Abstract
The activation of signalling pathways by ligand engagement with transmembrane receptors is responsible for determining many aspects of cellular function and fate. While these outcomes are initially determined by the nature of the ligand and its receptor, it is also essential that intracellular enzymes, adaptor proteins and transcription factors are correctly assembled to convey the intended response. In recent years, it has become evident that proteins that regulate the amplitude and duration of these signalling responses are also critical in determining the function and fate of cells. Of these, the Cbl family of E3 ubiquitin ligases and adaptor proteins has emerged as key negative regulators of signals from many types of cell-surface receptors. The array of receptors and downstream signalling proteins that are regulated by Cbl proteins is diverse; however, in most cases, the receptors have a common link in that they either possess a tyrosine kinase domain or they form associations with cytoplasmic PTKs (protein tyrosine kinases). Thus Cbl proteins become involved in signalling responses at a time when PTKs are first activated and therefore provide an initial line of defence to ensure that signalling responses proceed at the desired intensity and duration.
Keywords: Cbl, E3 ligase, receptor tyrosine kinase, T-cell receptor, thymocyte, ubiquitin
Abbreviations: APS, adaptor molecule containing pleckstrin homology and Src homology 2 domains; BLNK, B-cell linker; BMMC, bone-marrow-derived mast cell; CIN85, Cbl-interacting protein of 85 kDa; CTLA-4, cytotoxic T-lymphocyte-associated protein 4; DP, double positive; EGF, epidermal growth factor; FcεRI, high-affinity IgE receptor; GLUT4, glucose transporter 4; GRAIL, gene related to anergy in lymphocytes; HECT, homologous with E6AP C-terminus; hSpry2, human Sprouty 2; IL-2, interleukin 2; KPD, Komeda diabetes-prone; LAT, linker for activation of T-cells; MAIDS, murine AIDS; NFAT, nuclear factor of activated T-cells; PDGF, platelet-derived growth factor; PI3K, phosphoinositide 3-kinase; PKC, protein kinase C; PLC, phospholipase C; PTB, phosphotyrosine-binding; PTK, protein tyrosine kinase; RIP-gp, rat insulin promoter glycoprotein; RTK, receptor tyrosine kinase; SEM-5, sex muscle abnormal 5; SH, Src homology; SLAP, Src-like adaptor protein; Sli-1, suppressor of lineage defect-1; SLP-76, SH2 domain-containing leucocyte protein of 76 kDa; TCR, T-cell receptor; TKB, tyrosine-kinase-binding; UBA, ubiquitin-associated; Ubc, ubiquitin-conjugating; WASP, Wiskott–Aldrich syndrome protein; ZAP-70, ζ-associated protein of 70 kDa
DOMAIN STRUCTURE AND FUNCTION OF CBL PROTEINS
The mammalian Cbl family of proteins consists of three homologues known as c-Cbl, Cbl-b and Cbl-3 (Figure 1). All three share highly conserved regions in their N-terminal halves which encompass their TKB (tyrosine-kinase-binding), linker and RING finger domains. In contrast, the C-terminal regions of these family members are less well conserved; however, they do share common roles with abilities to interact with a range of SH2- (Src homology 2) and SH3-domain-containing proteins. Importantly, each of the highly conserved TKB, linker and RING domains have key roles in enabling Cbl proteins to function as E3 ubiquitin ligases. The process of ubiquitylation involves multiple steps and requires at least three groups of proteins known as E1, E2 and E3 (reviewed in [1,2]). The E1 enzyme initiates the process by activating the 76-amino-acid ubiquitin protein through the formation of a thiol ester bond with its C-terminal glycine residue. An E2 or Ubc (ubiquitin-conjugating) enzyme then accepts ubiquitin from the E1, and, in the final step of the process, an E3 ubiquitin ligase catalyses the transfer of ubiquitin molecules from the E2 to the ε-amino group of a lysine residue of the substrate protein. As well as directing ubiquitin transfer, E3 ligases are also responsible for determining substrate specificity (reviewed in [1]).
Figure 1. Cbl protein family in mammals (c-Cbl, Cbl-b and Cbl-3), Drosophila (D-Cbl, long and short) and C. elegans (Sli-1).
All Cbl proteins share a high level of sequence conservation between their TKB, linker (L) and RING finger (RF) domains. c-Cbl and Cbl-b have extensive proline-rich regions (black) in their C-terminal halves that mediate interactions with numerous SH3-domain-containing proteins. The TKB domain is composed of three interacting regions comprising a four-helix bundle (4H), a calcium-binding EF hand and a variant SH2 domain that is connected to the RING finger by a short linker domain. The PR domain in the C-terminus of c-Cbl and Cbl-b refers to a PX(P/A)XXR motif that binds SH3 domains of the CIN85/RUK (regulator of ubiquitous kinase)/CD2AP (C2-associated protein) family of proteins. The UBA domain at the C-terminus of c-Cbl, Cbl-b and D-Cbl(long) refers to a region with homology to UBA domains. Conserved tyrosine residues are shown in purple.
TKB domain
The foremost function of the TKB domain is to help determine Cbl's substrate specificity, which it does by engaging specific phosphorylated tyrosine residues on proteins that are to be ubiquitylated by Cbl. However, as will be described below, this role is not carried out exclusively by the TKB domain, since some substrates, such as Src family kinases, can be targeted by domains in the C-terminal region of Cbl proteins [3].
The best-studied Cbl substrates targeted by the TKB domain are RTKs (receptor tyrosine kinases) and non-receptor PTKs (protein tyrosine kinases) of the Syk family. Interaction with the TKB domain is mediated by three distinct subdomains comprising a four-helix bundle, a calcium-binding EF hand and a variant SH2 domain, all three of which are functionally required to form a unique PTB (phosphotyrosine-binding) module [4]. The crystal structure of the TKB domain complexed with a phosphopeptide representing its binding site in the ZAP-70 (ζ-associated protein of 70 kDa) PTK shows that it binds in a manner similar to that of other SH2–phosphopeptide complexes [4]. The TKB domain is unique to Cbl proteins and is highly conserved in all species, thus a proposal to call this the Cbl homology (CH) region remains appropriate [5]. The discovery that the TKB domain determines Cbl substrate specificity by targeting activated RTKs was an important contribution in our understanding of the function of Cbl proteins. Indeed, analyses of a well-characterized loss-of-function mutation in the TKB domain [6,7] revealed that the TKB domain is indispensable for Cbl to promote the ubiquitylation of activated RTKs [8].
The TKB domain, however, does not appear to promote strong associations, and it is generally accepted that this is not the principal means by which Cbl is recruited to activated RTKs, e.g. recruitment to the EGF (epidermal growth factor) receptor requires Cbl to associate with the Grb-2 adaptor protein and proline sequences in its C-terminal region (Figure 2) [9]. Thus the crucial role of the TKB domain interaction with RTKs may primarily be to ensure the appropriate orientation of the receptor such that Cbl's E3 ligase activity can promote the transfer of ubiquitin (Figure 2e). Consistent with this, the large four-helix bundle of the TKB domain has been found to anchor the RING finger domain, and this may also help to orientate the E2 conjugating enzyme with respect to lysine residues of the substrate [10].
Figure 2. Cbl-directed internalization, multi-ubiquitylation and degradation of activated RTKs, and modulation by Sprouty.
(a) Growth factor (GF) binding induces RTK tyrosine phosphorylation and the recruitment of Cbl to the activated receptor by adaptor proteins such as Grb2, which is required to support receptor endocytosis [144]. This allows the TKB domain to engage a specific phosphotyrosine on the RTK [e.g. pTyr1045 on the EGF receptor (EGFR)]. Activation of Src kinases after GF binding induces the tyrosine phosphorylation of Cbl and other proteins, including Sprouty. The association of Sprouty with the RING finger domain initially inhibits Cbl's recruitment of Ubc enzymes (E2s), but tyrosine phosphorylation of Sprouty removes this inhibition by displacing it from the RING finger to now interact with the TKB domain (b). This allows the RING finger to recruit an E2 conjugating enzyme which promotes the polyubiquitylation of Sprouty (c) and its subsequent proteasomal degradation (d). The TKB domain is freed to target the receptor (pTyr1045 of the EGF receptor) and the E3 ligase function of Cbl can then catalyse the transfer of a ubiquitin molecule from the RING-finger-bound E2 to the RTK (e). This process has been found to be sufficient to mediate receptor internalization. Continued addition of ubiquitin moieties leads to multi-ubiquitylation which marks the RTK for intracellular trafficking to the lysosome, where the receptor is degraded. Tyrosine phosphorylation of Cbl also enhances the recruitment of a CIN85–endophilin complex through a novel proline-arginine motif (shown as PR). This protein complex helps to promote receptor internalization by causing the membrane to invaginate. Cbl tyrosine phosphorylation also recruits SH2-domain-containing proteins, such as Crk and p85 (not shown), and this recruitment can enhance signalling responses from the receptor. For simplicity, the Grb2–Cbl interaction is not shown in (e), although it still contributes to maintaining Cbl's association with the receptor. Ub, ubiquitin.
Dissociation of the TKB domain from its target, even when the adaptor-mediated association is retained, appears to be sufficient to terminate the ubiquitylation process. TKB domain interactions may therefore determine the number of ubiquitin molecules transferred to the substrate and thus regulate the extent to which activated RTKs are ubiquitylated. It will be of interest to determine if dephosphorylation of conserved tyrosine residues in the TKB domain is involved in this process, since a recent report demonstrated that glutamate point mutations of specific residues in the TKB and linker domains constitutively activate the E3 ligase activity of c-Cbl [11]. Thus the TKB domain appears to have at least two important roles in regulating E3 ligase activity, and, as such, it is functionally more complex than classical SH2 or PTB domains.
The recognition of specific sequences in proteins such as RTKs by the TKB domain has been studied extensively in recent years. A consensus recognition sequence [i.e. NXpY(S/T)XXP] was proposed based on sites within the EGF, LET-23, VEGF (vascular endothelial growth factor) and neurotrophin receptors as well as ZAP-70, Syk and Sprouty 2 [12–16]. Interestingly, a study investigating the TKB-domain-binding site in the Met receptor found no similarity to this consensus [16]. The Met receptor motif of DpYR is conserved within other Met family members Ron and Sea, as well as in Met orthologues in pufferfish.
