Abstract
Drosophila NURF is an ISWI-containing ATP-dependent chromatin remodeling complex that regulates transcription by catalyzing nucleosome sliding. To determine in vivo gene targets of NURF, we performed whole genome expression analysis on mutants lacking the NURF-specific subunit NURF301. Strikingly, a large set of ecdysone-responsive targets is included among several hundred NURF-regulated genes. Null Nurf301 mutants do not undergo larval to pupal metamorphosis, and also enhance dominant-negative mutations in ecdysone receptor. Moreover, purified NURF binds EcR in an ecdysone-dependent manner, suggesting it is a direct effector of nuclear receptor activity. The conservation of NURF in mammals has broad implications for steroid signaling.
Keywords: NURF, chromatin remodeling, chromatin, Drosophila, Ecdysteroid, ISWI
The profound morphological changes that accompany insect metamorphosis and molting are coordinated by the action of the steroid hormone ecdysone (for review, see Thummel 2001). In common with other steroid hormones, ecdysone exerts its effects through a nuclear receptor (NR) that regulates transcription. Ecdysone is the ligand for the ecdysone receptor, a heterodimer between EcR and Ultraspiracle (USP), the Drosophila retinoid X receptor homolog. Both components of the heterodimer are required to allow hormone binding and transcription activation (Thomas et al. 1993; Yao et al. 1993). Studies of vertebrate NRs reveal that hormone binding triggers a conformational switch that allows the receptor to bind transcriptional coactivators, leading to the assembly of the transcription initiation complex (for review, see Glass et al. 1997).
There is growing evidence that the actions of NRs, like other transcription factors, occur in the context of chromatin. The condensation of DNA in chromatin can have profound effects on gene expression by blocking gene regulatory elements and inhibiting access of the transcription machinery. As such, it offers a convenient, epigenetic way of encoding information in the genome. Without changing the underlying DNA content, changes in nucleosome positioning and structure can alter the way in which genes are expressed. These altered chromatin states can either be triggered through post-translational modification of histones (Turner 2002; Fischle et al. 2003) or through the actions of the energy-utilizing, ATP-dependent chromatin remodeling factors (Lusser and Kadonaga 2003).
Changes in chromatin organization are an integral part of NR-mediated gene activation. Studies of vertebrate NRs have shown that steroid binding is accompanied by increased histone acetylation caused by the displacement of histone deacetylase activities and recruitment of histone acetylases (Collingwood et al. 1999; Privalsky 2004). Recently, Mazo and colleagues (Sedkov et al. 2003) have shown that a critical coactivator of the Drosophila ecdysone receptor is a histone H3 methyltransferase activity. In addition to changes in histone tail modifications, recruitment of ATP-dependent chromatin remodeling enzymes is instrumental in steroid-mediated gene activation. Most studies have focused on functions of the SWI/SNF family of ATP-dependent chromatin remodeling factors (Yoshinaga et al. 1992; Muchardt and Yaniv 1993; Fryer and Archer 1998). However, there is also evidence that ISWI complexes may also be required for NR action (Di Croce et al. 1999; Dilworth et al. 2000).
The nucleosome remodeling factor (NURF) is the founding member of the ISWI family of ATP-dependent chromatin remodeling factors. Like other ATP-dependent chromatin remodeling enzymes, NURF is a large multi-subunit protein complex that uses the energy of ATP hydrolysis to change the dynamic properties of nucleosomes (Tsukiyama and Wu 1995; Xiao et al. 2001). To date, Drosophila and human NURF have been purified and shown to catalyze energy-dependent nucleosome sliding (Tsukiyama and Wu 1995; Hamiche et al. 1999; Barak et al. 2003). By sliding nucleosomes, NURF can expose or occlude transcription factor-binding sites. Consistent with this, Drosophila NURF can regulate transcription of a reconstituted chromatin template in vitro (Mizuguchi et al. 1997).
Although NURF is composed of four subunits, only the largest subunit (in Drosophila NURF301; in humans BPTF) is specific to NURF (Xiao et al. 2001; Barak et al. 2003). All of the other subunits found in both Drosophila and human NURF—ISWI (SNF2L) and NURF55 (pRbAb46/48)—can form part of other chromatin remodeling complexes. As the largest subunit is obligatory for the assembly of NURF (Xiao et al. 2001), studies of the large NURF subunit provide the most direct route to understand the functions of NURF.
