Abstract
The expression of most Staphylococcus aureus virulence factors is controlled by the agr locus, which encodes a two-component signaling pathway whose activating ligand is an agr-encoded autoinducing peptide (AIP). A polymorphism in the amino acid sequence of the AIP and of its corresponding receptor divides S. aureus strains into four major groups. Within a given group, each strain produces a peptide that can activate the agr response in the other member strains, whereas the AIPs belonging to different groups are usually mutually inhibitory. We investigated a possible relationship between agr groups and human S. aureus disease by studying 198 S. aureus strains isolated from 14 asymptomatic carriers, 66 patients with suppurative infection, and 114 patients with acute toxemia. The agr group and the distribution of 24 toxin genes were analyzed by PCR, and the genetic background was determined by means of amplified fragment length polymorphism (AFLP) analysis. The isolates were relatively evenly distributed among the four agrgroups, with 61 strains belonging to agr group I, 49 belonging to group II, 43 belonging to group III, and 45 belonging to group IV. Principal coordinate analysis performed on the AFLP distance matrix divided the 198 strains into three main phylogenetic groups, AF1 corresponding to strains of agr group IV, AF2 corresponding to strains of agr groups I and II, and AF3 corresponding to strains of agr group III. This indicated that the agr type was linked to the genetic background. A relationship between genetic background, agr group, and disease type was observed for several toxin-mediated diseases: for instance, agr group IV strains were associated with generalized exfoliative syndromes, and phylogenetic group AF1 strains with bullous impetigo. Among the suppurative infections, endocarditis strains mainly belonged to phylogenetic group AF2 and agr groups I and II. While these results do not show a direct role of the agr type in the type of human disease caused by S. aureus, the agr group may reflect an ancient evolutionary division of S. aureus in terms of this species’ fundamental biology.
Staphylococcus aureus is both a commensal and an extremely versatile pathogen in humans, causing three basic syndromes: (i) superficial lesions such as skin abscesses and wound infections; (ii) deep-seated and systemic infections such as osteomyelitis, endocarditis, pneumonia, and bacteremia; and (iii) toxemic syndromes such as toxic shock syndrome (TSS) and staphylococcal scarlet fever (both due to toxic shock syndrome toxin 1 [TSST-1] and staphylococcal enterotoxins [SEs]), staphylococcal scalded-skin syndrome (SSSS; due to exfoliatins), and staphylococcal food poisoning (due to SEs) (1, 18, 24). With the exception of toxemia, the molecular basis of S. aureus pathogenicity is multifactorial, depending on the expression of a large class of accessory gene products that comprise cell wall-associated and extracellular proteins (24). Expression of most virulence factors in S. aureus is controlled by the agr locus, which encodes a two-component signaling pathway whose activating ligand is a bacterial-density-sensing peptide (autoinducing peptide) also encoded by agr (24). A polymorphism in the amino acid sequence of the autoinducing peptide and of its corresponding receptor (AgrC) has been described. S. aureus strains can be divided into four major groups on this basis: within a given group, each strain produces a peptide that can activate the agr response in the other member strains, whereas the autoinducing peptides produced by the different groups are usually mutually inhibitory (14, 16). Links between a peculiar agr type and a specific staphylococcal syndrome have been shown for TSS and SSSS. TSST-1-producing isolates belong to agr specificity group III (16) and mostly belong to a single clone, as shown by multilocus enzyme electrophoresis (MLEE) (23) and pulsed-field gel electrophoresis (PFGE) (3). Most exfoliatin-producing strains responsible for SSSS belong to agr group IV, but the clonality of these strains has not been investigated (14). agr group I was prevalent in a collection of 192 S. aureus strains, most of which were methicillin resistant, but no clinical information was available in this study (29).
The aim of the present study was to further investigate a possible relationship between agr groups (alleles) and the pattern of S. aureus disease. We studied 198 methicillin-susceptible strains from the French National Reference Center for Staphylococcal Toxemia strain collection, in which all clinical syndromes are represented, to determine their agr type and the distribution of 24 toxin genes (by PCR), as well as to determine their genetic background (by amplified fragment length polymorphism [AFLP] analysis). We then sought to determine the relationships between these characteristics and the type of clinical disease syndrome.
MATERIALS AND METHODS
Staphylococcal strains and corresponding disease syndromes.
The French National Reference Center for Staphylococcal Toxemia (Lyon, France) collects more than 800 strains yearly from patients with toxemic and nontoxemic staphylococcal diseases throughout France. For this study we selected a subset of 198 S. aureus strains isolated between January 1985 and December 1999. They were isolated from nose swabs (n = 3) and vaginal swabs (n = 11) of 14 asymptomatic carriers and from clinical specimens of 66 patients with S. aureus suppurative infections (necrotizing pneumonia caused by Panton-Valentine leukocidin-producing strains [n = 11], furunculosis [n = 11], native valve endocarditis [n = 19], finger pulp infections [n = 9], osteitis [n = 8], cellulitis and/or myositis [n = 4], and arthritis [n = 4]), 4 patients with enterocolitis, and 114 patients with acute toxemia, including 35 cases of TSS, 33 cases of staphylococcal scarlet fever, and 46 cases of SSSS (20 cases of generalized exfoliative syndrome and 26 cases of bullous impetigo). The types of infection were defined according to published criteria (8, 18, 19). All infections were community acquired. All of the strains were collected from hospitals located throughout France and were identified as S. aureus by their ability to coagulate citrated rabbit plasma (bioMérieux, Marcy l’Etoile, France) and to produce a clumping factor (Staphyslide Test; bioMérieux).
S. aureus strains RN6390 (agr group I), RN6923 (agr group II), RN8462 (agr group III), A980740 (agr group IV), and RN6911 (agr null) were used as controls for agr group identification (14, 16). Control strains used for toxin gene detection are listed in Table 1.
TABLE 1.