In addition to PTKs, several adaptor and regulatory proteins have been identified as targets of the TKB domain. These include BLNK, a B-cell linker protein [17], APS (adaptor molecule containing pleckstrin homology and SH2 domains), a protein phosphorylated in PDGF (platelet-derived growth factor) and insulin-stimulated cells [18], and Sprouty2, an inducible antagonist of Cbl E3 ligase activity [14]. A novel TKB phosphotyrosine recognition motif has also recently been determined near the C-terminus of APS at Tyr618 which is highly conserved in two other members of this adaptor family, SH-B and Lnk [19]. Compared with conventional TKB sites, the APS motif extends from an arginine residue at position −6 to a serine or threonine residue at position +1, i.e. RA(V/I)XNQpY(S/T). The TKB–APS crystal structure reveals that three residues N-terminal to Tyr618 (Arg612, Ala613 and Asn616) make specific contacts with residues in the four-helix bundle of the TKB domain [19]. Site-directed mutagenesis of any of these three residues was as equally disruptive to TKB domain interaction as a Y618F mutation, providing further evidence that the four-helix bundle, as well as the variant SH2 domain, makes an important contribution to phosphotyrosine sequence recognition. As Peschard et al. [16] point out, there is no precedent for a given PTB or SH2 domain to associate with unrelated ‘consensus’ binding sites. This raises interesting questions of why the Cbl TKB domain has this unusual versatility, and whether binding strengths differ among the array of target sites.
In contrast with the activation-dependent interactions described above, the TKB domain has been found to form a constitutive association with the SLAP (Src-like adaptor protein), which is involved in the down-regulation of activated antigen receptors on T-cells and thymocytes [20,21]. Whether this interaction involves all regions of the TKB domain, and whether SLAP and Cbl proteins function synergistically to mediate antigen receptor down-regulation, are details that are yet to be determined.
An essential role for the TKB domain has also recently been described for c-Cbl binding to, and stabilization of, microtubules [22]. Importantly, mutation of the Cbl TKB-SH2 domain, which disrupts binding to phosphotyrosine, did not affect the ability of the TKB domain to bind microtubules. Thus the highly conserved TKB domain confers both PTB-dependent and -independent functions and highlights the need to investigate further the roles of the four-helix bundle and EF hand regions of the TKB domain.
Linker domain
The TKB domain also makes intramolecular contacts with the highly conserved linker α-helix that separates it from the RING domain, and these contacts are centred on conserved residues Tyr368 and Tyr371 in human c-Cbl [10]. A remarkable property of Tyr371 is that replacement with phenylalanine abolishes c-Cbl's E3 ligase activity, while replacement with glutamate constitutively promotes its activity [11,23]. The reason for these effects is not known, although structural studies predict that phosphorylation of Tyr371 would result in significant changes to the orientation of the linker helix and therefore to the contacts made with the TKB and RING finger domains [10]. Interestingly, deletion or phenylalanine substitutions of Tyr371 has the same effect as deletion or alanine substitution of cysteine residues in the RING finger domain in abolishing Cbl's E3 ligase activity; however, only the deletion of Tyr371 (and Tyr368) causes c-Cbl to become oncogenic [24,25]. Thus the loss of c-Cbl E3 ligase activity is in itself insufficient to induce oncogenicity. Molecular modelling data predict that deletion of Tyr368 or Tyr371 would disrupt the highly conserved α-helix of the linker domain [10], and it is likely that it is this structural alteration, in addition to loss of E3 activity, which is required to activate fully the oncogenic potential of Cbl proteins.
RING finger domain and protein ubiquitylation
The development of in vitro reconstituted ubiquitylation assays proved that the highly conserved Cbl RING finger has intrinsic E3 ligase activity and can independently recruit E2s for the transfer of ubiquitin to substrates (Figure 2) [23,26–28]. The structural integrity of the RING finger domain is therefore an absolute requirement for Cbl proteins to function as E3 ligases. Furthermore, the structure of c-Cbl bound to the E2 enzyme UbcH7 has been solved, identifying multiple contacts between c-Cbl's RING finger domain and UbcH7 [10]. These landmark studies resulted in the definition of Cbl proteins as RING-type E3 ubiquitin protein ligases that direct the ubiquitylation of activated RTKs.
There is now a rapidly growing appreciation of ubiquitylation as a means of regulating proteins by affecting their internalization, trafficking, enzymatic activity, protein associations and abundance (reviewed in [1]). This versatility appears to be determined by the ability of E3 ligases to promote different types of protein ubiquitylation (Figure 3; and reviewed in [29]). Indeed, until recently, it was assumed that the striking ubiquitin smear associated with activated EGF and PDGF receptors in cells overexpressing c-Cbl or Cbl-b was due to Cbl-directed polyubiquitylation of the activated RTKs. However, while polyubiquitylation normally commits a protein to proteasomal degradation, it had been known for some time that EGF receptors are targeted to lysosomes for destruction. An important breakthrough came from the Yarden and Dikic groups who showed that c-Cbl directs mono-ubiquitylation on multiple sites of the EGF and PDGF receptors [30,31] and that this marks the activated RTKs for internalization and trafficking to the lysosome where the receptors are degraded (Figure 2; and reviewed in [32,33]). This ubiquitylation process presumably holds for the many other members of the RTK family that are also down-regulated by Cbl proteins. However, whether multiple mono-ubiquitylation is the outcome for all Cbl substrates is unknown, as their fates do differ to that of RTKs. For example, there are some Cbl substrates, in particular those that are not surface receptors or PTKs, whose protein levels are not affected by ubiquitylation. These include the p85 regulatory subunit of PI3K (phosphoinositide 3-kinase), Vav, CrkL and PLCγ-1 (phospholipase Cγ-1) [34,35].
Figure 3. Different types of ubiquitylation determine alternative fates for substrates targeted by E3 ligases.
The substrates targeted by Cbl proteins can either be multi-ubiquitylated, such as the EGF receptor following activation by ligand binding, or poly-ubiquitylated, as is the case for Sprouty proteins. The multi-ubiquitylated EGFR receptor is destined for lysosomal degradation, whereas Sprouty proteins are degraded in the 26 S proteasome. The mechanisms that enable Cbl proteins to direct both types of ubiquitylation remain to be determined. Ubiquitin linkages formed through Lys48 of ubiquitin direct proteins for proteasomal degradation, whereas linkages through Lys63 and Lys29 result in differing substrate fates (reviewed in [29]).
The types of effects that ubiquitylation can have on the fate and function of proteins has recently been highlighted by an exciting proposal by Mosesson et al. [30], where the authors present evidence that the EGF receptor kinase domain is the initial target for Cbl-mediated mono-ubiquitylation (see Figure 2). This so-called initiating or founder ubiquitin not only triggers EGF receptor internalization and sorting, but also could be envisaged to suppress kinase activity. If proved, this would be an effective way to terminate RTK signalling at an early point following ligand engagement, particularly when excessive ligand may produce strong signals that could be detrimental to the organism. Indeed, impaired deactivation of RTK signalling has been strongly implicated in tumorigenesis, and it is therefore of great interest that a number of RTKs become oncogenic through mutations or deletions encompassing their Cbl TKB-binding sites which render them no longer subject to ubiquitylation and down-regulation by Cbl (reviewed in [36,37]).
The c-Cbl RING finger domain is also involved in the regulation of EGF receptor fate by its interaction with the N-terminal region of human Sprouty2 (hSpry2) [38,39]. Sprouty was originally identified in a genetic screen in Drosophila as an antagonist of FGF (fibroblast growth factor) and EGF signalling [40,41]. Interestingly, it was subsequently found that hSpry2 binding to the RING finger domain inhibits Cbl's interaction with UbcH7 [42], suggesting that Sprouty proteins can function to negatively regulate Cbl E3 ligase activity. This negative regulation is removed when hSpry2 is phosphorylated following EGF receptor activation, resulting in its displacement from the RING finger to the TKB domain where it is poly-ubiquitylated by c-Cbl and targeted for proteasomal degradation (Figure 2) [14,39,43]. Thus the opposing roles played by c-Cbl and Sprouty in determining EGF receptor fate provides an additional level of regulation that can affect the diverse outcomes in response to growth factor stimulation. It will be of much interest to see whether our knowledge of the interplay between Cbl and Sprouty can be harnessed to down-regulate constitutively active RTKs that promote tumorigenesis.
The inhibition of c-Cbl-directed degradation of the EGF receptor has also been shown to involve the Ras-related protein Cdc42 which, upon activation, sequesters c-Cbl and prevents its interaction with the receptor [44]. The association between c-Cbl and Cdc42 is indirect and requires the presence of p85Cool/b-Pix which functions as a linker between the two. The p85Cool/b-Pix interactions involve its SH3 domain binding to c-Cbl proline residues while it interacts with activated Cdc42 through its Rho-insert region. It is hypothesized that the binding of activated Cdc42 to the p85Cool/b-Pix–c-Cbl complex sterically interferes with c-Cbl's binding to the EGF receptor. When Cdc42 undergoes GTP hydrolysis and is converted into its inactive state, it dissociates from p85Cool/b-Pix, and c-Cbl can then interact with the EGF receptor and mediate its down-regulation. In future studies, it will be important to determine whether there is a crosstalk or synergy between Cdc42- and Sprouty-mediated regulation of c-Cbl function, or whether the two mechanisms function independently. Alternatively, it is feasible that the relative involvement of each mechanism may depend on the strength of the stimulatory signal or the cell type.
In addition to the two mechanisms described above, c-Cbl function can itself be negatively regulated by ubiquitylation [45], a process which is enhanced by activated Src [3,46]. Thus, in Src-transformed cells, c-Cbl undergoes tyrosine phosphorylation which promotes its ubiquitylation and proteasomal degradation. A consequence of eliminating c-Cbl is reduced lysosomal destruction of the EGF receptor and increased recycling. This finding has been proposed to explain the link between Src activation and EGF receptor overexpression in tumour cells [46]. However, it is still not known how tyrosine phosphorylation can be modulated such that it promotes c-Cbl E3 ligase activity, yet can also direct its own ubiquitylation and degradation. Interestingly, a recent study showed that Cbl-3 was markedly more effective than either c-Cbl or Cbl-b in suppressing v-Src-induced transformation [28]. It will be informative to determine whether this effect on Src transformation, which is dependent on Cbl-3 ligase activity, is due to an inability of activated Src to promote the down-regulation of Cbl-3.