This property has been exploited in genetic studies where NURF301 and BPTF have been ablated to show that Drosophila and human NURF regulate transcription in vivo (Badenhorst et al. 2002; Barak et al. 2003). In Drosophila, mutation of Nurf301 blocks activation of the homeotic selector genes Ultrabithorax and engrailed (Badenhorst et al. 2002). siRNA-mediated knock-down of BPTF in human cells also prevents the activation of engrailed-1 and engrailed-2, the human homologs of engrailed (Barak et al. 2003).
However, the larval lethal phenotypes of Nurf301 and Iswi mutants (Deuring et al. 2000; Badenhorst et al. 2002) suggest that NURF has additional transcriptional targets. To identify these targets, we have used microarrays to profile gene expression in Drosophila Nurf301 mutants. Our results indicate that a major, additional function of NURF is to coregulate targets of the ecdysone receptor to allow progression from larval to pupal stages.
Results and Discussion
Isolation and phenotypic characterization of Nurf301 mutants
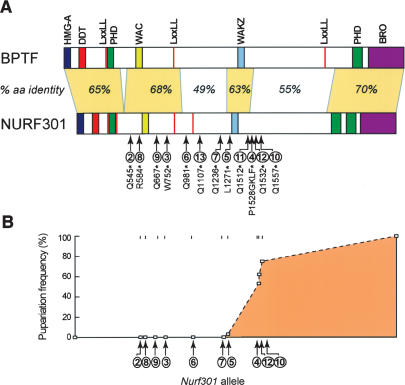
To characterize the physiological function of Drosophila NURF, we generated a series of EMS-induced lesions in the gene encoding the largest NURF subunit, Nurf301. We focused on NURF301, as the large subunit is the only NURF-specific subunit and is obligatory for the assembly of NURF (Xiao et al. 2001; Barak et al. 2003). Twelve EMS-induced lesions were isolated in Nurf301, all of which encode truncated NURF301 products (Fig. 1A; Supplementary Table 1). In addition to the previously reported male X-chromosome and melanotic tumor phenotypes, null Nurf301 mutants that truncate before the putative WAKZ motif (Xiao et al. 2001) displayed a non-pupariating phenotype (Fig. 1B). Mutants exhibit a slight developmental delay relative to heterozygous siblings, but larvae do not form pupae and can continue to survive in culture for up to 2 wk. In the few cases where white prepupae form, these retain an elongated larval form and fail to evert the anterior spiracles completely. Null Nurf301 mutants appear to undergo the larval molts normally. However, embryos contain a large dowry of maternally loaded Nurf301 mRNA, suggesting that Nurf301 mutants have sufficient NURF301 protein to support initial larval development, including the molt from L2 to L3.
Figure 1.
Analysis of Nurf301 mutants. (A) Domain structure of NURF301 and its human homolog BPTF. The location of EMS-induced lesions in 12 Nurf301 mutants is shown. (B) Pupariation frequency of hemizygous Nurf301 mutant animals. Null alleles of Nurf301 fail to pupariate.
In contrast to null Nurf301 alleles, Nurf301 mutations that truncate NURF301 after the WAKZ motif (Nurf3014, Nurf30110, Nurf30111, and Nurf30112 in Fig. 1) do pupariate, indicating that N-terminal fragments of NURF301 that extend beyond the WAKZ domain support pupariation. This agrees with in vitro data showing that these fragments contain sites of interaction with the other three NURF subunits and may be able to coordinate the assembly of a NURF complex (data not shown). Heteroallelic combinations of these alleles survive to adult stages. However, flies exhibit developmental abnormalities and are sterile (data not shown; see also Supplemental Material) indicating that the C-terminal regions of NURF301, while dispensable for pupariation, have functions.
Whole genome expression analysis of NURF mutants
To define gene targets of NURF that are required for pupariation, we compared whole genome expression profiles of null Nurf301 mutant (Nurf3012/Nurf3018) third instar larvae with those of wild-type larvae. We identified a set of 477 genes for which there was a statistically significant (P < 0.05) change in expression between mutant and wild-type samples. Of these, 274 genes were decreased at least threefold in Nurf301 mutants, while 203 exhibited at least threefold elevated levels of expression in the mutant samples, suggesting that NURF may function both as an activator and repressor of transcription.
Classification according to gene ontology (http://www.geneontology.org/) of the 274 genes that require NURF301 for expression revealed that a sizeable number correspond to ecdysone target genes (bracketed in Fig. 2A). An additional 30 of the 274 genes have no gene ontology classifications, but are known to be highly expressed at the larval/pupal transition (designated Yale-LP in Fig. 2A; Arbeitman et al. 2002) and may be additional ecdysone targets. Taken together, this indicates that NURF is required for expression of targets of the ecdysone receptor. Similar classifications of the genes that show increased expression in Nurf301 mutants indicated that many are immune-related genes. These will be discussed in greater detail elsewhere (P. Badenhorst and C. Wu, in prep.).
