Abstract
The only known niche of the human pathogen Helicobacter pylori is the gastric mucosa, where large fluctuations of pH occur, indicating that the bacterial response and resistance to acid are important for successful colonization. One of the few regulatory proteins in the H. pylori genome is a homologue of the ferric uptake regulator (Fur). In most bacteria, the main function of Fur is the regulation of iron homeostasis. However, in Salmonella enterica serovar Typhimurium, Fur also plays an important role in acid resistance. In this study, we determined the role of the H. pylori Fur homologue in acid resistance. Isogenic fur mutants were generated in three H. pylori strains (1061, 26695, and NCTC 11638). At pH 7 there was no difference between the growth rates of mutants and the parent strains. Under acidic conditions, growth of the fur mutants was severely impaired. No differences were observed between the survival of the fur mutant and parent strain 1061 after acid shock. Addition of extra iron or removal of iron from the growth medium did not improve the growth of the fur mutant at acidic pH. This indicates that the phenotype of the fur mutant at low pH was not due to increased iron sensitivity. Transcription of fur was repressed in response to low pH. From this we conclude that Fur is involved in the growth at acidic pH of H. pylori; as such, it is the first regulatory protein implicated in the acid resistance of this important human pathogen.
Helicobacter pylori, a gram-negative microaerophilic bacterium, is a gastric pathogen of humans. Infection always leads to chronic gastritis and may progress to peptic ulceration and gastric atrophy; it is associated with an increased risk for the development of gastric adenocarcinoma and mucosa-associated lymphomas (13). When H. pylori enters the stomach, it migrates from the acidic lumen into the gastric mucus layer, where the pH is thought to vary between 4.0 and 6.5 and where occasional acid shocks as low as pH 2 can occur (9, 25, 30). If left untreated, H. pylori infection usually lasts for decades or more, which indicates that the bacterium is well adapted to these acidic conditions. Survival of severe acid shock has been extensively studied and shown to depend on urease activity (10, 32). In contrast, little is known about the mechanisms that play a role in the growth of H. pylori at acidic pH, although several studies show that H. pylori grows in media with pHs ranging from 4 to 8 (22, 24, 31). During growth at acidic pH, urease-independent acid resistance mechanisms are essential (7), and 10 loci not related to urease are required in this process (8).
Acid resistance is an important trait for many bacteria and is a virulence determinant for various pathogens, like Salmonella enterica serovar Typhimurium, Vibrio cholerae, and Listeria monocytogenes (23, 27). Several (global) regulatory systems play a role in the orchestration of the response to acid stress in these organisms, like the OmpR/EnvZ, PhoP/Q, RpoS, and ferric uptake regulator (Fur) systems (3, 18, 21, 28). Remarkably, in the complete genome sequences of H. pylori strains J99 and 26695, homologues of genes that are involved in the regulation of acid resistance of other bacteria are absent, except for Fur (1, 35). In most organisms, Fur regulates the response to iron limitation (14, 26), but in S. enterica serovar Typhimurium Fur also takes part in the regulation of the acid resistance. The expression of several proteins that are part of the S. enterica serovar Typhimurium acid resistance system depends on Fur, and Fur-deficient mutants fail to mount an adaptive response to low pH and are thus acid sensitive (16, 17). Furthermore, the role of Fur in the acid tolerance response of S. enterica serovar Typhimurium is independent of the intracellular iron content and genetically separable from its role in iron acquisition (20). Although H. pylori Fur has less than 60% amino acid identity to other Fur proteins, it functions as an iron-dependent repressor of H. pylori genes involved in iron metabolism (4-6, 11, 15). The aim of this study was to determine whether the Fur homologue of H. pylori is involved in the acid resistance of this gastric human pathogen.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The H. pylori strains used were strains NCTC 11638 (National Collection of Type Cultures), 26695 (35), 1061 (19), and their isogenic fur mutants. Escherichia coli strain ER1793 was used as host for recombinant plasmids (New England Biolabs Inc., Beverly, Mass.). The plasmids used in this study were pBluescript, pGEM-Teasy (Promega, Madison, Wis.) for cloning PCR products, and pFUR3-CAT (5) for generation of H. pylori mutants of strains 1061 and NCTC 11638. E. coli was cultured either in liquid or on solid Luria-Bertani medium. For selection, E. coli media were supplemented with 20 mg of chloramphenicol per liter.