C-terminal Cbl sequences involved in SH2 and SH3 domain interactions
The sequence homology among Cbl proteins is less extensive C-terminal to the RING finger; however, with the exception of a short isoform found in Drosophila, they all have proline-rich regions that are involved in numerous SH3-domain interactions (Figure 1). These contribute to at least 15 and 17 potential SH3-domain-binding sites in c-Cbl and Cbl-b respectively, while Cbl-3 encodes five potential SH3-binding sites within the 84-amino-acid sequence C-terminal to the RING finger. c-Cbl and Cbl-b share an abundance of very similar motifs, including a Grb-2 SH3 consensus binding site of PPVPPR, a polyproline sequence of PPPPPP(E/D)R, as well as an atypical PX(P/A)XXR motif that binds SH3 domains of the CIN85 (Cbl-interacting protein of 85 kDa)/RUK (regulator of ubiquitous kinase)/CD2AP (C2-associated protein) family of proteins. The latter motif promotes ligand induced internalization of the EGF and Met receptors by recruiting endophilin via CIN85 and is further evidence that c-Cbl and Cbl-b have additional means other than ubiquitylation to down-regulate activated RTKs (Figure 2) [47,48]. Another key role that has been assigned to the proline-rich region in c-Cbl is its requirement for the ubiquitylation and proteasomal degradation of activated forms of Src [3]. Thus, in this case, the Cbl substrate is targeted by proline sequence interactions, rather than the TKB domain, although a TKB recognition site has been identified in the activation loop of Src at Tyr416 and is involved in the suppression of Src kinase activity [49].
c-Cbl and Cbl-b are prominent substrates of PTKs, and both are phosphorylated following the stimulation of a diverse array of cell-surface receptors. c-Cbl possesses 22 tyrosine residues, and Tyr700, Tyr731 and Tyr774 in the C-terminal region of human c-Cbl are the best-characterized phosphorylation sites. These residues are efficiently phosphorylated by Syk and the Src-family kinases Fyn, Yes and Lyn, but not by Lck or ZAP-70 [50]. Phosphorylation of Tyr700 and Tyr774 provides docking sites for the SH2 domain of Crk [51], an adaptor protein that forms prominent associations with c-Cbl and C3G, a guanine exchange factor for the GTPase Rap-1. In addition, phosphorylation of Tyr700 mediates an interaction with the SH2 domain of Vav1, a guanine exchange factor for the Rho family GTPases Rac and Cdc42 [52]. Cbl-b also possesses tyrosine residues with flanking sequences very similar to Tyr700 and Tyr774 which are found at positions 709 and 665 respectively [53]. Thus c-Cbl and Cbl-b are substrates for the same spectrum of PTKs and can recruit identical SH2-domain-containing proteins. A notable exception to this is Tyr731 which is unique to c-Cbl and does not share flanking sequence homology with either Tyr700 or Tyr774. Rather the Y731EAM motif provides a docking site for the SH2 domains of the p85 regulatory subunit of PI3K, thus enabling c-Cbl to function as a positive regulator by recruiting PI3K to the cell membrane [54–57]. The absence of this motif in Cbl-b highlights a potentially important divergence in the role and mode of action of these two highly similar regulatory proteins; however, this aspect has not been examined in depth with respect to growth factor or immune receptor signalling. That the adaptor binding properties of Cbl link them to proteins which provide a range of positive signalling functions is an interesting conundrum for a protein family that primarily functions to terminate activated signalling responses.
UBA (ubiquitin-associated) domain
c-Cbl and Cbl-b, but not Cbl-3 or Caenorhabditis elegans Cbl (known as Sli-1 for suppressor of lineage defect-1), have UBA domains at their C-terminal ends (Figure 1). UBA domains were originally identified as conserved motifs present in a range of proteins involved in ubiquitylation processes [58]. UBA domains have since been shown to interact with ubiquitin and ubiquitin-like domains of proteins such as Nedd8 (neuronal precursor of cell-expressed developmentally down-regulated gene 8). In addition, UBA domains can interact with each other, and this enables c-Cbl and Cbl-b to form homo- and hetero-dimers. Interestingly, the UBA domain of the DNA repair protein, Rad23, appears to inhibit polyubiquitin chain extension on substrate proteins [59] and, more recently, was shown to confer a stabilization signal that protects Rad23 from proteasomal degradation [60]. At present, the functional importance of the Cbl UBA domains are not known, since they are not required for the E3 ligase activity of either c-Cbl or Cbl-b [61]. Indeed the fact that Sli-1 does not possess a UBA domain supports this finding and provides an additional example of the evolution of Cbl proteins to undertake expanded functions in higher organisms. Notably, a recent study from the Lipkowitz laboratory showed that only the UBA domain from Cbl-b, and not c-Cbl, can interact with ubiquitin [61]. This is an intriguing finding, since it identifies an additional distinction between these two otherwise very similar proteins.
CBL PROTEIN FAMILY IN LOWER ORGANISMS
Cbl orthologues are present in C. elegans and Drosophila (Figure 1), and these genes and their products have been well characterized in numerous studies (reviewed in [62]). Indeed studies from the Sternberg laboratory first identified the C. elegans Cbl orthologue (Sli-1) as a negative regulator of RTK signalling [6,63], and it was this finding which directed mammalian studies to investigate the effects of Cbl proteins on regulating PTKs. The Sli-1 locus was originally defined by mutations that suppress partially defective forms of the LET-23 RTK, a C. elegans orthologue of the EGF receptor subfamily. Further analysis revealed that loss of Sli-1 function also suppresses a severe reduction-of-function allele of SEM-5 (sex muscle abnormal protein 5), an SH3–SH2–SH3 orthologue of mammalian Grb-2. In contrast, loss of Sli-1 function did not rescue loss-of-function LET-60 Ras mutations, suggesting that Sli-1/Cbl proteins function upstream of Ras. Thus the study of Sli-1 placed its activity at the level of the LET-23 receptor and its adaptor SEM-5 [6], and this conclusion has subsequently been confirmed in many studies of mammalian cells.
Cbl genes have also been identified in many other organisms, including sea squirts, mosquitoes, amphibians, fish and birds, but there is no evidence for Cbl genes in single-cell organisms or plants [64]. Thus it is likely that Cbl arose with the evolution of PTKs, and it is significant that neither is found in yeast. Amphibians and fish encode c-Cbl and Cbl-b homologues that are very similar to their mammalian counterparts, but they do not possess a Cbl-3 gene. Indeed, the amino acid sequences and intron/exon structure encompassing the TKB domain, linker and RING finger are almost identical between c-Cbl and Cbl-b from fish, mouse and humans, suggesting that they evolved by duplication from a common ancestor [64]. Thus, as multicellular organisms become more complex, Cbl proteins also evolved to expand their functional capabilities by increasing the spectrum of proteins with which they could associate. This added level of complexity of Cbl protein function has been investigated comprehensively in studies of the mammalian immune system (reviewed in [34,65–67]).
c-CBL AND CBL-B IN IMMUNITY: LESSONS FROM GENE KNOCKOUTS
The initial characterization of c-Cbl showed that it was ubiquitous, with the highest levels of expression detected in the thymus and testes [68]. Since the identification of c-Cbl as a prominent tyrosine kinase substrate in TCR-stimulated Jurkat T-cells [69,70], the role of Cbl proteins in T-cell signalling and function has been studied extensively. The importance of c-Cbl and Cbl-b in immunity and immune receptor signalling pathways has been demonstrated clearly by the phenotypes of the respective gene knockout mice. Consistent with a role for Cbl proteins as negative regulators, both c-Cbl−/− and Cbl-b−/− mice display hyperactive signalling downstream of the TCR (T-cell receptor). A characteristic feature of these mice is that loss of either Cbl protein lowers the activation threshold for signalling through the TCR. This not only results in hypersensitivity to low affinity/avidity engagement of the TCR, but also leads to activation of downstream signalling pathways without the normal requirement for co-receptor stimulation [71–75]. Interestingly, however, the predominant c-Cbl−/− and Cbl-b−/− phenotypes appear to be restricted to thymocytes and T-cells respectively [71–74]. This may reflect in part their relative tissue distribution, as c-Cbl is more highly expressed in the thymus compared with Cbl-b, while the reverse is true in peripheral T-cells [76]. In contrast, Cbl-3 is expressed primarily in epithelial cells of the gastrointestinal system, and loss of this protein does not promote an obvious immune phenotype [77].
Regulation of TCR signalling in Cbl−/− mice
In c-Cbl−/− mice, enhanced signalling is observed in CD4+CD8+ DP (double positive) thymocytes which show increased protein tyrosine phosphorylation that is in part a consequence of markedly elevated levels of the CD3, TCR and CD4 receptors, and of the Src family kinases, Lck and Fyn (Figures 4 and 5a) [71,72,78,79]. This is consistent with c-Cbl's role as an E3 ligase, and, indeed, similar increases are seen in a knockin mutant mouse carrying an inactivating mutation (C379A) in the c-Cbl RING finger domain (C. B. F. Thien and W. Y. Langdon, unpublished work), demonstrating that RING finger function is required for c-Cbl's ability to regulate levels of these proteins in vivo. The mechanism responsible for the high levels of CD3 and TCR on DP thymocytes from c-Cbl−/− and C379A RING finger mutant mice has not been determined fully; however, there does not appear to be a block in ligand-induced internalization (W. Y. Langdon and C. B. F. Thien, unpublished work). Whether enhanced receptor recycling in the DP thymocytes from c-Cbl mutant mice is responsible for the high levels of surface TCR and CD3 has yet to be investigated. Furthermore, loss of c-Cbl's E3 ligase activity towards the ζ-chain of the TCR could also have an effect on levels of the TCR complex, since transgenic overexpression of TCRζ increases total TCR surface expression [80,81].