Figure 2.
Whole genome expression analysis of Nurf301 mutants. (A) Gene ontology (GEO) classification of genes decreased in expression in Nurf301 mutants. Potential ecdysone target genes are bracketed. (B) Expression of known ecdysone target genes in Nurf301 mutants relative to wild type. Color bar denotes the magnitude and direction of change. Full microarray data sets are available at http://home.ccr.cancer.gov/badenhorst.
An involvement of NURF in EcR signaling agrees well with the pupariation defects we observe in Nurf301 mutants, which resemble the phenotypes of mutants in key downstream regulatory targets of the ecdysone receptor (Karim et al. 1993; Fletcher et al. 1995). To further confirm that NURF is required for ecdysone signaling, we examined the expression in Nurf301 mutants of all known ecdysone targets that had been annotated and included in the Affymetrix microarrays. Expression is shown schematically relative to the wild-type reference strain in Figure 2B. In Nurf301 mutants, expression of a significant majority of these genes was reduced by greater than fivefold. The few genes for which there were no relative differences in expression are genes that are predominantly expressed at the prepupal/pupal transition (after the point at which we sampled; for example, Edg84A and Eip63F-1 [Andres et al. 1993; Andres and Thummel 1995]) or whose induction would be difficult to detect in whole animals because of a high background of tissues in which expression is constitutive or even repressed by ecdysone—Eip71CD (Eip28/29) and Eip55E (Eip40) (Andres and Cherbas 1992).
Remarkably, although approximately equal numbers of genes exhibit elevated or reduced expression in Nurf301 mutants, among known ecdysone-responsive genes, Nurf301 mutation produced only decreases in expression. This suggests that NURF functions specifically as a coactivator in the ecdysone response. Finally, our microarray analysis shows that transcript levels of known ecdysone synthetic enzymes are unchanged in Nurf301 mutants (data not shown), indicating that NURF does not indirectly influence expression of responsive genes by affecting ecdysone levels.
Altered expression of EcR target genes in NURF mutants
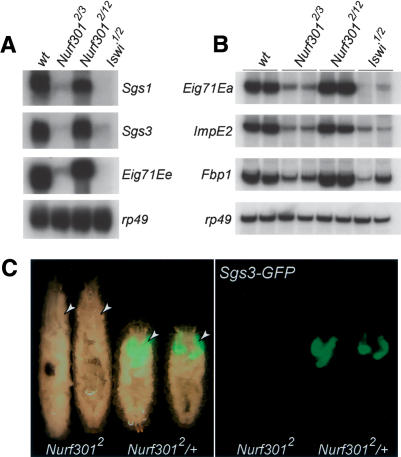
Next, we validated the altered expression of selected ecdysone targets by analyzing transcript levels using Northern analysis and semiquantitative RT-PCR. As shown in Figure 3A, Northern blotting confirms that Sgs1, Sgs3, and Eig71Ee are not expressed in null Nurf301 mutants (Nurf3012/Nurf3013 allelic combination) or Iswi mutants that lack the catalytic subunit of NURF. Similarly, semiquantitative RT-PCR confirms that expression of Eig71Ea, ImpE2, and Fbp1 is reduced in null Nurf301 or Iswi mutants (Fig. 3B). In contrast, transcript levels of EcR and usp are unchanged in Nurf301 mutants (Supplementary Fig. 1), indicating that the failure to express ecdysone target genes is not an indirect effect of reduced levels of the ecdysone receptor. As expected, allelic combinations with Nurf301 mutants that truncate after the putative WAKZ motif, and that are able to pupariate, do express ecdysone target genes (Nurf3012/Nurf30112 in Fig. 3A,B).
Figure 3.
NURF mutants do not express EcR target genes. Northern analysis of Sgs1, Sgs3, and Eig71Ee expression (A) and RT-PCR analysis of Eig71Ea, ImpE2, and Fbp1 expression (B) in Nurf301 and Iswi mutants relative to rp49. For brevity, the heteroallelic Nurf301 mutant combinations Nurf3012/Nurf3013 and Nurf3012/Nurf30112 are denoted by Nurf3012/3 and Nurf3012/12. The Iswi1/Iswi2 mutant combination is denoted by Iswi1/2. Using the null Nurf3012/Nurf3013 allelic combination in the Northern and RT-PCR analyses to validate microarray data (obtained using the null Nurf3012/Nurf3018 allelic combination) provides assurance against the potential influence of second site mutations in these strains. In B, data from two independent samples for each genotype are shown. (C) Expression of an Sgs3-GFP reporter transgene (Biyasheva et al. 2001) in Sgs3-GFP; Nurf3012/Nurf3012 homozygous mutants (-/-) and Sgs3-GFP; Nurf3012/TM6B, Tb1 siblings (-/+). Arrowheads label salivary glands.
