Abstract
Genetic studies in yeast have shown that the translation initiation factor eIF5 plays an important role in the selection of the AUG start codon. In order to ensure translation fidelity, the hydrolysis of GTP bound to the 40S preinitiation complex (40S·Met-tRNAi·eIF2·GTP), promoted by eIF5, must occur only when the complex has selected the AUG start codon. However, the mechanism that prevents the eIF5-promoted GTP hydrolysis, prior to AUG selection by the ribosomal machinery, is not known. In this work, we show that the presence of initiation factors eIF1, eIF1A and eIF3 in the 40S preinitiation complex (40S·eIF1·eIF1A·eIF3·Met-tRNAi·eIF2·GTP) and the subsequent binding of the preinitiation complex to eIF4F bound at the 5′-cap structure of mRNA are necessary for preventing eIF5-promoted hydrolysis of GTP in the 40S preinitiation complex. This block in GTP hydrolysis is released upon AUG selection by the 40S preinitiation complex. These results, taken together, demonstrate the biochemical requirements for regulation of GTP hydrolysis and its coupling to the AUG selection process during translation initiation.
Keywords: GTP hydrolysis, ribosomes, translation initiation, translation initiation factor eIF5
Introduction
The translation initiation process is mediated by a series of partial reactions each of which requires the participation of a large number of protein factors collectively called eukaryotic translation initiation factors (eIFs). According to the currently accepted view of translation initiation, an obligatory intermediate step involves the binding of the initiator Met-tRNA, as the Met-tRNAi·eIF2·GTP ternary complex, to a 40S ribosomal subunit containing bound initiation factor eIF3. This interaction leads to the formation of a 40S preinitiation complex (40S·eIF3·Met-tRNAi·eIF2·GTP), which is then recruited to the 5′-capped end of the mRNA via protein–protein interaction between the cap-bound initiation factor eIF4F and the 40S complex-bound eIF3. The 40S complex then scans the mRNA in a 5′ → 3′ direction until it is positioned at the initiating AUG codon where it forms the 40S initiation complex (40S·eIF3·mRNA·Met-tRNAi·eIF2·GTP) (also called 48S initiation complex). Subsequently, another initiation factor, eIF5, promotes hydrolysis of the GTP bound to eIF2 in the 40S initiation complex. This leads to release of the inactive eIF2·GDP (and possibly of other bound initiation factors) from the 40S complex, thereby allowing the eIF5B-mediated joining of the 60S ribosomal subunit to the 40S complex to form an 80S initiation complex (80S·Met-tRNAi·mRNA) that is competent to form the first peptide bond (Hershey and Merrick, 2000; Dever, 2002; Kapp and Lorsch, 2004).
Detailed characterization of the action of eIF5 has shown that in the translation initiation pathway, eIF5 acts as a GTPase activating protein (GAP) (Das and Maitra, 2001; Das et al, 2001). eIF5, by itself, does not hydrolyze either free GTP or GTP bound to free Met-tRNAi·eIF2·GTP ternary complex. Rather, eIF5-promoted hydrolysis of GTP occurs only when the ternary complex is bound to the 40S initiation complex (Chakrabarti and Maitra, 1991; Chaudhuri et al, 1994). Additionally, as with typical GAPs, eIF5 forms a complex with eIF2, the GTP-binding protein (Chaudhuri et al, 1994). This interaction is essential for eIF5 to function as a GAP, thereby activating the intrinsic GTPase activity of eIF2 (Das et al, 1997; Asano et al, 1999; Das and Maitra, 2000).
Hydrolysis of eIF2-bound GTP is a critical step in maintaining the overall fidelity of translation initiation in eukaryotes. Elegant genetic experiments in yeast have suggested that GTP hydrolysis should occur only after the 40S ribosomal machinery has selected the AUG start codon. Indeed, mutations in either eIF2 or eIF5 that result in premature GTP hydrolysis cause translation to be initiated from a non-AUG codon (Donahue, 2000). Biochemical assays, however, show that selection of the AUG codon by the 40S preinitiation complex, per se, is not a prerequisite for eIF5-promoted GTP hydrolysis. We have previously demonstrated that a 40S preinitiation complex (40S·Met-tRNAi·eIF2·GTP) can be formed in the presence of the initiation factor eIF1A alone and in the absence of an AUG codon (Chaudhuri et al, 1997b, 1999). We now observe that although such a ‘minimal' 40S preinitiation complex is an efficient substrate for eIF5-mediated GTP hydrolysis (see Figure 1), the resulting 80S complex formed is an ‘abortive' complex that is inactive in peptidyl transfer (see Figure 1). Additionally, biochemical studies in yeast cell-free extracts, free of 40S ribosomes, show that eIF5 is part of a multifactor complex consisting of eIF3, eIF1, eIF2 and stoichiometric amounts of tRNAimet (Asano et al, 2000). This implies that eIF5 is recruited to 40S ribosomes at an earlier stage of initiation prior to mRNA binding.
Figure 1.
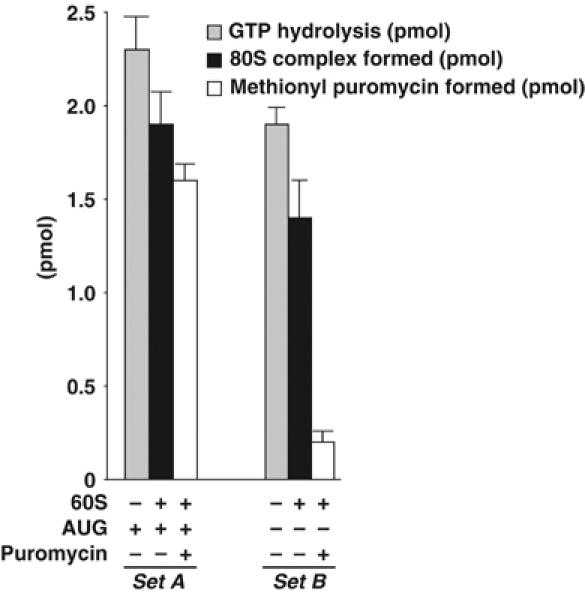
Methionyl puromycin reactivity of 40S complexes. The [3H]Met-tRNAi·eIF2·[γ-32P]GTP ternary complex was formed in reaction mixtures similar to those described in the section Isolation of the 40S preinitiation and initiation complexes. Two sets (sets A and B) of reaction mixtures (each set containing three separate reactions) were prepared. In each reaction, 3 pmol of the ternary complex was incubated with eIF1A (32 pmol) and 0.3A260 unit of 40S ribosomal subunits either in the presence (Set A) or absence (Set B) of an AUG triplet to form the 40S initiation complex (2.5 pmol) or the 40S preinitiation complex (2 pmol), respectively, as described previously (Chaudhuri et al, 1997b). Subsequently, reaction mixtures in each set were treated as follows. eIF5 (0.5 pmol) was added to one reaction mixture and hydrolysis of GTP was measured directly by 32Pi release (▒) as described in the section eIF5-mediated GTPase assay. In the second reaction, the Mg2+ concentration was raised to 5 mM, followed by the addition of eIF5 (0.5 pmol) and 60S ribosomal subunits, and analyzed through sucrose gradient centrifugation to determine the amount of 80S complex formed (▪). The third reaction of each set was treated with puromycin (1 mM final concentration) following 80S complex formation and assayed for methionyl puromycin (□), as described previously (Chakrabarti and Maitra, 1991). For each reaction, a parallel control reaction was carried out in which eIF5 was omitted and the values obtained (<0.1 pmol of either 32P released or 80S complex or methionyl puromycin formed) were subtracted.