Figure 4. c-Cbl preferentially regulates TCR and Lck levels in CD4/CD8 DP thymocytes and negatively regulates tyrosine kinase signalling following anti-CD3/4 cross-linking.
(a) CD4+CD8+ DP thymocytes and T-cells from lymph nodes of wild-type (+/+), c-Cbl−/− (c−/−) and Cbl-b−/− (b−/−) mice were analysed by flow cytometry for surface expression of TCRβ and intracellular levels of Lck. The results show that, in thymocytes, the negative regulation of surface TCR levels and intracellular Lck levels is carried out by c-Cbl and not by Cbl-b, and that this regulation by c-Cbl is not evident in peripheral T-cells. (b) Thymocytes from the above mice were left unstimulated or stimulated for 5 min at 37 °C with anti-CD3 and anti-CD4 antibodies, and total cell lysates were immunoblotted with anti-phosphotyrosine antibodies (4G10 monoclonal). The results show that c-Cbl, but not Cbl-b, has a marked effect on negatively regulating the activity of ZAP-70 and the tyrosine phosphorylation of its substrates SLP-76 and LAT. Molecular-mass sizes are shown in kDa.
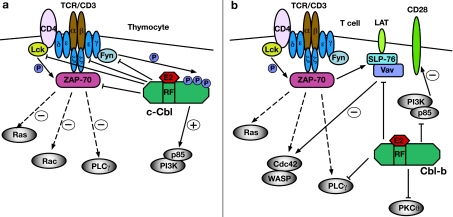
Figure 5. Distinct mechanisms of negative regulation of TCR signalling by c-Cbl in thymocytes (a) and Cbl-b in peripheral T-cells (b).
(a) In CD4/CD8 DP thymocytes, c-Cbl negatively regulates the surface expression levels of the TCRα/β and CD3ε/γ/δ receptors, as well as the intracellular levels of CD3ζ and the Src family kinases Lck and Fyn. c-Cbl also negatively regulates the activity of ZAP-70; however, the mechanism of this regulation has not been determined, as it does not involve the TKB or RING finger domains. c-Cbl also plays a positive role by enhancing PI3K activity through the recruitment of the p85 regulatory subunit to the cell membrane. (b) In peripheral T-cells, Cbl-b negatively regulates a distinct set of signalling proteins to those targeted by c-Cbl in thymocytes. Cbl-b ubiquitylates Vav, which suppresses its tyrosine phosphorylation and activity as a GDP/GTP-exchange factor for Cdc42. This inhibits the interaction of Cdc42 with WASP, thus reducing the extent of actin reorganization and TCR clustering. Cbl-b also ubiquitylates p85, which inhibits its recruitment to CD28 and TCRζ, therefore suppressing the activation of PI3K. PLCγ1 and PKCθ are ubiquitylated by Cbl-b, and their reduced activity suppresses calcium mobilization and the activation of transcription factors that lead to IL-2 production. It is important to note that none of the above substrates show evidence of reduced protein levels following Cbl-b directed ubiquitylation. (a) and (b) Broken arrows represent pathways downstream of ZAP-70 leading to Ras, Rac, PLCγ or Cdc42 activation, but where intermediate effectors have not been shown. Plus or minus signs indicate the resultant activating or inhibitory effect on various pathways as a consequence of Cbl protein activity towards ZAP-70, Vav or PI3K. RF, RING finger.
In contrast with the c-Cbl−/− mouse, the Cbl-b−/− mouse does not exhibit a thymic phenotype, and there are no increases in levels of cell-surface receptors or intracellular PTKs (Figure 4a). As a consequence, the PTK signal is not enhanced following CD3 and CD4 engagement (Figure 4b) [73,74]. However, peripheral T-cells from Cbl-b−/− mice exhibit a hyperproliferative phenotype that is uncoupled from a requirement for CD28 co-stimulation. Thus, in Cbl-b−/− T-cells, CD3 stimulation alone is sufficient to induce receptor clustering, raft aggregation, T-cell proliferation and IL-2 (interleukin-2) production at levels comparable with that seen by CD3+CD28 co-stimulation of wild-type cells [73,74]. Phosphorylation of proteins such as Vav1 is also sustained, and deregulation of the Vav/WASP (Wiskott–Aldrich syndrome protein) pathway in the absence of Cbl-b leads to enhanced formation of the immunological synapse and increased signalling. However, Cbl-b seems to selectively regulate the functions of Vav1, as loss of Cbl-b in the Vav1−/− mouse does not compensate for all CD28-Vav-mediated functions, such as IL-2 production [75,82].
Role of Cbl-b in T-cell anergy and autoimmunity
The tight regulation of immune responses and the prevention of autoimmunity are dependent on a number of checkpoint mechanisms that are employed by the organism. These include the deletion of autoreactive T-cells in the thymus (negative selection), control of peripheral T-cell activation by regulatory T-cells and the induction of anergy [83]. Autoreactive T-cells which escape deletion in the thymus undergo clonal expansion upon interaction with self-antigens in the periphery, but this ‘tolerizing’ signal normally renders them inactive or unresponsive to further stimulation, thereby preventing autoimmunity. Anergy is the mechanism of immunotolerance where the T-cell is maintained in a hypo- or un-responsive state following initial antigen challenge. Defects in any of these regulatory processes can have devastating consequences for the organism as the failure to properly induce tolerance can lead to the development or enhanced susceptibility to autoimmune diseases.
Cbl-b regulates autoimmunity
Studies of Cbl-b mutant mice and rats have clearly demonstrated a critical role for Cbl-b in regulating the development of autoimmune diseases. Cbl-b−/− mice are highly susceptible to the development of experimentally and spontaneously induced autoimmune diseases such as arthritis and diabetes [35,73–76,84]. An example of this is a recent study which showed that all Cbl-b−/− mice, but no wild-type mice, succumbed to late-onset arthritis in a non-adjuvant immunization protocol, indicating greatly exacerbated autoimmune responses in the absence of Cbl-b [35]. Indeed, repeated antigen challenge results in death due to the massive induction of cytokines causing multiple organ failure. In another study, Gronski et al. [84] showed that ablation of Cbl-b increased the incidence of diabetes in the RIP-gp (rat insulin promoter glycoprotein) transgenic mouse model to 100%, compared with only 50% of wild-type mice, when the mice were challenged with a low-affinity ligand. This study was an elegant demonstration of how Cbl-b negatively regulates the progression to diabetes by governing the intensity of TCR responsiveness to different affinity ligands.
Cbl-b has also been identified as a major susceptibility gene for insulin-dependent (Type I) diabetes in the KPD (Komeda diabetes-prone) rat [85], and a single nucleotide polymorphism in exon 12 was shown recently to be significantly associated with Type I diabetes [86]. The same study also noted genetic interaction between Cbl-b and CTLA-4 (cytotoxic T-lymphocyte-associated protein 4), a negative regulator of CD28 signalling, in Type I diabetes susceptibility. Interestingly Cbl-b's involvement in the development of Type I diabetes does not involve a perturbation to the translocation of the GLUT4 glucose transporters [87,88], as would be predicted from studies demonstrating a positive role for Cbl in GLUT4 translocation [89]. Therefore the likely cause of diabetes in the KPD rat and RIP-gp mouse models is through the development of autoimmunity. Indeed, both models show lymphocyte infiltration into pancreatic islets and other tissues such as the thyroid gland [84,85]. Interestingly, the Cbl-b mutation found in the KPD rat results in the expression of a C-terminally truncated protein that only retains the TKB, linker and RING finger domains. This finding is a clear demonstration that sequences downstream of the RING finger are of great importance for Cbl's regulation of immune receptor signalling.
Cbl-b and anergy
Effective T-cell activation, and subsequent IL-2 production, normally requires engagement of both the TCR and the co-stimulatory molecule CD28 on the naïve T-cell with the antigen-presenting cell. In contrast, stimulation of the TCR alone promotes an anergic response and induces tolerance. Loss of Cbl-b was found to remove the dependence on CD28 for T-cell activation [74]. This, and the increased susceptibility of Cbl-b mutant mice to autoimmunity, naturally led to studies investigating the role of Cbl-b in the development and maintenance of T-cell anergy. Cbl-b−/− mice demonstrate marked resistance to anergy induction or tolerizing signals mediated through engagement of the TCR alone, and their peripheral T-cells remain highly responsive to CD3+CD28 restimulation after anergizing treatment [35].
Interestingly, Wohlfert et al. [90] demonstrated that it is the effector T-cells which are functionally altered in Cbl-b−/− mice, whereas the regulatory T-cells are not affected. This was an important finding, as the ability of CD4+CD25+ regulatory T-cells to suppress the activation of effector cells plays a role in the prevention of autoimmunity in vivo [91–94]. Intriguingly, exposure to stronger activating signals, such as would exist in the Cbl-b−/− mouse, correlates with decreased suppressive activity by regulatory T-cells, as well as reduced suppressibility of effector T-cells [95]. Consistent with this, the activation and proliferation of Cbl-b−/− CD4+CD25− effector T-cells were found to be resistant to suppression by CD4+CD25+ regulatory T-cells and their downstream mediator TGF-β (transforming growth factor β) [90]. Importantly, however, this abnormality was specific for effector cells, as Cbl-b−/− regulatory T-cells did not differ from wildtype cells in functional in vitro assays.
