Abstract
The amyloid precursor protein (APP) plays a central role in Alzheimer's disease, but its physiological function and that of its mammalian paralogs, the amyloid precursor-like proteins 1 and 2 (APLPs), is still poorly understood. APP has been proposed to form dimers, a process that could promote cell adhesion via trans-dimerization. We investigated the dimerization and cell adhesion properties of APP/APLPs and provide evidence that all three paralogs are capable of forming homo- and heterocomplexes. Moreover, we show that trans-interaction of APP family proteins promotes cell–cell adhesion in a homo- and heterotypic fashion and that endogenous APLP2 is required for cell–cell adhesion in mouse embryonic fibroblasts. We further demonstrate interaction of all the three APP family members in mouse brain, genetic interdependence, and molecular interaction of APP and APLPs in synaptically enriched membrane compartments. Together, our results provide evidence that homo- and heterocomplexes of APP/APLPs promote trans-cellular adhesion in vivo.
Keywords: Alzheimer's disease, APP, cell adhesion, dimerization
Introduction
Research on the amyloid precursor protein (APP) has been focused on generation of the amyloid-β (Aβ) peptide, which is proteolytically derived from its precursor APP by consecutive cleavages by BACE-1 and γ-secretase (Aguzzi and Haass, 2003). Despite the wealth of studies regarding the physiological role of APP, there is little consensus about its function in vivo. Among other proposed functions, APP was found to promote neurite outgrowth, cell adhesion, and cell proliferation (Annaert and De Strooper, 2002). However, genetic studies have shown that APP knockout mice are viable, exhibiting only a mild phenotype including reduced forelimb grip strength and locomotor activity (Zheng et al, 1995; Li et al, 1996). Two paralogs of APP are known in mammals, termed amyloid precursor-like proteins 1 and 2 (APLP1 and APLP2). Mice with single knockouts of APLP1, APLP2, or double knockouts of APP/APLP1 are viable, while combinations of APLP2 and APP or APLP1 knockouts are lethal shortly after birth (von Koch et al, 1997; Heber et al, 2000), suggesting functional redundancy. Interestingly, triple knockout mice lacking all the three APP family members exhibit cortical dysplasia with ectopic neurons resembling a human type II lissencephaly phenotype (Herms et al, 2004). The observed phenomenon is in accordance with a redundant function of APP family proteins in neuronal cell adhesion. Redundancy of APP/APLPs is also reflected in a similar protein domain structure, where all the three family members share high homology of intra- and extracellular regions (Coulson et al, 2000). Moreover, all APP family members are processed in a similar manner by the same protease activities (Eggert et al, 2004), and it has also been shown that they interact with the Notch pathway during Drosophila development (Merdes et al, 2004). Despite the high degree of conservation between APP, APLP1, and APLP2, several differences can be noted. The expression of APLP1 is limited to the nervous system (Lorent et al, 1995), whereas APP and APLP2 are ubiquitously expressed (Tanzi et al, 1988; Slunt et al, 1994). Additionally, biochemical analyses suggested that APLP1 accumulates at the postsynapse (Kim et al, 1995), while APP (Ferreira et al, 1993) and APLP2 (Lyckman et al, 1998) are transported to presynaptic sites.
On the basis of its structural features, APP has been proposed to have a receptor-like function and it binds to different extracellular matrix proteins, such as heparin and collagen (Small et al, 1992; Beher et al, 1996). In this context, it is of interest that APP forms dimers (Scheuermann et al, 2001), which is reminiscent of classical receptor dimerization described for the EGF receptor (Schlessinger, 2002). However, it is unclear whether APP dimerization could also occur in an intercellular manner, as described for different cell adhesion molecules (CAMs), such as cadherins and nectins (Takai et al, 2003). This possibility is supported by the recent crystal structure of the APP extracellular carbohydrate domain (E2), which crystallized in the form of a trans-dimer (Wang and Ha, 2004). These intrinsic properties of APP would suggest a role in cell–cell adhesion via trans-dimerization. In order to address this hypothesis, we investigated homo- and heterointeraction of APP family proteins and analyzed their ability to promote intercellular adhesion.
Here we report for the first time that homo- and heterocomplexes of APP family proteins promote cell adhesion via trans-cellular interaction. We further demonstrate the existence of endogenous heterocomplexes of APP family proteins in mouse brain and synaptic compartments, suggesting a role for APP family proteins in cell–cell interaction.
Results
Homo- and heterointeraction of APP, APLP1, and APLP2
To address the question whether APP and its paralogs can interact with each other in a cellular system, we performed coimmunoprecipitation analyses of myc- and HA-tagged APP family proteins transiently expressed in COS7 cells. We tested if HA-tagged APP can be coimmunoprecipitated with the corresponding myc-tagged APP construct by using specific anti-myc and anti-HA antibodies (Figure 1A). As a control, the analysis was also carried out with cells expressing myc-tagged APP and vector only. Additionally, in order to exclude artificial postlysis aggregation, extracts from transfected cells expressing myc- or HA-tagged APP only were mixed after lysis and examined by immunoprecipitation as well (Figure 1A, lanes ‘mix').
Figure 1.
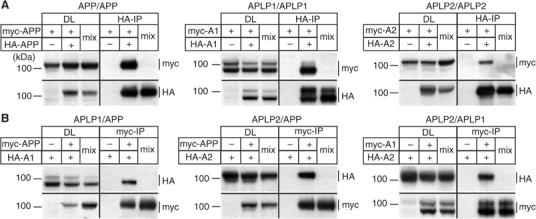
Homo- and heterointeraction of APP family proteins. (A) Homointeraction of myc- and HA-tagged APP, APLP1, and APLP2. Pairs of myc- and HA-tagged APP (myc-APP and HA-APP), APLP1 (myc-A1 and HA-A1), or APLP2 (myc-A2 and HA-A2) were expressed in COS7 cells. In all, 1/25 of each lysate was used as an input control (DL). HA-tagged APP/APLPs were immunoprecipitated and immunoblotted for myc- and HA-tagged constructs. As controls, cells transfected with the corresponding myc-tagged APP family member or postlysis mixtures (lanes ‘mix') of separately transfected myc- and HA-tagged APP/APLPs were immunoprecipitated. (B) Heterointeraction of myc- and HA-tagged APP, APLP1, and APLP2. Heterotypic pairs of myc- and HA-tagged APP, APLP1, or APLP2 were transfected into COS7 cells as indicated. In all, 1/25 of the each lysate was loaded as input control (DL). Myc-tagged APP/APLPs were immunoprecipitated and immunoblotted for myc- and HA-tagged APP/APLPs. Controls were performed as above.
Under these conditions, myc-tagged APP was specifically interacting with HA-tagged APP in cotransfected cells, but not in the control or after postlysis mixing (Figure 1A). With the same experimental setup, APLP1 and APLP2 were also found to specifically interact in a homophilic fashion to a similar extent as observed for APP (Figure 1A). Interestingly, we observed interaction of both mature and immature forms of APP/APLPs.
To prove the specificity of our approach, we performed the same experiment in reverse by immunoprecipitating the myc-tagged APP/APLP constructs. Again, we detected the coimmunoprecipitated corresponding HA-tagged constructs at comparable levels as in the experiments above (Supplementary Figure 1). These results show that APP/APLPs specifically interact in a homophilic fashion.