H. pylori was routinely cultured on Columbia agar (Oxoid, Basinstoke, United Kingdom) supplemented with 7% lysed horse blood and Dent H. pylori selective supplement (Oxoid; Dent plates). Bacteria were grown on plates or in liquid medium for 48 h at 37°C in an O2-CO2-N2 (5:10:85, vol/vol/vol) atmosphere. When appropriate, the Dent plates were supplemented with 20 mg of chloramphenicol per liter (Dent-chlor plates). After sterilization and the addition of blood and supplement to the medium, the pH of these Dent plates was adjusted with HCl where appropriate. The pH of the medium was monitored with a surface pH electrode (model L39A; Schott Geräte, Hofheim, Germany) and did not change more than 0.2 unit during the course of the experiments.
For liquid growth, H. pylori was cultured in brucella broth (Oxoid) supplemented with 3% (for strain 1061 and 26695) or 10% (for strain NCTC11638) newborn calf serum (Gibco, Paisley, Scotland) and Dent supplement. The differences in serum concentrations were necessary to obtain similar growth rates for all strains. The broth was adjusted to the desired pH with HCl after the addition of newborn calf serum and Dent supplement and subsequently was filter sterilized. To ensure that all strains were in the same growth phase, the bacteria were first grown to an optical density at 600 nm (OD600) of approximately 1 in pH 7 broth and subsequently diluted 1:100 into the test media to give a starting OD600 of 0.05. Bacterial growth was monitored by measuring the OD600 for 48 h, after which no increase in OD was observed. The pH of the medium was monitored by pH electrode (Schott Geräte) and did not change more than 0.5 unit during the course of the experiments, except when urea was added to the broth. Iron-restricted conditions were achieved by supplementing the brucella broth with 20 μM desferal (desferrioxamine-B; Sigma, St. Louis, Mo.). Iron-rich conditions were achieved by supplementing the iron-restricted medium with 100 μM FeCl3 (15). The iron content of the brucella broth was determined by inductively coupled plasma mass spectrometry (33).
Construction of fur mutants by allelic replacement.
The fur mutants of strains NCTC 11638 and 1061 were constructed by marker exchange mutagenesis using plasmid pFUR3-CAT (5). For generation of the 26695 fur mutant, a 431-bp fragment of the fur gene of strain 26695 was PCR amplified and cloned into pBluescript. Subsequently, the Campylobacter coli chloramphenicol resistance cassette (37) was inserted into the unique BclI restriction site and introduced into strain 26695 by natural transformation. In all strains the fur gene was successfully replaced, as determined by PCR amplifications performed on DNA isolated from these mutants with primers FurF (5"-GGTTTCTTCTCAAGGCATAG-3") and FurR (5"-GGGTATTTGGGCAAGACTTC-3"), which are located outside the fur gene fragment of pFUR3-CAT, and with outward facing primers (CatL [5"-TAGTGGTCGAAATACTCTTTTCGTG-3"] and CatR [5"-CCCTTATCGATTCAAGTGCATCATG-3"]) located within the chloramphenicol cassette. Mutation of the fur gene in all three strains resulted in the loss of iron regulation of the pfr and frpB1 genes (data not shown), which corresponds to the previously published phenotypes of H. pylori fur mutants (4, 11).
General genetic manipulations.
All standard DNA manipulations were performed as described previously (29). Plasmid DNA was isolated with the Miniprep spin kit (Qiagen Gmbh, Hilden, Germany). All enzymes were from New England Biolabs and were used as specified by the manufacturer.
Determination of urease activity.
The urease activity of fresh lysates was determined by measuring ammonia production from urea hydrolysis with the Berthelot reaction, as described previously (36).
Acid shock experiments.