While the studies described above provide some links as to how hyperactive T-cell signalling could lead to impaired peripheral tolerance, the biochemical mechanisms underlying this and the development of anergy and autoimmunity in Cbl-b−/− mice are not fully understood and still require further examination. However, it is thought that E3 ubiquitin ligases such as Cbl-b, GRAIL (gene related to anergy in lymphocytes) and Itch/Nedd4 affect T-cell activation thresholds by regulating the ubiquitylation and degradation of downstream effectors of T-cell tolerance/anergy (reviewed in [34,67]). In particular, targets such as the transcription factors NF-κB (nuclear factor κB) and NFAT (nuclear factor of activated T-cells) are known to control the expression of genes that are responsible for maintaining the anergic state, while ubiquitylation of PKC (protein kinase C) θ and PLC-γ1 by Cbl-b and Itch is thought to contribute to the early disintegration of the immunological synapse, thereby preventing further signalling [35,96]. In addition, the ubiquitylation of PLC-γ1 by Cbl-b has a profound effect on suppressing the tyrosine phosphorylation of PLC-γ, which in turn leads to a marked reduction in calcium mobilization [35]. Thus Cbl-b E3 ligase activity sets a threshold for TCR-induced calcium responses that are central for determining the anergic or responsive state of T-cells. Interestingly, the HECT (homologous with E6AP C-terminus)-type E3 ligases, Itch and Nedd4, can also target Cbl-b for ubiquitylation and degradation [45], suggesting that maintenance of Cbl-b levels is finely balanced in the regulation of anergy. Indeed, this was shown recently to be the case where Cbl-b levels were found to be up-regulated in T-cells after tolerizing conditions [35].
Role of Rap and Cbl proteins in T-cell signalling
A role for Cbl proteins in immunotolerance had previously been postulated when it was found that c-Cbl is hyperphosphorylated in anergic T-cells [97]. Both c-Cbl and Cbl-b possess tyrosine phosphorylation sites which can recruit Crk proteins [51,98], and it was suggested that increased Fyn activation and c-Cbl–CrkL formation contributes to the anergic state as anergized T-cells show increased Rap1 activation [97]. However, contrary to this study, which suggested a positive role for c-Cbl in regulating Rap1 activation, recent work from the Liu group examining c-Cbl- and Cbl-b-deficient mice suggest that Cbl proteins act to negatively regulate CrkL-mediated Rap1 activation [99,100]. They suggest that the absence of CrkL ubiquitylation by c-Cbl promotes increased membrane translocation of C3G, through its enhanced association with CrkL, and that this results in elevated guanine-nucleotide-exchange activity of C3G for Rap1. Consistent with the c-Cbl−/− thymocyte phenotype, expression of a T-cell-specific activated form of Rap1 has been shown to promote positive selection [101]. Similarly, Cbl-b also exerts the same effect on CrkL/C3G association and Rap1 activation in peripheral T-cells [100]. In both cases, CrkL localization and protein association, rather than its stability, are affected. Importantly, the effect was specific for C3G, as Cbl-b ubiquitylation of CrkL did not alter its association with ZAP-70 [100]. Whether these findings also reflect the compartmentalized functioning of c-Cbl and Cbl-b is not known, as corresponding analyses were not performed with c-Cbl−/− T-cells and Cbl-b−/− thymocytes. The reason for the discrepancy with earlier studies that showed a positive regulatory role for Cbl was not addressed, although it has been suggested that changes in Rap1 localization could be involved [67]. It is not clear then how to reconcile the role of Rap1 in T-cell anergy, nonetheless these studies demonstrate that Cbl proteins are prominent negative regulators of Rap1 and that this is important for integrin-mediated adhesion in thymocytes and T-cells [99,100]. Indeed, enhanced integrin affinity could contribute to stabilizing the formation of the immunological synapse and promote sustained TCR signalling in Cbl-b−/− T-cells [102].
Enhanced TCR signalling in thymocytes from c-Cbl mutant mice
While Cbl-b appears to be critical for peripheral T-cell responses, it is c-Cbl which modulates signalling in thymocytes. c-Cbl−/− thymocytes exhibit greatly increased protein tyrosine phosphorylation and signalling downstream of the TCR, and the requirement for CD4 co-stimulation is greatly reduced [71,78]. This is exemplified by the PTKs Lck and ZAP-70, which can be activated fully by CD3 stimulation alone in the c-Cbl−/−, whereas wild-type thymocytes require the engagement of both CD3 and CD4 [71,78]. The down-regulation of ZAP-70 was also affected, resulting in prolonged activity as measured by phosphorylation of substrates LAT (linker for activation of T-cells) and SLP-76 (SH2-domain-containing leucocyte protein of 76 kDa) [71,78]. In vitro studies identifying the negative regulatory site Tyr292 of ZAP-70 as a binding site for the TKB domain of c-Cbl [4,12,103,104] suggested that this could be the mechanism underlying ZAP-70 hyperactivation in c-Cbl−/− thymocytes. However, a c-Cbl(G304E) knockin mouse carrying a loss-of-function mutation in its TKB domain did not show similar deregulation of ZAP-70 activity, neither were levels of CD3, TCR, Fyn and Lck affected [79]. The TKB domain therefore is not involved in the negative regulation of these proteins in vivo. Indeed, the finding that many of the phenotypic changes seen in the c-Cbl−/− mouse are recapitulated in the C379A, but not the G304E, knockin mouse indicates that the RING finger, rather than the TKB, domain is central to many of c-Cbl's functions in thymocytes ([79]; and C. B. F. Thien and W. Y. Langdon, unpublished work).
Unlike the Src PTKs, ZAP-70 protein levels are not altered in the c-Cbl−/− mouse [78]. The finding by Mosesson et al. [30] that the initial ubiquitylation mediated by c-Cbl targeted residues within the EGF receptor kinase domain raised the intriguing possibility that this modification could regulate protein kinase activity directly. However, while ZAP-70 can mediate c-Cbl's association with, and ubiquitylation of, the TCRζ chain in vitro [80], to date, there is little evidence that ZAP-70 is a substrate for c-Cbl-mediated ubiquitylation and/or degradation. Indeed, analysis of the C379A knockin mouse shows that loss of the c-Cbl RING function also does not appear to regulate ZAP-70 activity directly (W. Y. Langdon and C. B. F. Thien, unpublished work). It remains to be determined whether other c-Cbl domains, such as proline sequences that interact with the ZAP-70 regulator Sts-2 [105–107], are involved. Thus the mode of c-Cbl's effect on ZAP-70 in vivo is yet to be resolved; however, the data from the c-Cbl−/− mouse does indicate that c-Cbl is able to regulate ZAP-70 activity by mechanisms other than by affecting protein levels.
Thymocyte development and selection
The strength and kinetics of responses downstream of the TCR are key factors determining whether thymocytes survive, or are actively deleted, by positive and negative selection respectively [108–110]. The amplitude and duration of these signalling responses are determined initially by the affinity of the TCR for antigen–MHC complexes and the total number of receptor interactions. It is therefore surprising, in light of the markedly enhanced TCR signal leading to the activation of ZAP-70, that c-Cbl−/− thymi develop with apparent normality, as judged by criteria such as size and developmental progression. However, experiments with MHC II-restricted TCR transgenic mice showed that positive selection of CD4+ thymocytes is enhanced in the absence of c-Cbl, i.e. a TCR-directed signal that would normally be too weak for positive selection is amplified without c-Cbl-mediated attenuation [72]. A similar effect was also observed in SLAP−/− mice which show increased surface TCR and CD3 expression, and up-regulation of CD5 and CD69 activation markers [21]. Thus the c-Cbl knockout suggests that c-Cbl acts at a later stage of thymocyte maturation by modulating thresholds of TCR signalling leading to MHC II-restricted positive selection [72]. However, c-Cbl deficiency can also partly rescue thymic development in SLP-76−/− or LAT−/− mice, although this process is still TCR-dependent, as no rescue is observed on a RAG-2−/− (recombination activating gene 2) background [111]. This provides evidence for a SLP-76- and LAT-independent pathway for T-cell development that is normally negatively regulated by c-Cbl. A role for c-Cbl in pre-TCR signalling has also been suggested by the finding that c-Cbl is required for pre-TCRα allelic exclusion [81]. Thus c-Cbl can regulate different stages of T-cell development, maturation and selection processes.
Localization of Cbl proteins
It is intriguing that a number of reports suggest that the extent to which immunological activation clusters are formed and sustained may determine or reflect positive and negative selection of thymocytes [112,113]. Interestingly, a recent study showed by multicolour confocal microscopy that Cbl-b co-localizes with CD3 and a number of tyrosine-phosphorylated and ubiquitylated proteins in immunological synapses formed after TCR triggering [114]. This suggests that recruitment of Cbl-b's E3 ligase activity may be critical to early signalling processes from the TCR.
c-Cbl has also been shown to localize to immunological synapses in T-cells [115]. Within these immunological synapses, however, c-Cbl appears to reside in the non-lipid raft/detergent-soluble fractions [116,117]. Lipid rafts are cholesterol- and glycosphingolipid-enriched membrane microdomains that are important in regulating the initiation and duration of TCR signalling through their ability to spatially organize and concentrate components of the immune receptor signalling complex, including immunoreceptors, palmitoylated Src family kinases and transmembrane adaptor proteins such as LAT, while excluding negative regulators such as CD45 (reviewed in [118]). In contrast with a study by Hawash et al. [116], a study by Xavier et al. [119] reported finding c-Cbl in rafts in T-cells; however, the reason for this discrepancy is not known, as similar fractionation methods were used by the two groups. Importantly, by analysis of immortalized c-Cbl−/− T-cells and Jurkat T-cells overexpressing c-Cbl, Hawash et al. [116] demonstrate that c-Cbl can sequester Lck from rafts, which would represent an additional mechanism of negative regulation. Similarly, it has been suggested that Cbl-b's negative regulation of p85 recruitment to CD28 may involve Cbl-b binding to, and thus retention of, p85 away from lipid rafts [120]. Differences in raft composition have been noted between immature and mature T- and B-cells, naïve and effector T-cells, and even between Th1 and Th2 T-cell subsets, where the same molecule can show different raft residence properties (reviewed in [118]). Thus it is possible that, while c-Cbl is recruited to lipid rafts after IgE triggering of FcεRIs (high-affinity IgE receptors) in mast cells [121] and in insulin-stimulated adipocytes [122], it may be completely or partially excluded in T-cells/thymocytes. However, further examination of Cbl protein localization is needed, particularly in thymocytes, as the ability to aggregate lipid rafts and cluster signalling proteins at the site of TCR activation appears to differ between thymocytes and T-cells [123]. Indeed, lipid rafts form inefficiently in thymocytes and may not play as important a role in TCR signalling as in mature cells.
c-Cbl and Cbl-b: differences in function and regulation
The molecular mechanisms by which c-Cbl and Cbl-b control thymocyte and T-cell activation and signalling are yet to be fully elucidated. Nevertheless, from these analyses, it is clear that both c-Cbl and Cbl-b primarily function to achieve the same goal by regulating the activation thresholds of signalling pathways downstream of immune receptors. In so doing, the action of Cbl proteins can be envisaged as a ‘fine-tuning’ mechanism or ‘rheostat’ that governs the strength of the ensuing response and the development of a functional T-cell repertoire. Indeed, the fact that c-Cbl and Cbl-b deficient mice are viable and fertile but that the loss of both is embryonic lethal before E10.5 (embryonic day 10.5), suggests that they have important overlapping functions [76], as might be expected from two such homologous proteins. Analysis of Cbl-b−/− mice in which c-Cbl has also been deleted in a T-cell-specific fashion demonstrate that both proteins contribute to the regulation of receptor levels and PTK-dependent signalling in thymocytes and peripheral T-cells [76]. The T-cell-restricted double Cbl knockout also exhibited other phenotypic characteristics of both the individual knockouts, including splenomegaly, enhanced extramedullary haemopoiesis and susceptibility to autoimmunity. This raises the question: is the reason then for the differing T-cell phenotype of c-Cbl−/− compared with Cbl-b−/− mice simply a reflection of their primary tissue distribution, or are there real functional differences between the two proteins? In addition, are these phenotypes due primarily to loss of E3 ubiquitin ligase activity or do they reflect other functional responsibilities of Cbl proteins?