We further asked whether APP/APLPs are capable of forming heterocomplexes as well. For this purpose, we expressed all possible combinations of myc- and HA-tagged APP/APLPs, and immunoprecipitated the according myc-tagged constructs as described above (Figure 1B). We found that all APP family proteins were efficiently coimmunoprecipitated in double-transfected cells, while no interaction was detected after postlysis mixing of single transfected cells. Comparable amounts of APP/APLP1, APP/APLP2, and APLP1/APLP2 heterocomplexes were recovered under these conditions independent of the type of tag used for the according APP/APLP construct (see Supplementary Figure 1).
Together, these results demonstrate that APP family proteins specifically form homo- and heterocomplexes, suggesting a strong tendency for dimerization/multimerization of APP/APLPs in a cellular context.
Interaction of APP/APLPs depends on the E1 domain
To characterize the site of interaction, we generated deletion constructs of APP/APLPs lacking the N-terminal growth factor-like and copper-binding domains (ΔE1), the carbohydrate domain (ΔE2), the entire ectodomain (ΔEC), or the intracellular domain (ΔCT) (Figure 2A). Both HA- and myc-tagged deletion constructs of all the three homologs were made and analyzed in coimmunoprecipitation experiments.
Figure 2.
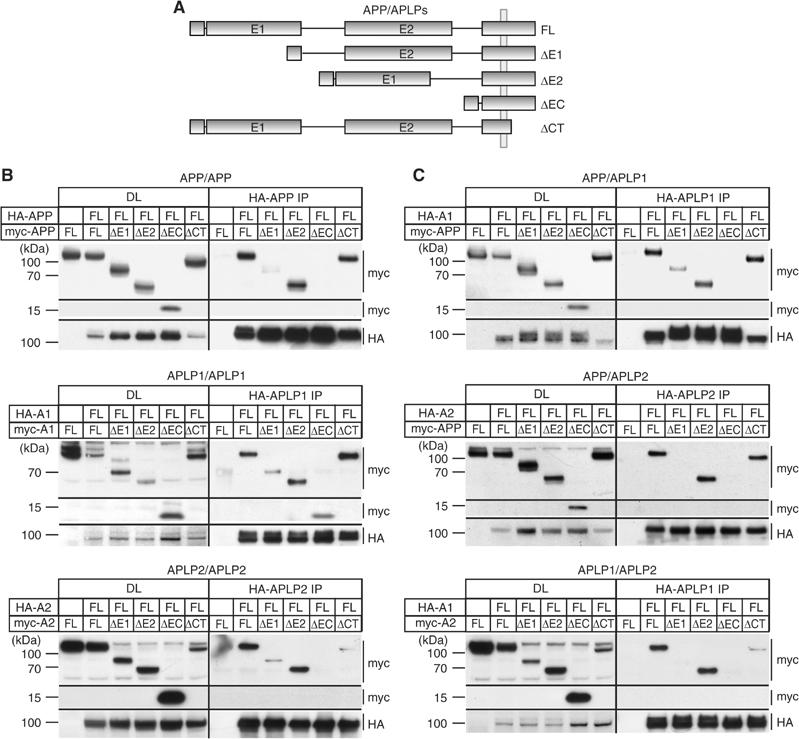
Mapping of APP/APLP homo- and heterointeraction. (A) Schematic drawing of APP/APLP constructs used. HA- and myc-tagged APP/APLP constructs lacking either the E1 domain (ΔE1), the E2 domain (ΔE2), the entire ectodomain (ΔEC), or the cytoplasmic domain (ΔCT) were analyzed. (B) Homointeraction of full-length HA-tagged APP, APLP1, and APLP2 with different deletion constructs in COS7 cells. HA-tagged APP (HA-APP FL) was coexpressed with vector only, myc-tagged APPΔE1 (ΔE1), APPΔE2 (ΔE2), APPΔEC (ΔEC), or APPΔCT (ΔCT). In all, 1/25 of each lysate was used for the DL. HA-tagged APP was immunoprecipitated from cell extracts and immunoblotted for myc- and HA-tagged constructs. The identical setup was used for APLP1 (HA-A1) and APLP2 (HA-A2) homointeraction with the corresponding deletion constructs. Lower levels of APLP2ΔCT interaction are due to lower expression levels of this construct. (C) Heterointeraction of APP, APLP1, and APLP2 with different deletion constructs in COS7 cells. As for the mapping of homointeractions, a HA-tagged APP family member was coexpressed with vector only or different APP/APLP deletion constructs and immunoprecipitated (as indicated). In all, 1/25 of each lysate was used as an input control (DL).
To assess homotypic interaction, HA-tagged full-length APP was coexpressed with the different myc-tagged APP deletion constructs and immunoprecipitated as described above. We found comparable amounts of coimmunoprecipitated myc-tagged full-length APP, APPΔE2, and APPΔCT (Figure 2B). Intriguingly, APP constructs lacking the E1 domain or the entire ectodomain (ΔEC) displayed poor or no detectable interaction, respectively. We also did not observe postlysis aggregation of the different deletion constructs (data not shown).
Similarly, little or no interaction of full-length APLP2 with APLP2ΔE1 or APPΔEC constructs was observed, respectively (Figure 2B). For APLP1, consistently lower, but detectable, amounts of myc-tagged APLP1ΔE1 or APLP1ΔEC were coimmunoprecipitated, while interaction levels of APLP1ΔE2 and APLP1ΔCT remained unchanged (Figure 2B).
These data suggest the E1 domain as the major region required for homodimerization of all three homologs. Additionally, the transmembrane (TM)/juxtamembrane region, especially of APLP1, might be involved in mediating homotypic binding.
Analogously, we performed a systematic mapping of heterotypic interaction of APP family proteins. APP/APLPs lacking the E2 domain or the intracellular domain were still interacting to a similar extent as the full-length proteins. In contrast, like already seen for homointeraction, all mutant APP/APLP1 lacking the E1 domain or the entire ectodomain were weakly, or not, coimmunoprecipitated, respectively (Figure 2C).
Taken together, these results show that the E1 domain is the major interface for homo- as well as heterotypic interaction of APP/APLPs.
Dimerization of APP, APLP1, and APLP2 at the cell surface
We further asked whether interaction of APP/APLPs also occurs at the cell surface, which would be prerequisite for dimerization-induced cell–cell adhesion. Therefore, we used a crosslinking approach utilizing the membrane impermeable crosslinker DTSSP to covalently link cell surface APP/APLPs in intact cells. COS7 cells expressing APP, APLP1, or APLP2 were labeled with [35S]methionine and incubated with DTSSP at 4°C to prevent endocytosis. Afterwards, cells were lysed and the according APP family members were immunoprecipitated with specific antibodies. As a control, equally treated cells without addition of DTSSP were analyzed in parallel. For all the three APP family members, specific bands corresponding to the size of homodimers (200–220 kDa) could be detected in DTSSP-treated cells, but not in control cells (Figure 3). To assess the nature of the crosslinked protein species, the crosslinking products were excised from the gel and the contained proteins were extracted. The samples were then denatured under reducing conditions in order to dissociate the crosslinker. After SDS–PAGE and autoradiography, we found that the recovered major bands were corresponding to the size of the APP/APLPs monomers (100–120 kDa) (Figure 3, lanes ‘el.d.', eluted dimers), indicating that all APP family proteins form homodimers at the cell surface.
Figure 3.