The acid shock was performed essentially as described previously (10). Bacteria were grown for 48 h, resuspended in phosphate-buffered saline (PBS) (pH 7), and centrifuged for 10 min at 3,000 × g. The pellets were resuspended in 1 ml of PBS, and aliquots of ∼1.25 × 109 bacteria, as determined by measuring the OD600, were resuspended in 5 ml of PBS of different pHs (7, 4.8, and 3.5). The pH of the PBS had been previously adjusted by addition of either HCl or NaOH. The bacteria were incubated for 60 min at 37°C under microaerobic conditions in 15-ml tubes with loose-fitting caps. The cells were then centrifuged at 3,000 × g for 10 min, and the pellet was resuspended in PBS (pH 7.2). Serial dilutions were made, plated on pH 7 Dent plates, and incubated for 5 days at 37°C under microaerobic conditions, and the CFU per milliliter were counted.
RNA analysis.
Total RNA was isolated from 4 × 109 bacteria grown for 24 h on RNeasy spin columns (Qiagen), as specified by the manufacturer. RNA samples were normalized to 16S and 23S rRNA band intensity. RNA was separated on 2% formaldehyde-1.5% agarose gels in 20 mM sodium phosphate buffer (pH 7), transferred to positively charged nylon membranes (Roche Diagnostics, Hoffmann-LaRoche, Basel, Switzerland), and covalently bound to these membranes with a UV cross-linker (Merck, Darmstadt, Germany). Transfer of RNA was confirmed by methylene blue staining, and the sizes of hybridizing RNA species were calculated from comparisons with a digoxigenin DIG-labeled RNA marker (RNA marker I; Roche Diagnostics). A fur-specific probe was amplified with primers FurF2 (5"-TCCATTTTAGAGCGCTTGAG-3") and Fur-R-T7 (5"-ctaatacgactcactatagggagaTTAACGACTTCATTCTGGCGGTT-3"). The resulting PCR fragment contained a T7 promoter sequence (indicated in lowercase type) and was used for the creation of an antisense RNA probe labeled with DIG by in vitro transcription with T7 RNA polymerase (Roche Diagnostics). Northern hybridization and stringency washes were done at 68°C. The hybridized RNA probe was detected by using the DIG-labeling and detection kit (Roche Diagnostics) and visualized using the chemiluminescent substrate CPD-Star (Amersham Pharmacia, Uppsala, Sweden).
Quantification of the chloramphenicol acyltransferase reporter protein.
The production of Cat reporter protein by the fur mutant of strain 1061 was measured with the Cat-ELISA system (Roche Diagnostics) (4). Bacteria were grown at pH 7 and 6 and were harvested by centrifugation. Lysis and determination of the Cat protein were performed with the Cat-ELISA system. The amount of Cat was calculated from a standard curve prepared with purified Cat protein from E. coli and standardized to the total protein content.
RESULTS
H. pylori fur mutants are not affected in their ability to survive acid shock.
To determine whether mutation of fur affects survival of acid shock, the 1061 fur mutant and the parent strain were submitted to acid shocks of pH 3.5 and 4.8 and to pH 7. The fur mutant and the parent strain did not show significant differences in survival of acid shocks at pH 3.5 and 4.8 (results not shown). This indicates that mutation of fur does not affect the ability of H. pylori to survive acid shocks.
H. pylori fur mutants are affected in their ability to grow at acidic pH.
To determine whether the inactivation of the fur gene affected the growth of H. pylori under acidic conditions, the H. pylori 1061 wild-type and fur mutant strains were grown on plates with different pHs. On pH 7 plates the fur mutant and its wild-type strain gave similar colony sizes. The fur mutant grew on pH 4.8, 5.0, 5.3, and 5.5 plates, but their colonies were smaller than those of the wild-type strain, even after more than 5 days of incubation. This difference between wild-type and fur mutant was most pronounced at pH 5.3.