In vitro, c-Cbl and Cbl-b appear to have equal capacity to act as E3 ubiquitin ligases towards a similar range of substrates. However, while the E3 ligase activity is critical for many Cbl functions, this appears to have differing consequences for c-Cbl compared with Cbl-b substrates. Indeed, unlike c-Cbl, ubiquitylation by Cbl-b does not often result in degradation of the substrate protein. Loss of Cbl-b does not alter expression levels of signalling molecules such as Vav or p85, known ubiquitylation substrates of Cbl-b (Figure 5b). Other signalling molecules or cell-surface receptors also show no change in protein levels in the absence of Cbl-b (Figure 4a) [75]. Rather, Cbl-b mediated ubiquitylation appears to affect protein localization and associations. For example, ubiquitylation of p85 by Cbl-b inhibits its association with CD28, an effect that is dependent on both the RING finger and p85 SH3 domains [124]. Thus Cbl-b's negative regulation of Vav phosphorylation and T-cell activation is thought to be mediated by its ability to alter recruitment of p85 to both the TCR (via TCRζ) and CD28 in a RING-finger-dependent manner. These studies were the first to reveal a proteolysis-independent function of the Cbl-b E3 ubiquitin ligase.
Liu and Gu [125] suggest that different T-cell phenotypes of c-Cbl and Cbl-b mice is in part due to differential targeting of substrates for ubiquitylation in vivo. This is a difficult hypothesis to test in primary mouse tissues, and it is still likely that the phenotypic differences between the two knockouts are partly due to variable expression levels in the thymus and peripheral T-cells. Indeed, by using quantitative reverse transcription–PCR, we have estimated an 8-fold higher expression level of c-Cbl mRNA in the thymus compared with Cbl-b (C. B. F. Thien and W. Y. Langdon, unpublished work). However, as noted already, the UBA domains of c-Cbl and Cbl-b do show different capabilities for dimerizing with other protein partners [61], so it is tempting to speculate that alternative modes of protein interaction could underlie some of the phenotypic differences between the two knockout mice. For example, c-Cbl's association with p85 is inducible and is largely dependent on the phosphorylation of Tyr731 (Tyr737 in mouse c-Cbl) and its recognition by the SH2 domains of p85 [54,55,57]. This tyrosine motif is absent in Cbl-b, and it is the C-terminal proline sequences that mediate its constitutive interaction with, and subsequent ubiquitylation of, the p85 subunit of PI3K [126]. The role of tyrosine phosphorylation in differential c-Cbl and Cbl-b function is an aspect that has not been studied extensively to date. However, this is particularly relevant as c-Cbl is such a prominent phosphotyrosine substrate, while Cbl-b shows negligible tyrosine phosphorylation upon TCR triggering (Figure 4b) [71,73]. Furthermore, tyrosine phosphorylation can modulate the E3 ligase activity of c-Cbl [11,23], so it will be interesting to determine whether Cbl-b E3 activity is similarly regulated. The lymphocyte-specific protein tyrosine phosphatase, Lyp, has been shown to constitutively associate with c-Cbl in thymocytes and T-cells, and its overexpression in Cos-7 cells reduces c-Cbl tyrosine phosphorylation [127]. However, additional studies are needed to investigate the role of Lyp in regulating c-Cbl tyrosine phosphorylation in vivo.
A number of studies have also revealed opposing effects of these highly similar proteins in other haemopoietic cells. For example, c-Cbl and Cbl-b have different roles in regulating mast cell responses, although their relative expression levels have not been determined. Loss of Cbl-b results in greatly increased FcεRI-induced protein tyrosine phosphorylation, particularly of Syk and PLCγ2, leading to increased Ca2+ flux, and histamine release in BMMCs (bone-marrow-derived mast cells) [128], and these effects appear to be dependent on the Cbl-b RING finger [129]. In contrast, c-Cbl deficiency does not greatly affect mast cell responses [128], despite in vitro studies showing that it can also be recruited and co-localized with FcεRIs in lipid rafts, and that it can target Syk for ubiquitylation [121,130,131].
Likewise, in B-cells, c-Cbl and Cbl-b have been found to show different functions, although, as with BMMCs, their relative expression levels have not been determined. c-Cbl inhibits PLCγ2 activation by blocking BLNK–PLCγ2 interaction [17], whereas loss of Cbl-b has no effect on this interaction. Rather Cbl-b appears to promote Btk–BLNK association and positively regulate Btk-mediated activation of PLCγ2 [132]. Thus there is not only the dichotomy of Cbl function in T- and B-cells, but also the question of how two such similar proteins can have opposing effects in the same cell type. A similar phenomenon was observed in Jurkat T-cells, where c-Cbl overexpression was found to suppress, while Cbl-b enhanced, TCR-induced activation of NFAT [133]. Thus different phenotypic effects exerted by c-Cbl and Cbl-b do not always reflect relative expression levels; however, the subtleties of these functional differences are yet to be fully understood.
c-Cbl and Cbl-b also appear to differ in the way that their protein levels are regulated. A number of studies suggest that Cbl-b activity, particularly with respect to anergy, is regulated at the level of its protein expression. For example, CD28 and CTLA-4 have opposing roles in modulating T-cell activation thresholds, and stimulation of these receptors leads to degradation or increased expression of Cbl-b expression respectively [134]. Similarly, induction of anergy mediated through calcineurin increases mRNA and protein levels of Cbl-b, as well as other E3 ubiquitin ligases, Itch and GRAIL, that are involved in anergic responses [35,96]. Consistent with a functional consequence of changes in Cbl-b protein levels, T-cells from Cbl-b−/− and Itch−/− mice are resistant to anergy induction, and CTLA-4−/− mice also show reduced expression of Cbl-b, which can be restored by inhibition of CD28 activation [134]. Notably, the ability of Cbl-b to negatively regulate PLCγ1 phosphorylation is only observed in activated/tolerized T-cells where Cbl-b expression is normally induced [35]. Thus Cbl-b expression levels may reset the activation threshold for antigen receptor stimulation in tolerized cells. Interestingly, the HECT E3 ligases, Itch and Nedd4, can also target Cbl-b for ubiquitylation and degradation [45], suggesting that maintenance of Cbl-b levels is finely balanced in the regulation of anergy.
Cbl-b has also been reported to undergo self-ubiquitylation and degradation upon TCR and CD28 engagement [134,135]. Thus T-cell activation appears to require the inactivation of Cbl-b, consistent with its role as a negative regulator and modulator of activation threshold. Interestingly, however, prolonged T-cell activation by CD3 or CD3+CD28 stimulation increases Cbl-b expression [100], perhaps acting as a novel negative-feedback mechanism. In contrast, no change in c-Cbl levels was observed under the same conditions. Indeed, with the exception of a report showing that EGF-activated Src can induce the tyrosine phosphorylation, ubiquitylation and subsequent degradation of c-Cbl [46], there is little evidence that c-Cbl function in haemopoietic cells is regulated in a similar manner.
Altering the balance of c-Cbl and Cbl-b protein levels within a cell could also conceivably modulate cellular responses. We have already postulated that T-cell phenotypes of c-Cbl−/− and Cbl-b−/− mice are likely to reflect their relative abundance in thymocytes and peripheral T-cells respectively. It is also interesting to note that microarray analysis of chronic myeloid leukaemia patients showed that responsiveness to the specific Bcr/Abl inhibitor, imatinib, correlated with an approx. 2-fold increase in levels of c-Cbl in peripheral blood [136]. A similar, but smaller, study by Kaneta et al. [137] did not cite changes to c-Cbl, but instead found Cbl-b to be one of the genes that is down-regulated in patients responsive to imatinib. Thus, in both cases, conditions which favour greater expression c-Cbl or reduced Cbl-b expression contribute to a phenotype of drug responsiveness. This is interesting given that c-Cbl and Cbl-b are differentially regulated and form distinctive signalling complexes in Bcr/Abl-transformed cells [138]. Indeed, c-Cbl appears to act as a positive mediator of Bcr/Abl signalling through its ability to interact with CrkL and PI3K, while Cbl-b negatively regulates cell migration of Bcr/Abl-transformed cells [138].
In a further example, immunosuppression induced by HIV infection was found to correlate with greatly increased Cbl-b, but not c-Cbl, protein expression in peripheral blood [139]. However, infection with MAIDS (murine AIDS) virus resulted in the downregulation of c-Cbl protein levels in T- and B-lymph node cells, although effects on Cbl-b expression were not examined [140]. Loss of c-Cbl was presumed to be through ubiquitin-mediated degradation processes which are known to be induced upon MAIDS infection. Thus virally induced immunosuppression appears to engage mechanisms which increase the relative abundance of Cbl-b to c-Cbl.