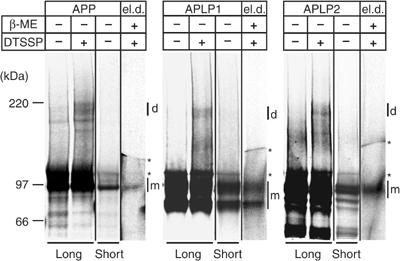
Dimerization of APP family proteins at the cell surface. [35S-Met]-labeled COS7 cells expressing APP, APLP1, or APLP2 were incubated either with or without the membrane-impermeable crosslinker DTSSP as indicated. APP/APLPs were immunoprecipitated with anti-APP (22734), anti-APLP1 (57), or anti-APLP2 (D2-II) antibodies, respectively. The samples were denatured without β-mercaptoethanol (β-ME) and analyzed on 3–8% Tris-acetate gels by autoradiography. Long and short exposures are shown as indicated to visualize crosslinked dimeric (d) or the monomeric (m) forms of APP/APLPs, respectively. Longer exposures show the crosslinked dimers compared to control cells without crosslinker. The crosslinking products (d) were subsequently extracted from the gel, denatured under reducing conditions (el.d.: eluted dimers), and analyzed on 3–8% Tris-acetate gels. The asterisks indicate unspecific signals present in all samples and controls.
Homophilic intercellular interaction of APP, APLP1, and APLP2
Our experiments so far have shown that mature, but also immature, APP/APLP forms are interacting in COS7 cells, suggesting that the large intracellular pool of APP family proteins forms lateral cis-dimers. However, to exclusively address trans-interactions at the cell surface, we asked if APP and its homologs are able to promote cell adhesion. Drosophila Schneider (S2) cells are a powerful tool to investigate trans-cellular interaction of cell adhesion proteins (Islam et al, 2004) and receptor–ligand binding (Klueg and Muskavitch, 1999). Expression of two interacting proteins in two separated pools of S2 cells causes coclustering after mixing of both cell pools, thus giving a direct readout for interaction of the two proteins.
We examined the properties of APP and its mammalian paralogs APLP1 and APLP2 to induce homotypic cell clustering. For this purpose, we transfected S2 cells with APP, APLP1, APLP2, or different extracellular deletion constructs thereof (ΔE1, ΔE2, or ΔEC; Figure 4A). Transfected single-cell suspensions were aggregated for 2 h and analyzed by immunocytochemistry. The transfection efficiency was within a comparable range of 20–25%, with high cell surface expression of all constructs (Figure 4A). Expression of APP/APLP full-length or ΔE2 constructs caused homotypic cell clustering, whereas no specific aggregation was observed for cells expressing constructs lacking the E1 domain or the entire ectodomain (Figure 4A). Recombinant expression of the full-length Notch receptor or GFP only did not induce homotypic cell clustering (data not shown), confirming the specificity of our assay. Quantification revealed that 6% of all APP- or APPΔE2-expressing cells were clustered, showing statistically significant interaction compared to APPΔE1- or APPΔEC-expressing cells (Figure 4A; P<0.05). Strikingly, 32 and 36% of APLP1- and APLP2-expressing cells were clustered, respectively (Figure 4A). The interaction of APLP1- or APLP2-transfected cells was again strictly dependent on the presence of the E1 domain, but not of the E2 domain, as APLP1ΔE1- and APLP2ΔE1-expressing cells were not clustered, respectively (Figure 4A; P<0.001). Consistent with these data, confocal microscopy studies of APLP1-expressing aggregated cells revealed that APLP1 strongly accumulated at sites of cell–cell contact transfected cells (Figure 4A), but not at contact sites of nontransfected cells (Figure 4A).
Figure 4.
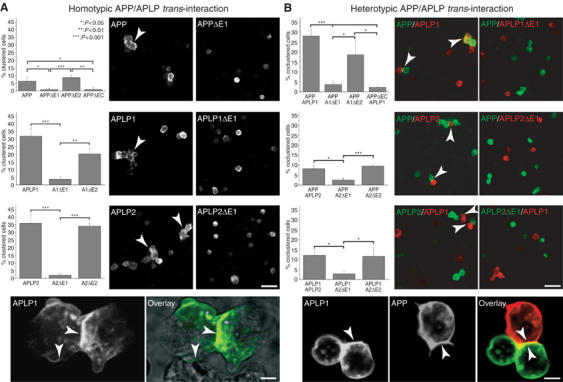
Homo- and heterotypic intercellular interactions of APP, APLP1, and APLP2. (A) Quantification and immunostainings of homotypic S2 cell clusters (indicated by arrows) expressing APP/APLPs or the corresponding deletion constructs. The percentage of clustered transfected cells from at least three independent experiments is shown for the different constructs as indicated (n⩾3, ±s.d., t-test, scale bar=20 μm). Lower panel: confocal analysis of APLP1 expressing S2 cells stained with an anti-myc antibody (APLP1) and overlaid with the transmission image (scale bar=3 μm). (B) Quantification and immunostainings of heterotypic cell contacts (indicated by arrows) of mixed pools of S2 cells expressing APP/APLPs or the corresponding extracellular deletion constructs as indicated. Direct heterotypic cell contacts were quantified from at least three independent experiments, and is given as the percentage of total transfected cells (n⩾3, ±s.d., t-test, scale bar=20 μm). Lower panel: confocal analysis of immunostained mixed pools of S2 cells expressing APLP1 (anti-myc) and APP (40090, scale bar=3 μm).
These data demonstrate that APP family proteins present at the cell surface can stabilize cell–cell interactions by homophilic trans-interaction via the E1 domain.
Heterotypic intercellular interaction of APP, APLP1, and APLP2
We further investigated heterotypic interactions between APP family proteins as our coimmunoprecipitation experiments already suggested a high degree of heterointeraction. Again, S2 cells were transiently transfected with APP, APLP1, APLP2, or the corresponding extracellular deletion constructs. Pairs of differentially transfected single-cell suspension pools were mixed, aggregated, and analyzed by immunocytochemistry. Intriguingly, 28% of mixed cell populations expressing APP and APLP1 displayed heterotypic cell contacts, suggesting a high degree of hetero-trans-interaction (Figure 4B). Similar results were obtained for mixed pools of APP and APLP2, or APLP1- and APLP2-expressing S2 cells, where heterocluster formation was counted for 8 or 12% of transfected cells, respectively (Figure 4B). Additionally, no interaction of GFP-expressing cells with APP/APLP-transfected cells was observed (Supplementary Figure 3), strongly suggesting that the observed cell contact formation requires direct interaction of APP family proteins. This is further supported by the finding that cells expressing APP/APLPs lacking the ΔE1 domain were not incorporated into clusters of APP-, APLP1-, or APLP2-expressing cells, respectively (Figure 4B). Consistently, confocal microscopy studies revealed that heterotypic aggregates of APP- and APLP1-expressing cells displayed accumulated APLP1 immunoreactivity at cell–cell contact sites, not only adjacent to APLP1-expressing cells as observed for homotypic clusters, but also at contact sites of APP-expressing cells (Figure 4B). APP, in turn, was also localized at cell contact sites and cellular protrusions framing adjacent APLP1-expressing cells (Figure 4B).
Together, these results demonstrate that APP, APLP1, and APLP2 have homo- and hetero-trans-interaction properties resulting in cell–cell adhesion, which specifically depend on E1 domain association.