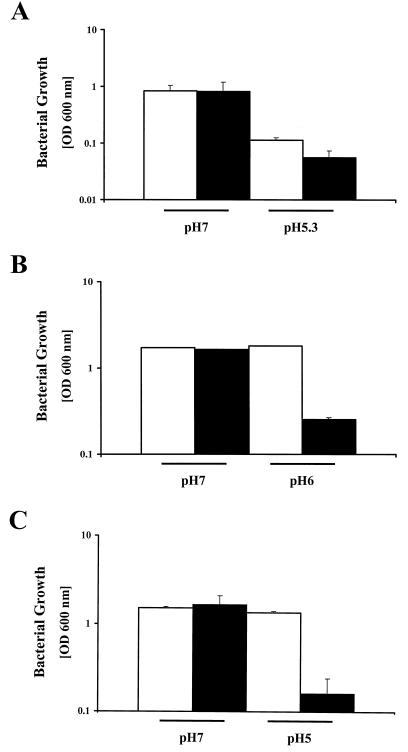
To quantify the growth defect of the fur mutant at low pH, strain 1061 and the 1061 fur mutant were grown in brucella media with pHs of 5.3 and 7 until the stationary phase was reached (after 48 h). The parent strain and the fur mutant have similar growth rates at pH 7 (Fig. 1A), but growth of the fur mutant was impaired at pH 5.3 (Fig. 1A) compared to the parent strain.
FIG. 1.
H. pylori fur mutants are defective in growth under acidic conditions. (A) Growth of H. pylori 1061 (white bars) and its fur mutant (black bars) after 48 h (stationary phase) in broth adjusted to pH 7 or 5.3. Note the logarithmic scale of the y axis. Results are the means of three independent experiments and standard errors of the mean. At pH 5.3, the difference between the parent strain and the mutant was significant, as determined by Student's t test (P < 0.05). (B) Growth of H. pylori NCTC 11638 (white bars) and its fur mutant (black bars) after 48 h (stationary phase) in broth adjusted to pH 7 or 6. Neither the parent strain nor the fur mutant grew at pH 5.3 (data not shown). Results are the means of three independent experiments and standard errors of the mean; due to the small standard errors, most of the error bars are not visible. At pH 6, the difference between the parent strain and mutant was significant (P < 0.05) as determined by Student's t test. (C) Growth of H. pylori 26695 (white bars) and its fur mutant (black bars) after 48 h (stationary phase) at pH 7 or 5. This experiment was repeated five times. Large experimental variation occurred; therefore, the results shown are a representative example of the means and standard errors of the mean of two independent experiments performed at the same day.
Since H. pylori strains are genetically diverse, we tested whether the fur mutants of strain 26695 and NCTC 11638 showed the same impairment in growth at acidic pH. NCTC 11638 is acid sensitive, and neither the parent strain nor the fur mutant grew at pH 5.3; therefore, the growth experiments were performed at pH 6 and 7 (Fig. 1B). Growth of the NCTC 11638 fur mutant was significantly reduced at pH 6 compared to that of the NCTC 11638 wild-type strain. Experiments with H. pylori strain 26695 were performed at pH 5, since the growth difference between the wild-type and fur mutant strains was most pronounced at this pH. Again, growth of the 26695 fur mutant was significantly reduced at pH 5 compared to the 26695 wild-type strain (Fig. 1C).
Mutation of fur does not affect urease-dependent acid resistance.
Urease activity has previously been shown to be important for survival of acid shocks and for growth at low pH (10, 31, 32). The urease activities of H. pylori 1061 and its fur mutant were 6.4 ± 2.1 and 8.4 ± 2.9 μmol of urea/min/mg of protein, and this difference was not significant (P = 0.15, n = 4; Student's t test). This excludes the possibility that the growth defect of the fur mutants at acidic pH is due to reduced urease activity.
To investigate whether urease was still fully functional at pH 5.3 in the fur mutant, the 1061 fur mutant and its parent strain were grown in broth at pH 7 and 5.3 supplemented with 5 mM urea. Growth of the fur mutant and the parent strain were identical at both pHs (data not shown).
Iron toxicity is not the cause of the growth defect of the fur mutant at pH 5.