FUTURE DIRECTIONS
Although recent discoveries have provided us with a clearer understanding of many aspects of how Cbl proteins regulate signalling responses, the intricacy of Cbl protein functions still cloud some of our interpretations. These complexities arise primarily from the vast number of proteins that can interact with Cbl proteins [at least 150 according to I. Dikic (personal communication)]. However, exciting new efforts are now being made to accurately reconstruct large-scale signalling networks by providing a framework for the application of mathematical methods to allow quantitative modelling of biochemical events as they occur in time and space [141]. Thus it is hoped that it will soon be possible to account for all the interconnective and functional relationships when analysing a signalling response that is regulated by Cbl proteins.
In future studies, it will also be important to try to utilize the knowledge that has been gained about Cbl protein function, with the aim of harnessing its negative regulatory activities towards disease-causing proteins. This could provide a means to suppress and down-regulate the activity of proteins such as oncogenic PTKs, as well as proteins involved in the development of autoimmune diseases. It is clear that Cbl E3 ligase activity is central to the regulation of many disease-causing proteins, so drugs that could enhance this activity may provide new therapeutic strategies for limiting or counteracting their constitutive signalling capabilities. Thus there may be potential therapeutic benefits by taking an alternative approach of indirectly targeting causative proteins through the enhancement of the natural restraining power of Cbl.
One possible avenue would be to screen for molecules that could mimic the effects of the phosphorylation of Tyr371, which, in human c-Cbl, is known to promote constitutive E3 activity, while retaining EGF receptor binding [11]. Thus there may be potential therapeutic benefits by taking an alternative approach of targeting causative proteins through the enhancement of the natural restraining power of Cbl. On the other side of the coin, a recent exciting finding has shown that a correlation exists between high c-Cbl expression and reduced Ron RTK expression in multiple sclerosis patient brains [142]. This is a significant finding since Ron-deficient mice exhibit greater neurological disease severity compared with normal littermates during experimental autoimmune encephalomyelitis [142]. Since Ron is down-regulated by c-Cbl-directed ubiquitylation [143], Ron expression levels could potentially be enhanced in multiple sclerosis patients by the development of small molecules that block E2 interactions with the c-Cbl RING finger domain.
From an experimental point of view, an area that has not been investigated adequately is the role of Cbl proteins in embryonic development. It is clear that Cbl proteins are essential for the survival of complex organisms, yet no studies are available that describe the causes of early embryonic lethality in the c-Cbl/Cbl-b double knockout mouse. Interestingly, we have found that it is possible to generate Cbl-b−/−/c-Cbl+/− mice at far greater frequencies than mice with a genotype of Cbl-b+/−/c-Cbl−/−. This breeding data suggested that c-Cbl is more essential than Cbl-b for embryonic development, and indeed, over the years, we have always found c-Cbl−/− mice more difficult to breed than Cbl-b−/− mice. Similarly, we can more readily produce Cbl-b−/−/c-Cbl+/G304E than Cbl-b+/−/c-CblG304E/G304E mice, which suggests further that retention of a functional c-Cbl TKB domain is more critical than Cbl-b function for embryonic survival (C. B. F. Thien and W. Y. Langdon, unpublished work).
As described in a number of the preceding sections, mice deficient for c-Cbl and Cbl-b have been invaluable in demonstrating the important roles played by these proteins in fine-tuning signalling thresholds in thymocytes and T-cells respectively. Analyses of knockout mice have already demonstrated that loss of c-Cbl and Cbl-b result in quite distinct phenotypes, and it is hoped that additional Cbl mutant mice will more adequately reveal the specific roles of c-Cbl and Cbl-b in various cell types. In particular, the generation of a greater range of Cbl knockin mice with well-defined loss-of-function or gain-of-function mutations will be of immense value in testing further many aspects of Cbl function in whole animals. Finally, a greater emphasis on the study of non-haemopoietic cells and tissues from these mice is also likely to be a future direction for testing the many aspects of RTK regulation that have previously only been rigorously studied in vitro. Thus, over the next few years, the analyses of Cbl mutant mice are likely to yield both new and confirmatory findings, with the anticipation of a better understanding of how Cbl proteins orchestrate the functional activity and fate of many partners to produce the desired intensity of a signalling response.
Acknowledgments
This work was supported by grants from NHMRC (National Health and Medical Research Council) (Canberra) and MHRIF (Medical and Health Research Infrastructure Fund) (Perth).
References
- 1.Weissman A. M. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 2.Passmore L. A., Barford D. Getting into position: the catalytic mechanisms of protein ubiquitylation. Biochem. J. 2004;379:513–525. doi: 10.1042/BJ20040198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yokouchi M., Kondo T., Sanjay A., Houghton A., Yoshimura A., Komiya S., Zhang H., Baron R. Src-catalyzed phosphorylation of c-Cbl leads to the interdependent ubiquitination of both proteins. J. Biol. Chem. 2001;276:35185–35193. doi: 10.1074/jbc.M102219200. [DOI] [PubMed] [Google Scholar]
- 4.Meng W., Sawasdikosol S., Burakoff S. J., Eck M. J. Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase. Nature (London) 1999;398:84–90. doi: 10.1038/18050. [DOI] [PubMed] [Google Scholar]
- 5.Bustelo X. R., Crespo P., López-Barahona M., Gutkind J. S., Barbacid M. Cbl-b, a member of the Sli-1/c-Cbl protein family, inhibits Vav-mediated c-Jun N-terminal kinase activation. Oncogene. 1997;15:2511–2520. doi: 10.1038/sj.onc.1201430. [DOI] [PubMed] [Google Scholar]
- 6.Yoon C. H., Lee J., Jongeward G. D., Sternberg P. W. Similarity of sli-1, a regulator of vulval development in C. elegans, to the mammalian proto-oncogene c-cbl. Science. 1995;269:1102–1105. doi: 10.1126/science.7652556. [DOI] [PubMed] [Google Scholar]
- 7.Thien C. B. F., Langdon W. Y. EGF receptor binding and transformation by v-cbl is ablated by the introduction of a loss-of-function mutation from the Caenorhabditis elegans sli-1 gene. Oncogene. 1997;14:2239–2249. doi: 10.1038/sj.onc.1201193. [DOI] [PubMed] [Google Scholar]
- 8.Lill N. L., Douillard P., Awwad R. A., Ota S., Lupher M. L., Jr, Miyake S., Meissner-Lula N., Hsu V. W., Band H. The evolutionarily conserved N-terminal region of Cbl is sufficient to enhance down-regulation of the epidermal growth factor receptor. J. Biol. Chem. 2000;275:367–377. doi: 10.1074/jbc.275.1.367. [DOI] [PubMed] [Google Scholar]
- 9.Waterman H., Katz M., Rubin C., Shtiegman K., Lavi S., Elson A., Jovin T., Yarden Y. A mutant EGF receptor defective in ubiquitylation and endocytosis unveils a role for Grb2 in negative signalling. EMBO J. 2002;21:303–313. doi: 10.1093/emboj/21.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng N., Wang P., Jeffrey P. D., Pavletich N. P. Structure of a c-Cbl–UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 11.Kassenbrock K. C., Anderson S. M. Regulation of ubiquitin protein ligase activity in c-Cbl by phosphorylation-induced conformational change and constitutive activation by tyrosine to glutamate point mutations. J. Biol. Chem. 2004;279:28017–28027. doi: 10.1074/jbc.M404114200. [DOI] [PubMed] [Google Scholar]
- 12.Lupher M. L., Songyang Z., Shoelson S. E., Cantley L. C., Band H. The Cbl phosphotyrosine-binding domain selects a D(N/D)XpY motif and binds to the TyrP292 negative regulatory phosphorylation site of ZAP-70. J. Biol. Chem. 1997;272:33140–33144. doi: 10.1074/jbc.272.52.33140. [DOI] [PubMed] [Google Scholar]
- 13.Lupher M. L., Jr, Rao N., Lill N. L., Andoniou C. E., Miyake S., Clark E. A., Druker B., Band H. Cbl-mediated negative regulation of the Syk tyrosine kinase: a critical role for Cbl phosphotyrosine-binding domain binding to Syk phosphotyrosine 323. J. Biol. Chem. 1998;273:35273–35281. doi: 10.1074/jbc.273.52.35273. [DOI] [PubMed] [Google Scholar]
- 14.Rubin C., Litvak V., Medvedovsky H., Zwang Y., Lev S., Yarden Y. Sprouty fine-tunes EGF signaling through interlinked positive and negative feedback loops. Curr. Biol. 2003;13:297–307. doi: 10.1016/s0960-9822(03)00053-8. [DOI] [PubMed] [Google Scholar]
- 15.Ohrt T., Mancini A., Tamura T., Niedenthal R. c-Cbl binds to tyrosine-phosphorylated neurotrophin receptor p75 and induces its ubiquitination. Cell. Signalling. 2004;16:1291–1298. doi: 10.1016/j.cellsig.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 16.Peschard P., Ishiyama N., Lin T., Lipkowitz S., Park M. A conserved DpYR motif in the juxtamembrane domain of the Met receptor family forms an atypical c-Cbl/Cbl-b tyrosine kinase binding domain site required for suppression of oncogenic activation. J. Biol. Chem. 2004;279:29565–29571. doi: 10.1074/jbc.M403954200. [DOI] [PubMed] [Google Scholar]
