Abstract
To promote faithful propagation of the genetic material during sexual reproduction, meiotic chromosomes undergo specialized morphological changes that ensure accurate segregation of homologous chromosomes. The molecular mechanisms that establish the meiotic chromosomal structures are largely unknown. We describe a mutation in a recently identified Histone H2A kinase, nhk-1, in Drosophila that leads to female sterility due to defects in the formation of the meiotic chromosomal structures. The metaphase I arrest and the karyosome, a critical prophase I chromosomal structure, require nucleosomal histone kinase-1 (NHK-1) function. The defects are a result of failure to disassemble the synaptonemal complex and to load condensin onto the mutant chromosomes. Embryos laid by nhk-1-/- mutant females arrest with aberrant polar bodies and mitotic spindles, revealing that mitosis is affected as well. We analyzed the role of Histone H2A phosphorylation with respect to the histone code hypothesis and found that it is required for acetylation of Histone H3 and Histone H4 in meiosis. These studies reveal a critical role for histone modifications in chromosome dynamics in meiosis and mitosis.
Keywords: Karyosome, synaptonemal complex, condensin, histone acetylation, chromosome structure
Chromosomes have three primary functions: to store genetic information, to enable gene expression, and to ensure faithful propagation of the genetic material to future generations. These diverse functions impose unique structural requirements and are reflected by the dramatic structural changes that the chromosomes undergo. In interphase, the chromosomes are decondensed, corresponding to open chromatin accessible for transcription. During mitosis, the chromosomes are condensed into rigid rods amenable to efficient transport to daughter cells. The molecular mechanisms that regulate chromosome dynamics are complex and not completely understood.
Sexual reproduction introduces additional challenges to chromosomes. Meiosis is a specialized cell division program that produces haploid gametes (oocytes and sperm) from diploid progenitors (for recent reviews, see Champion and Hawley 2002; Marston and Amon 2004). This is accomplished through two chromosomal segregation events, meiosis I and meiosis II, without an intervening DNA replication. To accommodate the specialized processes that occur during gametogenesis, meiotic chromosomes undergo specific morphological changes. In prophase of meiosis I, recombination occurs during which homologous chromosomes exchange genetic material. The synaptonemal complex (SC) is a protein lattice that runs along the length of the homologous chromosomes and stabilizes their pairing (Page and Hawley 2004). During recombination, double-strand break (DSB) formation and repair generate hybrid chromosomes, contributing to genetic diversity and allowing proper chromosome segregation by providing a physical link between homologs via chiasmata.
Subsequent to recombination in all metazoans, the oocytes remain in a prolonged prophase I that facilitates coordination of meiosis with development (Spradling 1993; Orr-Weaver 1995). During this time, the oocytes grow and accumulate nutrients essential to the embryo following fertilization. In the prophase I arrest in Drosophila, the chromosomes partially condense and form a characteristic structure within the oocyte nucleus, called a karyosome. Although the function of the karyosome is not clear, similar prophase I chromosomal arrangements are conserved among diverse organisms and may keep the chromosomes in close proximity to each other within the large volume of the oocyte nucleus (Gruzova and Parfenov 1993). The exact role of the karyosome and the mechanism by which it is formed remain exciting unanswered problems.
Another meiosis-specific chromosomal arrangement is the arrest at metaphase of either meiosis I (in insects) or meiosis II (in vertebrates) in preparation for fertilization. In Drosophila, upon release of the prophase I arrest, the oocyte chromosomes organize a spindle, remaining aligned at the metaphase I plate until ovulation (Theurkauf and Hawley 1992; Page and Orr-Weaver 1997). Upon ovulation, the metaphase I arrest is released, and meiosis resumes and proceeds to completion as the egg is fertilized and deposited. Proper completion of meiosis results in the formation of the polar body and establishment of the initial mitotic spindle in the embryos. The polar body, also called a rosette due to the characteristic arrangement of the chromosomes, forms from the condensed, unused meiotic products. Rosette formation is an excellent diagnostic for proper completion of meiosis. The fourth meiotic product, the female pronucleus, migrates to the male pronucleus, forming the first mitotic spindle and the zygotic genome. Proper regulation of meiosis-specific chromosomal dynamics is essential to the formation of viable gametes and offspring. Yet the molecular mechanisms that govern meiotic chromosome dynamics and architecture are poorly understood.
Major contributors to the diversity of chromosome structure are the chromosomal protein components, including the histones (Kornberg and Lorch 1999). Histones are subject to a variety of post-translational modifications, primarily at their N-terminal tails (Zhang and Reinberg 2001; Carrozza et al. 2003; Fischle et al. 2003a,b). Histone modifications affect all aspects of chromosome biology, including transcription, replication, recombination, and condensation, by affecting chromosome structure and by recruiting specific chromatin-binding proteins. The correlation between histone acetylation and transcriptional activity is one of the better understood functions of histone modifications (Eberharter and Becker 2002). Histone H1 and Histone H3 phosphorylation during prophase and metaphase of mitosis is associated with chromosome condensation through an unknown molecular mechanism (Hsu et al. 2000). Methylation of Histone H3 Lys 9 recruits chromodomain-containing proteins, such as heterochromatin protein 1 (HP1) (Lachner et al. 2001), while acetylation of histones recruits bromodomain-containing proteins (Dhalluin et al. 1999; Winston and Allis 1999; Zeng and Zhou 2002). Finally, phosphorylation of the histone variant, Histone H2A.X, or Histone H2Av in Drosophila, on a conserved serine is associated with formation and repair of DSBs during recombination and DNA damage (Rogakou et al. 1999; Mahadevaiah et al. 2001).
Additional histone modifications, including ones in regions outside the canonical N-terminal tails, have been identified more recently. Phosphorylation of Thr 119 in the C terminus of Drosophila Histone H2A (H2AT119ph) was identified biochemically (Aihara et al. 2004), but the biological functions of this modification are unknown. The Thr 119 residue is conserved among Histone H2A homologs, suggesting that it plays an important function. Nucleosomal histone kinase-1 (NHK-1) was shown to be specifically responsible for phosphorylating Histone H2A on Thr 119. NHK-1 is conserved among metazoans, indicating that NHK-1 homologs may function as histone kinases in other systems. Histone H2AT119 phosphorylation and NHK-1 protein were detected in embryos, consistent with a function in mitosis (Aihara et al. 2004).
In this study, we describe the first mutation in the nhk-1 gene. Through phenotypic analysis of the mutant, we determined the functional requirements for this histone modification in female meiosis. NHK-1 function is required during meiosis, and a mutation in nhk-1 leads to altered architecture of the oocyte chromosomes, including failure to assemble a karyosome, a metaphase I spindle, and a normal polar body. The SC formed normally and DSBs were formed and repaired. Strikingly, the SC failed to disassemble, and condensin failed to load on the oocyte chromosomes in the mutant. Therefore, by starting with a female-sterile mutation in Drosophila, we have gained further insight both into the requirements for chromosomal structure during meiosis and into the function of a histone modification.
Results
Identification of a mutation in nhk-1
The molecular mechanisms of meiosis are poorly understood. To identify new genes required for meiosis, we screened a collection of female-sterile mutants (Koundakjian et al. 2004) and identified Z3-0437 because the eggs laid by mutant females were arrested prior to completion of the first mitotic division.
The two chromosomal structures normally present in early embryos had aberrant morphologies in the arrested embryos. In wild-type embryos, the three unused meiotic products condense into a polar body, or rosette (Fig. 1A), surrounded by a mesh of microtubules (Fig. 1B). The polar bodies in the embryos laid by mutant females had aberrant configurations of chromosomes and microtubules. In some arrested embryos, the microtubules surrounding the polar body were normal (Fig. 1D), but the chromosomes within the polar body failed to assemble into a normal rosette (Fig. 1C). Alternatively, both the chromosomes and the microtubules of the polar body had aberrant morphologies, more closely resembling mitotic spindles, but lacking spindle asters (Fig. 1E,F).
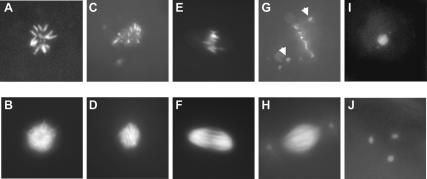
Figure 1.
