Abstract
Actinobacillus pleuropneumoniae is the causative agent of porcine pleuropneumonia, a disease characterized by pulmonary necrosis and hemorrhage caused in part by neutrophil degranulation. In an effort to understand the pathogenesis of this disease, we have developed an in vivo expression technology (IVET) system to identify genes that are specifically up-regulated during infection. One of the genes that we have identified as being induced in vivo is ohr, encoding organic hydroperoxide reductase, an enzyme that could play a role in detoxification of organic hydroperoxides generated during infection. Among the 12 serotypes of A. pleuropneumoniae, ohr was found in only serotypes 1, 9, and 11. This distribution correlated with increased resistance to cumene hydroperoxide, an organic hydroperoxide, but not to hydrogen peroxide or to paraquat, a superoxide generator. Functional assays of Ohr activity demonstrated that A. pleuropneumoniae serotype 1 cultures, but not serotype 5 cultures, were able to degrade cumene hydroperoxide. In A. pleuropneumoniae serotype 1, expression of ohr was induced by cumene hydroperoxide, but not by either hydrogen peroxide or paraquat. In contrast, an ohr gene from serotype 1 cloned into A. pleuropneumoniae serotype 5 was not induced by cumene hydroperoxide or by other forms of oxidative stress, suggesting the presence of a serotype-specific positive regulator of ohr in A. pleuropneumoniae serotype 1.
Actinobacillus pleuropneumoniae is the causative agent of porcine pleuropneumonia, a disease characterized by massive lung necrosis and pulmonary hemorrhage. This necrosis is due in part to the influx of host immune cells and the release of neutrophil lysosomal contents that include oxygen radicals, which can destroy the invading bacteria as well as the host tissue (27). In an effort to understand the pathogenesis of this respiratory disease and the effect of the host immune response on A. pleuropneumoniae, we have developed an in vivo expression technology (IVET) system to identify genes that are expressed during infection but not during growth in vitro on laboratory media (13). One of the genes that we have identified with this selection shows homology to ohr (organic hydroperoxide reductase), which has been described previously for Xanthomonas campestris (24), Pseudomonas aeruginosa (29), Enterococcus faecalis (33), and Bacillus subtilis (12). Ohr has been implicated in resistance to and detoxification of the organic peroxides, such as cumene hydroperoxide (CHP) (24, 29).
Bacteria are frequently exposed to reactive oxygen species during the course of infection. Oxygen radicals in the form of superoxides, hydrogen peroxides, and organic hydroperoxides can result from release of lysosomal contents within inflammatory cells or can be generated by bacterial cellular metabolism (3, 38). During infection of the porcine lung, A. pleuropneumoniae is exposed to oxygen radicals in the form of superoxides and peroxides generated by the neutrophil oxidative burst (27). A third class of oxygen radicals, organic hydroperoxides, can be generated either directly within the phagosome or as a consequence of oxygen radicals interacting with the bacterial cell membrane (reviewed in the work of Miller and Britigan [23]). To survive and protect its cellular metabolism within this dangerous milieu, A. pleuropneumoniae may require enzymes capable of inactivating these oxygen species (20).
A. pleuropneumoniae has been shown previously to contain catalase and two distinct superoxide dismutases, SodA and SodC, which can relieve a portion of the oxidative stress that occurs during infection (21). Enzymes that could detoxify the third category of oxidative stress reagents, the organic hydroperoxides, have not been previously identified in A. pleuropneumoniae. In this work, we identify another potentially protective gene, ohr, which is specifically induced during infection and produces a protein that is capable of detoxifying organic peroxides encountered by A. pleuropneumoniae during infection of the porcine lung.
MATERIALS AND METHODS
Bacterial strains.
The A. pleuropneumoniae strains that were used in this study and the plasmids propagated in these strains are listed in Table 1. A. pleuropneumoniae strains were cultured at 35°C on either brain heart infusion (BHI) (Difco, Detroit, Mich.) or heart infusion (Difco) medium supplemented with V factor (β-NAD) added at 10 μg/ml (BHIV or HIV, respectively). Riboflavin, when needed for maintenance of APP233, was added at 200 μg/ml. Medium was supplemented with ampicillin at 50 μg/ml to propagate plasmids or at 20 μg/ml to recover A. pleuropneumoniae from porcine lungs and to select for transformants after electroporation. Escherichia coli strain XLI-Blue mRF′ (Stratagene, La Jolla, Calif.) was used for construction and propagation of plasmids. E. coli was cultured on Luria-Bertani medium (Difco) supplemented with ampicillin at 100 μg/ml.
TABLE 1.
Characteristics of bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristic(s) | Source (reference) |
|---|---|---|
| Strains | ||
| APP225 | A. pleuropneumoniae ATCC 27088, serotype 1A, passaged through pigs | Fuller et al. (15) |
| APP227 | A. pleuropneumoniae ISU178, a serotype 5 field isolate, passaged through pigs | Fuller et al. (14) |
| APP233 | A double-crossover riboflavin auxotroph of APP225 | Fuller et al. (15) |
| ATCC 27089 | A. pleuropneumoniae serotype 2 | ATCCa |
| ATCC 27090 | A. pleuropneumoniae serotype 3 | ATCC |
| ATCC 33378 | A. pleuropneumoniae serotype 4 | ATCC |
| ATCC 33590 | A. pleuropneumoniae serotype 6 | ATCC |
| ISU53 | A. pleuropneumoniae serotype 7 | V. Rapp, Iowa State University |
| 405 | A. pleuropneumoniae serotype 8 | R. Nielsen, Denmark |
| CVJ1326 | A. pleuropneumoniae serotype 9 | R. Nielsen, Denmark |
| 13039 | A. pleuropneumoniae serotype 10 | R. Nielsen, Denmark |
| 56513 | A. pleuropneumoniae serotype 11 | R. Nielsen, Denmark |
| 1096 | A. pleuropneumoniae serotype 12 | R. Nielsen, Denmark |
| Plasmids | ||
| pTF86 | A. pleuropneumoniae IVET vector containing promoterless luxAB and ribBAH genes downstream of a unique BamHI cloning site | Fuller et al. (13) |
| piviK | pTF86 containing 801-bp insert with 294 bp of ohr in BamHI site | This study |
| pGEM-T | Apr TA cloning vector for PCR fragments | Promega |
| pinKE | pGEM-T containing 2.5-kb insert with ohr inverse PCR fragment | This study |
| pGZRS18 | AprA. pleuropneumoniae-E. coli shuttle vector | West (44) |
| pGeohr | pGZRS18 containing full-length ohr gene with 360 bp of upstream sequence | This study |
ATCC, American Type Culture Collection, Manassas, Va.
Molecular manipulations.
Genomic DNA from A. pleuropneumoniae was prepared according to the lysis-proteinase K method described in the work of Silhavy (36). Plasmid DNA was purified using Qiagen spin columns (Qiagen Inc., Valencia, Calif.). DNA-modifying enzymes were obtained from Roche (Roche Molecular Biochemicals, Indianapolis, Ind.) and used according to the manufacturer's specifications.