Lastly, as an additional demonstration that NURF is required for Sgs3 transcription, we examined expression of an Sgs3-GFP reporter transgene in null Nurf301 mutant animals. Sgs3-GFP is expressed in the salivary glands of Nurf3012/+ heterozygous animals but is not expressed in homozygous mutant Nurf3012 animals (Fig. 3C, arrowheads).
NURF physically associates with EcR
The failure of ecdysone-responsive genes to be expressed in NURF mutants indicates that NURF is a coactivator of the Drosophila ecdysone receptor (EcR). Thus, we tested whether NURF could physically interact with EcR. As shown by a pull-down assay (Fig. 4B), in vitro-translated EcR isoforms, EcR-A and EcR-B2, interacted with Flag-tagged recombinant NURF. These interactions were dependent on the presence of added ecdysone. No pull-down was observed in the absence of 20-hydroxyecdysone (Fig. 4B, lanes 2,5).
Figure 4.
NURF binds EcR in an ecdysone-dependent manner. (A) Structure of EcR isoforms and derivatives tested for NURF binding. (A/B) Variable-length N-terminal domain; (C) DNA-binding domain; (E) ligand-binding domain; (F) variable-length C-terminal domain; (GAL4 DBD) GAL4 DNA-binding domain; (AF1) activation function 1; (AF2) activation function 2 (amino acids 636-655 of the LBD). Full details of the GAL4 DBD-EcR fusions used in NURF pull-down are listed in the Supplemental Material. (B) Purified Flag-tagged recombinant NURF binds in vitro-translated EcR-A and EcR-B2 isoforms, and GBT-EL but not GBT-EL650 or GBT-EL/F645A. Binding is only detected in the presence (+) of 10 μM 20-hydroxyecdysone. Addition of solvent, ethanol, alone (-) cannot induce binding of NURF to EcR isoforms.
The ligand dependency of the NURF-EcR interaction implies that NURF functions as a coactivator for the ecdysone receptor. Like other NRs, EcR has two transcriptional activation function (AF) domains, the conserved AF2 located within the ligand-binding domain and isoform-specific AF1s at the N terminus (Fig. 4A). We observed that NURF is able to pull down, in a liganddependent manner, a minimal construct that contains the entire AF2 domain (GBT-EL in Fig. 4B). However, inactivation of AF2 either by C-terminal truncation (GBT-EL650 in Fig. 4B), or by mutation of F645, a conserved residue critical for interaction of mammalian NRs with coactivators (GBT-EL/F645A in Fig. 4B), blocks interaction with NURF. Our results extend the repertoire of transcription factors shown to interact with NURF. These data indicate that purified NURF is a coactivator that binds to the AF2 region of EcR, in addition to previously demonstrated interactions with the GAGA factor, GAL4-VP16, and HSF (Xiao et al. 2001).
NURF mutants enhance phenotypes caused by targeted blockade of EcR
To confirm further that NURF functions in ecdysone signaling in vivo, we assayed genetic interactions between EcR and components of NURF. A dominant-negative EcR mutant (EcR-F645A) was expressed in follicle cells in the developing egg chamber of female flies. EcR-F645A is defective in transcriptional activation (Hu et al. 2003) and interferes with ecdysone signaling, leading to a number of embryo defects including malformed dorsal appendages (see Fig. 5A). Decreasing the titer of a coactivator has been shown to enhance this phenotype (Cherbas et al. 2003). We observed that mutation of a single copy of any of three NURF subunits increases the frequency and severity of these aberrations (Fig. 5), consistent with NURF functioning as a coactivator for the ecdysone receptor.
Figure 5.