The nhk-1Z3-0437 mutation leads to aberrant chromosomal arrangements in the polar body, the mitotic spindle, and the metaphase I spindle. (A) Normal polar body, or rosette, in a control embryo stained with DAPI. (B) Normal microtubule arrangement in a control embryo. (C) The chromosomes in some embryos laid by nhk-1Z3-0437 mutant females formed aberrant rosettes. (D) Microtubules surrounding the abnormal rosette in C had a normal appearance. (E) The chromosomes in other embryos laid by nhk-1Z3-0437 mutant females appeared aligned on a spindle plate. (F) Microtubules surrounding the chromosomes in E formed a spindle that lacked asters. (G) Embryos laid by nhk-1Z3-0437 mutant females had one or two additional chromosomal arrangements that represented the female and male pronuclei with the majority of chromosomes at a mitotic spindle plate. Some chromosomes in these spindles failed to align at the plate and were found near the spindle poles (arrows). (H) The microtubules surrounding the aberrant mitotic chromosomes appeared normal and had asters, suggesting that the embryos were fertilized. (I) Normal metaphase I chromosome configuration in control stage 14 oocytes.(J) The chromosomes in nhk-1Z3-0437 mutant stage 14 oocytes failed to align at a metaphase I spindle and were found in three distinct foci.
In wild-type embryos, the fourth female meiotic product, the female pronucleus, migrates to the male pronucleus to establish the mitotic spindle, characterized by spindle asters. Most of the arrested embryos had a single mitotic spindle with asters (Fig. 1H). However, some chromosomes within the spindle were aberrantly localized to the poles, either due to failure to congress to the spindle plate or due to premature migration to the poles (Fig. 1G, arrows). We did not observe anaphase or interphase figures in the embryos, indicating a terminal arrest at metaphase. These results suggest that the mutant affects mitosis, or that defects in meiosis result in inability to proceed through the mitotic division in the early embryo.
The presence of a spindle with asters is consistent with the embryos being fertilized because the centrioles that nucleate the asters are provided by the sperm. We confirmed this possibility by mating the mutant females to males that carry a protein component of the sperm tail fused to green fluorescent protein (GFP) (Santel et al. 1997). We observed GFP-containing sperm tails in the embryos laid by mutant females, showing that the embryos were fertilized (data not shown). Fertilization suggests that the developmental program of the oocyte is unaffected in the mutant and that the phenotype is due specifically to defects in the cell division program. Taken together, the embryonic phenotypes reveal that the gene product mutated in this strain affects proper chromosomal dynamics.
Z3-0437 is a mutation in a histone kinase
To better understand the molecular function of the mutated gene, we determined the chromosomal location of the mutation to the CG6386/bällchen/nhk-1 gene (see Materials and Methods). The nhk-1 gene encodes a protein kinase with homologs in humans, mouse, Xenopus, and Caenorhabditis elegans (Aihara et al. 2004). In the Z3-0437 mutant, a single base pair substitution resulted in an amino acid change from Pro 117 to leucine (Fig. 2A). Pro 117 is in the kinase domain of NHK-1 and is highly conserved among kinases, suggesting that a mutation in that residue is likely to compromise the kinase activity of NHK-1. The Z3-0437 mutant is the first known mutation in nhk-1 in any organism. In addition to the female-sterile phenotype described here, this allele is also male-sterile, raising the possibility that nhk-1 has a role in the male germline. NHK-1 was previously detected in embryos (Aihara et al. 2004), suggesting that other alleles could cause lethality.
Figure 2.
nhk-1Z3-0437 is a mutation in a histone kinase. (A, top) A schematic of the NHK-1 protein structure. The black bar represents the kinase domain and the dark gray bar represents the basic-acidic-basic (BAB) domain conserved among the closest homologs of the NHK-1 (Aihara et al. 2004). The approximate position of the Z3-0437 Pro 117-to-leucine mutation is indicated. (Bottom) The protein sequence of a portion of the kinase domain of NHK-1 and its homologs, showing the Pro 117 (bold) mutated in the Z3-0473 mutant and just upstream of the catalytic loop important for kinase activity. (B) nhk-1 transcript is present in wild-type Drosophila ovaries—signal from antisense-strand probe. (C) The sense-strand probe shows the level of background signal. (D, top) Ovarian extracts were probed with anti-NHK-1 antibodies. The left lane is an extract from nhk-1Z3-0437/+ heterozygous control ovaries and shows that NHK-1 protein is present in the ovaries. The right lane is from nhk-1Z3-0437/Df mutant ovaries and shows a slight reduction in the NHK-1 protein levels due to the mutation. (Bottom) The same blot was stripped and reprobed with anti-α-tubulin antibodies as a control for equal loading of the extracts.
We used two methods to establish ovarian expression expected for a gene that plays a role in female meiosis. First, in situ hybridization with a probe against the antisense strand showed that the nhk-1 transcript is present in the ovaries at times of egg chamber development, during which the protein would be required for meiosis (Fig. 2B). The control, sense-strand probe showed a background level of signal (Fig. 2C). Second, we used an antibody against the NHK-1 protein (Aihara et al. 2004) to probe ovarian extracts and found that it is expressed in the ovaries, as would be expected for a protein with a function during meiosis (Fig. 2D, top panel). The NHK-1 protein levels in ovarian extracts from nhk-1Z3-0437/Df mutant females showed a slight reduction as compared with control nhk-1Z3-0437/+ extracts (Fig. 2D); thus, the P117L mutation also appears to lead to destabilization of the protein. The bottom panel in Figure 2D shows equal loading using an anti-α-tubulin antibody.
Given the specificity of NHK-1 in phosphorylating Histone H2AT119 (Aihara et al. 2004), the nhk-1Z3-0437 mutant presented a unique opportunity to dissect the role of a novel histone modification. Therefore, we analyzed its mutant phenotype in detail.
NHK-1 is required during meiosis
To determine whether NHK-1 is required during meiosis, we analyzed mutant ovaries by staining with DNA dye. We used three genetic backgrounds in our analysis: (1) nhk-1Z3-0437/nhk-1+, (2) nhk-1Z3-0437/nhk-1Z3-0437, and (3) nhk-1Z3-0437/Df(3R)Tl-I. We identified the Df(3R)Tl-I deficiency in the course of mapping the Z3-0437 mutation, due to its failure to complement the female sterility of Z3-0437 (see Materials and Methods). The deficiency is an important additional genetic tool because it completely removes the nhk-1 gene in an unrelated genetic background, confirming that the observed phenotypes are due to a mutation in nhk-1 and providing a loss-of-function allele. Because the deficiency removes other genes in addition to nhk-1, homozygous animals die early in development, and the deficiency was useful only in trans to the nhk-1 point mutation.
We observed two striking mutant phenotypes. First, we observed a defect in the metaphase I arrest in late-stage oocytes. In wild-type ovaries, the chromosomes in the oocytes at stage 14 align on the metaphase I spindle, with the nonexchange fourth chromosomes being pulled to the spindle poles (Fig. 1I). We observed normal metaphase I spindles in 50% of the mutant stage 14 oocytes. In the remaining 50% of the oocytes, the chromosomes were in three distinct foci (Fig. 1J), suggesting a failure to establish or maintain the metaphase I spindle. This phenotype was identical in both nhk-1Z3-0437/Z3-0437 and nhk-1Z3-0437/Df genetic backgrounds. These results show that NHK-1 function is required for establishment or maintenance of the metaphase I arrest.
Second, we observed a defect in formation of the karyosome in the nhk-1 mutant. The karyosome is a chromosomal structure that forms within the oocyte nucleus in prophase I (Fig. 3A). In the nhk-1Z3-0437/Z3-0437 mutant, a karyosome formed only in the early stage of oogenesis (stage 3), while in the nhk-1Z3-0437/Df background a karyosome failed to form at any stage. These observations suggest that the Z3-0437 allele is not a complete loss-of-function. To assist in identification of the oocyte chromosomes in the mutant, we stained the ovaries with antibodies against the cell cycle regulator Cyclin E that fills the oocyte nucleoplasm during prophase I. We found that the chromosomes in the mutant ovaries were at the periphery of the oocyte nucleus and had a beaded appearance (Fig. 3B). These results imply that NHK-1 function is required for formation and maintenance of the karyosome in the oocyte.