Oligonucleotide primers and reference strains used for toxin gene detection
| Toxin | Gene | GenBank accession no. | Primer(s) | Sequence (5′-3′) | Size of amplified product (bp) | Control strain |
|---|---|---|---|---|---|---|
| SEA | sea | M18970 | SEA-1 | GAAAAAAGTCTGAATTGCAGGGAACA | 560 | ATCC 13566 |
| SEA-2 | CAAATAAATCGTAATTAACCGAAGGTTC | |||||
| SEB | seb | M11118 | SEB-1 | ATTCTATTAAGGACACTAAGTTAGGGA | 404 | ATCC 13566 |
| SEB-2 | ATCCCGTTTCATAAGGCGAGT | |||||
| SEC | sec | X05815 | mpSEC-1 | GTAAAGTTACAGGTGGCAAAACTTG | 297 | ATCC 19095 |
| mpSEC-2 | CATATCATACCAAAAAGTATTGCCGT | |||||
| SED | sed | M28521 | SED-1 | GAATTAAGTAGTACCGCGCTAAATAATATG | 492 | FRI-1151m |
| SED-2 | GCTGTATTTTTCCTCCGAGAGT | |||||
| SEE | see | M21319 | SEE-1 | CAAAGAAATGCTTTAAGCAATCTTAGGC | 482 | ATCC 27664 |
| SEE-2 | CACCTTACCGCCAAAGCTG | |||||
| SEG | seg | AF064773 | SEG-1 | AATTATGTGAATGCTCAACCCGATC | 642 | A900322 |
| SEG-2 | AAACTTATATGGAACAAAAGGTACTAGTTC | |||||
| SEH | seh | U11702 | SEH-1 | CAATCACATCATATGCGAAAGCAG | 376 | ATCC 51811 |
| SEH-2 | CATCTACCCAAACATTAGCACC | |||||
| SEI | sei | AF064774 | SEI-1 | CTCAAGGTGATATTGGTGTAGG | 576 | A900322 |
| SEI-2 | AAAAAACTTACAGGCAGTCCATCTC | |||||
| SEJ | sej | AF053140 | mpSEJ-1 | TAACCTCAGACATATATACTTCTTTAACG | 300 | FRI-1151m |
| mpSEJ-2 | AGTATCATAAAGTTGATTGTTTTCATGCAG | |||||
| SEN | sen | AF285760 | mpSEN-1 | ATGAGATTGTTCTACATAGCTGCAAT | 680 | A900322 |
| mpSEN-2 | AACTCTGCTCCCACTGAAC | |||||
| SEO | seo | AF285760 | mpSEO-1 | AGTTTGTGTAAGAAGTCAAGTGTAGA | 180 | A900322 |
| mpSEO-2 | ATCTTTAAATTCAGCAGATATTCCATCTAAC | |||||
| SEM | sem | AF285760 | mpSEM-1 | CTATTAATCTTTGGGTTAATGGAGAAC | 300 | A900322 |
| mpSEM-2 | TTCAGTTTCGACAGTTTTGTTGTCAT | |||||
| TSST-1 | tst | J02615 | TST-1 | TTCACTATTTGTAAAAGTGTCAGACCCACT | 180 | FRI-1169 |
| TST-2 | TACTAATGAATTTTTTTATCGTAAGCCCTT | |||||
| ETA | eta | M17347 | mpETA-1 | ACTGTAGGAGCTAGTGCATTTGT | 190 | TC-7 |
| mpETA-3 | TGGATACTTTTGTCTATCTTTTTCATCAAC | |||||
| ETB | etb | M17348 | mpETB-1 | CAGATAAAGAGCTTTATACACACATTAC | 612 | TC-146 |
| mpETB-2 | AGTGAACTTATCTTTCTATTGAAAAACACTC | |||||
| PVL components | lukS-PV– | AB006796 | PVL-1 | ATCATTAGGTAAAATGTCTGGACATGATCCA | 433 | ATCC 49775 |
| S and F | lukF-PV | NPVL-2 | GCATCAASTGTATTGGATAGCAAAAGC | |||
| LukE-LukD | lukE-lukD | Y13225 | LUKDE-1 | TGAAAAAGGTTCAAAGTTGATACGAG | 269 | FRI-913 |
| LUKDE-2 | TGTATTCGATAGCAAAAGCAGTGCA | |||||
| LukM | lukM | D42144 | LUKM-1 | TGGATGTTACCTATGCAACCTAC | 780 | ATCC 31890 |
| LUKM-2 | GTTCGTTTCCATATAATGAATCACTAC | |||||
| Alpha-hemolysin | hla | M90536 | HLA-1 | CTGATTACTATCCAAGAAATTCGATTG | 209 | FRI-913 |
| HLA-2 | CTTTCCAGCCTACTTTTTTATCAGT | |||||
| Beta-hemolysin | hlb | S72497 | HLB-1 | GTGCACTTACTGACAATAGTGC | 309 | NCTC 7428 |
| HLB-2-2 | GTTGATGAGTAGCTACCTTCAGT | |||||
| Delta-hemolysin | hld | AF288215 | HLD-1 | AAGAATTTTTATCTTAATTAAGGAAGGAGTG | 111 | NCTC 9393 |
| HLD-2 | TTAGTGAATTTGTTCACTGTGTCGA | |||||
| Gamma-hemolysin | hlg | L01055 | mpHLG-1 | GTCAYAGAGTCCATAATGCATTTAA | 535 | ATCC 49775 |
| components A, B, and C | mpHLG-2 | CACCAAATGTATAGCCTAAAGTG | ||||
| Gamma-hemolysin | hlg-2 | D42143 | mpHLG2-1 | GACATAGAGTCCATAATGCATTYGT | 390 | RIMD 31092 |
| variant | mpHLG2-2 | ATAGTCATTAGGATTAGGTTTCACAAAG | ||||
| EDIN | edin | M63917 | EDIN-1 | GAAGTATCTAATACTTCTTTAGCAGC | 625 | E-1 |
| EDIN-2 | TCATTTGACAATTCTACACTTCCAAC |
Culture and DNA extraction.
Strains were grown on brain heart infusion agar or in the same broth at 37°C overnight. Genomic DNA used as target for PCR and AFLP assays was extracted by using a standard phenol-chloroform procedure (26), and the concentration of DNA was estimated spectrophotometrically (26).
Identification of agr alleles.