NURF mutants enhance dominant-negative EcR phenotypes. Embryos derived from females overexpressing a dominant-negative EcR variant (UAS-EcR-F645A) under the control of slbo-GAL4 show a low frequency (23%) of abnormal dorsal appendages. Removing one copy of a NURF subunit (by introducing either a Nurf301, Iswi, or Nurf38 mutant allele into the slbo-GAL4::UAS-EcR-F645A background) increases the frequency and severity of these abnormalities. (A, left) Dorsal appendages of a wild-type egg. Dorsal appendages of embryos from GAL4::UAS-EcR-F645A; Nurf301/+ females show malformation (arrowheads) and fusion (arrows). (B) Frequency of abnormal dorsal appendages in embryos from slbo-GAL4::UAS-EcR-F645A females and slbo-GAL4::UAS-EcR-F645A females containing one copy of either a Nurf301, Iswi, or Nurf38 mutant allele. (n) The number of embryos scored.
ATP-dependent chromatin remodeling and nuclear receptor function
Our results provide one of the first demonstrations of a biological requirement for an ISWI-containing chromatin remodeling enzyme (NURF) in steroid hormone-dependent transcriptional activation. To date, most studies of ATP-dependent chromatin remodeling during NR transactivation have focused on the SWI/SNF family of chromatin remodeling complexes. It has been shown that SWI/SNF enzymes are required for activation by the retinoid receptor heterodimer (RAR/RXR), glucocorticoid receptor, and estrogen receptor (Yoshinaga et al. 1992; Muchardt and Yaniv 1993; Fryer and Archer 1998; Dilworth et al. 2000). Moreover, interaction studies reveal that SWI/SNF remodeling complexes can be targeted to NRs through interactions with the noncore subunits BAF250, BAF57, and BAF60a (Nie et al. 2000; Belandia et al. 2002; Hsiao et al. 2003).
However, there are many families of ATP-dependent chromatin remodeling complexes, including those based on the SWI2/SNF2, ISWI, INO80, and CHD1 catalytic subunits. Each group of remodeling enzymes has distinct mechanisms of operation (for review, see Korbe and Horz 2004; Langst and Becker 2004). For example, SWI/SNF enzymes increase chromatin accessibility by DNA looping, whereas ISWI enzymes induce nucleosome sliding and the SWR1 category catalyzes histone exchange. It is important to determine whether NRs exclusively employ the SWI2/SNF2 branch or use a much wider repertoire of remodeling enzymes to exert their functions.
Here we provide evidence that the ISWI-containing chromatin remodeling enzyme NURF is a coactivator of a Drosophila NR, the ecdysone receptor. In addition to previous demonstrations of direct interactions between NURF and the GAGA factor and HSF (Xiao et al. 2001), we show that purified NURF binds to EcR in an ecdysone-dependent manner, suggesting it is a direct effector of NR activity. Our conclusions are broadly consistent with two previous studies that indicated that ISWI complexes are required for NR-dependent transactivation in vitro. Di Croce et al. (1999) observed that addition of recombinant ISWI stimulates activation of a chromatinized MMTV promoter by progesterone receptor. Dilworth et al. (2000) suggested that ATP-dependent chromatin remodeling by an ISWI-containing remodeling complex (present in chromatin assembly extracts) was required to stabilize the binding of RAR/RXR to targets in chromatin.
Inspection of the NURF301 coding sequence, and of the corresponding rat and human homologs, reveals that NURF301 contains two conserved NR (LxxLL) boxes (Fig. 1A). These motifs can mediate interaction between NRs and transcriptional coactivators (Darimont et al. 1998) and suggest a mechanism by which NURF can interact with, and be recruited by, EcR. We have as yet, despite numerous efforts, been unable to generate a suitable antibody that allows us to directly visualize recruitment of NURF to ecdysone-responsive promoters. However, chromatin immunoprecipitation (ChIP) using anti-ISWI antibodies shows that an ISWI-containing complex binds to the ecdysone response element of the hsp27 promoter. This ISWI ChIP signal is lost in Nurf301 mutants, suggesting that it is due to NURF recruitment (Supplementary Fig. 2).
Separation of function among ISWI-containing complexes
Analyses of Drosophila Iswi mutants have provided critical insights into the functions of ISWI-containing chromatin remodeling enzymes. These studies provided the first demonstration that ISWI chromatin remodeling complexes are required to maintain male X-chromosome morphology, for homeotic gene expression and metamorphosis (Deuring et al. 2000). However, Drosophila ISWI is the catalytic ATPase subunit of at least three complexes: ACF, CHRAC, and NURF (for review, see Corona and Tamkun 2004). As such, analysis of Iswi mutants alone does not allow the relative functions of these complexes to be discriminated. By focusing our investigations on the NURF-specific subunit NURF301, we have been able to define specific functions of NURF. The pupariation defects seen in Nurf301 mutants, and the reduced ecdysone target gene expression noted in Nurf301 mutants, highlights the critical function of NURF in EcR-dependent activation. As expected, defects seen in Nurf301 mutants are also observed in Iswi mutants. However, mutations in subunits specific to ISWI complexes other than NURF, for example, the ACF1 subunit of ACF and CHRAC, do not affect EcR function. Acf1 null mutants are semi-lethal but are able to produce viable adults (Fyodorov et al. 2004).