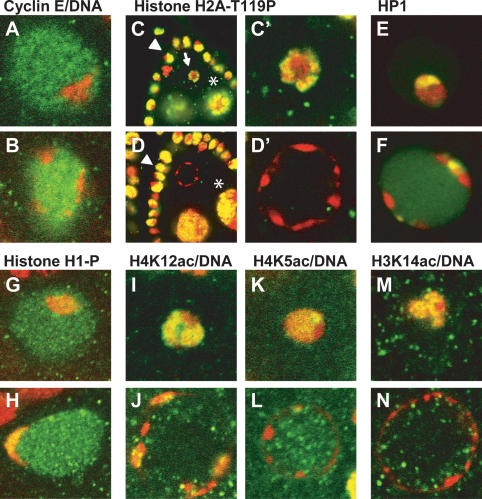
Figure 3.
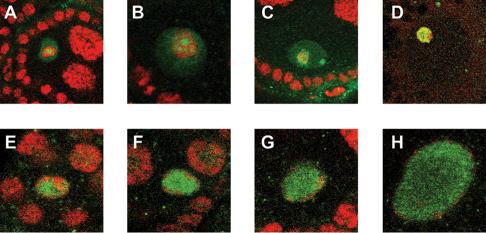
NHK-1 and the histone code. (A) The karyosome (stained with propidium iodide, red) is a chromosomal structure that forms within the prophase I nucleus (stained with Cyclin E, green) in control nhk-1Z3-0437/+ oocytes. (B) nhk-1Z3-0437/Z3-0437 mutant oocytes do not form a karyosome. Instead, the DNA (red) is found at the periphery of the oocyte nucleus (green). (C) Histone H2A-T119ph (green) is present in nurse cells (⋆), follicle cells (arrowhead), and the karyosomes (arrow) in control nhk-1Z3-0437/+ heterozygous ovaries, and it colocalizes with the DNA (red). Yellow denotes overlap between the signals. (C′) The control karyosome at a higher magnification showing the colocalization of Histone H2AT119ph with the DNA. (D) Histone H2A-T119ph persists in nurse cells (⋆) and follicle cells (arrowhead) in the mutant nhk-1Z3-0437/Z3-0437 ovaries. However, the DNA in the nhk-1Z3-0437/Z3-0437 mutant oocyte nucleus lacks Histone H2AT119ph. (D′) A higher magnification showing the absence of H2AT119ph in the mutant oocyte nucleus. (E) Heterochromatin protein 1 (HP1, green) localizes to the karyosome (red) in control nhk-1Z3-0437/+ oocytes. (F) HP1 colocalizes with the DNA in the nhk-1Z3-0437/Z3-0437 mutant oocytes. (G) Histone H1 is phosphorylated in the control nhk-1Z3-0437/+ karyosomes (DNA, red). (H) Phosphorylated Histone H1 colocalizes with the DNA (red) in the nhk-1Z3-0437/Z3-0437 mutant. (I) Histone H4K12 is acetylated in the control nhk-1Z3-0437/+ karyosomes (green, Histone H4K12ac; red, DNA; yellow, overlap between signals). (J) Histone H4K12ac (green) colocalizes with the DNA (red) in the nhk-1Z3-0437/Z3-0437 mutant oocytes (yellow, overlap between signals). (K) Histone H4K5 is acetylated in the control nhk-1Z3-0437/+ karyosomes (green, Histone H4K5ac; red, DNA; yellow, overlap between signals). (L) The DNA (red) in the nhk-1Z3-0437/Z3-0437 mutant oocytes is largely devoid of Histone H4K5ac (green). (M) Histone H3K14 is acetylated in the control karyosomes nhk-1Z3-0437/+ (green, Histone H3K14ac; red, DNA; yellow, overlap between signals). (N) The DNA (red) in the nhk-1Z3-0437/Df mutant oocytes is largely devoid of Histone H3K14ac (green).
Drosophila egg chambers are composed of three cell types: the somatically derived follicle cells, the germline-derived nurse cells, and the oocyte that undergoes meiosis (Spradling 1993). Having established that NHK-1 is required in the oocyte, it was of interest to analyze the follicle and nurse cells for any phenotypes. The follicle cells have a characteristic cell division program, starting with several mitotic divisions, followed by endocycles to increase the DNA content, and ending with amplification of four genomic loci (Claycomb and Orr-Weaver 2005). These characteristic chromosomal dynamics can be visualized by staining the ovaries with BrdU (Calvi et al. 1998; Calvi and Lilly 2004). We found that the follicle cells in the nhk-1Z3-0437/Z3-0437 mutant ovaries under-went the transitions from mitosis to endocycles to gene amplification in a manner and timing indistinguishable from their sibling controls (data not shown), suggesting that NHK-1 is not required in the follicle cells. Similarly, the chromosomes in the nurse cells undergo a distinctive transition from polyteny to polyploidy (King 1970). This transition was likewise unaffected in the nhk-1Z3-0437/Z3-0437 mutant ovaries (data not shown). We conclude that the nhk-1Z3-0437 mutation affects only the oocyte, providing an opportunity to analyze the function of NHK-1 specifically in meiosis.
Histone H2A is phosphorylated on T119 in meiosis
NHK-1 was shown to specifically phosphorylate Histone H2AT119 in vitro in the context of the nucleosome (Aihara et al. 2004). Therefore, we hypothesized that Histone H2AT119 is also a substrate of NHK-1 in meiosis. To test this hypothesis, we stained control nhk-1Z3-0437/+ Drosophila ovaries with an antibody specific to Histone H2AT119ph (Aihara et al. 2004). We found that this antibody stained the nuclei of all three ovarian cell types: the follicle cells, the nurse cells, and the oocyte (Fig. 3C,C′). Importantly, Histone H2AT119ph staining of the oocyte chromosomes was strongest at the time of SC disassembly, indicating that the peak of NHK-1 activity coincides with the first defect in the nhk-1 mutant (see below). Analysis of nhk-1Z3-0437/Df mutant ovaries with the anti-Histone H2AT119ph antibody showed that the staining persisted in the follicle cells and the nurse cells (Fig. 3D), but did not overlap with the DNA in the oocyte nucleus (Fig. 3D′). This suggests that the Z3-0437 allele specifically affects Histone H2A-T119ph in the oocyte and is consistent with the failure to observe mutant phenotypes in the nurse and follicle cells.
NHK-1 and the histone code
Covalent, post-translational modifications of histones affect chromosome dynamics by altering the pattern of proteins that bind to chromatin, and by affecting other histone modifications (Jenuwein and Allis 2001). The nhk-1 mutant provided the opportunity to analyze the consequences of lack of Histone H2AT119ph on other histone modifications and on binding of proteins to chromatin.
To explore the chromosomal architecture in the nhk-1Z3-0437 mutant oocytes, we analyzed the status of a panel of histone modifications (Table 1). First, localization of Histone H1ph is relevant because it occurs in early prophase, when we observed a phenotype in the nhk-1Z3-0437 mutant. Second, the localization pattern of HP1 is of interest because HP1 is recruited to chromatin by Histone H3K9me. Therefore, by analyzing HP1 localization, we monitored both the recruitment of proteins to chromatin and, indirectly, the methylation of Histone H3. We found that the karyosomes in the control nhk-1Z3-0437/+ Drosophila oocytes stained with HP1 (Fig. 3E) and Histone H1ph (Fig. 3G). In addition, we observed localization of HP1 (Fig. 3F) and Histone H1ph (Fig. 3H) to the nuclear periphery in the nhk-1Z3-0437/Z3-0437 mutant oocytes, coincident with the chromosomes. These results suggest that Histone H1ph, localization of HP1, and, indirectly, Histone H3K9me are not affected by a mutation in nhk-1. Therefore, despite the failure to form a karyosome, the chromosomes in the nhk-1Z3-0437 mutant oocytes are accessible to some chromatin-modifying proteins.
Table 1.