The primers Pan-1 (5′-ATG CAC ATG GTG CAC ATG CA-3′) and Pan-2 (5′-CAT AAT CAT GAC GGA ACT TGC TGC GCA-3′) (Eurogentec, Seraing, Belgium) were designed from agr group I to IV sequences (GenBank accession numbers M21854, AF001782, AF001783, and AF288215, respectively) to amplify a 1,234-bp agr fragment encompassing the 3′ end of agrB, all of agrD, and the 5′ end of agrC. Amplification was carried out on a PE-9600 thermocycler (Perkin-Elmer Corp., Norwalk, Conn.) under the following conditions: an initial 5-min denaturation step at 95°C; followed by 30 cycles of 1 min of denaturation at 94°C, 1 min of annealing at 55°C, and 1 min of extension at 72°C; and a final extension step at 72°C for 10 min. The PCR products were purified by using the High Pure kit (Boehringer Mannheim) and sequenced with the primers used for PCR (Genome Express, Grenoble, France). The 198 strains were assigned to one of the four agr groups by comparing the predicted product of AgrD and the N-terminal half of AgrC with those of the four control strains (see above).
Toxin gene detection.
Sequences specific for sea-e, seg-j, sem-o, tst, eta, etb, lukS-PV-lukF-PV, lukE-lukD, lukM, hla, hlb, hld, hlg, hlg-2, and edin, encoding SEA-E; SEG-J; SEM-O; TSST-1; ETA; ETB; PVL components S and F; LUKE-LUKD; LUKM; the alpha-, beta-, delta-, gamma-, and gamma variant hemolysins; and EDIN, respectively, were detected by PCR on a PE-9600 thermocycler (Perkin-Elmer ) as previously described (15, 19) with the primers shown in Table 1 (Eurogentec). Amplification of gyrA was used to confirm the quality of each DNA extract and the absence of PCR inhibitors (5). All PCR products were analyzed by electrophoresis through 1% agarose gels (Sigma, Saint Quentin Fallavier, France). The distribution of the 24 toxin genes among the 198 strains is available on-line (ftp://pbil.univ-lyon1.fr/pub/datasets/statox.txt).
AFLP.
The Perkin-Elmer AFLP Microbial Fingerprinting Kit was used according to the manufacturer’s recommendations, except that the TaqI primers and adapter were as described by Vos et al. (30). The following two AFLP conditions were used, with one selective nucleotide added at the 3′ extremity of each primer: EcoRI + A/TaqI + C (condition A/C) and EcoRI + T/TaqI + G (condition T/G). Touchdown PCR cycling was done as recommended by the manufacturer, in a PE-9600 thermocycler (Perkin-Elmer). Processed DNA samples were loaded in pools of two with fluorescent dyes in 6% (wt/vol) denaturing polyacrylamide gels for electrophoresis (ABI Prism 373 Sequencer; Perkin-Elmer).
AFLP data processing.
Perkin-Elmer GeneScan analysis software was used to extract data from electropherograms. Assignation of fragments to discrete categories, transformation of data into tabular binary matrices (LecPCR program), and calculation of genomic distance (DistAFLP program), with or without bootstrap resamplings, were done as previously described (22). The LecPCR and DistAFLP programs are freely available on the ADE-4 web server (http://pbil.univ-lyon1.fr/ADE-4/microb). DistAFLP provides output files in the ADE-4 binary format suitable for multivariate analysis (28). Genomic distances are estimated rates of nucleotide substitution over whole genomes by using the Dice similarity index and Jukes-Cantor correction (22).
Statistical analysis.