Preparation of electrocompetent A. pleuropneumoniae serotype 1 and electroporation of these cells with plasmid DNA prepared from either E. coli or A. pleuropneumoniae serotype 1 was performed as previously described (15). A. pleuropneumoniae serotype 5 (14) was made competent by the method of Ward et al. (43) and electroporated using the same electroporation conditions as those for serotype 1 but using plasmid prepared from E. coli. E. coli transformation was performed by the Hanahan method (16).
Southern blotting.
Genomic DNA from all 12 A. pleuropneumoniae serotypes (Table 1) was digested to completion using EcoRI and separated on a 0.8% agarose gel. DNA fragments were transferred to a Nytran (Schleicher & Schuell, Keene, N.H.) membrane by the method of Southern (35). The membrane was hybridized with a digoxigenin-labeled probe generated by PCR amplification of the complete ohr gene using primers (MM150, 5"-GACAAGAATTCAACAAGGACAATATTATG-3", and MM151, 5"-CCTAAATCGTCCCAGATCTGGTAGG-3") that flank the open reading frame (ORF) (Roche PCR DIG synthesis kit). For high-stringency hybridization, blots were incubated for 16 h at 42°C with the probe diluted in a hybridization buffer that contained 50% formamide, 5× SSC (1× SSC is 0.15 M sodium chloride plus 0.015 M sodium citrate), and 2% blocking reagent (Roche). High-stringency washes were performed at 68°C with 0.1× SSC-0.1% sodium dodecyl sulfate. For low-stringency hybridization, blots were incubated for 16 h at 42°C with the probe diluted in an aqueous solution containing 6× SSC and 1% blocking reagent, with no formamide added. Low-stringency blots were washed for 10 min at room temperature with 2× SSC-0.1% sodium dodecyl sulfate. Hybridizing bands were detected using alkaline phosphatase-tagged antidigoxigenin and the CDP* chemiluminescent substrate (Roche).
Infection model.
The infection of pigs with either APP233/piviK or APP233/pTF86 was performed as previously described (13). Briefly, each clone was inoculated into HIV broth containing 200 μg of riboflavin/ml, 50 μg of ampicillin/ml, and 5 mM calcium chloride and grown at 35°C to an optical density at 520 nm (OD520) of 0.8. The bacteria were washed with saline and resuspended in 10 ml of saline containing 200 μg of riboflavin/ml at a final concentration of 7 × 108 CFU. The bacterial suspension was inoculated, via an endotracheal catheter, into the lungs of 6-week-old pigs. The pigs were monitored for development of clinical signs of pleuropneumonia as previously described (18) and in conjunction with animal use approval. Sterile lung samples were collected for bacterial isolation and photography following necropsy. Lung samples were examined for in situ luciferase activity by using a Hamamatsu C1966 photonic microscope system (13). All experimental protocols for animal experiments were reviewed by the Michigan State University All-University Committee on Animal Use and Care, and all procedures conformed to university and U.S. Department of Agriculture regulations and guidelines.
Cloning of the intact ohr gene.
An inverse PCR technique was designed to clone the intact ohr gene (28). A 2.5-μg quantity of A. pleuropneumoniae serotype 1 genomic DNA was digested to completion with EcoRI, followed by a self-ligation using T4 DNA ligase (Roche) to form closed circular fragments. PCR amplification with AmpliTaq (Roche) was performed with 0.04 μg of ligated DNA using primers (MM139, 5"-AACCAAGTGAACCGTCATCTACTC-3", and MM140, 5"-GTGGCAAAGTCGGCACAAACC-3") designed from the known sequence of the iviK clone, which contained the promoter region and partial ORF of the ohr gene. These primers bound within the coding region of ohr and were oriented to PCR amplify the flanking regions. The resulting 2.5-kb PCR fragment was isolated from an agarose gel using the Qiaex II kit (Qiagen) and was cloned into the pGEM-T vector (Promega, Madison, Wis.) to form pinKE. This clone was sequenced using an ABI100 Model 377 automated sequencer (Applied Biosystems, Foster City, Calif.) and a primer (MM141, 5"-CTGTAGGCGTGGGAATCGGTC-3") internal to the known ohr sequence. A complete ORF was identified by comparison of the sequence obtained from the iviK clone and the sequence that resulted from the inverse PCR clone pinKE. The complete ohr ORF with the upstream promoter region was amplified from A. pleuropneumoniae serotype 1 genomic DNA using Pfu polymerase (Promega) and a primer pair (MM138, 5"-GGCTACGAAATATTGGACACG-3", and MM151) that binds 360 bp upstream of the start codon and 50 bp downstream of the stop codon. The PCR product was cloned into SmaI-cut pGZRS18 to form pGeohr (44). This plasmid was transformed into both A. pleuropneumoniae serotype 1 and A. pleuropneumoniae serotype 5.
Oxidative stress growth inhibition assay.
Hydrogen peroxide (Sigma, St. Louis, Mo.), CHP (Sigma), and the superoxide generator paraquat (Sigma) were used as oxidative stress reagents. Disk inhibition assays were used to analyze bacterial sensitivity to these reagents (38). Briefly, 100 μl of an overnight bacterial liquid culture was added to 3 ml of BHIV top agar (0.7%) and poured onto BHIV plates. Filter paper disks (10 mm, Whatman no. 1; Whatman Paper Ltd., Maidstone, England) saturated with 10 μl of 0.88 M hydrogen peroxide, 200 mM CHP, or 0.074 M paraquat were placed onto the hardened top agar (38). Diameters of the zones of growth inhibition were recorded after 22 h of incubation at 35°C under 5% CO2. Statistical analysis of zone diameter significance within each treatment group for serotypes containing ohr compared to serotypes lacking the gene was evaluated by a two-tailed Student t test. The Analysis ToolPak in Microsoft Excel (2000) was used to perform the t test under homoscedastic and heteroscedastic conditions.
Oxidative stress and measurement of lux expression.
Induction of oxidative stress in broth cultures was performed as follows. A. pleuropneumoniae strains were grown in 25 ml of BHIV broth containing 50 μg of ampicillin/ml, at 35°C and with shaking at 150 rpm, to an OD520 of 0.8 and were then dispensed into a 96-well microtiter plate in 200-μl aliquots. This was followed by a 30-min period of incubation at 35°C under 5% CO2 prior to addition of the stress reagent to allow for acclimation. For stress induction, stress reagents were added to a final sublethal concentration of 125 μM, 300 μM, or 1 mM CHP; 56 μM hydrogen peroxide; or 50 μM paraquat (38). Aliquots were taken for luminometric assays and primer extension at designated intervals.
Quantitative luciferase assays were performed using a Turner Model 20e luminometer as previously described (13). Briefly, 20 μl of culture was mixed with 20 μl of N-decyl aldehyde in a polypropylene luminometer cuvette. The sample was read in full integral, autoranging mode with a predelay of 0 s, a delay of 10 s, and an integration time of 30 s. The N-decyl aldehyde substrate was made by sonicating 20 mg of Essentially Fatty Acid Free Bovine Serum Albumin (Sigma)/ml with 1 μl of N-decyl aldehyde/ml in a sonicating water bath at room temperature. Luminometer readings were normalized to relative light units (RLU) per OD520 unit.
