding pocket in the large subunit rRNA. This region of 5S rRNA directly contacts helix 42, which is connected to the large subunit’s “GTPase associated center‘’. Thus, 5S rRNA may influence the positioning/function of this center through this interaction. This region of 5S rRNA also contacts helices 39 and 89. Helix 89 is located parallel to helix 91, and the tips of their loops are connected by a basepair. The opposite site of the loop-end of helix 91 also interacts with the sarcin–ricin loop, the second elongation factor binding site. Interestingly, this contact was affected by the “pure” mutations at A20C and U81C, causing a moderate decrease in the reactivity of G3027 (see Fig. 4a). Thus, these findings suggest that 5S rRNA may also influence the structure of both elongation factor binding sites. The observation that the pure mutants promoted weak protection of G3013 is also potentially very significant. This base is located in the lower part of the helix 95, in close vicinity to ribosomal protein L3, which was implicated in a proposed communication pathway between the peptidyltransferase center and the sarcin-ricin loop (Petrov et al. 2004).
The semi-dominant effects of the “mixed” mutants are also useful for linking structure with function. For example, alleles of this type that promoted increased +1 PRF were located in the region where 5S rRNA is sandwiched between ribosomal proteins L5 and L11 (bacterial proteins L18 and L5, respectively; Fig. 8b, red spheres), in the vicinity of the interaction between the peptidyl-tRNA T-loop and L11 (bacterial L5) (Yusupov et al. 2001). Previously, we showed that increased +1 PRF promoted by a mutant of ribosomal protein L5 was due to decreased affinity of ribosomes for peptidylt RNA (Meskauskas and Dinman 2001), and we suggest that this is also the case for those 5S rRNA mutants that map to this region. Conversely, all of the semi-dominant mutants that promoted decreased +1 PRF mapped to the region of the molecule that abuts the A-site (Fig. 8b, blue spheres). Ty1 mediated +1 PRF is driven by ribosomal pausing at a rare 0-frame AGG codon, which corresponds to the low abundance Arg-tRNA(CCU) (Kawakami et al. 1993). One possible explanation for the effects of this cluster of mutants could be that it is due to defects in the ability of ribosomes to distinguish between cognate and near- or non-cognate aa-tRNAs. Promiscuous misreading of the 0-frame AGG-Arg codon in the Ty1 slippery site would reduce ribosomal pause times at this frameshift signal, promoting a decrease in the frequency of +1 PRF. The observation of the allele specificity of effects on −1 PRF in the mixed mutants is less informative. We hypothesize that this may be because −1 PRF efficiencies can be affected by multiple factors. These include changes in rates of peptidyltransfer (Dinman et al. 1997; Meskauskas et al. 2003a, b), in affinities of ribosomes for peptidyland aa-tRNAs (Meskauskas et al. 2003a; Petrov et al. 2004), and in the interactions between ribosomes and elongation factors (Kinzy et al. 2002).
Lastly, why do multiple 5S rDNA alleles appear to be retained in eukaryotic genomes? Our demonstration that at least one of the naturally occurring yeast alleles has semi-dominant effects on L–A directed −1 PRF is the first demonstration that a naturally occurring allele of a ribosomal component can affect translational fidelity. We hypothesize that 5S rRNA sequence variants may have been evolutionarily selected to allow for fine tuning of gene expression at the post-transcriptional level. Interestingly, the 5S rRNA-ribosomal protein L5 complex is assembled onto large subunits late in ribosome biogenesis, and this complex can be dissociated from and re-associated onto core large subunits (Deshmukh et al. 1993, 1995; Brow and Geiduschek 1987; Yeh and Lee 1995). Thus, different versions of 5S rRNA could be added onto newly synthesized core 60S subunits, allowing cells to rapidly change the performance of their ribosomes. The studies using the yeast/Xenopus hybrids lend support to this suggestion. Another intriguing question is why so few of the RDN5 alleles affected frameshifting efficiencies. One potential reason for why more dramatic results were not observed could stem from our use of viral frameshift signals. Both the L–A virus and the Ty 1 retrotransposable element are endogenous to yeast, and their propagation can be severely affected by changes in PRF efficiencies (reviewed in Dinman 1995; Harger et al. 2002). Thus, it is possible that viral frameshift signals have evolved to be relatively immune to the effects of the naturally occurring 5S rRNA alleles. We have recently identified a large number of functional −1 PRF signals in chromosomally encoded yeast genes (J.D. Dinmanm, unpublished); it will be interesting to determine whether any of these are affected by any of the natural 5S rDNA alleles.