The table of toxin gene detection in the 198 isolates was analyzed by using classical principal component analysis (PCA). Only 19 toxin genes were used, since four were either always present (hla and hld) or always absent (edin and lukM). The AFLP results (distance matrix between the 198 strains) were analyzed by using principal coordinate analysis (PCO) (11). This method provides a description of the main structures of distance matrices in the form of factor maps, in the same way as PCA. PCA of the toxin genes and PCO of the AFLP results gave independent results, and the corresponding graphics cannot be compared. However, these two analyses can be linked by co-inertia analysis (CIA) (7), so that the results can be compared and the factor maps can be superimposed. CIA gives co-inertia axes that have the maximum possible covariance with the variables in each of the two data sets. By using the covariance instead of the correlation (as in canonical correlation analysis), CIA maximizes the product of the correlation by the projected variances, ensuring that co-inertia axes will have both a good correlation with the initial variables and real meaning for each of the two data sets (7).
RESULTS
Phylogenetic distribution of the clinical S. aureus strains.
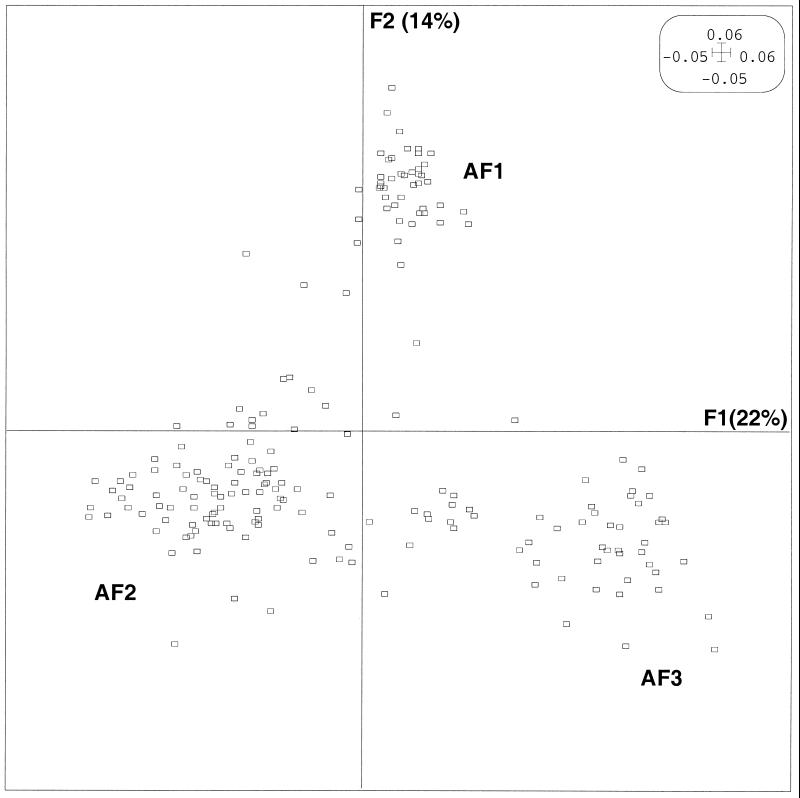
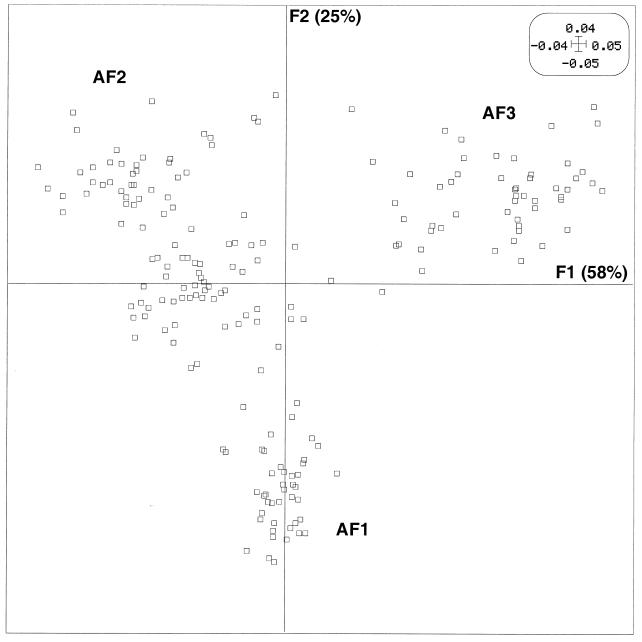
Since any relationship between the agr group and disease type would have to be interpreted according to the strain’s genetic background, we first conducted AFLP analysis of the 198 strains. PCO was performed on the distance matrix of the 198 isolates obtained by using the AFLP results to calculate genome divergence (i.e., the rate of nucleotide substitutions over whole genomes) between pairs of isolates. Figure 1 shows the factor map of the AFLP PCO. Since the first axis, F1, accounted for 22% of the variance, and the second axis, F2 (orthogonal to F1), accounted for the largest part (14%) of the variance not accounted for by F1 (data not shown), the results are expressed as the projections of each strain on a plane defined by these two axes, which were conserved for further analysis. This analysis divided the strains into three main phylogenetic groups, namely, AF1, a group with positive values on the F2 axis; AF2, a group with negative values on both the F1 and F2 axes; and AF3, a group distinguished from AF1 by negative values on the F2 axis.
FIG. 1.
PCO factor map (F1 × F2) of the 198 strains of S. aureus based on the AFLP data. The 198 strains are projected in the F1/F2 plane. This plane, obtained by computation, is defined by the two principal axes of the analysis: F1 explains most of the variance, and the second axis, F2 (orthogonal to F1), explains most of the remaining variance. Phylogenetic groups AF1, AF2, and AF3 are indicated. For clarity, when several strains are projected on the same point, only one is represented.
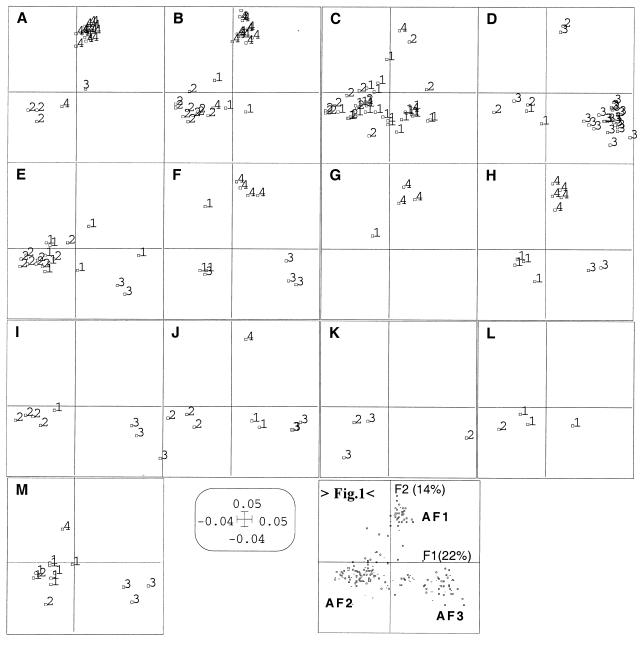
To evaluate the possible relationship between the disease type and the bacterial genetic background, the same AFLP PCO factor map was split according to the different diseases (classified in 13 disease types designated A to M, Table 2) for a better understanding of this relationship (Fig. 2). This analysis shows that the strains involved in scalded skin syndrome (disease type A) and bullous impetigo (disease type B) mainly belonged to phylogenetic group AF1 (27 of 46 strains). Enterotoxin-producing strains associated with scarlet fever and TSS (disease type C) mainly belonged to phylogenetic group AF2 (35 of 40 strains). The strains involved in TSST-1-associated disease (scarlet fever and menstrual and nonmenstrual TSS) (disease type D) belonged principally to phylogenetic group AF3 (21 of 28 strains). In contrast to toxin-mediated diseases, strains associated with suppurative infections (group E to M) did not seem to be specifically related to a particular AFLP cluster. Only endocarditis strains (disease type E) were mainly related to phylogenetic group AF2 (16 of 19 strains). The strains associated with diseases I, J, K, and L rarely belonged to group AF1. Interestingly, the strains associated with necrotizing pneumonia (disease F) and furuncles (disease H), which are both caused by Panton-Valentine leukocidin-producing strains (19), belonged to three phylogenetic groups, and the two graphics were highly superimposable.