Our studies on Drosophila NURF have important implications for NR function in mammals. Homologs of ISWI and the large subunit NURF301 exist in mouse and humans (Jones et al. 2000; Lazzaro and Picketts 2001). Moreover, human NURF has been purified and exhibits identical biochemical properties as Drosophila NURF (Barak et al. 2003). The human homolog of a Drosophila NURF target, engrailed (Deuring et al. 2000; Badenhorst et al. 2002), is also a target of human NURF (Barak et al. 2003). Given this conservation of function, it will be of interest to determine if gene targets of mammalian NRs related to Drosophila EcR also require NURF for expression.
Materials and methods
Genetics and Drosophila strains
Flies were raised at 25°C. All strains are as described by FlyBase (http://flybase.bio.indiana.edu). Full details of Nurf301 mutants and genetic crosses are in the Supplemental Material.
Whole genome expression analysis
RNA was isolated from Nurf3012/Nurf3018 mutant third instar larvae and from larvae of the w1118 isogenic line in which the Nurf301 EMS lesions were generated. Culture conditions, larval staging, and RNA isolation are described in the Supplemental Material. RNA was labeled and hybridized to Affymetrix Drosophila Genome Arrays according to the manufacturer's instructions. Microarrays were washed using an Affymetrix GeneChip Fluidics Station 400 at the University of Iowa microarray facility. Image data were captured using an Affymetrix GeneChip array scanner and converted to numerical output using Microarray Analysis Suite version 5.0. Further analysis and comparison was using Genespring software. Comparisons between wild-type and Nurf301 microarray data were made using the averaged signals over all samples.
Northern analysis and RT-PCR
Northern hybridizations were performed as described previously (Badenhorst et al. 2002). RT-PCR was performed using an Access RT-PCR kit (Promega). Before the amplification step, 0.2 μCi [α-32P]-deoxyadenosine 5′-triphosphate (Perkin Elmer, specific activity 6000Ci/mmol) was added as a tracer. Samples were run on 6% native PAGE gels, dried, and visualized using a Typhoon phosphorimager (Amersham Biosciences).
Protein interaction studies
35S-labeled EcR isoforms and derivatives (Hu et al. 2003) were in vitro translated using the TNT Quick-coupled transcription/translation system (Promega) in the presence of 35S-methionine. Flag-tagged NURF was generated by coexpression of all four recombinant NURF subunits in SF9 cells and purified as described (Xiao et al. 2001). Flag-tagged NURF exhibits potent nucleosome sliding activity. Purified NURF bound to anti-Flag agarose beads (Sigma) was incubated with 35S-labeled EcR in the presence of in vitro-translated USP for 30 min on ice. Beads were washed several times with ACB buffer (20 mM Hepes at pH 7.6, 2 mM MgCl2, 10% glycerol, 0.02% NP-40, 100 mM KCl) and bound proteins analyzed by SDS-PAGE followed by autoradiography. Ecdysone dependence of interactions was tested by addition of 10 μM 20-hydroxyecdysone (Sigma).
Enhancement of targeted EcR blockade
Newly eclosed adult females of the genotypes w1118; UAS-EcR-F645A/slbo-GAL4 UAS-GFP; Nurf301/+ w1118; UAS-EcR-F645A, Nurf38/slbo-GAL4 UAS-GFP; +/+ w1118; UAS-EcR-F645A, Iswi/slbo-GAL4 UAS-GFP; +/+ were mated to w1118 males and allowed to age for 1-2 wk at 20°C. Ovaries were then dissected, squashed, and observed by phase microscopy. Eggs were scored as either wild type or defective based on dorsal appendage morphology. Further details of the analysis are described in the Supplemental Material.
Acknowledgments
The following people generously provided flies and reagents for which we are grateful: David Bevan, Jim Kennison, Alban Moon, Yikang Rong, and John Tamkun. This work was supported by grants from the NSF to P.C., from the Wellcome Trust and Royal Society to P.B., and by the Intramural research program of the National Cancer Institute.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1342605.
Corresponding authors.