Acknowledgments
We wish to thank the members of the Dinman and Dontsova laboratories, with special thanks to Sarah Fraser, Ewan Plant, Steve Hutcheson and Alexey Bogdanov for their advice and support. This work was supported by grants to JDD from the National Institutes of Health (GM62143), and to JDD and OAD from the Fogarty International Center (TW005787), and to OAD from HHMI 55000303 and RFBR.
Footnotes
Sergey Kiparisov and Alexey Petrov contributed equally to this work
References
- Altschul SF, Gish W, Miller E, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Betzel C, Lorenz S, Furste JP, Bald R, Zhang M, Schneider TR, Wilson KS, Erdmann VA. Crystal structure of domain A of Thermus flavus 5S rRNA and the contribution of water molecules to its structure. FEBS Lett. 1994;351:159–164. doi: 10.1016/0014-5793(94)00834-5. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Xu H, Fink GR. A general method for the chromosomal amplification of genes into yeast. Science. 1988;239:280–282. doi: 10.1126/science.2827308. [DOI] [PubMed] [Google Scholar]
- Bogdanov AA, Dontsova OA, Dokudovskaya SS, Lavrik IN. Structure and function of 5S rRNA in the ribosome. Biochem Cell Biol. 1995;73:869–876. doi: 10.1139/o95-094. [DOI] [PubMed] [Google Scholar]
- Brow DA, Geiduschek EP. Modulation of yeast 5S rRNA synthesis in vitro by ribosomal protein YL3. J Biol Chem. 1987;262:13953–13958. [PubMed] [Google Scholar]
- Carroll K, Wickner RB. Translation and M1 dsRNA propagation: MAK18 = RPL41B and cycloheximide curing. J Bacteriol. 1995;177:2887–2891. doi: 10.1128/jb.177.10.2887-2891.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Yeast. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Deshmukh M, Tsay YF, Paulovich AG, Woolford JL., Jr Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol Cell Biol. 1993;13:2835–2845. doi: 10.1128/mcb.13.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh M, Stark J, Yeh LC, Lee JC, Woolford JL., Jr Multiple regions of yeast ribosomal protein L1 are important for its interaction with 5S rRNA and assembly into ribosomes. J Biol Chem. 1995;270:30148–30156. doi: 10.1074/jbc.270.50.30148. [DOI] [PubMed] [Google Scholar]
- Dinman JD. Ribosomal frameshifting in yeast viruses. Yeast. 1995;11:1115–1127. doi: 10.1002/yea.320111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman JD, Wickner RB. Ribosomal frameshifting efficiency and Gag/Gag-pol ratio are critical for yeast M1 doublestranded RNA virus propagation. J Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman JD, Wickner RB. 5S rRNA is involved in fidelity of translational reading frame. Genetics. 1995;141:95–105. doi: 10.1093/genetics/141.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman JD, Icho T, Wickner RB. A −1 ribosomal frameshift in a double-stranded RNA virus forms a Gag-pol fusion protein. Proc Natl Acad Sci USA. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman JD, Ruiz-Echevarria MJ, Czaplinski K, Peltz SW. Peptidyl transferase inhibitors have antiviral properties by altering programmed −1 ribosomal frameshifting efficiencies: development of model systems. Proc Natl Acad Sci USA. 1997;94:6606–6611. doi: 10.1073/pnas.94.13.6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokudovskaya S, Dontsova O, Shpanchenko O, Bogdanov A, Brimacombe R. Loop IV of 5S ribosomal RNA has contacts both to domain II and to domain V of the 23S RNA. RNA. 1996;2:146–152. [PMC free article] [PubMed] [Google Scholar]
- Farabaugh PJ. Programmed translational frameshifting. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford PJ, Southern EM. Different sequences for 5S RNA in kidney cells and ovaries of Xenopus laevis. Nat New Biol. 1973;241:7–12. doi: 10.1038/newbio241007a0. [DOI] [PubMed] [Google Scholar]
- Frank J. Electron microscopy of functional ribosome complexes. Biopolymers. 2003;68:223–233. doi: 10.1002/bip.10210. [DOI] [PubMed] [Google Scholar]