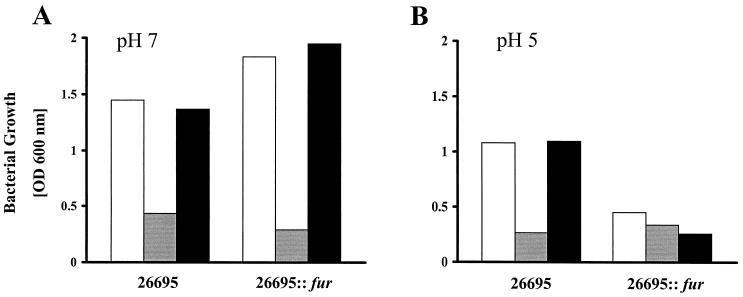
The total iron concentrations in the brucella media at pH 7 and 5.3 were similar (13.9 and 15 μM, respectively), which is far below the toxic level (>1 mM FeCl3) for H. pylori and even for a fur mutant (4). However, iron is more soluble at low pH, and mutation of fur in H. pylori results in an increase of iron uptake; therefore the amount of iron in the growth medium could be toxic under acidic conditions. Therefore, the growth defect of the fur mutants at low pH might be caused by iron toxicity instead of proton toxicity. To test this hypothesis, strain 26695 and its isogenic fur mutant were grown at pH 5 and 7 in medium supplemented with 20 μM desferal (iron restricted) and iron-restricted medium supplemented with 100 μM FeCl3 (iron rich). Changing the iron availability did not affect the acid sensitivity of the fur mutant at pH 5 or the growth at pH 7, which indicates that the acid-sensitive phenotype of the fur mutant in the growth medium we used is not caused by a defect in iron homeostasis (Fig. 2).
FIG. 2.
Influence of iron on fur-mediated acid sensitivity. The parent strain 26695 and its fur mutant were grown at pH 7 (A) or pH 5 (B) for 48 h in regular medium (white bars) or in iron-restricted (20 μM desferal; gray bars) or iron-rich (100 μM FeCl3; black bars) medium. The results shown are a representative example of the means of two independent experiments.
Transcription of fur is repressed under acidic conditions.
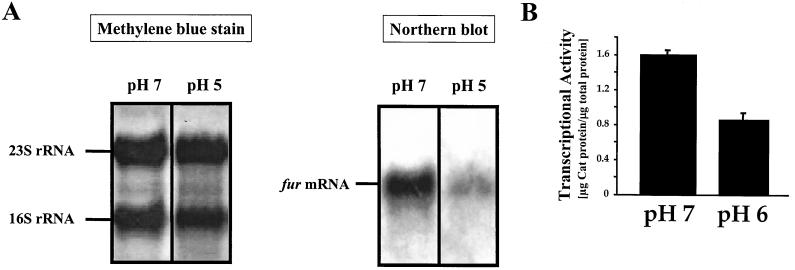
To investigate whether the function of Fur in acid resistance of H. pylori is mediated by a change in the expression of fur under different pH conditions, total RNA of the parent strain 1061 grown at pH 7 and 5.3 for 24 h was isolated and hybridized with a fur-specific probe. This analysis revealed that the fur gene is transcribed as a monocistronic unit and that transcription of fur was reduced at pH 5.3 (Fig. 3A). Similar effects were observed with fur in strains 26695 and NCTC 11638 (data not shown). Repression of fur under acidic conditions was independently confirmed by quantitating the levels of the Cat reporter protein in the 1061 fur mutant grown at pH 6 and 7 (Fig. 3B). To exclude the possibility that the observed reduction of fur transcription was the result of a general repression of transcription due to growth at acidic pH, we hybridized the same RNA samples with a probe specific for the catalase gene katA. Transcription of katA was not altered in response to pH (data not shown). Therefore, the environmental pH clearly affects the transcription of fur.
FIG. 3.
Transcriptional analysis of the fur gene under acidic conditions. (A) The left panel shows a methylene blue stain of transferred RNA prior to hybridization. The positions of 16S and 23S RNA are indicated. The right panel shows total RNA isolated from strain 1061 grown at pH 7 or 5.3 and analyzed by Northern hybridization with DIG-labeled fur antisense RNA. The band corresponding to the 0.5-kb fur mRNA is indicated. The experiment was repeated three times, with similar results. (B) Expression of the fur::cat reporter gene fusion under acidic conditions. The fur mutant of H. pylori strain 1061, where the promoterless cat gene is transcriptionally fused to the fur gene, was grown at pH 7 or 6, and the expression of the Cat protein was assayed by a Cat-specific ELISA. Results shown are the means of four experiments and standard errors of the mean.