NHK-1 and the histone code in prophase I oocytes
| Modification | Control oocyte | Mutant oocyte |
|---|---|---|
| H2AT119pha | + | − |
| H1pha | + | + |
| HP1 (H3me)a | + | + |
| H3aca | + | − |
| H3K9acb | NDc | ND |
| H3K14acb | + | − |
| H4acb | + | + |
| H4K5aca | + | − |
| H4K8aca | ND | ND |
| H4K12aca | + | + |
| H4K16acb | ND | ND |
| H3pha | ND | ND |
| H2AS10pha | ND | ND |
| H2Auba | ND | ND |
Ovaries from control nhk-1Z3-0437/+ and mutant females were stained with antibodies specific to the protein or the histone modification listed in the first column. (+) Colocalization of the antibody and DNA was observed in the prophase I oocyte nucleus; (−) DNA in the oocyte nucleus failed to colocalize with the antibody.
Mutant oocytes were from nhk-1Z3-0437/Z3-0437.
Mutant oocytes were from nhk-1Z3-0437/Df.
The available antibodies to these modifications did not stain Drosophila ovaries reliably to permit evaluation of the mutant.
To address the third common histone modification, we analyzed acetylation of Histone H3 and Histone H4 by staining for residue-specific (Histone H3 K9 and K14, and Histone H4 K5, K8, K12, and K16) and general Histone H3 and Histone H4 acetylation. Antibodies against Histone H3K9ac, Histone H4K8ac, and H4K16ac failed to stain the control ovaries (Table 1), indicating that either the acetylations are absent from Drosophila ovaries or that the antibodies are incompatible with the tissue. Histone H4ac (Table 1) and Histone H4K12ac (Fig. 3I,J; Table 1) were present in both control and mutant karyosomes, suggesting that Histone H2AT119ph does not affect these modification in meiosis. Strikingly, we found that Histone H4K5 and Histone H3K14 were acetylated in the karyosomes in the control oocytes (Fig. 3K,M, respectively), but not in the nhk-1 mutant (Fig. 3L,N, respectively), implying that Histone H2AT119ph is a prerequisite for acetylation of these residues in meiosis.
The SC fails to disassemble in the nhk-1Z3-0437 mutant
Failure of the mutant oocytes to form a karyosome implicated Histone H2A-T119ph in the formation of this structure. As little is known about the mechanisms of karyosome formation, we sought to explore whether the chromosomes in the nhk-1Z3-0437 mutant perform the meiosis-specific functions essential to formation of the egg, starting with an examination of homologous recombination early in prophase I.
To address whether recombination occurs normally, we analyzed formation of the SC and of DSBs. We used an antibody against the C(3)G protein, a component of the transverse element of the SC (Page and Hawley 2001). C(3)G is an excellent marker for SC formation and prophase I progression in the oocyte because of its characteristic localization pattern. Early in oocyte development, C(3)G is progressively restricted to the oocyte, where it localizes in a ribbon-like pattern along the chromosomes (Fig. 4A; Page and Hawley 2001). We found that the initial restriction and the ribbon-like localization of C(3)G in the oocyte nucleus were normal in the nhk-1Z3-0437/Df mutant (Fig. 4D). Following completion of recombination in wild-type oocytes, C(3)G dissociates from the chromosomes and is found diffusely throughout the oocyte nucleus, at the time when the chromosomes condense into the karyosome (Fig. 4B,C). Strikingly, we observed that C(3)G did not dissociate from the chromosomes in the later oocyte stages in the mutant. Instead, C(3)G retained the ribbon-like structure and migrated to the periphery of the oocyte nucleus (Fig. 4E,F). We obtained similar results with an antibody against another SC component, C(2)M (data not shown). These results suggest that Histone H2AT119ph by NHK-1 is required for disassembly of the SC in Drosophila.
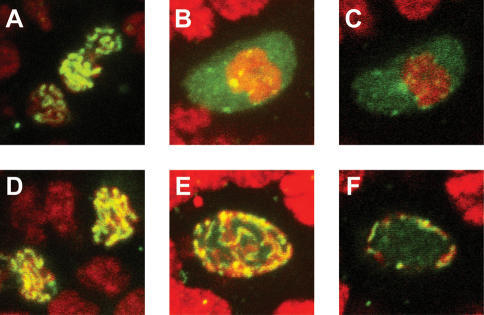
Figure 4.
The SC fails to dissociate from the chromosomes in the nhk-1 mutant oocytes. (A) The C(3)G protein (green) localizes to the oocyte nucleus in control nhk-1Z3-0437/+ early egg chambers. (B) C(3)G is localized diffusely throughout the oocyte nucleus in later egg chamber stages. This panel shows a projection of the entire oocyte nucleus. (C) A single slice through the middle of the oocyte nucleus showing that C(3)G is present throughout the nucleoplasm. (D) The C(3)G protein localizes to the oocyte nucleus in the nhk-1Z3-0437/Df mutant oocytes at early stages of oogenesis, and forms the expected ribbon pattern. (E) C(3)G fails to delocalize from the DNA in the nhk-1Z3-0437/Df mutant at later stages of oogenesis. A projection of the entire oocyte nucleus shows the persistence of the ribbon structure. (F) A single slice through the middle of the mutant oocyte nucleus shows that the persistent C(3)G ribbon is at the periphery of the nucleus and that the nucleoplasm is largely devoid of C(3)G in the mutant.
One possibility for failure in SC disassembly is a defect in DSB repair. Persistence of DSBs has been observed in mutants with aberrant karyosomes (Ghabrial et al. 1998; Ghabrial and Schupbach 1999). To determine whether the defect in SC disassembly in the nhk-1Z3-0437 mutant is due to failure to repair the DSBs, we analyzed the phosphorylation of Histone H2Av. The Histone H2Av variant is specifically phosphorylated on Ser 139 immediately following DSB formation (Madigan et al. 2002). We found that DSBs formed in the nhk-1Z3-0437/Df mutant with frequencies comparable to those in sibling controls (Fig. 5A,C, arrows). Egg chambers in which recombination is completed no longer show a signal with the anti-Histone H2AvS139ph antibody (Fig. 5B, arrow; Jang et al. 2003). Strikingly, we did not observe persistence of Histone H2Avph in the oocyte nucleus in the nhk-1Z3-0437/Df mutant (Fig. 5D, arrow), suggesting that the DSBs are repaired. We observed staining in the nurse cell and follicle cell nuclei in both mutant and control egg chambers, indicating that lack of signal is not due to failure of the antibody to enter the egg chambers. We conclude that the defect in SC disassembly in the nhk-1Z3-0437 mutant is not due to a failure to repair DSBs and propose that NHK-1 is required for SC disassembly at a step subsequent to DSB repair.
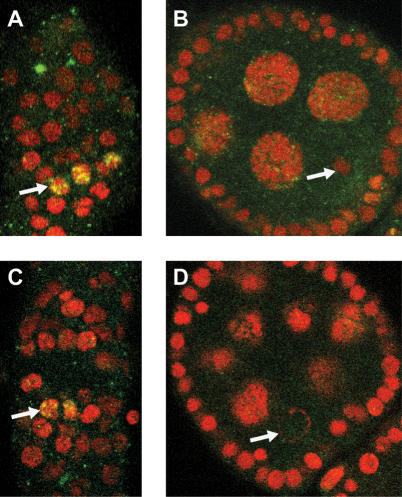
Figure 5.
Double-strand breaks are repaired in the nhk-1Z3-0437/Df mutant oocytes. (A) Control nhk-1Z3-0437/+ germarium shows phosphorylated Histone H2AvS139 (green) in a subset of nuclei (arrow) (red, DNA). (B) A karyosome (arrow) in a control nhk-1Z3-0437/+ egg chamber at a later stage of development did not stain with the anti-Histone H2AvS139ph antibody, suggesting that the double-strand breaks have been repaired. (C) The anti-Histone H2AvS139ph signal in the mutant nhk-1Z3-0437/Df germarium was indistinguishable from the control and appeared in a subset of nuclei (arrow), suggesting that double-strand breaks were formed properly in the mutant. (D) The oocyte DNA in the nhk-1Z3-0437/Df mutant at later stages of development (arrow) did not stain with the anti-Histone H2AvS139ph antibody, suggesting that the double-strand breaks were repaired and did not persist in the mutant.