Taken together, these genetic and biochemical studies indicate that eIF5-mediated GTP hydrolysis should be tightly regulated during the process of translation initiation. Since all the components required for GTP hydrolysis, namely eIF2, eIF5 and the 40S ribosome, are present in the 40S preinitiation complex, there must exist a mechanism by which premature GTP hydrolysis and therefore aberrant initiation is prevented prior to start codon selection by the 40S ribosomal machinery.
In this study, we have addressed the mechanism by which eIF5-promoted GTP hydrolysis is regulated during translation initiation. Specifically, we have investigated the in vitro requirements for the coupling of the two processes, namely AUG selection by the 40S preinitiation complex and eIF5-promoted GTP hydrolysis, using purified mammalian translation initiation factors. We show that when a 40S preinitiation complex is formed, containing bound initiation factors eIF1, eIF1A, eIF3, and cap-bound eIF4F, eIF5-promoted GTP hydrolysis is nearly completely prevented. In contrast, scanning of the mRNA by the 40S preinitiation complex and positioning of the complex at the AUG codon in the presence of the factors eIF4A, eIF4B and ATP restores eIF5-promoted GTP hydrolysis.
Results
Effect of AUG trinucleotide codon on eIF5-promoted GTP hydrolysis
We have previously shown that formation of the 40S initiation complex (recognition of the AUG codon by the 40S preinitiation machinery) is preceded by the initial transfer of Met-tRNAi·eIF2·GTP ternary complex to the 40S subunit in a reaction catalyzed by eIF1A (Chaudhuri et al, 1997b). This 40S initiation complex served as an efficient substrate for eIF5-promoted hydrolysis of GTP. Subsequent work showed that eIF1A could also carry out the same transfer of the ternary complex to 40S subunits in the absence of AUG triplet leading to the formation of a 40S preinitiation complex (Chaudhuri et al, 1999). Since all the components required for eIF5-promoted GTP hydrolysis are present in the 40S preinitiation complex, we investigated whether such an isolated 40S preinitiation complex is a substrate for eIF5-promoted GTP hydrolysis in vitro. We observed that GTP bound to eIF2, both in the 40S preinitiation complex (no AUG present) and the 40S initiation complex (AUG present), was nearly completely hydrolyzed by the addition of eIF5 (Figure 1). In agreement with previous reports published from this laboratory (Chakrabarti and Maitra, 1991), eIF5 acts catalytically in this reaction. Addition of 0.5 pmol of eIF5 can promote the hydrolysis of 2–2.5 pmol of GTP bound in the 40S complex (Figure 1). Addition of 60S ribosomal subunits to each reaction led to the formation of an 80S complex (Figure 1). It should be noted that in such an AUG-dependent ‘minimal' system, eIF5B is not required for 60S subunit joining following eIF5-promoted GTP hydrolysis (Raychaudhuri et al, 1987; Pestova et al, 2000). We observed that the 80S complex formed from the 40S preinitiation complex (in the absence of AUG) was unreactive to puromycin, indicating formation of a nonfunctional (abortive) 80S complex. In contrast, the 80S initiation complex formed from the 40S initiation complex in the presence of AUG was completely puromycin reactive, as expected, indicating formation of an elongation-competent 80S initiation complex. Clearly, therefore, for ‘productive' translation initiation, eIF5 must be prevented from promoting hydrolysis of GTP prior to binding of the 40S preinitiation complex at the AUG codon. This can be achieved if other components involved in the formation of 40S preinitiation complex somehow physically prevent eIF5 from promoting GTP hydrolysis prior to recognition of the AUG codon by the 40S preinitiation complex.
Binding of initiation factors eIF1, eIF1A, eIF3 and eIF4F to the 40S preinitiation complex
We have previously demonstrated that three initiation factors eIF1, eIF1A and eIF3 were together required for the quantitative transfer of Met-tRNAi (as Met-tRNAi·eIF2·GTP ternary complex) to form a stable 40S preinitiation complex. Under these conditions, all three factors remain stably associated with the resulting 40S preinitiation complex (Majumdar et al, 2003). Since the 40S preinitiation complex is recruited to the 5′-capped end of an mRNA via interaction with the cap-binding initiation factor eIF4F, it was of interest to investigate whether eIF4F could associate with the in vitro-synthesized 40S preinitiation complex. Experiments were carried out in which a 40S preinitiation complex was formed by incubating [3H]Met-tRNAi·eIF2·GMPPNP ternary complex with 40S ribosomal subunits in the presence of purified eIF1, eIF1A, eIF3 and eIF4F and the products were subjected to sucrose density gradient centrifugation. As shown in Figure 2C and D, a fraction of eIF4F (measured by Western blot analysis using antibodies to the eIF4E and eIF4G subunits of eIF4F) cosedimented with Met-tRNAi bound to the 40S particle. In accord with our previous observations (Majumdar et al, 2003), the other initiation factors, eIF1 (Figure 2A), eIF1A (Figure 2E) and eIF3 (Figure 2F), also cosedimented with the 40S complex. The binding of these factors to the 40S preinitiation complex was specific since no initiation factors were detected in the 40S region when 40S ribosomal subunits were omitted from the reactions (Figure 2, right panel).
Figure 2.