Functional analysis of Ohr activity.
An assay to evaluate the degradation of CHP was adapted from procedures developed by Dringen et al. (9) and Ochsner et al. (29). A. pleuropneumoniae was grown in BHIV broth to early stationary phase (OD520 = 0.8 to 1.0) and diluted with fresh prewarmed medium. CHP was added to a final concentration of either 0, 125, 300, or 600 μM CHP. Residual CHP concentrations were determined at 5-min intervals by a xylenol orange-iron reaction. At each time point, 100 μl of the culture was pelleted and 20 μl of the cell-free supernatant was added to 80 μl of 25 mM sulfuric acid in a 96-well microtiter plate. When all samples had been collected, 100 μl of freshly prepared reaction buffer containing 200 μM xylenol orange (Sigma), 200 μM ammonium ferrous sulfate (Sigma), and 25 mM sulfuric acid was added to each sample. After 10 min of incubation at room temperature, absorbance was read at 540 nm using a Bio-Tek ELISA Plate Reader Model EL310 (Bio-Tek Instruments, Inc., Winooski, Vt.). Concentrations of CHP in each sample were determined by comparison to a CHP standard curve performed at the time of each assay. Ohr activity was measured as micromoles of CHP degraded per minute.
To evaluate the induction of Ohr activity by CHP, CHP was added to 1 ml of the freshly diluted culture at a final concentration of 0, 125, or 300 μM, and this mixture was held without shaking for 30 min at 35°C. A sample was collected to determine the residual CHP concentration. Fresh CHP was added, and Ohr activity was assayed as described above.
Primer extension.
Primer extension of the ohr gene was performed as previously described (8) using a primer (MM220, 5"-CGAGTATGACCATCACGACCGCCAACTGC-3") that bound 30 bp into the ohr coding region. Bacteria were incubated for 30 min in a 96-well microtiter plate under inducing conditions with 1 mM CHP, and without CHP as a noninduced control. The mRNA was isolated using a hot-phenol extraction method (45). For reverse transcription, 10 μg of RNA was incubated with 1 pM [32P]ATP (Amersham Pharmacia Biotech, Piscataway, N.J.)-labeled primer and avian myeloblastosis virus reverse transcriptase (Promega) for 1 h at 42°C. The samples were separated on an 8% denaturing polyacrylamide gel along with a 35S-dATP (Amersham) sequencing ladder. The sequencing ladder was prepared using the Sequenase version 2.01 kit (Amersham) and the same primer that was used for primer extension (8).
Nucleotide sequence accession number.
The sequence reported in this paper has been submitted to GenBank and assigned accession no. AF395877.
RESULTS
Identification of ohr as an in vivo-induced gene.
We previously developed an IVET screen to identify genes from A. pleuropneumoniae that were induced during infection of the porcine lung but had minimal expression during in vitro growth on laboratory media (13). This IVET system utilized a promoter trap vector (pTF86) with a cloning site for genomic DNA fragments upstream of promoterless copies of the luxAB and ribBAH genes. A library of random genomic DNA fragments from A. pleuropneumoniae serotype 1 was cloned into the pTF86 vector and subsequently electroporated into APP233, a virulent A. pleuropneumoniae serotype 1 strain that is unable to produce riboflavin due to a directed mutation within the ribGBAH operon (15). When a functional promoter was placed in the cloning site of pTF86, riboflavin was produced and complemented the riboflavin deficiency of the host strain, APP233, restoring full virulence. Without a functional promoter, riboflavin was not produced and APP233 failed to survive and cause disease (15). Thus, the initial portion of the IVET procedure utilized infection of the natural host to select clones containing promoters that were expressed during infection. Clones containing functional promoters were isolated from the pig lung and examined for in vitro expression of the luxAB genes both quantitatively and qualitatively. Clones that had in vitro expression levels in APP233 that were ≤200 RLU/OD520 unit were identified as ivi (in vivo-induced) clones.
Forty-two unique ivi clones were identified during this selection. One of these clones, iviK, contained an 801-bp insert that included a partial ORF of 98 amino acids fused to luxAB, as well as 507 bp of upstream noncoding sequence. When this ORF, which contained a start codon but lacked a stop codon, was used to search microbial sequence databases, it showed 56% similarity to Ohr from X. campestris, an enzyme responsible for protection against organic peroxides (24).
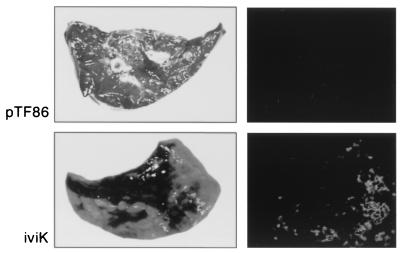
To confirm the in vivo induction of the iviK promoter, a pig was infected intratracheally with 7 × 108 CFU of APP233/piviK to monitor development and progression of the pulmonary disease. We have previously demonstrated that APP233 alone is avirulent at doses as high as 5 × 109 CFU in this animal infection model (13, 15). Within 6 h, the infected pig developed an increased respiratory rate and fever and showed depression and anorexia. The disease progressed to severe dyspnea by 9 h postinfection. At necropsy, 90 to 100% of the right lung lobes and the accessory lobe showed edema, hemorrhage, congestion, and regions of necrosis. These symptoms are consistent with peracute pleuropneumonia. A portion of the right caudal lung lobe from this pig was photographed by visible light camera and by photonic camera (Fig. 1). The visible light picture shows regions of severe necrosis and hemorrhage. The photonic camera picture of this same region of lung shows lux expression at the edges of this necrotic tissue, which is the region of active infection. In contrast to infection by this iviK clone, we have previously shown that infection with 5 × 109 CFU of a clone containing the pTF86 vector only does not result in disease symptoms in the pig (13). When lung tissue isolated from a pig infected with APP233/pTF86 was examined, there were no regions of necrosis or of lux expression (Fig. 1).
FIG. 1.
Pulmonary damage and in vivo lux expression resulting from infection of a swine lung with either APP233/pTF86 (top) or APP233/piviK (bottom). For each sample, on the left is an incidental light photograph of the lung specimen. On the right is a corresponding photograph, taken by photonic camera, showing bioluminescence of the same lung specimen. In the absence of a promoter, APP233/pTF86 showed no lung damage and no lux expression. In the presence of the piviK plasmid, significant lung damage was apparent and bioluminescence was seen.
Cloning and characterization of full-length ohr.