APP/APLPs promote intercellular adhesion of mouse embryonic fibroblasts (MEFs)
Having shown that overexpressed APP family proteins are capable of promoting cell adhesion via trans-cellular interaction, we asked if a lack of APP/APLPs would lead to loss of cell adhesion. To investigate this hypothesis, we used MEFs derived from APP/APLP knockout animals. We tested the cell adhesion properties of MEFs derived from wild-type (WT), APP or APLP2 knockout (APP−/−, APLP2−/−), and APP/APLP2 double knockout (Dko) mice in a modified cell aggregation assay under calcium- and magnesium-free conditions to avoid cadherin-mediated adhesion (Miura et al, 1992). MEFs were initially treated with 1 mM EDTA and single-cell suspensions were aggregated for 60 min at 80 r.p.m. As cell clusters are formed during aggregation, the number of particles (single cells or cell cluster are equal to one particle) decreases over time. Thus, stronger cell adhesion causes an increase in cell clustering, resulting in a decreased number of particles.
We detected solid and comparable cellular adhesion of WT and APP−/− MEFs shown by a decrease in particle number of approx. 40% (Figure 5A). Strikingly, cellular interaction of APLP2−/− and Dko MEFs was dramatically impaired, leading to significantly less cell aggregation (Figure 5A; P<0.01). However, after supplementation of calcium, APLP2−/− and Dko MEFs still exhibited similar aggregation properties as WT and APP−/− cells, indicating that the reduced amount of cellular interaction is not due to a lack of cadherin-mediated adhesion (Figure 5B). Since APP−/− MEFs were still showing strong cellular aggregation and MEFs do not express APLP1 endogenously (Supplementary Figure 4), we reasoned that APLP2 might be the essential family member for mediating cell–cell adhesion. To prove this hypothesis, we retransfected Dko cells with APLP2 (DkoAPLP2re). In addition, as we have shown strong trans-interaction of APLP1 in S2 cells, we also analyzed Dko cells stably expressing APLP1 (dkoAPLP1re) and APP−/− MEFs stably expressing APP (APP−/−APPre). The expression of APP family proteins in the different MEF lines was verified by Western blotting (Supplementary Figure 4). APP−/− and APP−/−APPre MEFs both showed strong cellular aggregation, with no significant difference (Figure 5A). Intriguingly, we observed strong cellular aggregation for DkoAPLP1re and DkoAPLP2re cell lines, showing that the cell adhesion properties observed in WT cells could be restored by APLP1 or APLP2 (Figure 5A; P<0.01).
Figure 5.
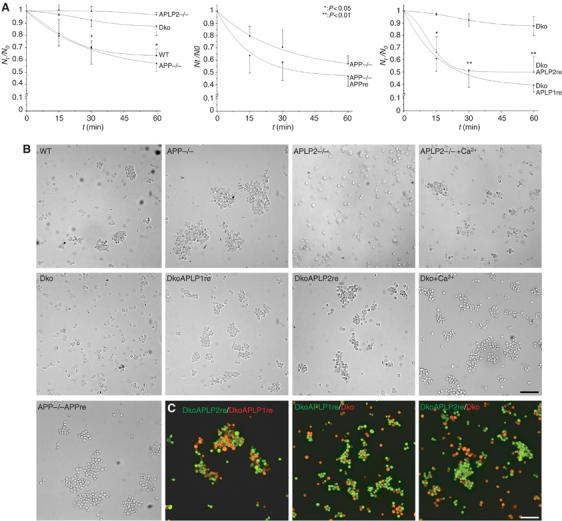
APP family proteins are required for cell adhesion. (A) Quantitative cellular aggregation of WT, APP knockout (APP−/−), APLP2 knockout (APLP2−/−), or APP/APLP2 double-knockout MEF cells (Dko). MEF cells were aggregated in suspension under calcium- and magnesium-free conditions for 1 h at 80 r.p.m. The number of particles (single cells and cell clusters) was counted at the indicated time points (Nt). The relative decrease in particle counts compared to t=0 (Nt/N0) is shown over time as a measure of aggregation of the different MEF cells (n⩾3, ±s.d., t-test). APP−/− MEFs were compared with APP retransfected cells (APP−/−APPre), and APLP1 (DkoAPLP1re) or APLP2 (DkoAPLP2re) rescued Dko cells were compared with parental Dko cells. (B) Typical micrographs of the different MEF lines after 60 min of aggregation. APLP2−/− and Dko cells were additionally aggregated in the presence of 1 mM Ca2+ to induce cadherin-mediated adhesion (scale bar=100 μm). (C) Equal amounts of differentially labeled Dko, DkoAPLP1re, and DkoAPLP12re cells (as indicated) were coaggregated for 60 min and analyzed by fluorescence microscopy (scale bar=100 μm).
We further assessed the specificity of APLP-induced cell adhesion by coaggregation experiments with the parental Dko cell line. For this purpose, calcein-red-labeled Dko cells were coaggregated with calcein-labeled dkoAPLP1re or dkoAPLP2re cells. We observed predominant homotypic aggregation of dkoAPLP1re and dkoAPLP2re MEFs, with only weak random coclustering of dko cells (Figure 5C). Coaggregation of labeled dkoAPLP1re and dkoAPLP2re cells, however, resulted in mixed cell aggregation, largely consisting of homotypic cell clusters (Figure 5C).
Taken together, our results clearly show that APLP2 is both required and sufficient for calcium-independent cell adhesion of MEFs. Moreover, APLP1 and APLP2 mediate MEF cell adhesion in a predominantly homotypic fashion, suggesting direct trans-cellular interaction.
Heterointeraction of APP/APLPs in vivo
We further aimed to investigate whether heterophilic interactions between APP family members exist in vivo. Therefore, we performed coimmunoprecipitations from WT mouse brain extracts and, to verify specificity, from APP−/−, APLP1−/−, and APLP2−/− mouse brain extracts (Figure 6).
Figure 6.

APP/APLPs form heterocomplexes in vivo. Brain extracts from WT, APP−/−, APLP1−/−, and APLP2−/− mice were subjected to immunoprecipitation and SDS–PAGE. In all, 20 μg of total extracts (DL) was loaded for comparison. (A) APP/APLP1 interaction was probed by immunoprecipitating with the anti-APLP1 antibody 57 and Western blot detection of APP (22C11), APLP1 (CT-11), and β-tubulin III (Tub). Note that only mature higher-molecular-weight forms of APP were coimmunoprecipitated with APLP1. The apparent size of the different APP bands is indicated. (B) APP/APLP2 interaction was probed by immunoprecipitating with the anti-APP antibody 22734 and Western blot detection of APLP2 (D2-13), APP (22C11), and β-tubulin III (Tub). The apparent size of the different APLP2 bands is indicated. (C) APLP1/APLP2 interaction was probed by immunoprecipitating with the anti-APLP1 antibody 57 and Western blot detection of APLP2 (D2-13), APLP1 (CT-11), and β-tubulin III (Tub).
To analyze APP/APLP1 interaction in vivo, APLP1 was immunoprecipitated with the C-terminal antibody 57 (Eggert et al, 2004). By this means, APP could be coimmunoprecipitated from WT and APLP2−/−, but not from APP−/− and APLP1−/− brain samples (Figure 6A), showing specific heterointeraction of APP and APLP1 in vivo. In mouse brain, at least three distinct forms of APP with molecular weights of 100, 108, and 120 kDa were detected (Figure 6A; as indicated), likely representing different post-translationally modified APP695 species (Sandbrink et al, 1997). Interestingly, only the mature, high-molecular-weight forms of APP with molecular weights of 108 and 120 kDa were coimmunoprecipitated with APLP1 (Figure 6A), suggesting that a particular pool of APP exists in complex with APLP1 in vivo. We never observed association of the major 100 kDa APP form with APLP1 in vivo making postlysis association highly unlikely.