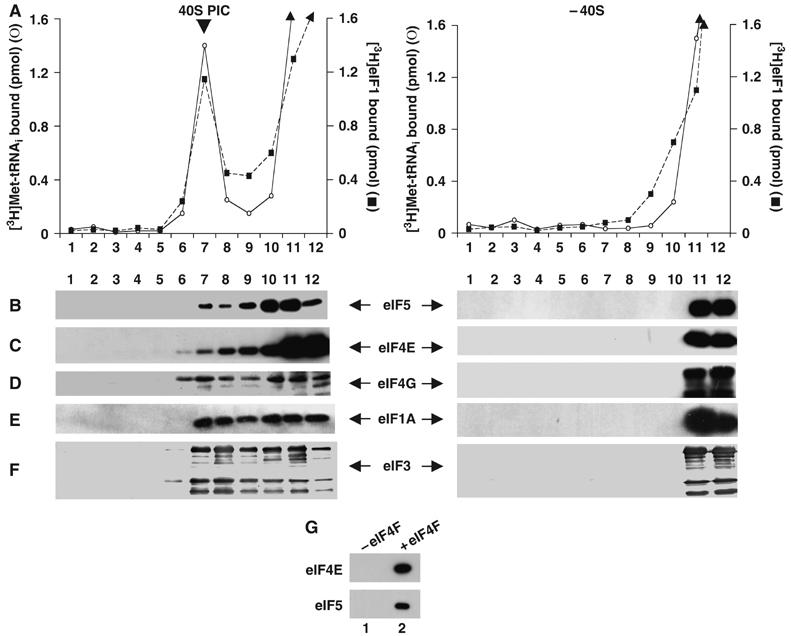
Binding of eIF4F and eIF5 to the 40S preinitiation complex. A 40S preinitiation complex was formed as described in Materials and methods in the presence of initiation factors eIF1A (16 pmol), eIF1 (16 pmol), eIF3 (10 pmol) and eIF4F (15 pmol) and [3H]Met-tRNAi. The 40S preinitiation complex was then incubated with recombinant eIF5 (5 pmol) at 25°C for 10 min and freed of unreacted components by sedimentation through sucrose density gradients. The sucrose gradient fractions (250 μl each) were subjected to TCA precipitation, followed by SDS–PAGE and Western blotting using antibodies specific for eIF5 (B), eIF4E (C), eIF4G (D), eIF1A (E) and eIF3 (F). Aliquots (10 μl) from each fraction were also monitored for 3H radioactivity prior to TCA precipitation to determine the position of the 40S preinitiation complex in the gradient (A). The binding of eIF1 (A) was monitored in a parallel reaction in which the 40S preinitiation complex was formed with unlabeled Met-tRNAi and with [3H]eIF1 instead of unlabeled eIF1 (Majumdar et al, 2003). Control reactions (right panel) in which 40S subunits were omitted were also analyzed. Under these conditions, none of the initiation factors were detected in the 40S region by Western blotting. It should be noted that the lower migrating bands observed in panel D are presumably due to degradation of eIF4G. In panel G, a 40S preinitiation complex was formed as described above, either in the absence (lane 1) or presence (lane 2) of eIF4F and purified by sucrose gradient centrifugation. Isolated complexes were incubated with m7GTP-Sepharose beads at 4°C for 1 h. The washed beads were analyzed by SDS–PAGE, followed by Western blotting using antibodies specific for eIF4E and eIF5.
Effect of initiation factors on eIF5-promoted GTP hydrolysis
We next investigated whether the association of the initiation factors with the 40S preinitiation complex affected the eIF5-mediated hydrolysis of bound GTP. An in vitro assay was carried out in which a preformed ternary complex, containing bound [γ-32P]GTP, was incubated with purified 40S ribosomal subunits and various combinations of the initiation factors eIF1, eIF1A, eIF3 and eIF4F. The 40S preinitiation complex formed in each case was purified through sucrose density gradients. Approximately 1 pmol of the purified 40S preinitiation complex, containing bound [γ-32P]GTP, was then incubated with purified recombinant eIF5 and the level of 32Pi released was measured (Figure 3). Under the conditions used, we observed that eIF5 promoted quantitative hydrolysis of the GTP bound to the 40S preinitiation complex formed with eIF1 and eIF1A (Figure 3, lane a). This indicates that eIF1 and eIF1A together cannot prevent premature hydrolysis of the 40S-bound GTP (the extent of GTP hydrolysis observed with the 40S preinitiation complex formed with eIF1 and eIF1A was taken as 100% and was used as a control in subsequent GTP hydrolysis reactions). However, the 40S preinitiation complex formed in the presence of either eIF3 alone (Figure 3, lane b) or eIF3 in combination with either eIF1A (Figure 3, lane c) or eIF1 (Figure 3, lane d), reduced the subsequent eIF5-mediated GTP hydrolysis by 40–50%. Preinitiation complexes formed in the presence of all three factors eIF3, eIF1 and eIF1A also prevented eIF5-promoted GTP hydrolysis by ∼50% (Figure 3, lane e). These results indicate that eIF3, which plays a key role in stabilizing the 40S preinitiation complex (Chaudhuri et al, 1999; Majumdar et al, 2003), may also regulate eIF5-mediated hydrolysis of GTP in the 40S preinitiation complex. It should be noted that although eIF1 has been shown to play an important role in AUG selection (Unbehaun et al, 2004; Maag et al, 2005), the factor does not appear to play a major role in preventing premature GTP hydrolysis.
Figure 3.
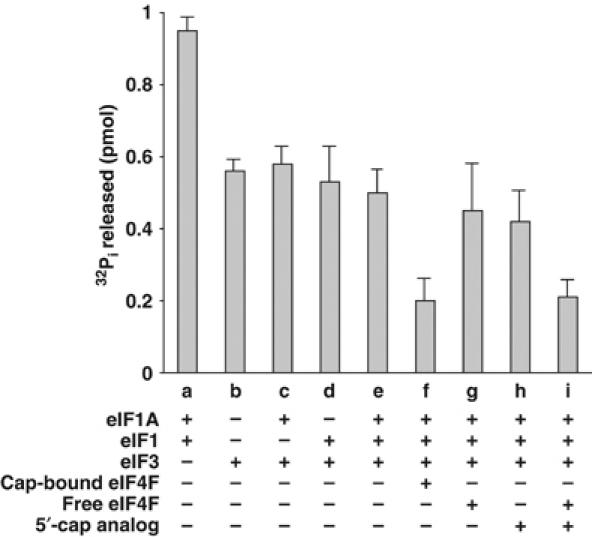
Prevention of eIF5-promoted hydrolysis of GTP bound in the 40S preinitiation complex by initiation factors. The 40S preinitiation complexes were formed at 1 mM Mg2+ using [γ-32P]GTP (5000 cpm/pmol) and [3H]Met-tRNAi (8000 cpm/pmol) in the presence of the indicated initiation factors. The amounts of factors used were as follows: 16 pmol eIF1, 16 pmol eIF1A, 14 pmol eIF3 and 16 pmol eIF4F (free and cap bound). Where indicated, 5′-cap analog (400 μM) was used. For the set containing eIF3 alone, the 40S preinitiation complex was formed at 5 mM Mg2+ to facilitate the transfer of the ternary complex to 40S ribosomal subunits in the absence of eIF1A and eIF1 (Chaudhuri et al, 1999). Each complex was isolated by sucrose gradient centrifugation as described in Materials and methods and approximately 1 pmol of the isolated complex containing bound [γ-32P]GTP was incubated with 0.5 pmol of recombinant eIF5 and the level of 32Pi released in each case was measured as described in the section eIF5-mediated GTPase assay. Control reactions were carried out for each set in which eIF5 was omitted and the values obtained (<0.05 pmol) were subtracted.
Since eIF4F could also associate with the 40S preinitiation complex (Figure 2C and D), we examined the effect of eIF4F on eIF5-promoted GTP hydrolysis. Accordingly, a cap analog-bound eIF4F was incubated with the 40S preinitiation complex formed in the presence of eIF1, eIF1A and eIF3. Interestingly, under these conditions, eIF5-promoted GTP hydrolysis was reduced by ∼80% (Figure 3, lane f). This reduction in GTP hydrolysis was specific for eIF4F containing the bound cap analog. Supplementation of the reaction with either eIF4F, free of the cap analog, or with the cap analog alone did not cause any additional inhibition of eIF5-promoted GTP hydrolysis over that observed without eIF4F (∼50% inhibition) (Figure 3, compare lanes g and h with lane e). An additional reduction of GTP hydrolysis (∼80% inhibition) was observed only when both the cap analog and eIF4F were added together to the 40S preinitiation complex (Figure 3, lane i). It should be noted that under these conditions, eIF5 was also bound to the 40S preinitiation complex (Figure 2B). To further confirm this interaction of eIF5 with the 40S preinitiation complex, we incubated the complex, isolated by sucrose gradient centrifugation, with m7GTP-Sepharose beads (which bind specifically to eIF4F). Analysis of the washed beads by SDS–PAGE, followed by Western blotting with antibodies specific for eIF5 and eIF4E, showed that binding of eIF5 to the beads was dependent on the presence of eIF4F in the 40S preinitiation complex (Figure 2G, compare lanes 1 and 2). These observations indicate that eIF4F and eIF5 were part of the same 40S preinitiation complex. Clearly, the inability of eIF5 to promote hydrolysis of GTP under these conditions was not due to a lack of association of eIF5 with the 40S preinitiation complex.