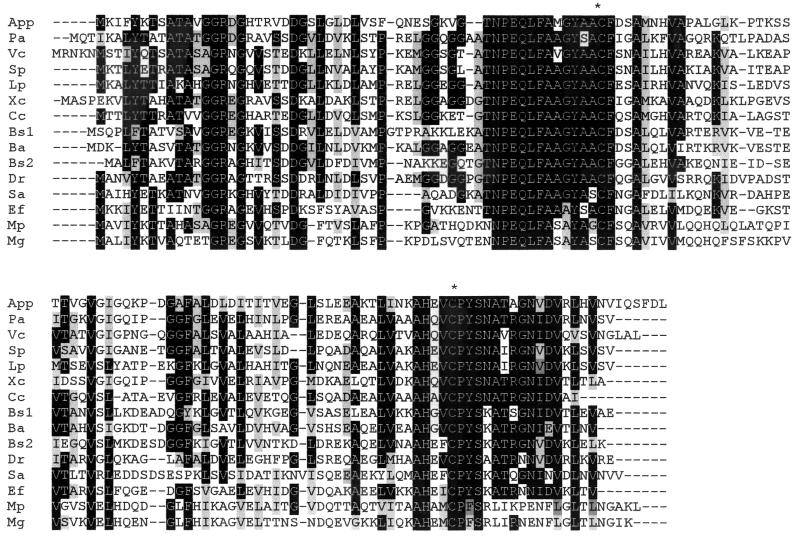
The nucleotide sequence of iviK was used to design inverse PCR primers to clone the full-length ohr gene. A 2.5-kb PCR product was obtained and cloned into pGEM-T to form pinKE. This insert was sequenced, and alignment of this sequence with that of iviK demonstrated a contiguous ORF of 432 bp encoding 143 amino acids. This ORF was identified based on sequence homology as ohr (Fig. 2). Primers were designed to amplify the entire ohr gene plus 360 bp of upstream sequence. This PCR product was cloned into the pGZRS18 vector to form pGeohr.
FIG. 2.
Protein sequence alignment of putative Ohr proteins from 14 bacterial species aligned with Boxshade v3.31 (http://biophysics.med.jhu.edu/prog/boxshade/PC__and__MAC/win16.zip) and ClustalX (41). Black-shaded regions indicate residues that are identical in the majority of species. Gray-shaded regions indicate residues that are functionally conserved in the majority of species. This alignment highlights the highly conserved regions surrounding the conserved cysteine residues (∗). Species abbreviations used are as follows: App, A. pleuropneumoniae; Pa, P. aeruginosa; Vc, Vibrio cholerae; Sp, Shewanella putrefaciens; Lp, Legionella pneumophila; Xc, X. campestris; Cc, Caulobacter crescentus; Bs1, B. subtilis YklA (OhrA); Ba, Bacillus anthracis; Bs2, B. subtilis YkzA (OhrB); Dr, Deinococcus radiodurans; Sa, Staphylococcus aureus; Ef, E. faecalis; Mp, Mycoplasma pneumoniae; Mg, Mycoplasma genitalium.
A potential ribosome binding site (AAGGA) at 12 bp upstream of the start codon was identified. Potential transcriptional terminators flanking the ORF were identified using the GCG Stemloop program (Genetics Computer Group [Madison, Wis.] program manual for the Wisconsin package, 8th ed.). The predicted protein sequence was compared to finished and partially finished bacterial genomes deposited in the Microbial Genomes BLAST databases using the BLAST programs (National Center for Biotechnology Information). The ORF had highest homology to Ohr from P. aeruginosa with 62% similarity and 48% identity when examined using BLAST pairwise homology (29, 40). Proteins with strong homology to Ohr were identified from 14 species of bacteria and were aligned to identify regions of identity and similarity (Fig. 2). This alignment shows two regions of high conservation that center around conserved cysteines, with the sequence outside of these regions lacking extensive similarity. Analysis of the protein using PSORT to predict cellular localization suggested that Ohr was a cytoplasmic protein (26).
The region upstream of Ohr was also used to search databases at both the nucleotide and protein levels using the BLAST programs (National Center for Biotechnology Information), and no matches of significant homology were detected. The upstream region contains several small ORFs, all of ≤50 amino acids, which lack significant homology to known proteins.
Distribution of ohr among serotypes correlates with organic peroxide resistance.
To determine if ohr was present in all 12 serotypes of A. pleuropneumoniae, Southern blotting was performed with EcoRI-digested genomic DNA from all 12 serotypes as the target and with the use of the full-length ohr gene from A. pleuropneumoniae serotype 1 to construct the probe. Hybridizing bands were seen with genomic DNA from A. pleuropneumoniae serotypes 1, 9, and 11 (Fig. 3A) under stringent conditions, whereas no hybridization was seen with genomic DNA from A. pleuropneumoniae serotypes 2 to 8, 10, or 12 even under relaxed conditions.
FIG. 3.
Correlation of ohr presence with resistance to organic hydroperoxides. The presence of the gene was determined by Southern blotting, under high-stringency conditions, of genomic DNA with the intact ohr gene as a probe (A). Molecular size standards (in kilobases) are shown on the left. Each lane in panel A indicates a different serotype. The resistance of each of the 12 A. pleuropneumoniae serotypes to oxidative stress agents was determined by measuring the diameter of the zone of growth inhibition due to CHP (B), hydrogen peroxide (C), and paraquat (D). The data presented are the averages of three experiments, with the standard deviations shown by error bars. The differences in the zone diameters, after incubation with CHP, seen with serotypes 1, 9, and 11, in comparison to the remaining serotypes, are statistically significant (P < 0.0001).
Sensitivity to oxidative stress reagents for each of the 12 A. pleuropneumoniae serotypes was examined using a disk inhibition assay. Overnight cultures were added to top agar and overlaid with disks saturated with hydrogen peroxide, CHP, or the superoxide generator paraquat. After 22 h, the zones of growth inhibition were recorded as a measure of the sensitivity of each serotype to the oxidative stress imposed. Of the 12 serotypes, 11 showed equivalent sensitivities to hydrogen peroxide and paraquat (Fig. 3C and D). The exception was serotype 6, which showed a significantly larger zone diameter (P < 0.01) with all three forms of oxidative stress, suggesting a decreased resistance to oxidative stress reagents in general for this serotype. Serotypes 2 to 8, 10, and 12 showed sensitivity to CHP similar to that seen with hydrogen peroxide and paraquat (Fig. 3B to D). In contrast, serotypes 1, 9, and 11 showed a significantly reduced zone of growth inhibition upon incubation with CHP, reflecting an increased resistance to oxidative stress caused by organic peroxides (Fig. 3B). This increased resistance of serotypes 1, 9, and 11 to CHP was highly statistically significant (P < 0.0001), and no statistically significant difference was seen for these serotypes in response to incubation with hydrogen peroxide or paraquat. This increased resistance to CHP, but not to hydrogen peroxide or paraquat, correlated with the presence of the ohr gene as shown by Southern blotting (Fig. 3A).
Induction of ohr in response to oxidative stress.