The condensin SMC4 does not localize to chromosomes in the nhk-1Z3-0437 mutant
Following completion of recombination, the chromosomes in the oocyte nucleus undergo partial condensation to form the karyosome. The molecular mechanism of chromosome condensation during meiotic prophase I is not well understood, although recently the condensin complex was shown to play a role in meiosis in C. elegans (Chan et al. 2004). Condensin is a conserved protein complex that consists of five subunits: two structural maintenance of chromosomes (SMC) proteins (SMC2 and SMC4) and three chromosome-associated proteins (CAP-D2, CAP-G, and CAP-H/Barren) (Swedlow and Hirano 2003).
We asked whether the condensin complex plays a role in meiotic chromosome condensation in Drosophila by analyzing the localization of the Drosophila ortholog of SMC4 (Steffensen et al. 2001). We found that SMC4 localizes to the oocyte chromosomes, and its localization suggests a role in karyosome formation. In the early egg chambers, SMC4 localized to a single focus within the presumptive karyosome (Fig. 6A). Subsequently, SMC4 spread to several foci within the karyosome (Fig. 6B). In later egg chambers, the overlap between the DNA and the SMC4 signal was progressively more extensive (Fig. 6C) and finally complete (Fig. 6D).
Figure 6.
The condensin SMC4 does not localize to chromosomes in the nhk-1Z3-0437 mutant. (A-D) Control nhk-1Z3-0437/+ oocyte nuclei at progressively later stages of development show the pattern of SMC4 localization during Drosophila meiosis. (A) SMC4 (green) was found throughout the nucleoplasm and at the center of the presumptive karyosome (red). (B) SMC4 localized to several foci within the karyosome. (C) Nearly complete overlap between the karyosome and SMC4. (D) Complete colocalization of SMC4 and the karyosome. (E-H) Four examples of nhk-1Z3-0437/Df mutant oocyte nuclei at progressively later stages of development. In all cases, the DNA (red) was at the periphery of the oocyte nucleus, and SMC4 was throughout the nucleoplasm and did not overlap with the DNA, showing that SMC4 does not colocalize with the DNA.
Failure of the nhk-1Z3-0437 mutant to form karyosomes suggested a defect in chromosome condensation. To test this possibility, we analyzed the localization pattern of SMC4 in nhk-1Z3-0437/Df mutant ovaries. Strikingly, SMC4 was diffusely distributed throughout the oocyte nucleus in the nhk-1Z3-0437/Df mutant oocytes. SMC4 did not colocalize with the DNA at the periphery of the nucleus, suggesting that SMC4 does not to load onto the chromosomes in the nhk-1Z3-0437 mutant. Figure 6E-H, shows a series of nhk-1Z3-0437/Df mutant oocyte nuclei at progressively later stages of egg chamber development, illustrating the failure of SMC4 and DNA to colocalize throughout oogenesis. We observed similar results with antibodies against the condensin subunit dCAP-D2 (Savvidou et al. 2005; data not shown). These results suggest that the condensin complex plays a role in condensation of the prophase I chromosomes in meiosis, and that loading of the condensin complex requires phosphorylation of Histone H2A by NHK-1 and/or disassembly of the SC. Given that H3 and H4ac increase in the oocyte nucleus at this time, it is also possible that a histone code, dependent on H2AT119ph, triggers disassembly of the SC and assembly of the condensin complex.
Cohesin, another conserved protein complex, is required to hold sister chromatids together from S phase to anaphase (for reviews, see Hirano 2000; Lee and Orr-Weaver 2001). Cohesin consists of two SMC subunits (SMC1 and SMC3) and two non-SMC subunits (SCC1/Mcd1/Rad21 and SCC3). To determine whether cohesin binding is affected in the nhk-1 mutant, we stained the control and mutant ovaries with a cocktail of antibodies against the SMC1 and SMC3 (S. Bickel, pers. comm.). In both control oocytes and mutant nhk-1z3-0437/Df oocytes colocalization of cohesin with DNA was observed, suggesting that sister chromatid cohesion is not affected by loss of NHK-1 function (data not shown). Taken together, our findings that the SC persists and cohesin is found on the chromosomes in the mutant are consistent with the observation that loss of cohesion leads to premature disassembly of the SC (Webber et al. 2004).
Discussion
In this study, we explored the functional requirements for a newly identified histone kinase in meiosis and found that NHK-1 functions in meiotic progression. The phenotypes of the female-sterile nhk-1Z3-0437 mutant showed that NHK-1 is required for the establishment of several meiosis-specific chromosomal configurations, including the prophase I karyosome, the metaphase I spindle, and the polar body. Histone H2AT119ph, as well as Histone H3K14ac and Histone H4K5ac, were reduced in the mutant oocytes, whereas the other histone modifications examined were unaffected. Strikingly, disassembly of the SC and loading of condensin failed in the mutant. Therefore, we suggest that NHK-1 and Histone H2AT119ph are required specifically for proper chromosome architecture in meiosis.
Meiosis and the histone code
The histone code hypothesis conjectures functional interactions among histone modifications and between modifications and proteins that bind to them (Strahl and Allis 2000). It has been postulated that histones have signature modification profiles in meiosis (Kimmins and Sassone-Corsi 2005) to accommodate the meiosis-specific chromosomal events. However, most research in vertebrates has been limited to identifying the modifications and has not provided extensive insight into their function (Kim et al. 2003).
Our finding that NHK-1 is required for meiosis, presumably via its phosphorylation of Histone H2AT119 (Aihara et al. 2004), prompted us to examine the presence of other histone modifications in Drosophila oocytes. We found that Histone H1 is phosphorylated, HP1 is bound (indicative of histone H3 methylation), and Histone H3 and H4 are acetylated during prophase I of meiosis in Drosophila. The presence of these histone modifications in oocyte nuclei suggests that they play a role in chromosome dynamics during meiosis. It is of interest to note that Histone H4 and H3 were shown to be acetylated in mouse oocytes in prophase I (Kim et al. 2003), suggesting an evolutionary conservation. Future analysis of mutations in histone modifying enzymes may shed light on the functions of these modifications in meiosis.
One stipulation of the histone code is that histone modifications affect each other (Strahl and Allis 2000). To test this hypothesis with respect to Histone H2AT119ph, we examined the panel of histone modifications in the nhk-1Z3-0437 mutant ovaries. Histone H1ph, HP1 binding, and Histone H4K12ac were unaffected in the mutant oocytes, consistent with Histone H2AT119ph being downstream or independent of them in Drosophila meiosis. In contrast, Histone H3K14ac and Histone H4K5ac were absent specifically from the chromosomes in the nhk-1Z3-0437 mutant oocytes, indicating that Histone H2AT119ph is a prerequisite for acetylation of these residues. Although the significance of these acetylations is unclear at present, it is intriguing to speculate that they play an important role in meiosis.
Another stipulation of the histone code is that modifications recruit or exclude proteins from binding to chromatin (Strahl and Allis 2000). One possibility is that Histone H2AT119ph is required for function of the histone acetyltransferases for Histone H3K14 and Histone H4K5, the residues that lack acetylation in the mutant oocytes. There is a precedent for such a cascade of dependencies in transcription: Phosphorylation of H3 Ser 10 leads to acetylation of H3 Lys 14 (Jenuwein and Allis 2001). Histone H2AT119 phosphorylation may also be a prerequisite for proper chromosomal associations of SMC4 and for dissociation of the SC.
In conclusion, Histone H2AT119 phosphorylation appears to affect both other histone modifications and the binding of proteins to chromatin as predicted by the histone code hypothesis. Histone H2AT119 phosphorylation by NHK-1 may, therefore, be a key component of a meiotic histone code in oocytes.
How does the karyosome form?
The Drosophila karyosome is a subnuclear organelle comprised of the prophase I chromosomes. It is the best studied example of a family of similar structures found throughout evolution (Gruzova and Parfenov 1993). Although the exact function of the karyosome is unclear, several Drosophila mutants that disrupt karyosome structure (this study; see below) lead to female sterility, suggesting that karyosome formation is required for fertility. In addition, it has been postulated that retaining the oocyte chromosomes in close proximity within the large oocyte nucleus (the germinal vesicle) facilitates proper chromosome segregation during the meiotic divisions (Gruzova and Parfenov 1993; Homer et al. 2005). Therefore, understanding the molecular mechanisms that regulate karyosome formation is of great interest.