- Funari SS, Rapp G, Perbandt M, Dierks K, Vallazza M, Betzel C, Erdmann VA, Svergun DI. Structure of free Thermus flavus 5S rRNA at 1.3 nm resolution from synchrotron X-ray solution scattering. J Biol Chem. 2000;275:31283–31288. doi: 10.1074/jbc.M004974200. [DOI] [PubMed] [Google Scholar]
- Harger JW, Dinman JD. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA. 2003;9:1019–1024. doi: 10.1261/rna.5930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harger JW, Dinman JD. Evidence against a direct role for the Upf proteins in frameshifting or nonsense codon readthrough. RNA. 2004;10:1721–1729. doi: 10.1261/rna.7120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harger JW, Meskauskas A, Dinman JD. An ’integrated model’ of programmed ribosomal frameshifting and posttranscriptional surveillance. Trends Biochem Sci. 2002;27:448–454. doi: 10.1016/s0968-0004(02)02149-7. [DOI] [PubMed] [Google Scholar]
- Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- Huber PW, Rife JP, Moore PB. The structure of helix III in Xenopus oocyte 5S rRNA: an RNA stem containing a two nucleotide bulge. J Mol Biol. 2001;312:823–832. doi: 10.1006/jmbi.2001.4966. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JL, Dinman JD. Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res. 2004;32:e160–e170. doi: 10.1093/nar/gnh157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Paned S, Faioa B, Moore DP, Boeke JD, Farabaugh PJ, Strathern JN, Nakamura Y, Garfinkel DJ. A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics. 1993;135:309–320. doi: 10.1093/genetics/135.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzy TG, Harger JW, Carr-Schmid A, Kwon J, Shastry M, Justice MC, Dinman JD. New targets for antivirals: the ribosomal A-site and the factors that interact with it. Virology. 2002;300:60–70. doi: 10.1006/viro.2002.1567. [DOI] [PubMed] [Google Scholar]
- Lorenz S, Perbandt M, Lippmann C, Moore K, DeLucas LJ, Betzel C, Erdmann VA. Crystallization of engineered Thermus flavus 5S rRNA under earth and microgravity conditions. Acta Crystallogr D Biol Crystallogr. 2000;56:498–500. doi: 10.1107/s0907444900001736. [DOI] [PubMed] [Google Scholar]
- Meskauskas A, Dinman JD. Ribosomal protein L5 helps anchor peptidyl-tRNA to the P-site in Saccharomyces cerevisiae. RNA. 2001;7:1084–1096. doi: 10.1017/S1355838201001480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskas A, Baxter JL, Carr EA, Yasenchak J, Gallagher JEG, Baserga SJ, Dinman JD. Delayed rRNA processing results in significant ribosome biogenesis and functional defects. Mol Cell Biol. 2003a;23:1602–1613. doi: 10.1128/MCB.23.5.1602-1613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskas A, Harger JW, Jacobs KLM, Dinman JD. Decreased peptidyltransferase activity correlates with increased programmed −1 ribosomal frameshifting and viral maintenance defects in the yeast Saccharomyces cerevisiae. RNA. 2003b;9:982–992. doi: 10.1261/rna.2165803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF, Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Lancaster L, Dallas A, Fredrick K, Earnest TN, Cate JH. Structure of the ribosome at 5.5 Å resolution and its interactions with functional ligands. Cold Spring Harb Symp Quant Biol. 2001;66:57–66. doi: 10.1101/sqb.2001.66.57. [DOI] [PubMed] [Google Scholar]
- Oakes M, Aris JP, Brockenbrough JS, Wai H, Vu L, Nomura M. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J Cell Biol. 1998;143:23–34. doi: 10.1083/jcb.143.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Ohtake Y, Wickner RB. KRB1, a suppressor of mak 7-1 (a mutant RPL4A), is RPL4B, a second ribosomal protein L4 gene, on a fragment of Saccharomyces chromosome XII. Genetics. 1995a;140:129–137. doi: 10.1093/genetics/140.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake Y, Wickner RB. Yeast virus propagation depends critically on free 60S ribosomal subunit concentration. Mol Cell Biol. 1995b;15:2772–2781. doi: 10.1128/mcb.15.5.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S (1977) Inhibitors of protein synthesis. In: Weissbach H, Pestka S (eds) Molecular mechanisms of protein biosynthesis. Academic, New York, pp 467–553