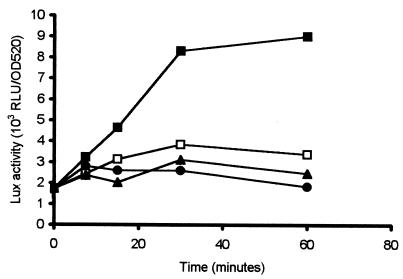
To characterize the expression of ohr in response to different oxidative stress reagents, induction studies were performed with wild-type A. pleuropneumoniae serotype 1. The analysis was performed with the wild-type strain in order to decrease the oxidative stress imposed on the cultures due to the presence of riboflavin in the medium that is necessary for growth of the APP233 strain. APP225/piviK, which contained the ohr promoter-luxAB fusion, was induced in microtiter plates with paraquat, CHP, or hydrogen peroxide. Addition of CHP, at 125, 300, or 1,000 μM, resulted in a rapid increase in lux expression in comparison to the noninduced control (Fig. 4). Neither paraquat nor hydrogen peroxide caused any induction, and the level of lux expression was equivalent to that seen in the absence of oxidative stress (Fig. 4). The concentrations of oxidative stress reagents were selected as the maximal sublethal dose (data not shown).
FIG. 4.
Induction of the cloned ohr promoter in A. pleuropneumoniae serotype 1 (APP225/piviK) by various oxidative stress reagents. CHP (1 mM; ▪), paraquat (50 μM; •), and hydrogen peroxide (56 μM; ▴) were added as oxidative stress reagents, and Lux activity, expressed as RLU per OD520 unit, was measured. Lux activity was also measured in control cultures to which no inducing agent was added (□). Data presented are from a representative experiment. Trends were identical in all experiments.
Comparison of induction of ohr in A. pleuropneumoniae serotype 1 and A. pleuropneumoniae serotype 5.
We compared the expression of the ohr::lux fusion by using APP225/piviK (serotype 1) and APP227/piviK (serotype 5) during normal growth in culture and under induction by CHP. A growth curve was determined for each serotype to evaluate the expression level of ohr during growth in an aerated broth culture. The expression of ohr for both A. pleuropneumoniae serotype 1 and A. pleuropneumoniae serotype 5, as measured by luciferase activity, remained constant during normal growth, with the increase over time directly correlated with the increase in total cell number (data not shown). The expression level was independent of serotype, with both serotypes showing low but equal expression levels during normal growth in the absence of inducers. This expression level of luciferase in the wild-type strains of A. pleuropneumoniae serotype 1 and A. pleuropneumoniae serotype 5, which averaged 1,000 RLU/OD unit, is greater than that seen for the A. pleuropneumoniae serotype 1 riboflavin mutant (APP233), which was never higher than 200 RLU/OD unit. Expression of luciferase from the pTF86 vector alone, with no insert, showed minimal expression of <50 RLU/OD unit in both serotypes (data not shown).
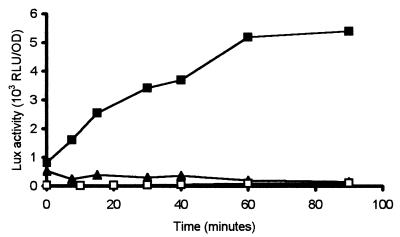
Expression of ohr under inducing conditions was examined for both A. pleuropneumoniae serotype 1 and A. pleuropneumoniae serotype 5. Induction of ohr in response to incubation with CHP was seen only in A. pleuropneumoniae serotype 1, not in A. pleuropneumoniae serotype 5 (Fig. 5). APP225/piviK showed rapid induction of expression, as measured by Lux assay with a twofold increase within 10 min post-exposure to CHP. Lux activity increased over time, with maximal levels detected between 30 and 60 min after induction. In contrast, APP227/piviK showed no increase in lux expression in response to CHP and maintained a level of expression slightly greater than that of the vector-only control. These data suggest that incubation with CHP does not cause induction of ohr in A. pleuropneumoniae serotype 5.
FIG. 5.
Expression of the ohr promoter is induced by 125 μM CHP in A. pleuropneumoniae serotype 1 and not in serotype 5. lux expression driven by the ohr promoter was measured over time in A. pleuropneumoniae serotype 1 (APP225/piviK) (▪) and in A. pleuropneumoniae serotype 5 (APP227/piviK) (▴) in comparison to a vector-only control, APP225/pTF86 (□). Data presented are from a representative experiment. Trends were identical in all experiments.
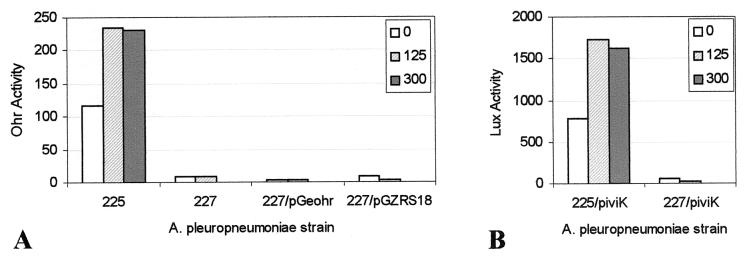
These induction data were confirmed through a functional assay of Ohr enzymatic activity. A xylenol orange colorimetric assay was used to determine the concentration of CHP after incubation of CHP with bacteria. Each serotype was grown in broth and then diluted into fresh media containing 125 or 300 μM CHP followed by incubation for 30 min to allow for induction. After this induction period, 125, 300, or 600 μM CHP was added and the rate of CHP degradation was measured. APP225, A. pleuropneumoniae serotype 1, showed significant Ohr activity, as measured by the rate of CHP degradation, in the absence of induction. Induction of A. pleuropneumoniae serotype 1 for 30 min with either 125 or 300 μM CHP resulted in an approximately twofold increase in enzymatic activity, which correlated well with the increase in ohr expression as measured by Lux activity (Fig. 5 and 6).
FIG. 6.
(A) Ohr activity, expressed as micromoles of CHP degraded per minute, is induced by CHP in A. pleuropneumoniae serotype 1 (APP225) but not in serotype 5 containing the intact ohr gene plus promoter region (APP227/pGeohr). Controls include APP227 with no plasmid and APP227 containing the pGZRS18 shuttle vector only. For induction, CHP was added at final concentrations of 0, 125, or 300 μM to 1-ml broth cultures, which were held for 30 min at 35°C. Ohr activity was measured as degradation of CHP. (B) Lux activity, measured as RLU per OD unit, is similarly induced in APP225/piviK but not in APP227/piviK. Induction conditions for these assays were identical to those described for panel A. Data presented are from a representative experiment. Trends were identical in all experiments.
In contrast, neither APP227 (A. pleuropneumoniae serotype 5), APP227/pGZRS18, nor APP227/pGeohr showed significant Ohr activity in the absence of induction, and no increase in activity was evident under inducing conditions. These results correlate with and confirm the lack of expression of ohr in APP227 as measured by Lux activity (Fig. 5 and 6). The assays performed with APP227 showed that CHP concentrations of ≥300 μM CHP were lethal to the cells, and thus, induction and assays were performed at 125 μM CHP.