The spindle mutants in Drosophila (spindle-B, spindle-D, and okra) are characterized by defects in karyosome formation, in establishment of the dorsal/ventral polarity of the oocyte and the egg, and defects in repair of the DSBs following recombination (Ghabrial et al. 1998; Ghabrial and Schupbach 1999). The nhk-1Z3-0437 mutant does not show either of the latter phenotypes, consistent with the NHK-1 histone kinase having a primary role in karyosome formation, whereas the spindle mutants may affect karyosome function as a secondary consequence of other defects. The specific requirement for NHK-1 implicates Histone H2AT119ph in formation of the karyosome. The conservation of the NHK-1 kinase and Thr 119 in Histone H2A among diverse species suggests that this histone modification plays a conserved role in chromosomal dynamics in meiosis.
Is NHK-1 required at multiple steps during meiosis?
The nhk-1Z3-0437 mutant showed several interesting phenotypes in the oocyte. First, all embryos laid by mutant females showed aberrant polar bodies and an arrest at metaphase of the first mitotic division. Second, we observed a defect in the metaphase I arrest in 50% of late-stage oocytes. Finally, the karyosome failed to form in all early oocytes. It is not clear how the dispersed chromosomes in the mutant karyosomes later form three masses at metaphase I, but it suggests that the chromosomes are not fragmented and that bivalent associations are retained.
These results can be interpreted in two ways. First, NHK-1 may be required continuously throughout meiosis for each of the events affected by the Z3-0437 mutation. Continuous requirement may be due to multiple substrates or due to dynamic Histone H2A phosphorylation and dephosphorylation. Second, NHK-1 function may be required specifically for karyosome formation, whereas the metaphase I and the embryonic phenotypes may be a consequence of the karyosome defect. This model suggests that Histone H2AT119ph serves as an epigenetic mark that contributes to proper completion of later events, such as establishment of the metaphase I arrest and progression through the first mitotic division. To test whether the early mitotic defects in the nhk-1 mutant were due to a failure to load condensin in mitosis, we examined the dCAP-D2 condensin subunit and found it localized onto chromosomes in embryos from nhk-1 mutant females (Savvidou et al. 2005; data not shown). Nevertheless, we think it likely that NHK-1 plays essential roles in mitosis that may be revealed by additional alleles of nhk-1.
Does NHK-1 function outside the oocyte?
Histone H2AT119 is phosphorylated in the follicle cells, the nurse cells, and the oocyte, suggesting that NHK-1 functions in all three ovarian cell types. However, we observed mutant phenotypes specifically in the oocyte. There are two reasons why Histone H2A phosphorylation persists in the mutant nurse and follicle cells and mutant defects were not observed. First, Histone H2A may be phosphorylated by another kinase in the nurse and follicle cells. Second, NHK-1 may phosphorylate Histone H2A in all cell types, but the oocyte may require higher levels of kinase activity for phosphorylation. The levels of Histone H2AT119ph may be reduced in the mutant nurse and follicle cells, but the concentration on these polyploid chromosomes could still permit detection by the antibody. We favor the second idea because partial loss-of-function alleles of essential genes often lead to female sterility. Our observation that the nhk-1Z3-0437/Df(3R)Tl-I allelic combination has a stronger mutant phenotype than the nhk-1Z3-0437/Z3-0437 allelic combination is consistent with nhk-1Z3-0437 being a partial loss-of-function allele. Stronger alleles of nhk-1 may, therefore, reveal additional requirements for this kinase in other cell types, including in male meiosis.
In conclusion, by using a female-sterile Drosophila mutant, we uncovered a key step in the formation of the karyosome, a conserved structure with an elusive function. Based on the analysis of the mutant phenotype, we propose the following model for karyosome formation: Following DSB repair and dephosphorylation of Histone H2AvS139, phosphorylation of Histone H2AT119 by NHK-1 leads to disassembly of the SC, loading of condensin onto the chromosomes, and subsequent condensation into a karyosome. In addition to providing insight into karyosome formation, the nhk-1Z3-0437 mutant is an excellent tool for elucidating the interactions of a specific histone modification within the histone code.
Materials and methods
Genetic and molecular mapping
To identify the gene mutated in the Z3-0437 line, we performed complementation analysis with a series of third-chromosome deficiencies (Bloomington Stock Center) and identified lines Df(3R)Tl-P and Df(3R)Tl-I that failed to complement the female sterility of Z3-0437. To refine the region, we employed site-specific male recombinational mapping (Chen et al. 1998) using the EP3619, EP1141, and EP3030 lines. The combination of these two methods localized the mutation to a region of 13 genes. DNA sequencing of exons in this region revealed a mutation in the CG6386/bällchen/nhk-1 gene.
Genomic DNA for sequencing was isolated from five males homozygous for the Z3-0437 mutation or a reference strain from the same screen (Koundakjian et al. 2004), essentially as previously described (Ballinger and Benzer 1989). Sequencing was performed at the Massachusetts Institute of Technology Center for Cancer Research, Howard Hughes Medical Institute Biopolymers Laboratory.
In situ hybridization
In situ hybridization (Tautz and Pfeifle 1989) was performed as previously described (Royzman et al. 1997). Ovaries from wild-type (y, w) fattened females were dissected in Grace's medium and hybridized at 55°C. Digoxygenin-labeled sense and anti-sense RNA probes were made according to the manufacturer's instructions (Roche Applied Sciences). The 1.6-kb CG6386/bällchen/nhk-1 ORF was amplified by PCR, using primers PR203 (5′-TGGCCCGGCATTTCG-3′) and PR205 (5′-CTATC GAATTCAGTAAATGTAGG-3′). The PCR product was cloned into the pCRII-TOPO vector (Invitrogen). The sense probe was transcribed using the T7 promoter in the vector, and the anti-sense probe was transcribed using the Sp6 promoter in the vector.
Protein extracts and immunoblot analysis
Drosophila ovarian protein extracts were prepared by an acid extraction procedure to ensure that chromosomal proteins were extracted from the chromosomes. Ovaries from ∼50 females were dissected in Grace's insect medium (Gibco) after 2 d at 25°C on protein-rich food. The ovaries were washed in 1 mL of PBS and were resuspended in 5 volumes of lysis buffer (10 mM HEPES at pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 1.5 mM PMSF). Following homogenization, sulfuric acid was added to 0.4 N final concentration and the homogenate was incubated on ice for 30 min. Following centrifugation for 10 min at 4°C, the supernatant was transferred to a Slide-A-Lyzer dialysis cassette (Pierce) and the acid was dialyzed according to the manufacturer's instructions.
The proteins were separated by SDS-PAGE followed by transfer to nitrocellulose. The anti-NHK-1 antibody was used at a 1:200 dilution (Aihara et al. 2004). Anti-α-tubulin antibodies (clones 1/2 and 1/34, Accurate Chemical and Scientific Corporation) were used at a 1:400 dilution. Secondary antibodies were from Jackson ImmunoResearch Laboratories and were used according to the manufacturer's recommendation.
Cytology and microscopy
Newly eclosed females were aged for 12-24 h on standard corn-based medium supplemented with dry yeast. Ovaries were dissected in Grace's medium. For staining with the DNA dye (DAPI), ovaries were fixed in 8% formaldehyde (Ted Pella, Inc.) in PBS for 10-15 min and washed (3 × 10 min) in PBS (Ashburner 1989; Theurkauf 1994). For immunofluorescence staining, ovaries were processed as described (Belmont et al. 1989; de Cuevas et al. 1996).
The anti-Cyclin E antibodies were made in guinea pigs against a GST fusion to the N-terminal 296 amino acids of the protein and were used at 1:500 dilution for ovary staining.
The anti-HP1 antibody (Developmental Studies Hybridoma Bank, University of Iowa), shown to specifically recognize the HP1 protein in cytological staining (James et al. 1989), was used at a 1:5 dilution. The antibodies against differentially modified histones were shown to specifically recognize the designated modification (Lin et al. 1989; Braunstein et al. 1993, 1996; Perry et al. 1993; Lu et al. 1994) and were used according to the manufacturer's recommendations (Upstate Biotechnology).