TABLE 2.
agr group and AFLP group distribution according to the disease type
| Disease type | Letter designation | No. of strains
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Total (n) |
agr group (n)
|
AFLP group (n)
|
|||||||
| I | II | III | IV | AF1 | AF2 | AF3 | |||
| Exfoliative toxin-mediated disease | |||||||||
| Generalized exfoliative syndrome | A | 20 | 0 | 3 | 1 | 16 | 15 | 5 | 0 |
| Bullous impetigo | B | 26 | 3 | 10 | 0 | 13 | 12 | 14 | 0 |
| Subtotal | 46 | 3 | 13 | 1 | 29 | 27 | 19 | 0 | |
| Enterotoxin-mediated disease | C | ||||||||
| TSS | 24 | 16 | 7 | 0 | 1 | 1 | 20 | 2 | |
| Scarlet fever | 16 | 10 | 5 | 0 | 1 | 2 | 15 | 0 | |
| Subtotal | 40 | 26 | 12 | 0 | 2 | 3 | 35 | 2 | |
| TSST-1-mediated disease | D | ||||||||
| Menstrual TSS | 5 | 0 | 0 | 5 | 0 | 1 | 0 | 4 | |
| Nonmenstrual TSS | 6 | 1 | 0 | 5 | 0 | 0 | 1 | 5 | |
| Scarlet fever | 17 | 1 | 4 | 12 | 0 | 1 | 4 | 12 | |
| Subtotal | 28 | 2 | 4 | 22 | 0 | 2 | 5 | 21 | |
| Suppurative infections | |||||||||
| Endocarditis | E | 19 | 8 | 9 | 2 | 0 | 0 | 16 | 3 |
| Necrotizing pneumonia | F | 11 | 3 | 0 | 4 | 4 | 5 | 3 | 3 |
| Cellulitis and/or myositis | G | 4 | 1 | 0 | 0 | 3 | 3 | 1 | 0 |
| Furunculosis | H | 11 | 4 | 0 | 2 | 5 | 5 | 4 | 2 |
| Osteitis and/or osteomyelitis | I | 8 | 1 | 4 | 3 | 0 | 0 | 5 | 3 |
| Finger pulp infections | J | 9 | 2 | 3 | 3 | 1 | 1 | 5 | 3 |
| Arthritis | K | 4 | 0 | 1 | 3 | 0 | 0 | 3 | 1 |
| Subtotal | 66 | 19 | 16 | 18 | 13 | 14 | 37 | 15 | |
| Enterocolitis | L | 4 | 3 | 1 | 0 | 0 | 0 | 3 | 1 |
| Colonization | M | ||||||||
| Nose | 3 | 0 | 1 | 2 | 0 | 0 | 1 | 2 | |
| Vagina | 11 | 8 | 1 | 1 | 1 | 1 | 9 | 1 | |
| Subtotal | 14 | 8 | 2 | 3 | 1 | 1 | 10 | 3 | |
| Overall total | 198 | 61 | 49 | 43 | 45 | 47 | 109 | 42 | |
FIG. 2.
AFLP PCO factor map (F1 × F2) of the 198 strains, split according to the 13 disease types designated A to M: A, generalized exfoliative syndrome; B, bullous impetigo; C, enterotoxin-mediated diseases; D, TSST-1-mediated diseases; E, endocarditis; F, necrotizing pneumonia; G, cellulitis and/or myositis; H, furunculosis; I, osteitis and/or osteomyelitis; J, finger pulp infections; K, arthritis; L, enterocolitis; M, colonization. Numbers 1 to 4 in panels A to M indicate the agr group of each strain. Panel “>Fig. 1<” is a reduction of Fig. 1 showing the axes (F1 and F2) and the positions of the phylogenetic groups (AF1, AF2, and AF3).
Relationships between agr groups, genetic background, and disease type.
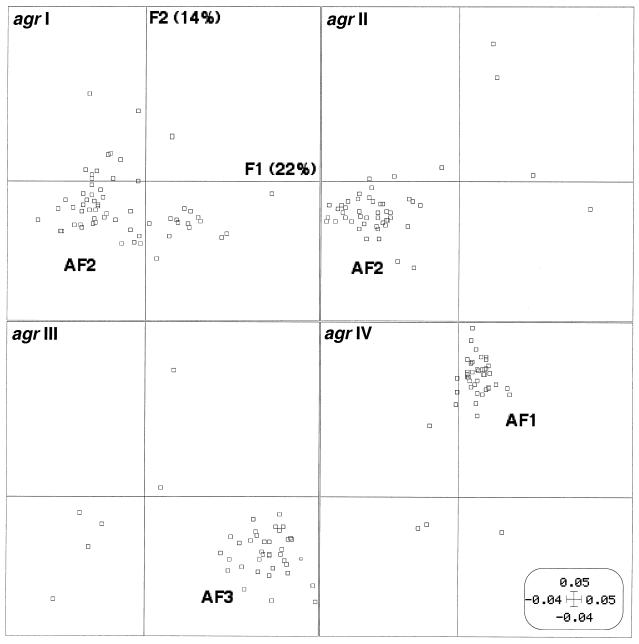
The agr group of the 198 S. aureus isolates was analyzed by amplification and sequencing. All isolates were classified as part of one of the four agr groups, and the distribution was relatively even, with 61 strains belonging to agr I, 49 belonging to agr II, 43 belonging to agr III, and 45 belonging to agr IV (Table 2). The AFLP PCO factor map for the 198 isolates was then split according to the four agr groups (Fig. 3). This clearly individualized strains from groups III and IV, while strains from groups I and II were partly superimposed. Thus, there was a very strong relationship between the AFLP cluster and the agr group in spite of the fact that a few isolates appeared to be outliers from the clusters of isolates belonging to the same agr group (four strains of agr group I, four strains of agr group II, six strains of agr group III, and three strains of agr group IV). This finding clearly indicated that agr types are associated with genetic backgrounds of strains. agr group labeling of each isolate on the AFLP PCO factor map showed a clear relationship between the genetic background, the agr group and the disease type, particularly for toxin-mediated diseases (Fig. 2, panels A to D). For instance, agr group IV strains involved in generalized exfoliative syndrome and bullous impetigo (disease groups A and B) were particularly associated with phylogenetic group AF1. Likewise, agr group III strains involved in TSST-1-mediated diseases (disease group D) were particularly associated with phylogenetic group AF3, and strains causing SE-mediated diseases (disease group C) belonged to agr group I or II and phylogenetic group AF2.
FIG. 3.
AFLP PCO factor map (F1 × F2) of the 198 strains of S. aureus as in Fig. 1 split according to the agr groups (agr I to IV). Phylogenetic groups AF1, AF2, and AF3 are indicated.
Relationships between toxin genes and genetic background.