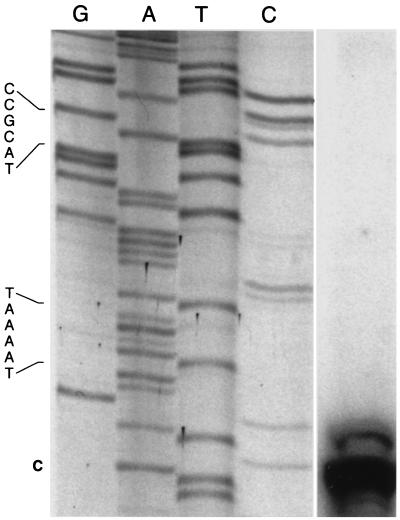
Evaluation of the mRNA start site in serotype 1 under CHP induction.
Primer extension was performed using mRNA isolated from APP225/piviK that had been induced with CHP. The major transcriptional start site was located 31 bp upstream of the ohr start codon (Fig. 7). A −10 region (TAAAAT) was identified 6 bp upstream of the transcription start site. However, no −35 region similar to that found in A. pleuropneumoniae housekeeping genes (TTRAA, where R is A or G) could be identified (8). In the region in which a −35 site would be expected to exist, a SoxS binding motif (ACCGCAT) was found (34). This proposed SoxS binding motif is an exact match for the previously published E. coli SoxS binding motif (AnnGCAY) (34). Primer extension under noninducing conditions was also performed using sixfold more RNA. A primer extension product could not be detected under these noninducing conditions (data not shown).
FIG. 7.
Primer extension analysis of APP225/piviK induced with 1 mM CHP. The sequencing reactions and the primer extension reaction were performed with an identical primer. The lane on the right contains the primer extension product. The transcriptional start site is labeled in bold (“C” at bottom). The −10 region (TAAAAT) and the potential SoxS binding motif (CCGCAT) are indicated on the left. An intervening region between the sequencing ladder and the primer extension product was removed, without alteration of alignment.
DISCUSSION
Using the IVET system that we developed to facilitate identification of A. pleuropneumoniae genes involved in the pathogenesis of disease, we identified 42 unique promoter clones that were specifically induced during infection of the natural swine host. In this study, we report the identification and characterization of one of these in vivo-induced clones, ohr, which encodes an organic hydroperoxide reductase that we hypothesize is involved in protection from oxidative stress encountered during infection.
Enzymes responsible for conferring enhanced resistance to oxidative stress encountered during infection of the respiratory tract are potentially important virulence factors for organisms that cause pneumonia, such as A. pleuropneumoniae (6, 46). The mechanisms by which A. pleuropneumoniae causes disease lead to an environment filled with oxygen radicals (21, 27). Upon infection of the porcine lung by A. pleuropneumoniae, the host immune response to bacterial cell components, such as lipopolysaccharide, triggers an influx of inflammatory cells, particularly neutrophils, which are intended to limit the bacterial infection. Within this environment, A. pleuropneumoniae produces hemolysins and cytotoxins, in the form of RTX (repeats in toxin) toxins. These pore-forming RTX toxins secreted by A. pleuropneumoniae insert themselves into eukaryotic cell membranes and cause lysis and cell death of neutrophils and macrophages, which in turn release phagocyte contents such as oxygen radicals in the form of peroxides and superoxides (7, 11, 34). To survive in this environment, A. pleuropneumoniae likely requires enzymes that allow the bacteria to evade or detoxify these oxygen radicals. A. pleuropneumoniae has previously been shown to produce several enzymes involved in response to oxidative stress, including catalase and two separate types of superoxide dismutase (19, 21). This is the first report that this pulmonary pathogen produces an additional oxidative stress protectant, an organic hydroperoxide reductase, and the first demonstration that this type of enzyme can be specifically induced in vivo during the course of infection.
Ohr enzymes have been recently described for X. campestris, P. aeruginosa, E. faecalis, and B. subtilis and have been shown to be important in the survival of these bacteria when exposed to oxidative stress in vitro, although Ohr has not been previously implicated in virulence (12, 24, 29, 33). The ohr gene from these organisms exhibits a pattern of expression similar to that of A. pleuropneumoniae ohr. In each of these organisms, ohr is induced specifically in response to organic hydroperoxides, with little or no induction in response to hydrogen peroxide and superoxide (12, 24, 29, 33). This pattern of induction is distinct from that seen with ahpC, which encodes Ahp (alkyl hydroperoxide reductase), a second class of organic hydroperoxide reductases found in many bacterial species, including E. coli, Salmonella enterica serovar Typhimurium, B. subtilis, and P. aeruginosa (1, 4, 30, 37). Ahp enzymes are induced by hydrogen peroxide and organic peroxides but not superoxides (30, 34). We identified putative Ohr sequences, based on homology to these five identified Ohr proteins, from nine additional species (Fig. 2), although not from any members of the family Enterobacteriaceae. The predicted Ohr proteins from these 14 species share two regions of strong homology that flank conserved cysteines, which may be responsible for the reduction of peroxides to the corresponding alcohol (10). Similar conserved cysteines are seen in Ahp enzymes, which are functionally similar to Ohr enzymes but not closely related at the DNA level or protein level.
When we examined the type strains of the 12 known serotypes of A. pleuropneumoniae for the presence of ohr by Southern blotting, we were able to detect an ohr homologue only in A. pleuropneumoniae serotypes 1, 9, and 11 and not in serotypes 2 to 8, 10, and 12. The intensity of the band in A. pleuropneumoniae serotype 1 was greater than that of either serotype 9 or serotype 11, suggesting that the gene in serotypes 9 and 11 was not completely homologous to that of serotype 1, from which the probe was prepared. This distribution correlates with what is known about the relatedness of A. pleuropneumoniae serotypes. Serotypes 1, 9, and 11 are closely related to one another, having essentially the same lipopolysaccharide O-antigen chain, the same complement of RTX toxins produced, and an identical genotype for one of these toxins, apxIA (17, 31). A. pleuropneumoniae serotypes 1 and 9 have also been shown previously to be closely related to one another and distinct from serotypes 2 to 8 by multilocus enzyme electrophoresis (25). The differential distribution of ohr among the serotypes may reflect an evolutionary relatedness of these serotypes.
The presence of ohr among the 12 serotypes of A. pleuropneumoniae correlates with resistance to oxidative stress reagents. Serotypes 2 to 8, 10, and 12, which do not contain an ohr gene, were equally sensitive to all types of oxidative stress agents tested, as judged by the zone of growth inhibition upon exposure to CHP, paraquat, and hydrogen peroxide (Fig. 3). In contrast, serotypes 1, 9, and 11 were significantly less sensitive to growth inhibition by CHP than were the other serotypes but were similar to the other serotypes in sensitivity to hydrogen peroxide and paraquat. A. pleuropneumoniae serotypes 1, 9, and 11 showed an increased resistance to CHP, but not to hydrogen peroxide or superoxide, that correlates with the presence of the ohr gene (Fig. 3).