The anti-C(3)G antibody (a kind gift of M. Lilly, National Institutes of Health, Bethesda, MD), shown to specifically recognize the C(3)G protein (Anderson et al. 2005), was used at a 1:1000 dilution. For staining using the anti-Histone H2AvS139ph antibody, ovaries were processed as described (Belmont et al. 1989). The anti-Histone H2AvS139ph antibody (a kind gift of R.L. Glaser, State University of New York, Albany, NY), shown to specifically recognize the phosphorylated form of the Drosophila Histone H2Av (Madigan et al. 2002), was used at a 1:100 dilution. The anti-SMC4 and dCAP-D2 antibodies (a kind gift of M. Heck, Edinburgh University, Edinburgh, Scotland), shown to specifically recognize the SMC4 and dCAP-D2 proteins in cytological staining (Steffensen et al. 2001; Savvidou et al. 2005), were used at 1:500 and 1:1000 dilutions, respectively. The anti-SMC1 and anti-SMC3 antibodies (a kind gift of S. Bickel, Dartmouth University, Hanover, NH), shown to specifically recognize the SMC1 and SMC3 proteins in cytological staining (S. Bickel, pers. comm.) were used at 1:2000 and 1:1000 dilutions, respectively. Secondary antibodies were from Jackson Immunoresearch Laboratories, Inc. The DNA was stained with propidium iodide (Sigma-Aldrich).
For analysis of the embryonic phenotype, embryos were collected for 2 h, dechorionated in 50% bleach for 3 min, and fixed in 4% formaldehyde for 15 min. The embryos were treated with RNase for 1 h and stained with YOYO (Molecular Probes) according to the manufacturer's recommendation. Embryo preparation for antibody localization was performed as previously described (Whittaker et al. 2000). Anti-α-tubulin antibodies (clones 1/2 and 1/34) were used at a 1:40 dilution. To determine fertilization, virgin nhk-1Z3-0437/Z3-0437 mutant females were mated to males carrying a transgene that fuses GFP to a spermtail protein (a kind gift of B. Wakimoto, University of Washington, Seattle, WA) (Santel et al. 1997; Fitch and Wakimoto 1998). Embryos were mounted in 70% glycerol in PBS.
Embryos and egg chambers were photographed on a Zeiss Axiophot microscope with Plan-Neofluar 25× Inn Korr or 40× oil objectives with a SPOT RT CCD camera and software. Alternatively, a Zeiss LSM510 Laser-scanning confocal microscope at the W.M. Keck Microscopy Facility at the Whitehead Institute was used.
Acknowledgments
The maternal-effect mutant collection was kindly provided by Charles Zuker's laboratory, University of California, San Diego. We thank Edmund Koundakjian, David Cowan, and Robert Hardy of the Zuker laboratory for establishing the collection of F3 viable lines, and Barbara Wakimoto, Dan Lindsley, and Mike McKeon for identifying the subset of female-sterile lines. We gratefully acknowledge the work of fellow lab members in jointly screening this collection: K. Dej (currently of McMaster University, Hamilton, ON, Canada), G. Bosco (currently of The University of Arizona, Tucson, AZ), L. Lee (currently of Vanderbilt University Medical Center, Nashville, TN), H. Kashevsky, D. Epstein, and J. Pesin. We thank Tamar Resnick for providing control embryos. The anti-Cyclin E antibodies were made by Helena Kashevsky in this laboratory. We thank R.L. Glaser for the anti-Histone H2AvS139 antibody, M. Heck for the anti-SMC4 and anti-dCAP-D2 antibodies, M. Lilly for the anti-C(3)G antibody, S. Bickel for the anti-SMC1 and anti-SMC3 antibodies, and Kim McKim for anti-C(2)M and anti-Histone H2AvS139 antibodies. Andrew Baltus, Gloria Brar, Andreas Hochwagen, Jana Koubova, Emmy Rogakou, and Dimitris Thanos provided helpful comments on the manuscript. I.I. was supported as a Special Fellow by the Leukemia and Lymphoma Society and by a National Research Service Award post-doctoral fellowship (GM63395) from the National Institutes of Health. This work was supported by a grant from the National Institutes of Health to T.O.-W. (GM39341).
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1348905.
References
- Aihara H., Nakagawa, T., Yasui, K., Ohta, T., Hirose, S., Dhomae, N., Takio, K., Kaneko, M., Takeshima, Y., Muramatsu, M., et al. 2004. Nucleosomal histone kinase-1 phosphorylates H2A Thr 119 during mitosis in the early Drosophila embryo. Genes & Dev. 18: 877-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L.K., Royer, S.M., Page, S.L., McKim, K.S., Lai, A., Lilly, M.A., and Hawley, R.S. 2005. Juxtaposition of C(2)M and the transverse filament protein C(3)G within the central region of Drosophila synaptonemal complex. Proc. Natl. Acad. Sci. 102: 4482-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. 1989. Drosophila. A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Ballinger D.G. and Benzer, S. 1989. Targeted gene mutations in Drosophila. Proc. Natl. Acad. Sci. 86: 9402-9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont A.S., Braunfeld, M.B., Sedat, J.W., and Agard, D.A. 1989. Large-scale chromatin structural domains within mitotic and interphase chromosomes in vivo and in vitro. Chromosoma 98: 129-143. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Rose, A.B., Holmes, S.G., Allis, C.D., and Broach, J.R. 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes & Dev. 7: 592-604. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Sobel, R.E., Allis, C.D., Turner, B.M., and Broach, J.R. 1996. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol. 16: 4349-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi B.R. and Lilly, M.A. 2004. Fluorescent BrdU labeling and nuclear flow sorting of the Drosophila ovary. Methods Mol. Biol. 247: 203-213. [DOI] [PubMed] [Google Scholar]
- Calvi B.R., Lilly, M.A., and Spradling, A.C. 1998. Cell cycle control of chorion gene amplification. Genes & Dev. 12: 734-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza M.J., Utley, R.T., Workman, J.L., and Cote, J. 2003. The diverse functions of histone acetyltransferase complexes. Trends Genet. 19: 321-329. [DOI] [PubMed] [Google Scholar]
- Champion M.D. and Hawley, R.S. 2002. Playing for half the deck: The molecular biology of meiosis. Nat. Cell Biol. 4 Suppl: s50-s56. [DOI] [PubMed] [Google Scholar]
- Chan R.C., Severson, A.F., and Meyer, B.J. 2004. Condensin restructures chromosomes in preparation for meiotic divisions. J. Cell Biol. 167: 613-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Chu, T., Harms, E., Gergen, J.P., and Strickland, S. 1998. Mapping of Drosophila mutations using site-specific male recombination. Genetics 149: 157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb J.M. and Orr-Weaver, T.L. 2005. Developmental gene amplification: Insights into DNA replication and gene expression. Trends Genet. 21: 149-162. [DOI] [PubMed] [Google Scholar]
- de Cuevas M., Lee, J.K., and Spradling, A.C. 1996. α-Spectrin is required for germline cell division and differentiation in the Drosophila ovary. Development 122: 3959-3968. [DOI] [PubMed] [Google Scholar]
- Dhalluin C., Carlson, J.E., Zeng, L., He, C., Aggarwal, A.K., and Zhou, M.M. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399: 491-496. [DOI] [PubMed] [Google Scholar]
- Eberharter A. and Becker, P.B. 2002. Histone acetylation: A switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 3: 224-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W., Wang, Y., and Allis, C.D. 2003a. Binary switches and modification cassettes in histone biology and beyond. Nature 425: 475-479. [DOI] [PubMed] [Google Scholar]
- ____. 2003b. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15: 172-183. [DOI] [PubMed] [Google Scholar]
- Fitch K.R. and Wakimoto, B.T. 1998. The paternal effect gene ms(3)sneaky is required for sperm activation and the initiation of embryogenesis in Drosophila melanogaster. Dev. Biol. 197: 270-282. [DOI] [PubMed] [Google Scholar]
- Ghabrial A. and Schupbach, T. 1999. Activation of a meiotic checkpoint regulates translation of Gurken during Drosophila oogenesis. Nat. Cell Biol. 1: 354-357. [DOI] [PubMed] [Google Scholar]
- Ghabrial A., Ray, R.P., and Schupbach, T. 1998. okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes & Dev. 12: 2711-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruzova M.N. and Parfenov, V.N. 1993. Karyosphere in oogenesis and intranuclear morphogenesis. Int. Rev. Cytol. 144: 1-52. [DOI] [PubMed] [Google Scholar]
- Hirano T. 2000. Chromosome cohesion, condensation, and separation. Annu. Rev. Biochem. 69: 115-144. [DOI] [PubMed] [Google Scholar]
- Homer H.A., McDougall, A., Levasseur, M., and Herbert, M. 2005. Restaging the spindle assembly checkpoint in female mammalian meiosis I. Cell Cycle 4: 650-653. [DOI] [PubMed] [Google Scholar]
- Hsu J.Y., Sun, Z.W., Li, X., Reuben, M., Tatchell, K., Bishop, D.K., Grushcow, J.M., Brame, C.J., Caldwell, J.A., Hunt, D.F., et al. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102: 279-291. [DOI] [PubMed] [Google Scholar]
- James T.C., Eissenberg, J.C., Craig, C., Dietrich, V., Hobson, A., and Elgin, S.C. 1989. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell Biol. 50: 170-180. [PubMed] [Google Scholar]
- Jang J.K., Sherizen, D.E., Bhagat, R., Manheim, E.A., and McKim, K.S. 2003. Relationship of DNA double-strand breaks to synapsis in Drosophila. J. Cell Sci. 116: 3069-3077. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. and Allis, C.D. 2001. Translating the histone code. Science 293: 1074-1080. [DOI] [PubMed] [Google Scholar]
- Kim J.M., Liu, H., Tazaki, M., Nagata, M., and Aoki, F. 2003. Changes in histone acetylation during mouse oocyte meiosis. J. Cell Biol. 162: 37-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmins S. and Sassone-Corsi, P. 2005. Chromatin remodelling and epigenetic features of germ cells. Nature 434: 583-589. [DOI] [PubMed] [Google Scholar]
- King R.C. 1970. Ovarian development in Drosophila melanogaster. Academic Press, New York.