Taken together, these results suggest that eIF4F must be bound to the 5′-cap structure in order to prevent premature eIF5-mediated GTP hydrolysis, although both the cap analog-bound eIF4F (Figure 2C and D) and eIF4F, free of cap analog (data not shown), can associate with the 40S preinitiation complex.
Effect of binding of the 40S preinitiation complex to the 5′-end of mRNA on eIF5-promoted hydrolysis of GTP
The observations described above prompted us to examine the influence of the interaction between the 40S preinitiation complex and the 5′-end of a bona fide mRNA on eIF5-promoted GTP hydrolysis. In particular, we were interested in evaluating whether restoration in eIF5-promoted GTP hydrolysis could be obtained under conditions that would allow the 40S preinitiation complex to scan the mRNA and reach the AUG start codon. For this purpose, a 200-nt-long 5′-capped mRNA, designated pCON-PK-wild type mRNA, was generated (see Materials and methods). This mRNA was labeled with 32P (Materials and methods) and incubated with [3H]Met-tRNAi (as a Met-tRNAi·eIF2·GTP ternary complex), 40S ribosomal subunits and eIF1, eIF1A, eIF3, and eIF4F (devoid of cap analog) in a standard initiation reaction mixture. The resulting 40S preinitiation complex was then subjected to sucrose gradient centrifugation. As shown in Figure 4A, both 3H and 32P radioactivity cosedimented with the 40S particles, indicating that the 40S preinitiation complex was bound to the mRNA.
Figure 4.
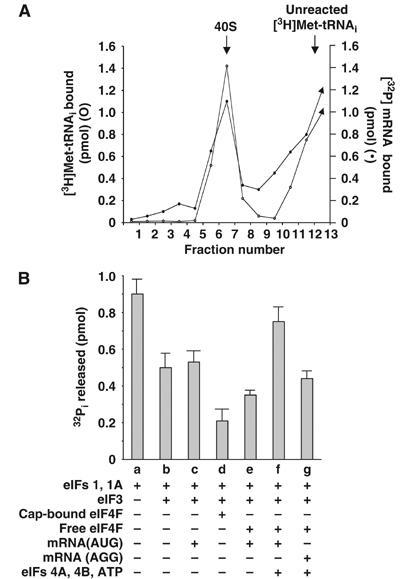
(A) mRNA binding by the eIF4F-containing 40S preinitiation complex. A 40S preinitiation complex was formed containing 5 pmol eIF3, 12 pmol eIF1A, 12 pmol eIF1, and 8 pmol eIF4F (devoid of 5′-cap analog) and 3 pmol of [3H]Met-tRNAi·eIF2·GTP ternary complex (8000 cpm/pmol). The complex was then incubated with 32P-labeled pCON-PK-wild type mRNA (1200 cpm/pmol) (Materials and methods). Following incubation, each reaction mixture was sedimented through a sucrose gradient (Materials and methods). Fractions were collected from the bottom of each tube and the amount of 32P or 3H radioactivity was determined by liquid scintillation counting. Control reaction mixtures (not shown), in which either 40S subunits or the ternary complex was omitted, were also analyzed. Under these conditions, 3H or 32P radioactivity was not detected in the 40S region. (B) Influence of pCON-PK-wild type mRNA on eIF5-mediated hydrolysis of GTP bound to the 40S preinitiation complex. A 40S preinitiation complex was formed, as described in Figure 3 legend, in the presence of the indicated factors. Where indicated, the preinitiation complexes were incubated with the pCON-PK-wild type mRNA (2 μg) either in the absence (lane e) or presence (lane f) of eIF4A (2 μg), eIF4B (2.5 μg) and ATP (1 mM). In lane g, a reaction similar to that in lane f was prepared except that the wild-type mRNA was replaced by pCON-PK-AGG mRNA. The 40S complexes were isolated by sucrose gradient centrifugation and eIF5-promoted GTP hydrolysis was measured by incubating 1 pmol of isolated complex with 0.5 pmol of recombinant eIF5. Control reactions without eIF5 were included in each case as described in Figure 3 legend.
The effect of (a) binding of the 40S preinitiation complex to the pCON-PK-wild type mRNA and (b) its subsequent positioning at the AUG start codon on eIF5-promoted GTP hydrolysis was then determined. For this purpose, the 40S preinitiation complex was formed in the presence of eIF1, eIF1A, eIF3, and eIF4F (free of cap analog) and incubated with the pCON-PK-wild type mRNA as described above. In parallel reactions, eIF4A and eIF4B as well as ATP were also included during incubation of the 40S preinitiation complex with the mRNA. In each case, the 40S complex formed was isolated by sucrose density gradient centrifugation and then used as a substrate for eIF5-promoted GTP hydrolysis. We observed that when the 40S complex was formed in the absence of eIF4A, eIF4B and ATP, there was only a marginal increase in the eIF5-promoted hydrolysis of bound GTP over that obtained with the 40S preinitiation complex containing cap-bound eIF4F (but no mRNA) (Figure 4B, compare lanes d and e). It should be noted that inclusion of the mRNA had no effect on GTP hydrolysis when eIF4F was omitted in the formation of the 40S preinitiation complex (Figure 4B, compare lanes b and c). In contrast, incubation of eIF5 with the 40S complex containing wild-type mRNA in the presence of eIF4A, eIF4B and ATP restored 70–80% of the level of GTP hydrolysis (Figure 4B, lane f). To examine whether the restored GTP hydrolysis required the presence of the AUG codon in the mRNA, we mutated the single AUG codon in the wild-type mRNA to AGG. Restoration of the eIF5-promoted GTP hydrolysis was substantially lower with the mutated AGG codon-containing mRNA than with the mRNA containing the AUG codon (Figure 4B, lane g).
Position of the 40S preinitiation complex on the mRNA during eIF5-promoted GTP hydrolysis
Next, we mapped the position of the 40S ribosomal complex on the mRNA under conditions where eIF5-promoted GTP hydrolysis was restored. For this purpose, we carried out a toe-print assay (Pestova et al, 1996; Kozak, 1997), which involves annealing of a 32P-labeled deoxyoligonucleotide primer to the 3′-end of an mRNA template containing the bound ribosomal complex, followed by reverse transcription of the annealed primer to generate complementary DNAs. The reverse transcriptase reaction is terminated at the leading edge of the bound complex, resulting in the generation of a cDNA of defined length, which can then be analyzed using a sequencing gel. It has been shown previously (Pestova et al, 1998) that, owing to the large size of ribosomal complexes, the reverse transcription reaction terminates about 17 nt downstream of the point at which the ribosomal complexes are arrested on the mRNA.