References
- Andres A.J. and Cherbas, P. 1992. Tissue-specific ecdysone responses: Regulation of the Drosophila genes Eip28/29 and Eip40 during larval development. Development 116: 865-876. [DOI] [PubMed] [Google Scholar]
- Andres A.J. and Thummel, C.S. 1995. The Drosophila 63F early puff contains E63-1, an ecdysone-inducible gene that encodes a novel Ca2+-binding protein. Development 121: 2667-2679. [DOI] [PubMed] [Google Scholar]
- Andres A.J., Fletcher, J.C., Karim, F.D., and Thummel, C.S. 1993. Molecular analysis of the initiation of insect metamorphosis: A comparative study of Drosophila ecdysteroid-regulated transcription. Dev. Biol. 160: 388-404. [DOI] [PubMed] [Google Scholar]
- Arbeitman M.N., Furlong, E.E., Imam, F., Johnson, E., Null, B.H., Baker, B.S., Krasnow, M.A., Scott, M.P., Davis, R.W., and White, K.P. 2002. Gene expression during the life cycle of Drosophila melanogaster. Science 297: 2270-2275. [DOI] [PubMed] [Google Scholar]
- Badenhorst P., Voas, M., Rebay, I., and Wu, C. 2002. Biological functions of the ISWI chromatin remodeling complex NURF. Genes & Dev. 16: 3186-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak O., Lazzaro, M.A., Lane, W.S., Speicher, D.W., Picketts, D.J., and Shiekhattar, R. 2003. Isolation of human NURF: A regulator of Engrailed gene expression. EMBO J. 22: 6089-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belandia B., Orford, R.L., Hurst, H.C., and Parker, M.G. 2002. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 21: 4094-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biyasheva A., Do, T.V., Lu, Y., Vaskova, M., and Andres, A.J. 2001. Glue secretion in the Drosophila salivary gland: A model for steroid-regulated exocytosis. Dev. Biol. 231: 234-251. [DOI] [PubMed] [Google Scholar]
- Cherbas L., Hu, X., Zhimulev, I., Belyaeva, E., and Cherbas, P. 2003. EcR isoforms in Drosophila: Testing tissue-specific requirements by targeted blockade and rescue. Development 130: 271-284. [DOI] [PubMed] [Google Scholar]
- Collingwood T.N., Urnov, F.D., and Wolffe, A.P. 1999. Nuclear receptors: Coactivators, corepressors and chromatin remodeling in the control of transcription. J. Mol. Endocrinol. 23: 255-275. [DOI] [PubMed] [Google Scholar]
- Corona D.F. and Tamkun, J.W. 2004. Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim. Biophys. Acta 1677: 113-119. [DOI] [PubMed] [Google Scholar]
- Darimont B.D., Wagner, R.L., Apriletti, J.W., Stallcup, M.R., Kushner, P.J., Baxter, J.D., Fletterick, R.J., and Yamamoto, K.R. 1998. Structure and specificity of nuclear receptor-coactivator interactions. Genes & Dev. 12: 3343-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuring R., Fanti, L., Armstrong, J.A., Sarte, M., Papoulas, O., Prestel, M., Daubresse, G., Verardo, M., Moseley, S.L., Berloco, M., et al. 2000. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell 5: 355-365. [DOI] [PubMed] [Google Scholar]
- Di Croce L., Koop, R., Venditti, P., Westphal, H.M., Nightingale, K.P., Corona, D.F., Becker, P.B., and Beato, M. 1999. Two-step synergism between the progesterone receptor and the DNA-binding domain of nuclear factor 1 on MMTV minichromosomes. Mol. Cell 4: 45-54. [DOI] [PubMed] [Google Scholar]
- Dilworth F.J., Fromental-Ramain, C., Yamamoto, K., and Chambon, P. 2000. ATP-driven chromatin remodeling activity and histone acetyl-transferases act sequentially during transactivation by RAR/RXR In vitro. Mol. Cell 6: 1049-1058. [DOI] [PubMed] [Google Scholar]
- Fischle W., Wang, Y., and Allis, C.D. 2003. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15: 172-183. [DOI] [PubMed] [Google Scholar]
- Fletcher J.C., Burtis, K.C., Hogness, D.S., and Thummel, C.S.1995. Drosophila E74 gene is required for metamorphosis and plays a role in the polytene chromosome puffing response to ecdysone. Development 121: 1455-1465. [DOI] [PubMed] [Google Scholar]
- Fryer C.J. and Archer, T.K. 1998. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature 393: 88-91. [DOI] [PubMed] [Google Scholar]