- Kornberg R.D. and Lorch, Y. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98: 285-294. [DOI] [PubMed] [Google Scholar]
- Koundakjian E.J., Cowan, D.M., Hardy, R.W., and Becker, A.H. 2004. The Zuker collection: A resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics 167: 203-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M., O'Carroll, D., Rea, S., Mechtler, K., and Jenuwein, T. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116-120. [DOI] [PubMed] [Google Scholar]
- Lee J.Y. and Orr-Weaver, T.L. 2001. The molecular basis of sister-chromatid cohesion. Annu. Rev. Cell Dev. Biol. 17: 753-777. [DOI] [PubMed] [Google Scholar]
- Lin R., Leone, J.W., Cook, R.G., and Allis, C.D. 1989. Antibodies specific to acetylated histones document the existence of deposition- and transcription-related histone acetylation in Tetrahymena. J. Cell Biol. 108: 1577-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M.J., Dadd, C.A., Mizzen, C.A., Perry, C.A., McLachlan, D.R., Annunziato, A.T., and Allis, C.D. 1994. Generation and characterization of novel antibodies highly selective for phosphorylated linker histone H1 in Tetrahymena and HeLa cells. Chromosoma 103: 111-121. [DOI] [PubMed] [Google Scholar]
- Madigan J.P., Chotkowski, H.L., and Glaser, R.L. 2002. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 30: 3698-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah S.K., Turner, J.M., Baudat, F., Rogakou, E.P., de Boer, P., Blanco-Rodriguez, J., Jasin, M., Keeney, S., Bonner, W.M., and Burgoyne, P.S. 2001. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27: 271-276. [DOI] [PubMed] [Google Scholar]
- Marston A.L. and Amon, A. 2004. Meiosis: Cell-cycle controls shuffle and deal. Nat. Rev. Mol. Cell Biol. 5: 983-997. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T.L. 1995. Meiosis in Drosophila: Seeing is believing. Proc. Natl. Acad. Sci. 92: 10443-10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S.L. and Hawley, R.S. 2001. c(3)G encodes a Drosophila synaptonemal complex protein. Genes & Dev. 15: 3130-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2004. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20: 525-558. [DOI] [PubMed] [Google Scholar]
- Page A.W. and Orr-Weaver, T.L. 1997. Activation of the meiotic divisions in Drosophila oocytes. Dev. Biol. 183: 195-207. [DOI] [PubMed] [Google Scholar]
- Perry C.A., Dadd, C.A., Allis, C.D., and Annunziato, A.T. 1993. Analysis of nucleosome assembly and histone exchange using antibodies specific for acetylated H4. Biochemistry 32: 13605-13614. [DOI] [PubMed] [Google Scholar]
- Rogakou E.P., Boon, C., Redon, C., and Bonner, W.M. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146: 905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royzman I., Whittaker, A.J., and Orr-Weaver, T.L. 1997. Mutations in Drosophila DP and E2F distinguish G1-S progression from an associated transcriptional program. Genes & Dev. 11: 1999-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A., Winhauer, T., Blumer, N., and Renkawitz-Pohl, R. 1997. The Drosophila don juan (dj) gene encodes a novel sperm specific protein component characterized by an unusual domain of a repetitive amino acid motif. Mech. Dev. 64: 19-30. [DOI] [PubMed] [Google Scholar]
- Savvidou E., Cobbe, N., Steffensen, S., Cotterill, S., and Heck, M.M. 2005. Drosophila CAP-D2 is required for condensin complex stability and resolution of sister chromatids. J. Cell Sci. 118: 2529-2543. [DOI] [PubMed] [Google Scholar]
- Spradling A.C. 1993. Developmental genetics of oogenesis. In The development of Drosophila melanogaster (eds. M. Bate and A. Martinez Arias), pp. 1-70. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Steffensen S., Coelho, P.A., Cobbe, N., Vass, S., Costa, M., Hassan, B., Prokopenko, S.N., Bellen, H., Heck, M.M., and Sunkel, C.E. 2001. A role for Drosophila SMC4 in the resolution of sister chromatids in mitosis. Curr. Biol. 11: 295-307. [DOI] [PubMed] [Google Scholar]
- Strahl B.D. and Allis, C.D. 2000. The language of covalent histone modifications. Nature 403: 41-45. [DOI] [PubMed] [Google Scholar]
- Swedlow J.R. and Hirano, T. 2003. The making of the mitotic chromosome: Modern insights into classical questions. Mol. Cell 11: 557-569. [DOI] [PubMed] [Google Scholar]
- Tautz D. and Pfeifle, C. 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98: 81-85. [DOI] [PubMed] [Google Scholar]
- Theurkauf W.E. 1994. Immunofluorescence analysis of the cytoskeleton during oogenesis and early embryogenesis. In Methods in cell biology (eds. L.S.B. Goldstein and E.A. Fyrberg), pp. 489-505. Academic Press, New York. [DOI] [PubMed]
- Theurkauf W.E. and Hawley, R.S. 1992. Meiotic spindle assembly in Drosophila females: Behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J. Cell Biol. 116: 1167-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber H.A., Howard, L., and Bickel, S.E. 2004. The cohesion protein ORD is required for homologue bias during meiotic recombination. J. Cell Biol. 164: 819-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker A.J., Royzman, I., and Orr-Weaver, T.L. 2000. Drosophila double parked: A conserved, essential replication protein that colocalizes with the origin recognition complex and links DNA replication with mitosis and the down-regulation of S phase transcripts. Genes & Dev. 14: 1765-1776. [PMC free article] [PubMed] [Google Scholar]
- Winston F. and Allis, C.D. 1999. The bromodomain: A chromatin-targeting module? Nat. Struct. Biol. 6: 601-604. [DOI] [PubMed] [Google Scholar]
- Zeng L. and Zhou, M.M. 2002. Bromodomain: An acetyl-lysine binding domain. FEBS Lett. 513: 124-128. [DOI] [PubMed] [Google Scholar]
- Zhang Y. and Reinberg, D. 2001. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes & Dev. 15: 2343-2360. [DOI] [PubMed] [Google Scholar]