Assembly of the 40S complex on the wild-type mRNA in the presence of eIF1, eIF1A, eIF3, eIF4F as well as eIF4A, eIF4B and ATP resulted in a strong toe-print at a position 17 nt downstream of the AUG start codon (Figure 5, lane 2). This indicates that the toe-print corresponded to the 40S complex positioned at the AUG codon to form the 40S initiation complex. This toe-print was absent in the control lane 1, which contained only the wild-type mRNA and lacked the initiation factors as well as the 40S ribosomal subunits, indicating that the toe-print in lane 2 was indeed due to the ribosomal complex assembled on the mRNA. However, when the complex was assembled on an mRNA in which the AUG was mutated to AGG, the intensity of the toe-print at the position of the 40S initiation complex was greatly diminished (Figure 5, lane 3). Furthermore, several weak bands appeared at the position of the 40S initiation complex in lane 3, which are indicative of stalling of the 40S complex at those positions. Such stalling may occur due to the sequences surrounding the AGG codon in the AGG-mRNA as well as the secondary structure that might form in the control mRNA (e.g., the AvaI site CCCCGGGC might pair with GCUGGGG downstream). At any rate, these results demonstrate that under conditions that support the eIF5-promoted GTP hydrolysis, the 40S complex was positioned at the initiation codon of the wild-type mRNA.
Figure 5.
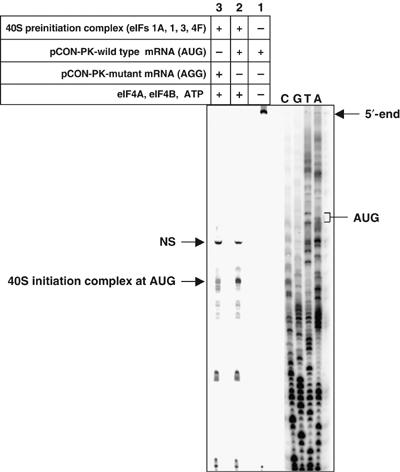
Toe-printing analysis of the position of 40S preinitiation complex on the mRNA during eIF5-promoted GTP hydrolysis. Capped mRNA transcripts, preannealed to a 32P-labeled primer, 5′-GGCATCGTAAAGAACATTTTGAG-3′ (∼4 pmol), were incubated under standard reaction conditions for primer extension assays, as described in Materials and methods either with preformed 40S preinitiation complexes in the presence of eIF4A (2 μg), eIF4B (2 μg) and ATP (1 mM) (lanes 2 and 3) or without these components (lane 1). PCON-PK-wild type mRNA (containing a single AUG codon) was used in lanes 1 and 2, while pCON-PK-mutant mRNA (in which the AUG codon is mutated to AGG) was used in lane 3. Primer extension was carried out at 25°C for 15 min with Superscript II reverse transcriptase (2 U/μl). Different cDNA products obtained in the reactions are indicated by arrows (NS, nonspecific cDNA product). Reference lanes A, T, G and C depict a dideoxynucleotide ladder obtained by primer extension with the pCON-PK-wild type mRNA in the presence of dideoxynucleotides.
Discussion
In this paper, we have examined the mode of action of eIF5 regarding its role in the selection of the AUG start codon by the 40S ribosomal preinitiation complex. Our interest in this problem stemmed from the observation of Donahue and his associates, who, using an elegant genetic approach, showed that in the yeast Saccharomyces cerevisiae, eIF2, a GTP-binding initiation factor, and eIF5, a GAP, (as well as eIF1) play essential roles in maintaining the fidelity of translation initiation (Donahue, 2000). Subsequent biochemical assays by this group demonstrated that mutations in the genes encoding the above protein factors lead to either an elevated level of GTP hydrolysis or aberrant dissociation of eIF2 from Met-tRNAi. This results in premature release of bound factors from the 40S preinitiation complex and presumably allows the translation machinery to initiate from a non-AUG codon (Donahue, 2000). These observations suggested that the GTP hydrolysis step, during translation initiation, is coupled to the ribosomal recognition of an AUG start codon. However, a question that remained unanswered is how is the eIF5-promoted hydrolysis of GTP in the 40S preinitiation complex prevented prior to the selection of the initiation codon?
Using in vitro biochemical studies, we observed that selection of an AUG codon is not a prerequisite for eIF5-promoted GTP hydrolysis. Rather, a 40S preinitiation complex, devoid of any bound initiation factors, is also an efficient substrate for eIF5-promoted GTP hydrolysis (Figure 1). However, the resulting 80S complex is nonfunctional and represents an ‘abortive' initiation complex (Figure 1). The possibility thus exists that, prior to AUG selection, eIF5-promoted GTP hydrolysis may be modulated by other initiation factors that normally bind the 40S preinitiation complex. Previously, we have shown that the binding of three initiation factors, eIF1, eIF1A and eIF3, to the 40S ribosomal subunits was necessary for the formation of a maximally stable 40S preinitiation complex (Majumdar et al, 2003). We now observe that in such a 40S preinitiation complex, eIF5-promoted GTP hydrolysis is prevented by ∼50% (Figure 3, lane e). However, in the absence of eIF3, the other two initiation factors, eIF1 and eIF1A, together could not prevent eIF5-promoted GTP hydrolysis (Figure 3, lane a). This finding can be explained by our previous observation that the stable association of eIF1 and eIF1A with the 40S preinitiation complex depended on the presence of eIF3 (Majumdar et al, 2003). Thus, it is possible that in the absence of eIF3, eIF1 and eIF1A cannot bind stably to the 40S preinitiation complex and thus cannot prevent eIF5 from promoting hydrolysis of the bound GTP. Clearly, eIF3 appears to play a central role in preventing premature eIF5-promoted GTP hydrolysis.
eIF3 is also involved in recruiting the 40S preinitiation complex to the 5′-capped end of mRNA by interacting with the cap-bound initiation factor eIF4F (Dever, 2002). Indeed, eIF4F is found associated with the 40S preinitiation complex containing bound eIF1, eIF1A and eIF3 (Figure 2C and D). The presence of a cap analog-bound eIF4F in the 40S preinitiation complex prevented premature eIF5-mediated GTP hydrolysis by as much as 80% (Figure 3, lane f). Surprisingly, although both cap analog-bound eIF4F and eIF4F devoid of the cap analog associate with the 40S preinitiation complex with comparable efficiency (Figure 2 and data not shown), eIF4F exerted its effect on eIF5-promoted GTP hydrolysis only when bound to the cap (Figure 3, lanes f and i). The eIF4E subunit of eIF4F has been shown to undergo marked conformational changes upon binding the 5′-capped mRNA structure (Marcotrigiano et al, 1997; Matsuo et al, 1997; Gross et al, 2003). Our results indicate that this specific cap-bound conformation of the protein is, perhaps, required to prevent eIF5-promoted GTP hydrolysis in the 40S preinitiation complex.