The increased resistance to organic peroxides but not to other forms of oxidative stress seen for A. pleuropneumoniae serotypes 1, 9, and 11 correlates well with data on the induction of the ohr promoter by various stress reagents in A. pleuropneumoniae serotype 1. Induction of ohr was measured by luciferase assays using the ohr promoter fused to luxAB reporter genes and by assay of Ohr enzymatic activity via colorimetric detection of CHP degradation. With both of these methods, ohr expression in A. pleuropneumoniae serotype 1 was induced by CHP but not by either hydrogen peroxide or paraquat (Fig. 4 and 6).
We cloned both the intact serotype 1 ohr gene plus promoter region and an ohr::luxAB gene fusion into A. pleuropneumoniae serotype 5, which lacks ohr. During growth in broth under noninducing conditions, serotype 1 and serotype 5 showed low but equivalent expression as assayed by lux expression. However, while A. pleuropneumoniae serotype 1 is rapidly induced upon exposure to CHP, this induction is not seen with A. pleuropneumoniae serotype 5, either as increased expression of luciferase or as increased Ohr enzymatic activity (Fig. 5 and 6). We conclude that A. pleuropneumoniae serotype 5 not only does not contain a wild-type ohr gene but also is unable to respond to exposure to CHP by induction of the cloned serotype 1 ohr gene. This suggests that A. pleuropneumoniae serotype 5 may lack not only the ohr gene itself but also an additional gene(s) necessary to increase the expression of ohr in A. pleuropneumoniae serotype 1.
Multiple regulators that respond to oxidative stress have been identified for other prokaryotes, but none have as yet been identified for A. pleuropneumoniae. Three of the most well-studied regulators of oxidative stress responses in bacteria are OxyR, PerR, and SoxR (2, 34). OxyR, which has been identified for many gram-negative bacteria, is activated by exposure to peroxide, induces expression of ahpC (alkyl hydroperoxide reductase) and catalase, and also represses its own expression (30, 42). In both X. campestris and P. aeruginosa, ohr expression was not altered by lack of OxyR (24, 29). In many gram-positive organisms, ahpC and catalase are regulated by PerR, a homologue of the ferric uptake regulator Fur, which is functionally analogous to OxyR (2, 5, 34). Both of these regulators have known binding motifs that were not found in the A. pleuropneumoniae ohr promoter region (5, 42). SoxR, a transcription factor that is activated by superoxide, induces expression of a second transcription factor, SoxS, which in turn induces expression of superoxide-regulated genes such as sodA. SoxRS has also been shown elsewhere to regulate ahpC in some organisms but has not been demonstrated to be activated by peroxides (22, 32, 34). In P. aeruginosa, ohr induction was not affected by mutations in SoxR (29).
Recently, a novel transcriptional regulator, ohrR, has been described for B. subtilis and X. campestris (12, 39). In both organisms, ohrR is located immediately upstream of the ohr gene and encodes a 17-kDa peptide that is a member of the MarR family of transcriptional repressors. Expression of B. subtilis ohrA and X. campestris ohr is induced by organic peroxides and repressed by OhrR (12, 39). In B. subtilis, a 15-bp inverted repeat sequence overlapping the −10 promoter element is required for OhrR-mediated repression of the ohrA gene (39). No such repeat was found in the X. campestris ohrR-ohr intragenic region.
Analysis of 507 bp upstream of A. pleuropneumoniae ohr showed no evidence for an ohrR homologue. When we examined the promoter region of A. pleuropneumoniae ohr for potential regulatory sequences, we identified a potential SoxS box but no PerR or OxyR binding sequences or potential inverted repeats, such as that seen with the B. subtilis OhrR binding motif. SoxS in other organisms does not respond to organic peroxides, and the A. pleuropneumoniae ohr gene was not induced by superoxide generators. Our results suggest that a novel regulator or regulatory sequence is responsible for induction of ohr in A. pleuropneumoniae and that this novel regulator exists in A. pleuropneumoniae serotype 1 and not in serotype 5. Further studies are in progress to identify this regulatory molecule and to evaluate the role of Ohr in pulmonary infection caused by A. pleuropneumoniae.
Acknowledgments
This work was supported by USDA CSREES grant 98-02202 awarded to M.H.M. R.J.S. is the recipient of a Harold Wetterberg Foundation fellowship.
We thank Scott Doree for assistance with nucleotide sequencing and primer extension procedures. We thank C. Oliver Duran and Bo Norby for assistance with the animal infection experiments. We thank Gerald Shea for assistance with statistical analysis.
Editor: V. J. DiRita
REFERENCES
- 1.Antelmann, H., S. Engelmnn, R. Schmid, and M. Hecker. 1996. General and oxidative stress responses in Bacillus subtilis: cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J. Bacteriol. 178:6571-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bsat, N., A. Herbig, L. Casillas-Martinez, P. Setlow, and J. D. Helmann. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29:189-198. [DOI] [PubMed] [Google Scholar]
- 3.Buettner, G. R. 1993. The pecking order of free radicals and antioxidants: lipid peroxidation, alpha-tocopherol, and ascorbate. Arch. Biochem. Biophys. 300:535-543. [DOI] [PubMed] [Google Scholar]
- 4.Chae, H. Z., K. Robison, L. B. Poole, G. Church, and G. Storz. 1994. Cloning and sequencing of thio-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thio-specific antioxidant define a large family of antioxidant enzymes. Proc. Natl. Acad. Sci. USA 91:7017-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, L., L. Keramati, and J. D. Helmann. 1995. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc. Natl. Acad. Sci. USA 92:8190-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Mello, R. A., P. R. Langford, and J. S. Kroll. 1997. Role of bacterial Mn-cofactored superoxide dismutase in oxidative stress responses, nasopharyngeal colonization, and sustained bacteremia caused by Haemophilus influenzae type B. Infect. Immun. 65:2700-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dom, P., F. Haesebrouck, and P. DeBaetselier. 1992. Stimulation and suppression of the oxygenation activity of porcine pulmonary alveolar macrophage by Actinobacillus pleuropneumoniae and its metabolites. Am. J. Vet. Res. 53:1113-1118. [PubMed] [Google Scholar]
- 8.Doree, S. M., and M. H. Mulks. 2001. Identification of an Actinobacillus pleuropneumoniae consensus promoter structure. J. Bacteriol. 183:1983-1989. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Dringen, R., L. Kussmaul, and B. Hamprecht. 1998. Detoxification of exogenous hydrogen peroxide and organic hydroperoxides by cultured astroglial cells assessed by microplate assay. Brain Res. Protoc. 2:223-228. [DOI] [PubMed] [Google Scholar]
- 10.Ellis, H. R., and L. B. Poole. 1997. Roles for the two cysteine residues of AhpC in catalysis of peroxide reduction by alkyl hydroperoxide reductase from Salmonella typhimurium. Biochemistry 36:13349-13356. [DOI] [PubMed] [Google Scholar]