DISCUSSION
Fur is a global regulator of the response to iron in many bacteria (14, 26). The genome of the gastric pathogen H. pylori contains a Fur homologue, whose function in several aspects of iron homeostasis has been the subject of several recent studies (4, 11, 15). As in other organisms, H. pylori Fur functions as an iron-responsive regulator of iron homeostasis (4, 11) but also in the nickel-responsive induction of urease expression (36). In this study we show that Fur also plays an important role in urease-independent growth at acidic pH, which, to our knowledge, is the first report of a regulator being involved in acid resistance of H. pylori. At acidic pH, the growth of H. pylori fur mutants in three different strain backgrounds was significantly impaired, even though the strains differed in their respective sensitivities to acidic pH. Furthermore, the use of pH 7 precultures to start the different pH cultures ensured that the wild-type and fur mutant strains were all in the same growth phase, which excludes the possibility that impaired growth at low pH was due to differences in growth phase.
The H. pylori 1061 and NCTC 11638 fur mutants contained a fur gene that was inactivated by a chloramphenicol cassette (cat) that lacks promoter and terminator sequences (37), making polar effects highly unlikely, although they cannot be excluded. However, it is unlikely that the insertion of cat affected the genes downstream of fur since they are in the opposite orientation. In addition, the genes directly flanking fur, encoding HP1026 (JHP0396), a conserved hypothetical helicase-like protein, and HP1028 (JHP0398), a hypothetical protein, are not expected to function in either iron metabolism or acid resistance (1, 35).
In H. pylori, Fur has been implicated in the regulation of urease expression (36). Since urease plays an important role in acid resistance of H. pylori (10, 31), the acid sensitivity of the fur mutant might be due to reduced urease activity. However, the urease activities of wild-type and fur mutant H. pylori are comparable, and addition of urea to the pH 5 growth medium restores growth of the fur mutant to levels similar to that of the wild-type strain. This indicates that the role of Fur in acid resistance of H. pylori is thus independent of urease and is mediated through a yet unknown mechanism.
Iron is more soluble under acidic conditions than under alkaline conditions. In addition, fur mutation derepresses iron uptake genes, which could result in an increased iron uptake by H. pylori fur mutants. Therefore, the growth defect that we observed at acidic pH in the fur mutant could also be due to iron toxicity. However, when we added an excess of iron to the parent strain at pH 5, this did not have a negative effect on its growth. Also, growth of the fur mutant under iron-restricted conditions did not abolish the growth defect, nor did an excess of iron abolish the growth at acidic pH. These results indicate that the role of Fur in the acid resistance of H. pylori is independent of its role in iron acquisition.
Interestingly, transcription of fur is repressed under acidic conditions. This repression was also apparent when fur transcription was determined by measuring the Cat production from the inserted promoterless cat cassette, indicating that it is not an effect of Fur autoregulation. This means that the expression of Fur is (partly) regulated by other proteins. The regulatory properties of Fur are based on the higher affinity for DNA of Fur containing iron than of Fur without iron. In H. pylori, fur transcription is regulated not only by pH but also by iron (34); this indicates that apart from the amount of free iron present in the cell, Fur-mediated regulation is controlled by changing the transcription of fur in H. pylori. It is unlikely that the pH-dependent repression of fur transcription is due to a general repression of transcription at acidic pH, since transcription of catalase was not altered at acidic pH. Furthermore, in studies using microarray hybridization, transcription of several H. pylori genes was induced under acidic conditions (2, 12). In both studies, the repression of fur transcription at low pH was not observed. This could be due to the use of solid media in those studies, whereas in this study we have used broth-grown bacteria; it could also be because fur transcription levels are near detection limits. Interestingly, transcription of HP1562 (ceuE), encoded by an open reading frame putatively involved in iron uptake, is induced under acidic conditions (2). Taken together, these results indicate that the relation between iron and pH homeostasis in H. pylori is more complex than was previously thought and that H. pylori is well adapted to the high bioavailability of iron that is presumed to exist in the acidic gastric environment in which that this bacterium thrives.