- Fyodorov D.V., Blower, M.D., Karpen, G.H., and Kadonaga, J.T. 2004. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes & Dev. 18: 170-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass C.K., Rose, D.W., and Rosenfeld, M.G. 1997. Nuclear receptor coactivators. Curr. Opin. Cell Biol. 9: 222-232. [DOI] [PubMed] [Google Scholar]
- Hamiche A., Sandaltzopoulos, R., Gdula, D.A., and Wu, C. 1999. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell 97: 833-842. [DOI] [PubMed] [Google Scholar]
- Hsiao P.W., Fryer, C.J., Trotter, K.W., Wang, W., and Archer, T.K. 2003. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol. Cell. Biol. 23: 6210-6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Cherbas, L., and Cherbas, P. 2003.Transcription activation by the ecdysone receptor (EcR/USP): Identification of activation functions. Mol. Endocrinol. 17: 716-731. [DOI] [PubMed] [Google Scholar]
- Jones M.H., Hamana, N., and Shimane, M. 2000. Identification and characterization of BPTF, a novel bromodomain transcription factor. Genomics 63: 35-39. [DOI] [PubMed] [Google Scholar]
- Karim F.D., Guild, G.M., and Thummel, C.S. 1993. Drosophila Broad-Complex plays a key role in controlling ecdysone-regulated gene expression at the onset of metamorphosis. Development 118: 977-988. [DOI] [PubMed] [Google Scholar]
- Korber P. and Horz, W. 2004. SWRred not shaken; mixing the histones. Cell 117: 5-7. [DOI] [PubMed] [Google Scholar]
- Langst G. and Becker, P.B. 2004. Nucleosome remodeling: One mechanism, many phenomena? Biochim. Biophys. Acta 1677: 58-63. [DOI] [PubMed] [Google Scholar]
- Lazzaro M.A. and Picketts, D.J. 2001. Cloning and characterization of the murine Imitation Switch (ISWI) genes: Differential expression patterns suggest distinct developmental roles for Snf2h and Snf2l. J. Neurochem. 77: 1145-1156. [DOI] [PubMed] [Google Scholar]
- Lusser A. and Kadonaga, J.T. 2003. Chromatin remodeling by ATP-dependent molecular machines. Bioessays 25: 1192-1200. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G., Tsukiyama, T., Wisniewski, J., and Wu, C. 1997. Role of nucleosome remodeling factor NURF in transcriptional activation of chromatin. Mol. Cell 1: 141-150. [DOI] [PubMed] [Google Scholar]
- Muchardt C. and Yaniv, M. 1993. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 12: 4279-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z., Xue, Y., Yang, D., Zhou, S., Deroo, B.J., Archer, T.K., and Wang, W. 2000. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell. Biol. 20: 8879-8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalsky M.L. 2004. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu. Rev. Physiol. 66: 315-360. [DOI] [PubMed] [Google Scholar]
- Sedkov Y., Cho, E., Petruk, S., Cherbas, L., Smith, S.T., Jones, R.S., Cherbas, P., Canaani, E., Jaynes, J.B., and Mazo, A. 2003. Methylation at lysine 4 of histone H3 in ecdysone-dependent development of Drosophila. Nature 426: 78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H.E., Stunnenberg, H.G., and Stewart, A.F. 1993. Heterodimerization of the Drosophila ecdysone receptor with retinoid X receptor and ultraspiracle. Nature 362: 471-475. [DOI] [PubMed] [Google Scholar]
- Thummel C.S. 2001. Molecular mechanisms of developmental timing in C. elegans and Drosophila. Dev. Cell 1: 453-465. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T. and Wu, C. 1995. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell 83: 1011-1020. [DOI] [PubMed] [Google Scholar]
- Turner B.M. 2002. Cellular memory and the histone code. Cell 111: 285-291. [DOI] [PubMed] [Google Scholar]
- Xiao H., Sandaltzopoulos, R., Wang, H.M., Hamiche, A., Ranallo, R., Lee, K.M., Fu, D.G., and Wu, C. 2001. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell 8: 531-543. [DOI] [PubMed] [Google Scholar]
- Yao T.P., Forman, B.M., Jiang, Z., Cherbas, L., Chen, J.D., McKeown, M., Cherbas, P., and Evans, R.M. 1993. Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature 366: 476-479. [DOI] [PubMed] [Google Scholar]
- Yoshinaga S.K., Peterson, C.L., Herskowitz, I., and Yamamoto, K.R. 1992. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science 258: 1598-1604. [DOI] [PubMed] [Google Scholar]