- Petes TD. Meiotic mapping of yeast ribosomal deoxyribonucleic acid on chromosome XII. J Bacteriol. 1979a;138:185–192. doi: 10.1128/jb.138.1.185-192.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes TD. Yeast ribosomal DNA genes are located on chromosome XII. Proc Natl Acad Sci USA. 1979b;76:410–414. doi: 10.1073/pnas.76.1.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov A, Meskauskas A, Dinman JD. Ribosomal protein L3: influence on ribosome structure and function. RNA Biol. 2004;1:59–65. doi: 10.4161/rna.1.1.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P (1990) Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning, a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
- Sigmund CD, Ettayebi M, Borden A, Morgan EA. Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol. 1988;164:673–690. doi: 10.1016/s0076-6879(88)64077-8. [DOI] [PubMed] [Google Scholar]
- Smith MW, Meskauskas A, Wang P, Sergiev PV, Dinman JD. Saturation mutagenesis of 5S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:8264–8275. doi: 10.1128/MCB.21.24.8264-8275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer SS, Wickner RB. Co-curing of plasmids affecting killer double-stranded RNAs of Saccharomyces cerevisiae: [HOK], [NEX], and the abundance of L are related and further evidence that M1 requires L. J Bacteriol. 1982;150:545–551. doi: 10.1128/jb.150.2.545-551.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz TA, Moore PB. RNA, the first macromolecular catalyst: the ribosome is a ribozyme. Trends Biochem Sci. 2003;28:411–418. doi: 10.1016/S0968-0004(03)00169-5. [DOI] [PubMed] [Google Scholar]
- Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. 5S Ribosomal RNA Database. Nucleic Acids Res. 2002;30:176–178. doi: 10.1093/nar/30.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velichutina IV, Hong JY, Mesecar AD, Cherno YO, Liebman SW. Genetic interaction between yeast Saccharomyces cerevisiae release factors and the decoding region of 18S rRNA. J Mol Biol. 2001;305:715–727. doi: 10.1006/jmbi.2000.4329. [DOI] [PubMed] [Google Scholar]
- Wai HH, Vu L, Oakes M, Nomura M. Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo. Nucleic Acids Res. 2000;28:3524–3534. doi: 10.1093/nar/28.18.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB. Double-stranded RNA replication in the yeast: the killer system. Annu Rev Biochem. 1986;55:373–395. doi: 10.1146/annurev.bi.55.070186.002105. [DOI] [PubMed] [Google Scholar]
- Wickner RB. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol Rev. 1996;60:250–265. doi: 10.1128/mr.60.1.250-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Leibowitz MJ. Two chromosomal genes required for killing expression in killer strains of Saccharomyces cerevisiae. Genetics. 1976;82:429–442. doi: 10.1093/genetics/82.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Porter-Ridley S, Fried HM, Ball SG. Ribosomal protein L3 is involved in replication or maintenance of the killer double-stranded RNA genome of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1982;79:4706–4708. doi: 10.1073/pnas.79.15.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN, Nierhaus KH. The ribosome through the looking Glass. Angew Chem Int Ed Engl. 2003;42:3464–3486. doi: 10.1002/anie.200200544. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Sundaralingam M. Two crystal forms of helix II of Xenopus laevis 5S rRNA with a cytosine bulge. RNA. 2000;6:1316–1324. doi: 10.1017/s135583820000090x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh LC, Lee JC. An in vitro system for studying RNA protein interaction: application to a study of yeast ribosomal protein L1 binding to 5S rRNA. Biochimie. 1995;77:167–173. doi: 10.1016/0300-9084(96)88121-1. [DOI] [PubMed] [Google Scholar]
- Yonath A, et al. Crystallographic studies on the ribosome, a large macromolecular assembly exhibiting severe nonisomorphism, extreme beam sensitivity and no internal symmetry. Acta Crystallogr A. 1998;54:945–955. doi: 10.1107/s010876739800991x. [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]