The 40S preinitiation complex, containing eIF1, eIF1A, eIF3 and eIF4F, efficiently binds to an mRNA (Figure 4A). However, eIF5-promoted hydrolysis of the bound GTP was restored only in the presence of the additional factors eIF4A, eIF4B and ATP (Figure 4B, lane f). Under these conditions, toe-printing analysis revealed that the 40S ribosomal complex was positioned at the AUG codon of the mRNA (Figure 5, lane 2). However, when the single AUG codon in the mRNA was mutated to AGG, both GTP hydrolysis (Figure 4B, lane g) and the intensity of the signal at the selective position of the 40S complex (Figure 5, lane 3) were markedly decreased. It should be noted that some amount of GTP hydrolysis was detected in the presence of the mutated AGG-mRNA. It is likely that during scanning, either eIF4F gets dissociated from the 40S complex or, in the absence of an AUG codon, the scanning complex probably stalls on the mRNA allowing eIF5 to promote hydrolysis of some of the bound GTP. Nevertheless, these results directly demonstrate that the process of AUG selection by the 40S preinitiation complex is linked to the hydrolysis of the bound GTP.
The role of eIF4A and ATP (along with eIF4B) in promoting mRNA-dependent GTP hydrolysis (Figure 4B, compare lanes e and f) is not completely clear. It should be noted that although eIF4A has been shown to possess RNA helicase activity, the exact role of eIF4A and ATP in the scanning reaction is still ill-defined (Gingras et al, 1999). In fact, it has been hypothesized that the helicase activity of eIF4A may act to rearrange secondary structures in the rRNA, causing a conformational change in the 40S ribosomal subunit (Gingras et al, 1999; Kapp and Lorsch, 2004). It seems likely that with the particular mRNA used in this study (which lacks secondary structure near the 5′-end), free eIF4A may be needed not to promote scanning, but to alter interactions among the bound initiation factors. Such structural alterations might then facilitate the essential eIF5–eIF2 interaction required for hydrolysis of the GTP bound in the 40S complex.
Several recent reports in the yeast S. cerevisiae have shown that the C-terminal of eIF5 that interacts with the β subunit of eIF2 (Das et al, 1997; Asano et al, 1999) is also involved in interaction with several other initiation factors. These factors include eIF1 (Asano et al, 1999; Asano et al, 2000), the Nip1 (eIF3c) subunit of eIF3 (Phan et al, 1998) and the eIF4G subunit of eIF4F (Asano et al, 2001). Interestingly, this eIF4G–eIF5 interaction was found to be mutually exclusive to the critical eIF5–eIF2β interaction, indicating that these two interactions are temporally separated in the translation initiation pathway (Asano et al, 2001). Moreover, a mutation in the HEAT domain of eIF4G moderately enhances translation from non-AUG codons (He et al, 2003), suggesting that formation of a complex between eIF4G and eIF5 is important for maintaining the integrity of the scanning ribosomal preinitiation complex.
The initiation factors that bind to the C-terminal region of eIF5 were shown, in this study, to also prevent premature eIF5-promoted GTP hydrolysis in the 40S preinitiation complex. We also found that eIF5 was stably associated with the 40S preinitiation complex (Figure 2B). This association suggests that binding of eIF1, eIF1A, eIF3 and eIF4F to the 40S preinitiation complex might sterically prevent eIF5 from interacting with eIF2, resulting in inhibition of aberrant GTP hydrolysis in the 40S preinitiation complex positioned at the 5′-capped end of an mRNA. However, it is possible that once the 40S complex scans the 5′-UTR to reach the AUG start codon, codon–anticodon base pairing between the initiator Met-tRNA and the AUG codon may lead to a conformational change in the 40S complex, resulting in the reorientation of the bound factors in the complex, or the release of one or more of the bound initiation factors. As a result, eIF5 may then be able to interact with eIF2β, which triggers the GAP activity of eIF5 and hydrolysis of the GTP bound in the 40S complex (Figure 6). In this context, it is interesting to note that Lorsch and his associates (Maag et al, 2005) recently detected a marked conformational change in the 40S complex upon its binding to the AUG codon of a synthetic mRNA. These investigators also used FRET analysis to show that eIF1 is released from the 40S complex upon AUG selection. However, in a parallel study using a natural mRNA, Pestova's group (Unbehaun et al, 2004) did not observe the release of bound initiation factors at the stage of AUG recognition. Rather, the release of eIF2·GDP was observed following the subsequent eIF5-promoted GTP hydrolysis, while the release of eIF1 and eIF3 was detected subsequent to 60S ribosomal subunit joining. Clearly, these issues involving the exact mechanism of the release of factors require further analysis.
Figure 6.
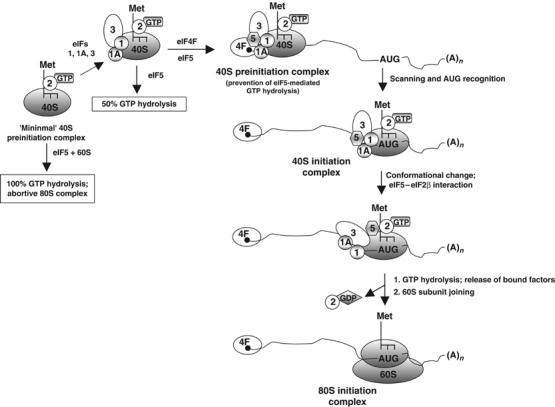
Proposed model for the regulation of GTP hydrolysis during translation initiation. Initiation factor eIF5 bound to the 40S preinitiation complex (positioned at the 5′-end of the mRNA) is prevented from interacting with eIF2 due to the presence of the other initiation factors eIF1, eIF1A, eIF3 and eIF4F. In the absence of this interaction, eIF5 cannot act as a GAP to promote hydrolysis of the GTP bound to the 40S preinitiation complex. Following scanning and positioning of the 40S complex at the AUG start codon, a conformational change and/or release of one or more of the bound initiation factors allows eIF5 to interact with eIF2 thereby leading to GTP hydrolysis. Although shown bound to the 5′-cap structure, whether eIF4F remains bound to the cap or is a part of the scanning 40S complex is not yet clear.
Materials and methods
Purified initiation factors and other reagents
The preparation of 3H-labeled rabbit liver initiator Met-tRNAi (8000 cpm/pmol) and 40S and 60S ribosomal subunits from Artemia salina eggs, purified eIF2 and eIF3 from rabbit reticulocyte lysates and bacterial-expressed recombinant rat eIF5, human eIF1A and human eIF1 protein (unlabeled or 3H-labeled) was described previously (Chaudhuri et al, 1997b; Das and Maitra, 2000; Majumdar et al, 2003). Initiation factor eIF4F was purified from 0.5 M KCl-wash proteins of rabbit reticulocyte polysomal pellets by an adaptation of the procedure described by Sonenberg and associates (Edery et al, 1983). The factor eIF4A was purified from rabbit reticulocyte lysates using an FPLC-Hi-Trap Blue column, while eIF4B was expressed as a His-tagged protein in Escherichia coli BL21 (DE3) cells and purified using Ni2+-NTA agarose column as described previously (Pause et al, 1994). Rabbit IgG antibodies specific for mammalian eIF1A and eIF5 and total IgY antibodies specific for mammalian eIF3 subunits were isolated as described (Chaudhuri et al, 1997a, 1997b).