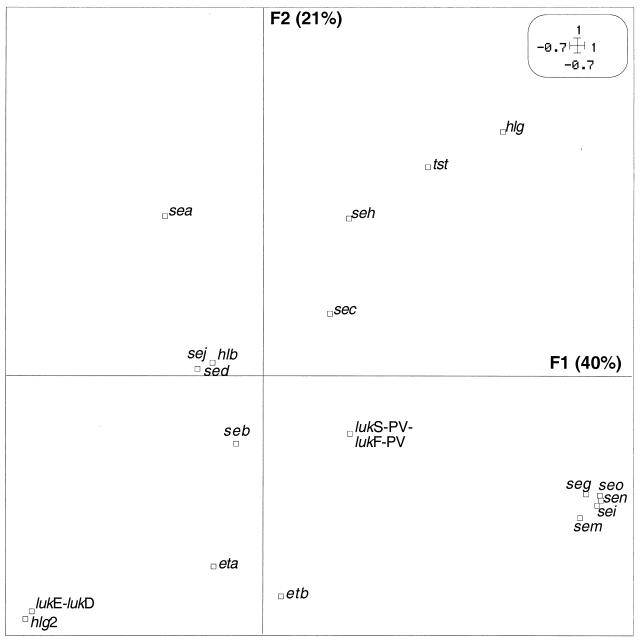
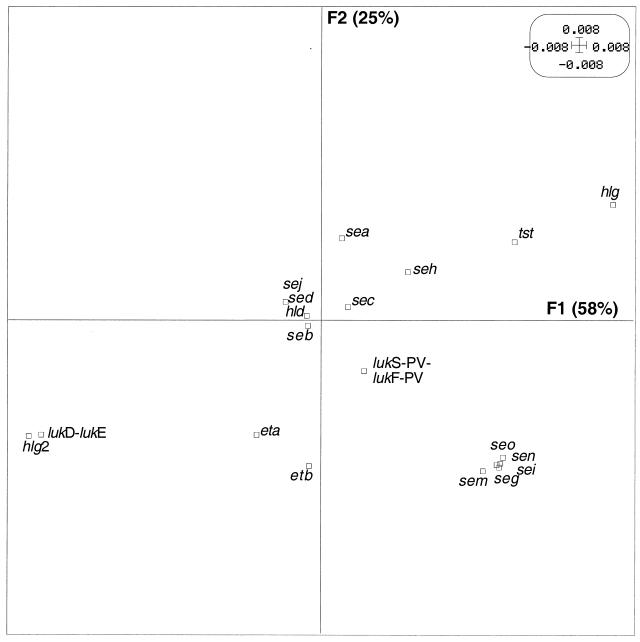
To determine whether the observed link between toxin-mediated diseases and genetic background was also found at the level of toxin genes, we analyzed the relationship between the distribution of 24 toxin genes and the genetic background. We first determined whether preferential combinations of toxin genes occurred among the 198 clinical strains, by means of PCA. Since the first axis, F1, and the second axis, F2 (orthogonal to F1), accounted for the largest part of the variance in PCA (40 and 21%, respectively), the results were expressed as the projections of each toxin gene on a plane defined by these two components (F1 and F2) (Fig. 4). On the first component, a group of five toxin genes (seg, sei, sen, seo, and sem, all encoded by the enterotoxin gene cluster) (15) was clearly individualized and slightly opposed to sea. On the second component there was an opposition between hlg-2 (together with lukE-lukD and, less strongly, eta and etb) and hlg (and, to a lesser extent, tst and seh). The other toxin genes (seb, sec, sed, sej, hlb, and lukS-PV-lukF-PV) were too near to the origin to be interpreted. The preferential combinations or exclusions of toxin genes suggested a nonrandom distribution of toxin genes in these strains.
FIG. 4.
PCA of 19 toxin genes in the 198 S. aureus strains. The variables are projected in the F1/F2 plane (defined as in Fig. 1). In each pair of axes, the variables located in a given direction relative to the origin can be considered positively associated, whereas the variables located in opposite directions can be considered antagonistic. Variables plotted near the origin cannot be interpreted. For example, the five gene toxins seg, sei, sen, sem, and seo can be considered associated with one another and negatively associated with sea. The toxin gene codes are given in Table 1.
To confirm the link between the toxin gene distribution and the genetic background of the strains, we coupled the AFLP clusters and the toxin gene analysis by using CIA (7). The PCA of the toxin gene table and the PCO of the AFLP results were coupled. Figure 5 shows the CIA factor map for the toxin genes, while Fig. 6 shows the CIA factor map for the AFLP patterns of the 198 isolates. These factor maps were very similar to the factor maps of the separate analyses (Fig. 3 and 4). The correlation coefficients between the co-inertia axes were 0.813 and 0.783 for the first and second axes, respectively. The percentages of variance derived from the co-inertia axes were 92 and 86% for the first axis in the toxin gene space and in the AFLP space, respectively. For the first two axes, the corresponding percentages were 98 and 83%. These very strong correlations and percentages of explained variance reflected a strong relationship between the toxin gene distribution and AFLP clusters. Hence, tst and hlg were associated with phylogenetic group AF3 and opposed to groups AF1 and AF2. The five genes belonging to the enterotoxin gene cluster (seg, sei, sem, sen, and seo) were associated with group AF1 and opposed to group AF2. lukD-lukE and hlg-2 were opposed to phylogenetic group AF3 and associated with groups AF1 and AF2. eta and etb were associated with phylogenetic group AF1 (Fig. 5 and 6). The distribution of the 24 toxin genes, the agr group, and the AFLP group among the 198 strains is available on-line (ftp://pbil.univ-lyon1.fr/pub/datasets/statox.txt).
FIG. 5.
First factor map (F1 × F2) of the CIA of the 19 toxin genes. This factor map is very similar to the PCA factor map (Fig. 4), with a few exceptions (sea and seb, for example). Genes with similar positions on this map have a similar profile of presence or absence among the 198 strains (as in Fig. 4), but they also belong to strains that have a similar AFLP profile (see Fig. 6).
FIG. 6.
First factor map (F1 × F2) of the CIA of the 198 S. aureus strains. This factor map is very similar to the PCO factor map (Fig. 1), except that the second (vertical) axis is inverted. Phylogenetic group AF1 is in the lower part of the graph, while groups AF2 and AF3 are in the upper part. Strains with similar positions on this map have genes present in the same direction on the map in Fig. 5.