Acknowledgments
We thank Frank Fassbinder for his valuable contribution to construction of fur mutants and E. Schmuck for performing the ICP at the Chemische Landesuntersuchungsanstalt Freiburg.
Part of this study was financially supported by Netherlands Organization for Scientific Research (NWO 901-14-206) to A.H.M.V.V., the Deutsche Forschungsgemeinschaft (DFG, Ki201/8-2) to M.K., and the Wellcome Trust to D.J.K.
Editor: J. T. Barbieri
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Ang, S., C. Z. Lee, K. Peck, M. Sindici, U. Matrubutham, M. A. Gleeson, and J. T. Wang. 2001. Acid-induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect. Immun. 69:1679-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bearson, S., B. Bearson, and J. W. Foster. 1997. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 147:173-180. [DOI] [PubMed] [Google Scholar]
- 4.Bereswill, S., S. Greiner, A. H. M. van Vliet, B. Waidner, F. Fassbinder, E. Schiltz, J. G. Kusters, and M. Kist. 2000. Regulation of ferritin-mediated cytoplasmic iron storage by the ferric uptake regulator homolog (Fur) of Helicobacter pylori. J. Bacteriol. 182:5948-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bereswill, S., F. Lichte, S. Greiner, B. Waidner, F. Fassbinder, and M. Kist. 1999. The ferric uptake regulator (Fur) homologue of Helicobacter pylori: functional analysis of the coding gene and controlled production of the recombinant protein in Escherichia coli. Med. Microbiol. Immunol. 188:31-40. [DOI] [PubMed] [Google Scholar]
- 6.Bereswill, S., F. Lichte, T. Vey, F. Fassbinder, and M. Kist. 1998. Cloning and characterization of the fur gene from Helicobacter pylori. FEMS Microbiol. Lett. 159:193-200. [DOI] [PubMed] [Google Scholar]
- 7.Bijlsma, J. J. E., M. M. Gerrits, R. Imamdi, C. M. J. E. Vandenbroucke-Grauls, and J. G. Kusters. 1998. Urease-positive, acid-sensitive mutants of Helicobacter pylori: urease-independent acid resistance involved in growth at low pH. FEMS Microbiol. Lett. 167:309-313. [DOI] [PubMed] [Google Scholar]
- 8.Bijlsma, J. J. E., A. L. Lie, I. C. Nootenboom, C. M. J. E. Vandenbroucke-Grauls, and J. G. Kusters. 2000. Identification of loci essential for the growth of Helicobacter pylori under acidic conditions. J. Infect. Dis. 182:1566-1569. [DOI] [PubMed] [Google Scholar]
- 9.Chu, S., S. Tanaka, J. D. Kaunitz, and M. H. Montrose. 1999. Dynamic regulation of gastric surface pH by luminal pH. J. Clin. Investig. 103:605-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clyne, M., A. Labigne, and B. Drumm. 1995. Helicobacter pylori requires an acidic environment to survive in the presence of urea. Infect. Immun. 63:1669-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delany, I., A. B. Pacheco, G. Spohn, R. Rappuoli, and V. Scarlato. 2001. Iron-dependent transcription of the frpB gene of Helicobacter pylori is controlled by the Fur repressor protein. J. Bacteriol. 183:4932-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong, Q., D. Hyde, C. Herra, C. Kean, P. Murphy, C. A. O'Morain, and M. Buckley. 2001. Identification of genes regulated by prolonged acid exposure in Helicobacter pylori. FEMS Microbiol. Lett. 196:245-249. [DOI] [PubMed] [Google Scholar]
- 13.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fassbinder, F., A. H. M. Van Vliet, V. Gimmel, J. G. Kusters, M. Kist, and S. Bereswill. 2000. Identification of iron-regulated genes of Helicobacter pylori by a modified fur titration assay (FURTA-Hp). FEMS Microbiol. Lett. 184:225-229. [DOI] [PubMed] [Google Scholar]
- 16.Foster, J. W. 1991. Salmonella acid shock proteins are required for the adaptive acid tolerance response. J. Bacteriol. 173:6896-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster, J. W., and H. K. Hall. 1992. Effect of Salmonella typhimurium ferric uptake regulator (fur) mutations on iron- and pH-regulated protein synthesis. J. Bacteriol. 174:4317-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster, J. W., Y. K. Park, I. S. Bang, K. Karem, H. Betts, H. K. Hall, and E. Shaw. 1994. Regulatory circuits involved with pH-regulated gene expression in Salmonella typhimurium. Microbiology 140:341-352. [DOI] [PubMed] [Google Scholar]