We also investigated APP/APLP2 interaction by immunoprecipitating APP with the APP-specific antibody 22734 (Figure 6B). We detected specifically coimmunoprecipitated APLP2 in WT and APLP1−/− brain extracts only (Figure 6B), showing endogenous interaction of APP/APLP2. Additionally, we could also coimmunoprecipitate APLP2 with the anti-APLP1 antibody 57 from WT and APP−/−, but not from APLP1−/− or APLP2−/− brain extracts (Figure 6C). Interestingly, for both APP/APLP2 and APLP1/APLP2 interactions, only the mature 120 kDa form of APLP2 was coimmunoprecipitated, which corresponds to the observed association of mature APP with APLP1 (Figure 6A).
These data further corroborate the interaction of APP/APLPs by showing the existence of common complexes of mature, higher-molecular-weight, but not immature, forms of APP family proteins in vivo.
Post-translational accumulation of APLPs in APP−/− mice
Unexpectedly, during our analysis, we noticed changes in the protein levels of APLP1 and APLP2 in APP−/− mice. To investigate this observation in more detail, different 4- or 8-months-old knockout mice were compared with age-matched WT mice (Figure 7A).
Figure 7.
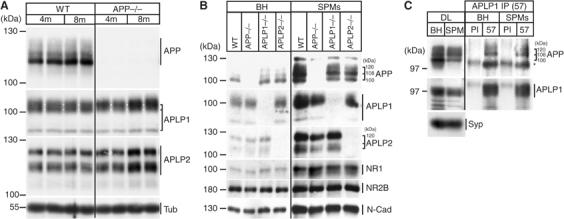
Accumulation of APLPs in APP−/− mouse brain and synaptic enrichment and interaction. (A) In all, 20 μg of total mouse brain lysates from 4- (4 m) or 8-months- (8 m) old WT and APP−/− mice were subjected to SDS–PAGE and immunoblotted for APP, APLP1, and APLP2, and β-tubulin III (Tub). The size of the different APP/APLP bands is indicated by brackets. Note that that the highest APLP1 band is specifically accumulating in APP−/− mice compared to WT (size is indicated by brackets and white lines). (B) Synaptic plasma membranes (SPMs) were prepared from BH of WT, APP−/−, APLP1−/−, and APLP2−/− mice. From each fraction, 20 μg of total protein was subjected to SDS–PAGE and immunoblotted with antibodies against APP (22C11), APLP1 (CT-11), and APLP2 (D2-II). Enrichment of synaptic compartments was confirmed with NMDA receptor 1 (NR1) and 2B (NR2B), and N-cadherin antibodies (N-Cad). (C) Total extracts (BH) and SPM preparations from WT mouse brains were directly loaded to the gel (DL) or immunoprecipitated either with PI or an anti-APLP1 antibody (57), followed by Western blot detection of APP (22C11), APLP1 (CT-11), and synaptophysin (Syp). DLs of BH and SPMs are shown in comparison. The asterisk indicates a signal of the antibody heavy chain.
Interestingly, we detected an age-dependent increase of APLP1 and APLP2 protein levels in APP−/− mice compared to WT. This was evident in brain extracts from 8-months-, but not 4-months-old APP−/− mice (Figure 7A), while neither APLP1−/− nor APLP2−/− mice displayed changes in APP/APLP protein levels (data not shown). Intriguingly, the signal pattern of APLP1 appeared different in APP−/− extracts, as the intensity of the highest-molecular-weight form of APLP1 was already increased in 4-months-old APP−/− mice, which was even more pronounced in the 8-months-old mice (Figure 7A).
Taken together, accumulation of APLP1 and APLP2 in APP−/− mice indicates deregulation of APLP1 and APLP2 metabolism, possibly due to the absence of APP as a binding partner.
APP, APLP1, and APLP2 are enriched in synaptic plasma membranes (SPMs) and APP interacts with APLP1 in synaptic compartments
It has been previously shown that the high-molecular-weight forms of APP with 108 and 120 kDa are particularly enriched in synaptic compartments (Beher et al, 1999). Therefore, we wanted to investigate whether the high-molecular-weight forms of APP and APLP2 found to interact might be identical to those enriched in synaptic compartments.
For this purpose, SPM fractions were isolated from total brain homogenates (BH) of WT, APP−/−, APLP1−/−, and APLP2−/− mice by Ficoll and Sucrose gradient ultracentrifugation as described previously (Beher et al, 1999). Consequently, we found a strong enrichment of 108 and 120 kDa forms of APP in SPMs compared to levels of the major 100 kDa form (Figure 7B). The synaptic enrichment of APP was comparable in samples of WT, APLP1−/−, or APLP2−/− fractions. Likewise, APLP1 and APLP2 strongly accumulated in SPMs to a comparable extent (Figure 7B), and, similarly to APP, we found enrichment especially of the highest-molecular-weight form of APLP1 and APLP2 in SPMs (Figure 7B). Enrichment of SPM compartments in the purified fractions was confirmed by the expected accumulation of N-cadherin and NMDA receptor 1 and 2B compared to total brain extracts (Figure 7B).
Since the 108 and 120 kDa APP species found to interact with APLP1 (Figure 6A) were indeed enriched in the synaptic preparations, we asked whether the APP/APLP1 heterocomplex exists in synaptic membrane compartments. Therefore, total extracts and SPMs from WT mouse brains were immunoprecipitated either with preimmune serum (PI) or the anti-APLP1 antibody 57, respectively (Figure 7C). We found that the 108 and 120 kDa forms of APP were specifically coimmunoprecipitated with APLP1 from SPMs to a similar extent as from total extracts (Figure 7C), as comparable amounts of high-molecular-weight forms of APP were present in direct loads (DLs) of both fractions (Figure 7C).
Taken together, these data show enrichment of all APP family proteins and the existence of an APP/APLP1 heterocomplex in synaptic membrane compartments.
Discussion
In our work, we were able to demonstrate homo- and heterointeraction of APP, APLP1, and APLP2. Moreover, we have identified the N-terminal E1 domain as the major interaction interface for dimerization of cellular APP/APLPs. APP dimerization has previously been shown to occur in vitro, as recombinant secreted APP is mainly recovered as a dimer, suggesting that homodimerization is an intrinsic property of APP (Beher et al, 1996; Scheuermann et al, 2001). In addition, we could now show that APP/APLP homodimers are present at the cell surface of COS7 cells. However, as APP family proteins expressed in COS7 cells displayed a mainly immature glycosylation pattern, the majority of APP/APLP interaction is most likely taking place in intracellular compartments. Therefore, our data indicate that this intracellular APP/APLP pool primarily forms cis-type interactions in COS7 cells. While we have found a major role of the E1 domain in this respect, recent reports have also implicated the transmembrane region as a site for cis-dimerization. The APP mutant K624C was shown to form constitutive dimers by disulfide bridging within the Aβ domain, suggesting close proximity and interaction of the APP juxtamembrane/TM region (Scheuermann et al, 2001). Moreover, the APP transmembrane domain efficiently dimerized in a bacterial assay (Vooijs et al, 2004). Although we observed a major relevance for dimerization of the transmembrane region only for APLP1, our coimmunoprecipitation conditions might have not allowed the detection of weaker interactions in the absence of the E1 domain.