DISCUSSION
We investigated a possible relationship between agr groups and the pattern of S. aureus disease and found a strong association between the agr types and certain diseases. However, in most cases the association reflected the link between the disease types, the pattern of toxin genes, and the genetic background of the strains. For instance, the strains causing SE-mediated diseases (disease group C) belonged to agr group I or II and phylogenetic group AF2. We thus concluded that, in most of the disease types considered (mainly toxin-mediated diseases), the agr alleles and toxin genes evolved contemporaneously with their parent strains and that horizontal transfer played only a marginal role. These findings confirm that specific bacterial pathogenicities are each essentially associated with a specific clone or group of clones.
The association of virulence factors with specific backgrounds has been discussed recently for several bacterial species such as Escherichia coli, in which a clonal distribution of virulence genes has been reported among clinical strains isolated from bloodstream infections (12, 20), neonatal meningitis (2), and extraintestinal infections (25). Regarding S. aureus, multilocus sequence typing comparison of isolates recovered from asymptomatic nasal carriers and from patients with severe diseases revealed that invasive diseases were primarily caused by a subset of genotypes unrepresentative of the carriage population as a whole (4). Another study of the genetic structure of S. aureus, involving MLEE, revealed that a single clone (designated ET41) of S. aureus producing TSST-1 causes most epidemiologically unrelated cases of urogenital TSS (23). The observation of a large proportion of asymptomatic female genital-tract carriers of this clone suggested that ET41 was highly adapted to the cervicovaginal tract. More recently, Booth et al. (3) used PFGE to analyze 405 clinical isolates of S. aureus and found that five phylogenetic lineages were highly prevalent and widely distributed, in contrast to 85 other lineages which occurred with frequencies of <2.5%. One of the five prevalent lineages (SAL1) comprised most TSST-1-producing strains, confirming the observation by Musser et al. (23). Booth et al. also found that SAL1 was enriched among the normal flora of the anterior nares and that lineage SAL4, which comprised 90% of methicillin-resistant S. aureus (MRSA) strains, was significantly associated with respiratory tract infections (3).
We studied a different population of methicillin-susceptible S. aureus clinical strains causing a broad spectrum of community-acquired infections. In agreement with the findings of Musser et al. (23) and Booth et al. (3), we confirmed the clonality of TSST-1-producing strains. In addition, we found that a number of diseases, such as SSSS, bullous impetigo, scarlet fever, TSS and, to a lesser extent, infective endocarditis, were preferentially associated with one of the three phylogenetic lineages that structured our strain population. In this respect, we fully agree with the concept developed by Falkow et al. that “the basic unit of bacterial pathogenicity is the clone or lineage that expands due to the possession of unique combinations of virulence genes” (10). With the exception of infective endocarditis, most of the diseases listed above are caused by specific toxins whose genetic determinants are frequently carried by potentially mobile elements such as plasmids (etb, seb, etc.), phages, or pathogenicity islands (tst, egc, lukFD-lukSE, etc.) (17). CIA, used to couple PCA of the toxin gene table (Fig. 4) and PCO of the AFLP results (Fig. 1), clearly indicated that the distribution of toxin genes was closely linked to the strain’s genetic background (Fig. 5 and 6). This suggested that the virulence determinants did not spread homogeneously among various genetic backgrounds or, at least, that the efficiency of such genetic exchanges between the three major lineages was low. Similarly, Booth et al. found that tst, cna (the collagen-binding protein gene), and hlb were associated with certain PFGE lineages and not with others, suggesting limited horizontal transfer among lineages (3). This is in accordance with the observation that, in E. coli, horizontal transfer generally does not disrupt the clonal structure of the species (6, 21, 27). A stable link between virulence and phylogeny could correspond to the necessity of having virulence determinants move into a particular genetic background for the emergence of a “virulent clone” (9, 10, 25). In contrast, PVL-associated diseases (necrotizing pneumonia and furunculosis) appeared to be caused by strains of the three lineages (Fig. 2). Since PVL is encoded by a bacteriophage, it is likely that in this case the bacteriophage has spread easily among the different backgrounds. With other suppurative diseases (groups I, J, K, and L [i.e., osteitis, finger pulp infections, arthritis, and enterocolitis, respectively]), the rarity of associations with phylogenetic group AF1 (Fig. 2) cannot yet be interpreted, since we do not know the virulence factors specifically involved in these infections. Microbial surface component-recognizing adhesive matrix molecules are likely play a role in these settings (13).
Considering the possible relation between the agr group and the disease type, we first postulated a relation between the agr group and the capacity to induce a specific disease. Ji et al. observed that the vast majority of menstrual toxic shock strains belonged to agr group III but that strains belonging to the other two groups had no apparent clinical specificity (16). We have previously shown that most ET-producing strains belong to agr group IV (14). In another study, agr group I was the most prevalent among 192 carrier and disease isolates, but 71% of the isolates were MRSA strains, which are known to be highly clonal (29). In our carefully selected collection of S. aureus clinical isolates (mainly causing community-acquired infections), the four agr types were relatively evenly distributed (Table 2). The agr group distribution correlated strongly with the genetic background of the strains and thus, indirectly, with certain disease profiles. The observed link between the agr group and genetic background was also found among coagulase-negative staphylococci in our laboratory: amplification-sequencing yielded 25 distinct agr variants among 13 staphylococcal species; the overall topology of the phylogenetic tree constructed from the DNA sequences of the 25 agr alleles was remarkably similar to that constructed from 16S rRNA loci of the 13 staphylococcal species, arguing against significant horizontal transfer of agr genes between populations of staphylococci (D. Dufour et al., unpublished data). As proposed by Novick, this agr grouping may represent the first subdivision of S. aureus based on the fundamental biology of the organism (24). Finally, though we cannot attribute a direct responsibility of the agr type in disease initiation, we can speculate that the preferential association between certain agr alleles, certain toxin genes, and a particular genetic background may make the activation of virulence factors more efficient. To paraphrase Falkow (10), we propose that the basic unit of bacterial pathogenicity would be the clone or lineage, which expands because it possesses particular combinations of virulence and regulatory genes in the appropriate genetic background.
Acknowledgments
We thank N. Violland, C. Courtier, and C. Gardon for technical assistance and D. Young for editing the manuscript.
This work was made possible by using the sequencing device facilities of the DTAMB at UCBL.