- 19.Goodwin, A., D. Kersulyte, G. Sisson, v. Z. Veldhuyzen, D. E. Berg, and P. S. Hoffman. 1998. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 28:383-393. [DOI] [PubMed] [Google Scholar]
- 20.Hall, H. K., and J. W. Foster. 1996. The role of fur in the acid tolerance response of Salmonella typhimurium is physiologically and genetically separable from its role in iron acquisition. J. Bacteriol. 178:5683-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall, H. K., K. L. Karem, and J. W. Foster. 1995. Molecular responses of microbes to environmental pH stress. Adv. Microb. Physiol. 37:229-272. [DOI] [PubMed] [Google Scholar]
- 22.Kangatharalingam, N., and P. S. Amy. 1994. Helicobacter pylori comb. nov. exhibits facultative acidophilism and obligate microaerophilism. Appl. Environ. Microbiol. 60:2176-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merrell, D. S., and A. Camilli. 1999. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol. Microbiol. 34:836-849. [DOI] [PubMed] [Google Scholar]
- 24.Morgan, D. R., R. Freedman, C. E. Depew, and W. G. Kraft. 1987. Growth of Campylobacter pylori in liquid media. J. Clin. Microbiol. 25:2123-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quigley, E. M., and L. A. Turnberg. 1987. pH of the microclimate lining human gastric and duodenal mucosa in vivo. Studies in control subjects and in duodenal ulcer patients. Gastroenterology 92:1876-1884. [DOI] [PubMed] [Google Scholar]
- 26.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 27.Riesenberg-Wilmes, M. R., B. Bearson, J. W. Foster, and R. Curtis. 1996. Role of the acid tolerance response in virulence of Salmonella typhimurium. Infect. Immun. 64:1085-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowbury, R. J. 1997. Regulatory components, including integration host factor, CysB and H-NS, that influence pH responses in Escherichia coli. Lett. Appl. Microbiol. 24:319-328. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Schade, C., G. Flemström, and L. Holm. 1994. Hydrogen ion concentration in the mucus layer on top of acid-stimulated and -inhibited rat gastric mucosa. Gastroenterology 107:180-188. [DOI] [PubMed] [Google Scholar]
- 31.Scott, D. R., D. Weeks, C. Hong, S. Postius, K. Melchers, and G. Sachs. 1998. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology 114:58-70. [DOI] [PubMed] [Google Scholar]
- 32.Stingl, K., E. M. Uhlemann Em, G. Deckers-Hebestreit, R. Schmid, E. P. Bakker, and K. Altendorf. 2001. Prolonged survival and cytoplasmic pH homeostasis of Helicobacter pylori at pH 1. Infect. Immun. 69:1178-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutton, K. L., and J. A. Caruso. 1999. Liquid chromatography-inductively coupled plasma mass spectrometry. J. Chromatogr. Ser. A 856:243-258. [DOI] [PubMed] [Google Scholar]
- 34.Szczebara, F., L. Dhaenens, S. Armand, and M. O. Husson. 1999. Regulation of the transcription of genes encoding different virulence factors in Helicobacter pylori by free iron. FEMS Microbiol. Lett. 175:165-170. [DOI] [PubMed] [Google Scholar]
- 35.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J.Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 36.Van Vliet, A. H. M., E. J. Kuipers, B. Waidner, B. J. Davies, N. de Vries, C. W. Penn, C. M. J. E. Vandenbroucke-Grauls, M. Kist, S. Bereswill, and J. G. Kusters. 2001. Nickel-responsive induction of urease expression in Helicobacter pylori is mediated at the transcriptional level. Infect. Immun. 69:4891-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, Y., and D. E. Taylor. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94:23-28. [DOI] [PubMed] [Google Scholar]