Above cis-dimerization, we provide for the first time evidence that APP forms trans-interactions at the cellular level, a property that is even more pronounced for APLP1 and APLP2. As for the coimmunoprecipitation studies, we found the E1 domain also to be essential for cellular interaction of APP/APLP-expressing S2 cells. The observed accumulation of APP/APLPs at sites of cell contact further indicates direct trans-cellular interaction. In contrast to the recently reported trans-dimerization of the APP E2 domain (Wang and Ha, 2004), we did not observe a major relevance of this domain for dimerization. However, as our analysis focused on the cellular proteins and not on isolated domains, we cannot exclude that E2 domain association plays a role in other processes, for example, for secreted APP/APLPs. Nevertheless, our data together with previous studies strongly suggest that both the transmembrane and the E1 domain are involved in lateral dimerization, while the formation of adhesive complexes only requires the E1 domain (Figure 8). In all APP family proteins, the E1 domain is connected to the further C-terminal domains by a highly charged acidic region. We propose that this presumably very flexible linker is needed to allow interaction of E1 domains in both lateral and trans-cellular dimers. With this model, cis- as well as trans-dimerization of APP/APLPs could be achieved with a single E1-binding mode (Figure 8).
Figure 8.
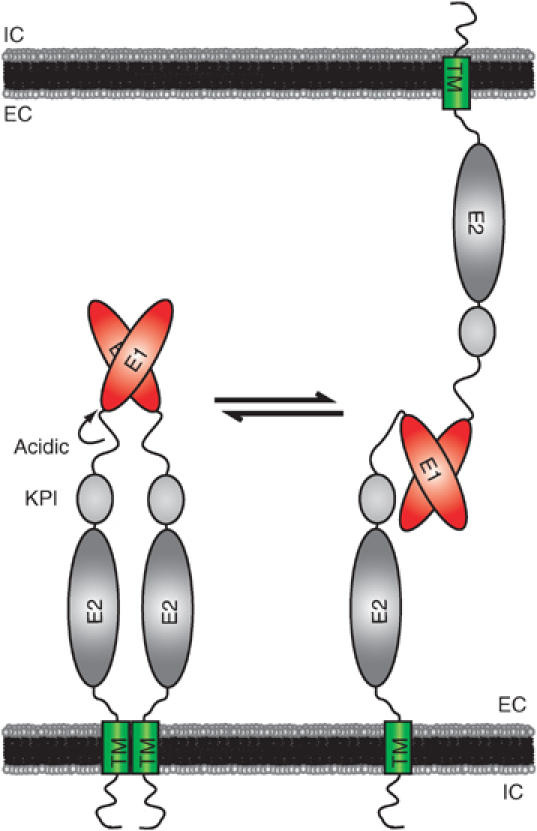
A model for cis- and trans-interaction of APP family proteins. The schematic model of APP/APLP domain organization and interaction is shown. The N-terminal E1 domain is linked to a highly flexible acidic region, followed by the alternatively spliced Kunitz-type protease inhibitor (KPI) domain (for APP and APLP2), the E2 domain, the juxtamembrane/TM region, and the cytosolic domain. Based on our results, we suggest that APP family proteins are capable of forming lateral and adhesive dimers in homo- and heterotypic fashions. The E1 domain is crucial for both cis- and trans-interactions, while the TM region could additionally contribute to lateral dimerization.
Cell–cell adhesion mediated by cell CAMs like cadherins and nectins is essential for tissue and synapse formation and executed by trans-dimerization of these proteins present at adjacent cells or at pre- and postsynaptic sites (Takai et al, 2003). In analogy, we could demonstrate that MEFs lacking endogenous APLP2 exhibit strongly reduced calcium-independent cell adhesion compared to WT cells. This defect could be rescued by exogenously expressing APLP2 or APLP1, showing that they directly induce cell adhesion. Moreover, we observed preferential homotypic cell aggregation of APLP1- and APLP2-expressing Dko MEFs. This finding is consistent with our S2 cell aggregation data showing strong homotypic interaction of APLP1 and APLP2 strictly depending on the E1 domain. Together, our data suggest that MEF cell–cell adhesion requires APLP2 and can be mediated by direct trans-interaction of APLP1 or APLP2, strongly supporting the view that APP family proteins have CAM-like trans-dimerization properties.
Our findings might play a role in neuronal cell adhesion as well, as we could show that mature endogenous APP/APLPs are associated as heterocomplexes in mouse brain. Our coimmunoprecipitation studies in COS7 cells indicate that, in principle, both mature and immature APP/APLPs can interact with similar efficiency. This observation is consistent with in vitro dimerization of recombinantly expressed soluble APP (Scheuermann et al, 2001). Additionally, both mature and immature human APP forms from APP23 transgenic mice coimmunoprecipitate with APLP1 (P Soba, unpublished results), showing that in vivo APP/APLP interaction is basically independent of glycosylation. However, we found that endogenous heterocomplexes exclusively contain mature APP/APLPs. Moreover, these interacting high-molecular-weight species of APP family proteins strongly accumulated in SPM fractions. In addition, the APP/APLP1 heterocomplex could be recovered from SPMs. As only mature APP is present at the cell surface in neurons (Yamazaki et al, 1997), we favor a model that endogenous APP/APLP heterointeraction in the brain is limited to the cell surface, which would allow trans-cellular binding. APP and APLP2 have previously been shown to be transported to presynaptic terminals (Yamazaki et al, 1995; Lyckman et al, 1998) and growth cones of neurons (Beher et al, 1999), while APLP1 has been reported to localize to the postsynapse (Kim et al, 1995). Interestingly, all APP family members exhibit developmentally increased expression levels correlating with postembryonic synaptogenesis (Moya et al, 1994; Sandbrink et al, 1994; Lorent et al, 1995), and recent progress revealed that APP and APLP2 are required for neuromuscular synaptogenesis (Wang et al, 2005). Together with our data showing not only strong homo-trans-interaction of APLP1 and APLP2 but also remarkable hetero-trans-interaction of APP with APLP1, these findings offer the intriguing possibility that trans-cellular APP/APLP interaction is involved in regulating synaptogenesis.
In summary, we have shown that APP, APLP1, and APLP2 have cis- and trans-interaction properties, which largely depend on the conserved E1 domain. Moreover, we have provided evidence that APP family proteins have an essential CAM-like role in cell–cell adhesion. Together with previous genetic studies of APP/APLP2 double-knockout and APP/APLP triple-knockout mice displaying defects in synaptogenesis (Wang et al, 2005) and neuronal migration (Herms et al, 2004), respectively, a picture of the molecular function of APP family proteins in control of cell adhesion is finally emerging. Further studies elucidating the interactions of APP, APLP1, and APLP2 could therefore advance our understanding, not only of their normal physiological function but also of the pathological role in Alzheimer's disease.
Materials and methods
Expression constructs
pCDNA3.1 and pMT (Invitrogen) constructs of N-terminally myc/HA-tagged human APP and APLP1, or C-terminally myc/HA-tagged APLP2, together with corresponding deletion constructs were generated by PCR or have been described previously (Paliga et al, 1997; Scheuermann et al, 2001). N-terminally myc-tagged APP, C-terminally myc-tagged APLP1 (Paliga et al, 1997), APLP2 (Eggert et al, 2004), and Notch (Loewer et al, 2004) were subcloned into the pUAST vector. Gal4 was PCR-amplified and cloned into the pMT/V5-His-TOPO vector (Invitrogen). pCEP4 APP695, APLP1, and APLP2-763 have been described (Paliga et al, 1997; Eggert et al, 2004). The identity of all PCR-amplified constructs was confirmed by sequencing.