Isolation of the 40S preinitiation and 40S initiation complexes
The 40S preinitiation complex containing [3H]Met-tRNAi and [γ-32P]GTP was isolated free of unreacted components as follows. In stage I of the reaction, reaction mixtures (50 μl) containing 20 mM Tris–HCl, pH 7.5, 100 mM KCl, 5 mM 2-mercaptoethanol, 60 μg of nuclease-free bovine serum albumin, 20 μM [γ-32P]GTP (8000–10 000 cpm/pmol), 50 pmol of [3H]Met-tRNAi (8000 cpm/pmol) and 8–10 μg of eIF2 were incubated at 37°C for 5 min to form a [3H]Met-tRNAi·eIF2·[γ-32P]GTP ternary complex. Based on microgram of eIF2 added, the efficiency of ternary complex formation, measured by nitrocellulose membrane filtration assay (Raychaudhuri et al, 1987), varies between 40 and 60%. In stage 2, another set of reaction mixtures (90 μl each) containing 20 mM Tris–HCl, pH 7.5, 100 mM KCl, 1 mM MgCl2, 2.5 mM 2-mercaptoethanol (Buffer T), 2.0A260 unit of 40S ribosomal subunits and, where indicated, 10 μg of eIF3, 300 ng of eIF1A, 250 ng of eIF1 and 5 μg eIF4F (bound to the cap analog m7GTP) were incubated at 37°C for 4 min and then supplemented with 50 μl of the stage 1 reaction mixture. The Mg2+ concentration of each reaction mixture (now 125 μl each) was adjusted to 1 mM, and the reaction mixtures were incubated at 37°C for 4 min, chilled in an ice-water bath, and then layered onto a 5 ml of 7.5–30% (w/v) sucrose density gradient containing Buffer T and centrifuged at 48 000 r.p.m. for 105 min in an SW 50.1 rotor. Fractions (200 μl) were collected from the bottom of the gradients, and aliquots (10 μl) of each fraction were counted in a liquid scintillation spectrometer to determine the radioactivity profile. The 40S preinitiation complex fractions containing bound [3H]Met-tRNAi and [γ-32P]GTP were pooled, divided into 1 pmol aliquots and stored at −135°C. The 40S initiation complex was isolated using the same reaction conditions as described for the isolation of 40S preinitiation complex, except that at stage II, mixtures (120 μl) were supplemented with 5 μg eIF4F (devoid of the m7GTP cap analog), 2 μg (∼20 pmol) wild-type or mutant mRNA, 1 μg eIF4A, 1.5 μg eIF4B and 1 mM ATP. Such reactions yielded a 40S preinitiation complex that could scan the mRNA for the presence of an initiating codon.
Construction of pCON-PK plasmids
For the generation of an mRNA, a T7 promoter-based plasmid, designated pCON-PK-wild type plasmid, was used. This plasmid was derived from the p-CON-0 plasmid (a kind gift from Marilyn Kozak) (Kozak, 1997). It consisted of the region between nucleotides 32 and 102 of p-CON-0 plasmid, which contained a single ATG codon followed by a pseudoknot (Kozak, 1998) and GGCATCGTAAAGAACATTTTGA. Transcription of this plasmid (linearized with EcoRI), using the mMESSAGE mMACHINE T7 Ultra coupled transcription-capping kit (Ambion), yielded a 200-nt-long 5′-capped pCON-PK-wild type mRNA, which included a 5′-capped 50-nt-long 5′-UTR, a single AUG codon and a 150-nt-long open reading frame, and terminated in a pseudoknot structure at the 3′-end. The pseudoknot was introduced in the mRNA to prevent the scanning ribosomal complex from falling off the mRNA. A mutant plasmid was constructed from the wild-type plasmid by mutating the ATG codon at position 58 of the wild-type plasmid to AGG. Transcription of this mutant plasmid yielded the pCON-PK-AGG mRNA in which the single AUG codon was changed to AGG. Capped mRNAs were purified from enzymatic reactions using the MEGAclear Purification Kit (Ambion) and quantified by measuring the absorbance at 260 nm. A 32P-labeled pCON-PK-wild type mRNA was generated using the coupled transcription-capping kit as described above, except that 1 μl of [α-32P]UTP (6000 Ci/mmol; Amersham) was included in the reaction mixture.
Primer extension assays
The oligonucleotide 5′-GGCATCGTAAAGAACATTTTGAG-3′ served as the primer for the reverse transcription reaction. It was labeled at the 5′-end with T4 polynucleotide kinase and [γ-32P]ATP (6000 Ci/mmol; Amersham) and preannealed to either pCON-PK-wild type mRNA or pCON-PK-AGG mRNA (0.25 μg or ∼3 pmol) by heating for 1 min at 65°C, followed by incubation at 37°C for 8 min in a buffer containing 40 mM Tris–HCl, pH 7.5, and 0.2 mM EDTA and then kept on ice for 5 min. A preformed ternary complex (eIF2·[3H]Met-tRNAi·GTP) was incubated with 40S ribosomal subunits (1.2A260 units) in the presence of eIF1 (200 ng), eIF1A (250 ng), eIF3 (7 μg), and eIF4F (5 μg), devoid of cap analog, at 37°C for 5 min in Buffer T to form the 40S preinitiation complex. The preinitiation complex was incubated with either pCON-PK-wild type mRNA or pCON-PK-AGG mRNA (preannealed to the 32P-labeled primer) at 37°C for 4 min in the presence of 100 U of RNaseOUT (Invitrogen) and, where indicated, 1 mM ATP, eIF4A (1 μg) and eIF4B (1 μg). To determine the position of the 40S complex on the mRNA, reaction mixtures (125 μl each) were incubated with all the four deoxynucleoside triphosphates (final concentration of 0.5 mM each) and 2 U/μl Superscript II reverse transcriptase (Invitrogen) in a buffer containing 50 mM Tris–HCl, pH 8.3, 75 mM KCl and 6 mM MgCl2 at 25°C for 15 min. Reactions were terminated by extraction with phenol–chloroform–isoamyl alcohol (24:23:1) and cDNA products were precipitated by the addition of an equal volume of isopropanol followed by overnight storage at −80°C. cDNA products were resuspended, mixed with formamide, heated at 90°C for 1 min and analyzed by electrophoresis through 8% polyacrylamide sequencing gels. The products were compared to a sequencing ladder obtained by primer extension of the pCON-PK-wild type mRNA in the presence of dNTPs and the avian myeloblastosis virus reverse transcriptase (Promega).
eIF5-mediated GTPase assay
For this assay, a 40S preinitiation complex (40S·[3H]Met-tRNAi·eIF2·[γ-32P]GTP) or 40S initiation complex (40S·mRNA·[3H]Met-tRNAi·eIF2·[γ-32P]GTP) was prepared and isolated free of unreacted components by sucrose gradient centrifugation as described above. An aliquot (80 μl) of isolated 40S preinitiation complex or 40S initiation complex, each containing 1 pmol of bound [γ-32P]GTP, was incubated with 25 ng of eIF5 at 22°C for 10 min. The release of 32Pi due to hydrolysis of [γ-32P]GTP bound to the 40S complex was measured using the ammonium phosphomolybdate method (Raychaudhuri et al, 1987) as follows. The reaction was terminated by the addition of 100 μl of 20 mM silicotungstic acid in 10 mM H2SO4 followed by centrifugation in an Ependorf microfuge. To the supernatant were added 200 μl of 2 mM KH2PO4 and 100 μl of 5% ammonium molybdate in 4 N H2SO4. The reaction tubes were incubated for 1 min at 37°C, followed by the addition of 500 μl of isobutanol:benzene (1:1). The tubes were stirred vigorously for 20 s on a vortex mixture followed by a brief (30 s) centrifugation in an Ependorf microfuge to separate the phases. A 200 μl aliquot from the upper organic phase from each tube was assayed for radioactivity by counting in Aquasol in a liquid scintillation spectrometer.