Editor: E. I. Tuomanen
REFERENCES
- 1.Arbuthnott, J. P., D. C. Coleman, and J. S. de Azavedo. 1990. Staphylococcal toxins in human disease. Soc. Appl. Bacteriol. Symp. Ser. 19:101S–107S. [DOI] [PubMed] [Google Scholar]
- 2.Bingen, E., B. Picard, N. Brahimi, S. Mathy, P. Desjardins, J. Elion, and E. Denamur. 1998. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J. Infect. Dis. 177:642–650. [DOI] [PubMed] [Google Scholar]
- 3.Booth, M. C., L. M. Pence, P. Mahasreshti, M. C. Callegan, and M. S. Gilmore. 2001. Clonal associations among Staphylococcus aureus isolates from various sites of infection. Infect. Immun. 69:345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Day, N. P., C. E. Moore, M. C. Enright, A. R. Berendt, J. M. Smith, M. F. Murphy, S. J. Peacock, B. G. Spratt, and E. J. Feil. 2001. A link between virulence and ecological abundance in natural populations of Staphylococcus aureus. Science 292:114–116. [DOI] [PubMed] [Google Scholar]
- 5.De Buyser, M. L., A. Morvan, F. Grimont, and N. El Solh. 1989. Characterization of Staphylococcus species by ribosomal RNA gene restriction patterns. J. Gen. Microbiol. 135:989–999. [DOI] [PubMed] [Google Scholar]
- 6.Desjardins, P., B. Picard, B. Kaltenbock, J. Elion, and E. Denamur. 1995. Sex in Escherichia coli does not disrupt the clonal structure of the population: evidence from random amplified polymorphic DNA and restriction-fragment-length polymorphism. J. Mol. Evol. 41:440–448. [DOI] [PubMed] [Google Scholar]
- 7.Doledec, S., and D. Chessel. 1994. Co-inertia analysis: an alternative method for studying species-environment relationships. Freshwater Biol. 31:277–294. [Google Scholar]
- 8.Durack, D. T., A. S. Lukes, and D. K. Bright. 1994. New criteria for diagnosis of infective endocarditis: utilization of specific echocardiographic findings. Duke Endocarditis Service. Am. J. Med. 96:200–209. [DOI] [PubMed] [Google Scholar]
- 9.Falkow, S. 1996. The evolution of pathogenicity in Escherichia coli, Shigella, and Salmonella, p.2723–2729. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 10.Falkow, S. 1997. What is a pathogen? ASM News 63:359–370. [Google Scholar]
- 11.Gower, J. C. 1966. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53:325–338. [Google Scholar]
- 12.Hilali, F., R. Ruimy, P. Saulnier, C. Barnabe, C. Lebouguenec, M. Tibayrenc, and A. Andremont. 2000. Prevalence of virulence genes and clonality in Escherichia coli strains that cause bacteremia in cancer patients. Infect. Immun. 68:3983–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Höök, M., and T. J. Foster. 2000. Staphylococcal surface proteins, p.386–391. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 14.Jarraud, S., G. J. Lyon, A. M. Figueiredo, G. Lina, F. Vandenesch, J. Etienne, T. W. Muir, and R. P. Novick. 2000. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J. Bacteriol. 182:6517–6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarraud, S., M. A. Peyrat, A. Lim, A. Tristan, M. Bes, C. Mougel, J. Etienne, F. Vandenesch, M. Bonneville, and G. Lina. 2001. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J. Immunol. 166:669–677. [DOI] [PubMed] [Google Scholar]
- 16.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027–2030. [DOI] [PubMed] [Google Scholar]
- 17.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Itao, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Uni, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225–1240. [DOI] [PubMed] [Google Scholar]
- 18.Lina, G., Y. Gillet, F. Vandenesch, M. E. Jones, D. Floret, and J. Etienne. 1997. Toxin involvement in staphylococcal scalded skin syndrome. Clin. Infect. Dis. 25:1369–1373. [DOI] [PubMed] [Google Scholar]
- 19.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128–1132. [DOI] [PubMed] [Google Scholar]
- 20.Maslow, J. N., T. S. Whittam, C. F. Gilks, R. A. Wilson, M. E. Mulligan, K. S. Adams, and R. D. Arbeit. 1995. Clonal relationships among bloodstream isolates of Escherichia coli. Infect. Immun. 63:2409–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milkman, R. 1997. Recombination and population structure in Escherichia coli. Genetics 146:745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mougel, C., S. Teysier, C. d’Angelo, K. Groud, M. Neyra, K. Sidi-Boumedine, A. Cloeckaert, M. Pelloille, S. Baucheron, E. Chaslus-Dancla, S. Jarraud, H. Meugnier, F. Forey, F. Vandenesch, G. Lina, J. Etienne, J. Thioulouse, C. Manceau, P. Robbe, R. Nalin, J. Briolay, and X. Nesme. Experimental and theoretical evaluation of typing methods based upon random amplification of genomic restriction fragments (AFLP) for bacterial population genetics. Genet. Sel. Evol., in press.
- 23.Musser, J. M., P. M. Schlievert, A. W. Chow, P. Ewan, B. N. Kreiswirth, V. T. Rosdahl, A. S. Naidu, W. Witte, and R. K. Selander. 1990. A single clone of Staphylococcus aureus causes the majority of cases of toxic shock syndrome. Proc. Natl. Acad. Sci. USA 87:225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novick, R. P. 2000. Pathogenicity factors and their regulation, p.392–407. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 25.Picard, B., J. S. Garcia, S. Gouriou, P. Duriez, N. Brahimi, E. Bingen, J. Elion, and E. Denamur. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Smith, J. M., N. H. Smith, M. O’Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thioulouse, J., D. Chessel, S. Dolédec, and J. M. Olivier. 1997. ADE-4: a multivariate analysis and graphical display software. Stat. Comp. 7:75–83. [Google Scholar]
- 29.van Leeuwen, W., W. van Nieuwenhuizen, C. Gijzen, H. Verbrugh, and A. van Belkum. 2000. Population studies of methicillin-resistant and -sensitive Staphylococcus aureus strains reveal a lack of variability in the agrD gene, encoding a staphylococcal autoinducer peptide. J. Bacteriol. 182:5721–5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, and M. Zabeau. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]