Antibodies
Antibodies against APP (22734 and 40090, gifts from Gerd Multhaup; 22C11 (Weidemann et al, 1989)), APLP1 (57 (Eggert et al, 2004); CT-11, Calbiochem), APLP2 (D2-II, Calbiochem), myc (A-14 and 9E10, Santa Cruz), HA (3F10, Roche), NR1 (Synaptic Systems), NR2B (Santa Cruz), N-cadherin (BD Biosciences), Synaptophysin (Sigma), and β-tubulin III (Sigma) were used. The monoclonal anti-APLP2 antibody D2-13 was generated against the C-terminal 15 amino acids of human APLP2 and verified for specificity. Secondary HRP-coupled secondary antibodies were from Promega. Alexa488 and Alexa594 fluorescent dye coupled goat secondary antibodies were from Invitrogen.
Coimmunoprecipitation of myc/HA-tagged APP family proteins
COS7 cells were transiently cotransfected with myc/HA-tagged pCDNA3.1-APP, -APLP1, -APLP2, or empty vector with Lipofectamine Plus (Invitrogen) according to the manufacturer's protocol. Cells lysates were incubated either with anti-myc-agarose (Sigma) or anti-HA-sepharose beads (Roche), washed, and denatured. Total lysates and immunoprecipitates were analyzed on 4–12% Bis–Tris gels (Invitrogen) and probed for HA- and myc-tagged APP family proteins.
Cell surface crosslinking of APP family proteins
COS7 cells stably expressing APP, -APLP1, or -APLP2 were labeled with 35S-methionine and incubated with or without 1 mM 3,3′-dithiobis[sulfosuccinimidyl propionate] (DTSSP, Pierce) to crosslink proteins present at the cell surface. APP, APLP1, and APLP2 were immunoprecipitated and denatured under nonreducing conditions and analyzed on 3–8% Tris-acetate gels (Invitrogen), followed by autoradiography. The crosslinked dimers were excised from the gel, extracted, denatured under reducing conditions, and re-analyzed on 3–8% Tris-acetate gels by autoradiography.
S2-cell aggregation assay
S2-cells were transiently transfected as described (Klueg et al, 2002) using Effectene (Qiagen). Consistent transfection efficiencies of 20–25% were achieved for all constructs analyzed. For analysis of homotypic interactions, 4 × 105 cells in single-cell suspension were aggregated in a 24-well plate for 2 h at 80 r.p.m. on a horizontal shaker. For heterotypic aggregation, two separately transfected pools of S2 cells were mixed (2 × 105 cells each) as single-cell suspensions and aggregated as described above. Cells were transferred to coverslips and immunostained for the overexpressed constructs according to standard procedures. For quantification, at least three low-magnification fields with equal cell densities were taken from each experiment in a blinded fashion and the immunostained cells were counted. For homotypic aggregation experiments, clusters of three or more transfected cells were scored as positive. For heterotypic aggregation experiments, transfected cells with direct heterotypic cell contact were scored as positive. In total, 300–800 transfected cells were counted per experiment, and the data from at least three independent experiments (n⩾3, ±s.d.) were evaluated. Statistical significance was tested with an unpaired t-test. Cell surface expression of APP, APLP1, and APLP2 was confirmed by immuno-EM labeling of transfected S2 cells with specific antbodies (see Supplementary Figure 2).
MEF cell aggregation assay
Cell aggregation of MEF cells was performed in calcium- and magnesium-free Hank's balanced salt solution (HBSS) essentially as described (Miura et al, 1992). For coaggregations, MEF cells were prelabeled with calcein or calcein red-orange (Invitrogen). For quantification, the number of cell particles was counted in duplicate per indicated timepoint and the ratio of particles after 15, 30, and 60 min (Nt) compared to starting conditions at t=0. N0 was calculated and plotted against time. Experiments were performed at least in triplicates (n⩾3, ±s.d.) and statistical significance was tested with an unpaired t-test.
Brain extraction, coimmunoprecipitation, and SPM preparation
Preparation of brain extracts and SPM was performed essentially as described (Beher et al, 1999). Mouse brain extracts were prepared from 6–9-months-old (for coimmunoprecipitation and SPM preparation) or 4- and 8-months-old (for age related APP/APLP levels) age-matched mice of the indicated genotype. For coimmunoprecipitation, BH or SPMs were incubated with the indicated antibodies overnight at 4°C and analyzed on 7 or 8% Tris-glycine gels.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Material and Methods
Acknowledgments
We thank Birgit Hub for excellent technical assistance in EM work, and Andreas Langer for providing MEF cells. We also thank Gerd Multhaup for antibodies 22734 and 40090, Eva Zinser and Tobias Hartmann for APP−/−APPre MEF cells, and Peter Becker for S2 cells. This work was supported by a Fritz Thyssen Stiftung Fellowship (to PS), the Alzheimer Forschungs Initiative e.V (to SK and GM), and the DFG priority program (to SK, UM, and KB). We apologize to all contributors whose work could not be cited or discussed due to space restrictions.
References
- Aguzzi A, Haass C (2003) Games played by rogue proteins in prion disorders and Alzheimer's disease. Science 302: 814–818 [DOI] [PubMed] [Google Scholar]
- Annaert W, De Strooper B (2002) A cell biological perspective on Alzheimer's disease. Annu Rev Cell Dev Biol 18: 25–51 [DOI] [PubMed] [Google Scholar]
- Beher D, Elle C, Underwood J, Davis JB, Ward R, Karran E, Masters CL, Beyreuther K, Multhaup G (1999) Proteolytic fragments of Alzheimer's disease-associated presenilin 1 are present in synaptic organelles and growth cone membranes of rat brain. J Neurochem 72: 1564–1573 [DOI] [PubMed] [Google Scholar]
- Beher D, Hesse L, Masters CL, Multhaup G (1996) Regulation of amyloid protein precursor (APP) binding to collagen and mapping of the binding sites on APP and collagen type I. J Biol Chem 271: 1613–1620 [DOI] [PubMed] [Google Scholar]
- Coulson EJ, Paliga K, Beyreuther K, Masters CL (2000) What the evolution of the amyloid protein precursor supergene family tells us about its function. Neurochem Int 36: 175–184 [DOI] [PubMed] [Google Scholar]
- Eggert S, Paliga K, Soba P, Evin G, Masters CL, Weidemann A, Beyreuther K (2004) The proteolytic processing of the amyloid precursor protein gene family members APLP-1 and APLP-2 involves alpha-, beta-, gamma-, and epsilon-like cleavages: modulation of APLP-1 processing by n-glycosylation. J Biol Chem 279: 18146–18156 [DOI] [PubMed] [Google Scholar]
- Ferreira A, Caceres A, Kosik KS (1993) Intraneuronal compartments of the amyloid precursor protein. J Neurosci 13: 3112–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber S, Herms J, Gajic V, Hainfellner J, Aguzzi A, Rulicke T, von Kretzschmar H, von Koch C, Sisodia S, Tremml P, Lipp HP, Wolfer DP, Muller U (2000) Mice with combined gene knock-outs reveal essential and partially redundant functions of amyloid precursor protein family members. J Neurosci 20: 7951–7963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herms J, Anliker B, Heber S, Ring S, Fuhrmann M, Kretzschmar H, Sisodia S, Muller U (2004) Cortical dysplasia resembling human type 2 lissencephaly in mice lacking all three APP family members. EMBO J 23: 4106–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam R, Kristiansen LV, Romani S, Garcia-Alonso L, Hortsch M (2004) Activation of EGF Receptor Kinase by L1-mediated Homophilic Cell Interactions. Mol Biol Cell 15: 2003–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Wu K, Xu JL, McAuliffe G, Tanzi RE, Wasco W, Black IB (1995) Selective localization of amyloid precursor-like protein 1 in the cerebral cortex postsynaptic density. Brain Res Mol Brain Res 32: 36–44 [DOI] [PubMed] [Google Scholar]