Acknowledgments
This research was supported by Grant GM15399 from the National Institutes of Health and by Cancer Core Support Grant P30CA13330 from the National Cancer Institute. We are indebted to Dr Marilyn Kozak of the Robert Wood Johnson Medical School, University of Medicine and Dentistry, New Jersey, Dr Michael Brenowitz and Somdeb Mitra of the Albert Einstein College of Medicine for considerable help in the toe-printing assays described in the text. We also thank Dr Jerard Hurwitz of the Sloan-Kettering Cancer Research Center, New York and Dr Jonathan R Warner of the Albert Einstein College of Medicine for critically reading the manuscript.
References
- Asano K, Clayton J, Shalev A, Hinnebusch AG (2000) A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNA(Met) is an important translation initiation intermediate in vivo. Genes Dev 14: 2534–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Krishnamoorthy T, Phan L, Pavitt GD, Hinnebusch AG (1999) Conserved bipartite motifs in yeast eIF5 and eIF2B epsilon, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J 18: 1673–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Shalev A, Phan L, Nielsen K, Clayton J, Valasek L, Donahue TF, Hinnebusch AG (2001) Multiple roles for the C-terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J 20: 2326–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A, Maitra U (1991) Function of eukaryotic initiation factor 5 in the formation of an 80 S ribosomal polypeptide chain initiation complex. J Biol Chem 266: 14039–14045 [PubMed] [Google Scholar]
- Chaudhuri J, Chakrabarti A, Maitra U (1997a) Biochemical characterization of mammalian translation initiation factor 3 (eIF3). Molecular cloning reveals that p110 subunit is the mammalian homologue of Saccharomyces cerevisiae protein Prt1. J Biol Chem 272: 30975–30983 [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Chowdhury D, Maitra U (1999) Distinct functions of eukaryotic translation initiation factors eIF1A and eIF3 in the formation of the 40 S ribosomal preinitiation complex. J Biol Chem 274: 17975–17980 [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Das K, Maitra U (1994) Purification and characterization of bacterially expressed mammalian translation initiation factor 5 (eIF-5): demonstration that eIF-5 forms a specific complex with eIF-2. Biochemistry 33: 4794–4799 [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Si K, Maitra U (1997b) Function of eukaryotic translation initiation factor 1A (eIF1A) (formerly called eIF-4C) in initiation of protein synthesis. J Biol Chem 272: 7883–7891 [DOI] [PubMed] [Google Scholar]
- Das S, Ghosh R, Maitra U (2001) Eukaryotic translation initiation factor 5 functions as a GTPase-activating protein. J Biol Chem 276: 6720–6726 [DOI] [PubMed] [Google Scholar]
- Das S, Maiti T, Das K, Maitra U (1997) Specific interaction of eukaryotic translation initiation factor 5 (eIF5) with the beta-subunit of eIF2. J Biol Chem 272: 31712–31718 [DOI] [PubMed] [Google Scholar]
- Das S, Maitra U (2000) Mutational analysis of mammalian translation initiation factor 5 (eIF5): role of interaction between the beta subunit of eIF2 and eIF5 in eIF5 function in vitro and in vivo. Mol Cell Biol 20: 3942–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Maitra U (2001) Functional significance and mechanism of eIF5-promoted GTP hydrolysis in eukaryotic translation initiation. Prog Nucleic Acid Res Mol Biol 70: 207–231 [DOI] [PubMed] [Google Scholar]
- Dever TE (2002) Gene-specific regulation by general translation factors. Cell 108: 545–556 [DOI] [PubMed] [Google Scholar]
- Donahue TF (2000) Genetic approaches to translation initiation in Saccharomyces cerevisiae. In Translational Control of Gene Expression, Sonenberg N, Hershey JWB, Mathews MB (eds) pp 487–502. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Edery I, Humbelin M, Darveau A, Lee KA, Milburn S, Hershey JW, Trachsel H, Sonenberg N (1983) Involvement of eukaryotic initiation factor 4A in the cap recognition process. J Biol Chem 258: 11398–11403 [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N (1999) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68: 913–963 [DOI] [PubMed] [Google Scholar]
- Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G (2003) Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115: 739–750 [DOI] [PubMed] [Google Scholar]
- He H, von der Haar T, Singh CR, Ii M, Li B, Hinnebusch AG, McCarthy JE, Asano K (2003) The yeast eukaryotic initiation factor 4G (eIF4G) HEAT domain interacts with eIF1 and eIF5 and is involved in stringent AUG selection. Mol Cell Biol 23: 5431–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey JWB, Merrick WC (2000) Pathway and mechanism of initiation of protein synthesis. In Translational Control of Gene Expression, Sonenberg N, Hershey JWB, Mathews MB (eds) pp 33–88. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratry Press [Google Scholar]
- Kapp LD, Lorsch JR (2004) The molecular mechanics of eukaryotic translation. Annu Rev Biochem 73: 657–704 [DOI] [PubMed] [Google Scholar]
- Kozak M (1997) Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J 16: 2482–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (1998) Primer extension analysis of eukaryotic ribosome–mRNA complexes. Nucleic Acids Res 26: 4853–4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D, Fekete CA, Gryczynski Z, Lorsch JR (2005) A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol Cell 17: 265–275 [DOI] [PubMed] [Google Scholar]
- Majumdar R, Bandyopadhyay A, Maitra U (2003) Mammalian translation initiation factor eIF1 functions with eIF1A and eIF3 in the formation of a stable 40 S preinitiation complex. J Biol Chem 278: 6580–6587 [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK (1997) Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89: 951–961 [DOI] [PubMed] [Google Scholar]
- Matsuo H, Li H, McGuire AM, Fletcher CM, Gingras AC, Sonenberg N, Wagner G (1997) Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat Struct Biol 4: 717–724 [DOI] [PubMed] [Google Scholar]
- Pause A, Methot N, Svitkin Y, Merrick WC, Sonenberg N (1994) Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J 13: 1205–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, Hellen CU (1998) Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394: 854–859 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU, Shatsky IN (1996) Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol 16: 6859–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU (2000) The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403: 332–335 [DOI] [PubMed] [Google Scholar]
- Phan L, Zhang X, Asano K, Anderson J, Vornlocher HP, Greenberg JR, Qin J, Hinnebusch AG (1998) Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol Cell Biol 18: 4935–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri P, Chevesich J, Ghosh S, Maitra U (1987) Characterization of eukaryotic initiation factor 5 from rabbit reticulocytes. Evidence that the initiation factor is a monomeric protein of Mr of about 58 000–62 000. J Biol Chem 262: 14222–14227 [PubMed] [Google Scholar]
- Unbehaun A, Borukhov SI, Hellen CU, Pestova TV (2004) Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon–anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev 18: 3078–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]