- 11.Frey, J. 1995. Virulence in Actinobacillus pleuropneumoniae and RTX toxins. Trends Microbiol. 3:257-261. [DOI] [PubMed] [Google Scholar]
- 12.Fuangthong, M., S. Atichartpongkul, S. Mongkolsuk, and J. D. Helmann. 2001. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 183:4134-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuller, T. E., R. J. Shea, B. J. Thacker, and M. H. Mulks. 1999. Identification of in vivo induced genes in Actinobacillus pleuropneumoniae. Microb. Pathog. 27:311-327. [DOI] [PubMed] [Google Scholar]
- 14.Fuller, T. E., B. J. Thacker, C. O. Duran, and M. H. Mulks. 2000. A genetically-defined riboflavin auxotroph of Actinobacillus pleuropneumoniae as a live attenuated vaccine. Vaccine 18:2867-2877. [DOI] [PubMed] [Google Scholar]
- 15.Fuller, T. E., B. J. Thacker, and M. H. Mulks. 1996. A riboflavin auxotroph of Actinobacillus pleuropneumoniae is attenuated in swine. Infect. Immun. 64:4659-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 17.Jansen, R., J. Briaire, E. M. Kamp, A. L. J. Gielkens, and M. A. Smits. 1993. Structural analysis of the Actinobacillus pleuropneumoniae-RTX-toxin I (ApxI) operon. Infect. Immun. 61:3688-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jolie, R. A., M. H. Mulks, and B. J. Thacker. 1995. Cross-protection experiments in pigs vaccinated with Actinobacillus pleuropneumoniae subtypes 1A and 1B. Vet. Microbiol. 45:383-391. [DOI] [PubMed] [Google Scholar]
- 19.Kilian, M., J. Nicolet, and E. L. Biberstein. 1978. Biochemical and serological characterization of Haemophilus pleuropneumoniae (Matthews and Pattison 1961) Shope 1964 and proposal of a neotype strain. Int. J. Syst. Bacteriol. 28:20-26. [Google Scholar]
- 20.Kuo, C. F., T. Mashino, and I. Fridovich. 1987. α,β-Dihydroxyisovalerate dehydratase. A superoxide-sensitive enzyme. J. Biol. Chem. 262:4724-4727. [PubMed] [Google Scholar]
- 21.Langford, P. R., B. M. Loynds, and J. S. Kroll. 1996. Cloning and molecular characterization of Cu,Zn superoxide dismutase from Actinobacillus pleuropneumoniae. Infect. Immun. 64:5035-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manchado, M., C. Michan, and C. Pueyo. 2000. Hydrogen peroxide activates the SoxRS regulon in vivo. J. Bacteriol. 182:6842-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, R. A., and B. E. Britigan. 1997. Role of oxidants in microbial pathophysiology. Clin. Microbiol. Rev. 10:1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mongkolsuk, S., W. Praituan, S. Loprasert, M. Fuangthong, and S. Chamnongpol. 1998. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J. Bacteriol. 180:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musser, J. M., V. J. Rapp, and R. K. Selander. 1987. Clonal diversity in Haemophilus pleuropneumoniae. Infect. Immun. 55:1207-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakai, K., and M. Kanehisa. 1991. Expert system for predicting protein localization sites in Gram-negative bacteria. Proteins 11:95-110. [DOI] [PubMed] [Google Scholar]
- 27.Nicolet, J. 1992. Actinobacillus pleuropneumoniae, p. 401-408. In A. D. Leman et al. (ed.), Diseases of swine, 7th ed. Iowa State University Press, Ames.
- 28.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ochsner, U. A., D. J. Hassett, and M. L. Vasil. 2001. Genetic and physiological characterization of ohr, encoding a protein involved in organic hydroperoxide resistance in Pseudomonas aeruginosa. J. Bacteriol. 183:773-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ochsner, U. A., M. L. Vasil, E. Alsabbagh, K. Parvatiyar, and D. J. Hassett. 2000. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 182:4533-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perry, M. B., E. Altman, J. R. Brisson, L. M. Beynon, and J. C. Richards. 1990. Structural characteristics of the antigenic capsular polysaccharides and lipopolysaccharides involved in the serological classification of Actinobacillus (Haemophilus) pleuropneumoniae strains. Serodiagn. Immunother. Infect. Dis. 4:299-308. [Google Scholar]
- 32.Pomposiello, P. J., M. H. J. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rince, A., J. Giard, V. Pichereau, S. Flahaut, and Y. Auffray. 2001. Identification and characterization of gsp65, an organic hydroperoxide resistance (ohr) gene encoding a general stress protein in Enterococcus faecalis. J. Bacteriol. 183:1482-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosner, J. L., and G. Storz. 1997. Regulation of bacterial responses to oxidative stress. Curr. Top. Cell. Regul. 35:163-177. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Silhavy, T. J. 1984. DNA extraction from bacterial cells, p. 137-139. In T. J. Silhavy et al. (ed.), Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Storz, G., F. S. Jacobson, L. A. Tartaglia, R. W. Morgan, L. A. Silveira, and B. N. Ames. 1989. An alkyl hydroperoxide reductase induced by oxidative stress in Salmonella typhimurium and Escherichia coli: genetic characterization and cloning of ahp. J. Bacteriol. 171:2049-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Storz, G., and M. B. Toledano. 1994. Regulation of bacterial gene expression in response to oxidative stress. Methods Enzymol. 236:196-207. [DOI] [PubMed] [Google Scholar]
- 39.Sukchawalit, R., S. Loprasert, S. Atichartpongkul, and S. Mongkolsuk. 2001. Complex regulation of the organic hydroperoxide resistance gene (ohr) from Xanthomonas involves OhrR, a novel peroxide-inducible negative regulator, and posttranscriptional modifications. J. Bacteriol. 183:4405-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tatusova, T. A., and T. L. Madden. 1999. Blast 2 sequences--a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. [DOI] [PubMed] [Google Scholar]
- 41.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toledano, M. B., I. Kullik, F. Trinh, P. T. Baird, T. D. Schneider, and G. Storz. 1994. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78:897-909. [DOI] [PubMed] [Google Scholar]
- 43.Ward, C. K., M. L. Lawrence, H. P. Veit, and T. J. Inzana. 1998. Cloning and mutagenesis of a serotype-specific DNA region involved in encapsulation and virulence of Actinobacillus pleuropneumoniae serotype 5a: concomitant expression of serotype 5a and 1 capsular polysaccharides in recombinant A. pleuropneumoniae serotype 1. Infect. Immun. 66:3326-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.West, S. E., M. J. Romero, L. B. Regassa, N. A. Zielinski, and R. A. Welch. 1995. Construction of Actinobacillus pleuropneumoniae-Escherichia coli shuttle vectors: expression of antibiotic resistance genes. Gene 160:81-86. [DOI] [PubMed] [Google Scholar]
- 45.Xiong, X., N. De La Cruz, and W. S. Reznikoff. 1991. Downstream deletion analysis of the lac promoter. J. Bacteriol. 173:4570-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yesilkaya, H., A. Kadioglu, N. Gingles, J. E. Alexander, T. J. Mitchell, and P. W. Andrew. 2000. Role of manganese-containing superoxide dismutase in oxidative stress and virulence of Streptococcus pneumoniae. Infect. Immun. 68:2819-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]