- Klueg KM, Alvarado D, Muskavitch MA, Duffy JB (2002) Creation of a GAL4/UAS-coupled inducible gene expression system for use in Drosophila cultured cell lines. Genesis 34: 119–122 [DOI] [PubMed] [Google Scholar]
- Klueg KM, Muskavitch MA (1999) Ligand-receptor interactions and trans-endocytosis of Delta, Serrate and Notch: members of the Notch signalling pathway in Drosophila. J Cell Sci 112 (Part 19): 3289–3297 [DOI] [PubMed] [Google Scholar]
- Li ZW, Stark G, Gotz J, Rulicke T, Gschwind M, Huber G, Muller U, Weissmann C (1996) Generation of mice with a 200-kb amyloid precursor protein gene deletion by Cre recombinase-mediated site-specific recombination in embryonic stem cells. Proc Natl Acad Sci USA 93: 6158–6162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer A, Soba P, Beyreuther K, Paro R, Merdes G (2004) Cell-type-specific processing of the amyloid precursor protein by Presenilin during Drosophila development. EMBO Rep 5: 405–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorent K, Overbergh L, Moechars D, De Strooper B, Van Leuven F, Van den Berghe H (1995) Expression in mouse embryos and in adult mouse brain of three members of the amyloid precursor protein family, of the alpha-2-macroglobulin receptor/low density lipoprotein receptor-related protein and of its ligands apolipoprotein E, lipoprotein lipase, alpha-2-macroglobulin and the 40,000 molecular weight receptor-associated protein. Neuroscience 65: 1009–1025 [DOI] [PubMed] [Google Scholar]
- Lyckman AW, Confaloni AM, Thinakaran G, Sisodia SS, Moya KL (1998) Post-translational processing and turnover kinetics of presynaptically targeted amyloid precursor superfamily proteins in the central nervous system. J Biol Chem 273: 11100–11106 [DOI] [PubMed] [Google Scholar]
- Merdes G, Soba P, Loewer A, Bilic MV, Beyreuther K, Paro R (2004) Interference of human and Drosophila APP and APP-like proteins with PNS development in Drosophila. EMBO J 23: 4082–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura M, Asou H, Kobayashi M, Uyemura K (1992) Functional expression of a full-length cDNA coding for rat neural cell adhesion molecule L1 mediates homophilic intercellular adhesion and migration of cerebellar neurons. J Biol Chem 267: 10752–10758 [PubMed] [Google Scholar]
- Moya KL, Benowitz LI, Schneider GE, Allinquant B (1994) The amyloid precursor protein is developmentally regulated and correlated with synaptogenesis. Dev Biol 161: 597–603 [DOI] [PubMed] [Google Scholar]
- Paliga K, Peraus G, Kreger S, Durrwang U, Hesse L, Multhaup G, Masters CL, Beyreuther K, Weidemann A (1997) Human amyloid precursor-like protein 1—cDNA cloning, ectopic expression in COS-7 cells and identification of soluble forms in the cerebrospinal fluid. Eur J Biochem 250: 354–363 [DOI] [PubMed] [Google Scholar]
- Sandbrink R, Masters CL, Beyreuther K (1994) APP gene family: unique age-associated changes in splicing of Alzheimer's betaA4-amyloid protein precursor. Neurobiol Dis 1: 13–24 [DOI] [PubMed] [Google Scholar]
- Sandbrink R, Monning U, Masters CL, Beyreuther K (1997) Expression of the APP gene family in brain cells, brain development and aging. Gerontology 43: 119–131 [DOI] [PubMed] [Google Scholar]
- Scheuermann S, Hambsch B, Hesse L, Stumm J, Schmidt C, Beher D, Bayer TA, Beyreuther K, Multhaup G (2001) Homodimerization of amyloid precursor protein and its implication in the amyloidogenic pathway of Alzheimer's disease. J Biol Chem 276: 33923–33929 [DOI] [PubMed] [Google Scholar]
- Schlessinger J (2002) Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell 110: 669–672 [DOI] [PubMed] [Google Scholar]
- Slunt HH, Thinakaran G, Von Koch C, Lo AC, Tanzi RE, Sisodia SS (1994) Expression of a ubiquitous, cross-reactive homologue of the mouse beta-amyloid precursor protein (APP). J Biol Chem 269: 2637–2644 [PubMed] [Google Scholar]
- Small DH, Nurcombe V, Moir R, Michaelson S, Monard D, Beyreuther K, Masters CL (1992) Association and release of the amyloid protein precursor of Alzheimer's disease from chick brain extracellular matrix. J Neurosci 12: 4143–4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai Y, Shimizu K, Ohtsuka T (2003) The roles of cadherins and nectins in interneuronal synapse formation. Curr Opin Neurobiol 13: 520–526 [DOI] [PubMed] [Google Scholar]
- Tanzi RE, McClatchey AI, Lamperti ED, Villa-Komaroff L, Gusella JF, Neve RL (1988) Protease inhibitor domain encoded by an amyloid protein precursor mRNA associated with Alzheimer's disease. Nature 331: 528–530 [DOI] [PubMed] [Google Scholar]
- von Koch CS, Zheng H, Chen H, Trumbauer M, Thinakaran G, van der Ploeg LH, Price DL, Sisodia SS (1997) Generation of APLP2 KO mice and early postnatal lethality in APLP2/APP double KO mice. Neurobiol Aging 18: 661–669 [DOI] [PubMed] [Google Scholar]
- Vooijs M, Schroeter EH, Pan Y, Blandford M, Kopan R (2004) Ectodomain shedding and intramembrane cleavage of mammalian Notch proteins is not regulated through oligomerization. J Biol Chem 279: 50864–50873 [DOI] [PubMed] [Google Scholar]
- Wang P, Yang G, Mosier DR, Chang P, Zaidi T, Gong YD, Zhao NM, Dominguez B, Lee KF, Gan WB, Zheng H (2005) Defective neuromuscular synapses in mice lacking amyloid precursor protein (APP) and APP-Like protein 2. J Neurosci 25: 1219–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ha Y (2004) The X-ray structure of an antiparallel dimer of the human amyloid precursor protein E2 domain. Mol Cell 15: 343–353 [DOI] [PubMed] [Google Scholar]
- Weidemann A, Konig G, Bunke D, Fischer P, Salbaum JM, Masters CL, Beyreuther K (1989) Identification, biogenesis, and localization of precursors of Alzheimer's disease A4 amyloid protein. Cell 57: 115–126 [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Koo EH, Selkoe DJ (1997) Cell surface amyloid beta-protein precursor colocalizes with beta 1 integrins at substrate contact sites in neural cells. J Neurosci 17: 1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Selkoe DJ, Koo EH (1995) Trafficking of cell surface beta-amyloid precursor protein: retrograde and transcytotic transport in cultured neurons. J Cell Biol 129: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Jiang M, Trumbauer ME, Sirinathsinghji DJ, Hopkins R, Smith DW, Heavens RP, Dawson GR, Boyce S, Conner MW, Stevens KA, Slunt HH, Sisoda SS, Chen HY, Van der Ploeg LH (1995) beta-Amyloid precursor protein-deficient mice show reactive gliosis and decreased locomotor activity. Cell 81: 525–531 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Material and Methods