- 17.Yasuda T., Maeda A., Kurosaki M., Tezuka T., Hironaka K., Yamamoto T., Kurosaki T. Cbl suppresses B cell receptor-mediated phospholipase C (PLC)-γ2 activation by regulating B cell linker protein–PLCγ2 binding. J. Exp. Med. 2000;191:641–650. doi: 10.1084/jem.191.4.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yokouchi M., Wakioka T., Sakamoto H., Yasukawa H., Ohtsuka S., Sasaki A., Ohtsubo M., Valius M., Inoue A., Komiya S., Yoshimura A. APS, an adaptor protein containing PH and SH2 domains, is associated with the PDGF receptor and c-Cbl and inhibits PDGF-induced mitogenesis. Oncogene. 1999;18:759–767. doi: 10.1038/sj.onc.1202326. [DOI] [PubMed] [Google Scholar]
- 19.Hu J., Hubbard S. R. Structural characterization of a novel Cbl phosphotyrosine recognition motif in the APS family of adapter proteins. J. Biol. Chem. 2005;280:18943–18949. doi: 10.1074/jbc.M414157200. [DOI] [PubMed] [Google Scholar]
- 20.Tang J., Sawasdikosol S., Chang J.-H., Burakoff S. J. SLAP, a dimeric adapter protein, plays a functional role in T cell receptor signaling. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9775–9780. doi: 10.1073/pnas.96.17.9775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sosinowski T., Killeen N., Weiss A. The Src-like adaptor protein downregulates the T cell receptor on CD4+CD8+ thymocytes and regulates positive selection. Immunity. 2001;15:457–466. doi: 10.1016/s1074-7613(01)00195-9. [DOI] [PubMed] [Google Scholar]
- 22.Teckchandani A. M., Birukova A. A., Tar K., Verin A. D., Tsygankov A. Y. The multidomain protooncogenic protein c-Cbl binds to tubulin and stabilizes microtubules. Exp. Cell Res. 2005;306:114–127. doi: 10.1016/j.yexcr.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 23.Levkowitz G., Waterman H., Ettenberg S. A., Katz M., Lavi S., Iwai K., Reiss Y., Ciechanover A., Lipkowitz S., Yarden Y. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell. 1999;4:1–20. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 24.Andoniou C. E., Thien C. B. F., Langdon W. Y. Tumour induction by activated abl involves tyrosine phosphorylation of the product of the cbl oncogene. EMBO J. 1994;13:4515–4523. doi: 10.1002/j.1460-2075.1994.tb06773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thien C. B. F., Walker F., Langdon W. Y. Ring finger mutations that abolish c-Cbl-directed polyubiquitination and downregulation of the EGF receptor are insufficient for cell transformation. Mol. Cell. 2001;7:355–365. doi: 10.1016/s1097-2765(01)00183-6. [DOI] [PubMed] [Google Scholar]
- 26.Joazeiro C. A. P., Wing S. S., Huang H.-K., Leverson J. D., Hunter T., Liu Y.-C. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 27.Ettenberg S. A., Magnifico A., Cuello M., Nau M. M., Rubinstein Y. R., Yarden Y., Weissman A. M., Lipkowitz S. Cbl-b-dependent coordinated degradation of the epidermal growth factor receptor signaling complex. J. Biol. Chem. 2001;276:27677–27684. doi: 10.1074/jbc.M102641200. [DOI] [PubMed] [Google Scholar]
- 28.Kim M., Tezuka T., Tanaka K., Yamamoto T. Cbl-c suppresses v-Src-induced transformation through ubiquitin-dependent protein degradation. Oncogene. 2004;23:1645–1655. doi: 10.1038/sj.onc.1207298. [DOI] [PubMed] [Google Scholar]
- 29.Hicke L. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 30.Mosesson Y., Shtiegman K., Katz M., Zwang Y., Vereb G., Szollosi J., Yarden Y. Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J. Biol. Chem. 2003;278:21323–21326. doi: 10.1074/jbc.C300096200. [DOI] [PubMed] [Google Scholar]
- 31.Haglund K., Sigismund S., Polo S., Szymkiewicz I., Di Fiore P. P., Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 2003;5:461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- 32.Dikic I., Giordano S. Negative receptor signaling. Curr. Opin. Cell Biol. 2003;15:128–135. doi: 10.1016/s0955-0674(03)00004-8. [DOI] [PubMed] [Google Scholar]
- 33.Dikic I., Szymkiewicz I., Soubeyran P. Cbl signaling networks in the regulation of cell function. Cell. Mol. Life Sci. 2003;60:1805–1827. doi: 10.1007/s00018-003-3029-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y. C. Ubiquitin ligases and the immune response. Annu. Rev. Immunol. 2004;22:81–127. doi: 10.1146/annurev.immunol.22.012703.104813. [DOI] [PubMed] [Google Scholar]
- 35.Jeon M.-S., Atfield A., Venuprasad K., Krawczyk C., Sarao R., Elly C., Yang C., Arya S., Bachmaier K., Su L., et al. Essential role of the E3 ubiquitin ligase Cbl-b in T cell anergy induction. Immunity. 2004;21:167–177. doi: 10.1016/j.immuni.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 36.Bache K. G., Slagsvold T., Stenmark H. Defective downregulation of receptor tyrosine kinases in cancer. EMBO J. 2003;23:2707–2712. doi: 10.1038/sj.emboj.7600292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peschard P., Park M. Escape from Cbl-mediated downregulation: a recurrent theme for oncogenic deregulation of receptor tyrosine kinases. Cancer Cell. 2003;3:519–532. doi: 10.1016/s1535-6108(03)00136-3. [DOI] [PubMed] [Google Scholar]
- 38.Wong E. S. M., Lim J., Low B. C., Chen Q., Guy G. R. Evidence for direct interaction between Sprouty and Cbl. J. Biol. Chem. 2001;276:5866–5875. doi: 10.1074/jbc.M006945200. [DOI] [PubMed] [Google Scholar]
- 39.Fong C. W., Leong H. F., Wong E. S. M., Lim J., Yusoff P., Guy G. R. Tyrosine phosphorylation of Sprouty2 enhances its interaction with c-Cbl and is crucial for its function. J. Biol. Chem. 2003;278:33456–33464. doi: 10.1074/jbc.M301317200. [DOI] [PubMed] [Google Scholar]
- 40.Hacohen N., Kramer S., Sutherland D., Hiromi Y., Kasnow M. A. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92:253–263. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- 41.Casci T., Vinòs J., Freeman M. Sprouty, an intracellular inhibitor of Ras signaling. Cell. 1999;96:655–665. doi: 10.1016/s0092-8674(00)80576-0. [DOI] [PubMed] [Google Scholar]
- 42.Wong E. S. M., Fong C. W., Lim J., Yusoff P., Low B. C., Langdon W. Y., Guy G. R. Sprouty2 attenuates epidermal growth factor receptor ubiquitylation and endocytosis, and consequently enhances Ras/Erk signalling. EMBO J. 2003;21:4796–4808. doi: 10.1093/emboj/cdf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall A. B., Jura N., DaSilva J., Jang Y. J., Gong D., Bar-Sagi D. hSpry2 is targeted to the ubiquitin-dependent proteasome pathway by c-Cbl. Curr. Biol. 2003;13:308–314. doi: 10.1016/s0960-9822(03)00086-1. [DOI] [PubMed] [Google Scholar]
- 44.Wu W. J., Tu S., Cerione R. A. Activated Cdc42 sequesters c-Cbl and prevents EGF receptor degradation. Cell. 2003;114:715–725. doi: 10.1016/s0092-8674(03)00688-3. [DOI] [PubMed] [Google Scholar]
- 45.Magnifico A., Ettenberg S. A., Yang B., Mariano J., Tiwari S., Fang S., Lipkowitz S., Weissman A. M. WW domain HECT E3s target Cbl RING finger E3s for proteasomal degradation. J. Biol. Chem. 2003;278:43169–43177. doi: 10.1074/jbc.M308009200. [DOI] [PubMed] [Google Scholar]
- 46.Bao J., Gur G., Yarden Y. Src promotes destruction of c-Cbl: implications for oncogenic synergy between Src and growth factor receptor. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2438–2443. doi: 10.1073/pnas.0437945100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soubeyran P., Kowanetz K., Szymkiewicz I., Langdon W. Y., Dikic I. Cbl–CIN85–endophilin complex mediates ligand-induced downregulation of EGF receptors. Nature (London) 2002;416:183–187. doi: 10.1038/416183a. [DOI] [PubMed] [Google Scholar]
- 48.Petrelli A., Gilestro G. F., Lanzardo S., Comoglio P. M., Migone N., Giordano S. The endophilin–CIN85–Cbl complex mediates ligand-dependent downregulation of c-Met. Nature (London) 2002;416:187–190. doi: 10.1038/416187a. [DOI] [PubMed] [Google Scholar]
- 49.Sanjay A., Houghton A., Neff L., DiDomenico E., Bardelay C., Antoine E., Levy J., Gailit J., Bowtell D., Horne W. C., Baron R. Cbl associates with Pyk2 and Src to regulate Src kinase activity, αvβ3 integrin-mediated signaling, cell adhesion, and osteoclast motility. J. Cell Biol. 2001;152:181–195. doi: 10.1083/jcb.152.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsygankov A. Y., Mahajan S., Fincke J. E., Bolen J. B. Specific association of tyrosine-phosphorylated c-Cbl with Fyn tyrosine kinase in T cells. J. Biol. Chem. 1996;271:27130–27137. doi: 10.1074/jbc.271.43.27130. [DOI] [PubMed] [Google Scholar]
- 51.Andoniou C. E., Thien C. B. F., Langdon W. Y. The two major sites of cbl tyrosine phosphorylation in abl-transformed cells select the crkL SH2 domain. Oncogene. 1996;12:1981–1989. [PubMed] [Google Scholar]
- 52.Marengére L. E. M., Mirtsos C., Kozieradzki I., Veillette A., Mak T. W., Penninger J. M. Proto-oncoprotein Vav interacts with c-Cbl in activated thymocytes and peripheral T cells. J. Immunol. 1997;159:70–76. [PubMed] [Google Scholar]
- 53.Keane M. M., Rivero-Lezcano O. M., Mitchell J. A., Robbins K. C., Lipkowitz S. Cloning and characterization of cbl-b: a SH3-binding protein with homology to the c-cbl proto-oncogene. Oncogene. 1995;10:2367–2377. [PubMed] [Google Scholar]
- 54.Hunter S., Burton E. A., Wu S. C., Anderson S. M. Fyn associates with Cbl and phosphorylates tyrosine 731 in Cbl, a binding site for phosphatidylinositol 3-kinase. J. Biol. Chem. 1999;274:2097–2106. doi: 10.1074/jbc.274.4.2097. [DOI] [PubMed] [Google Scholar]
- 55.Ueno H., Sasaki K., Honda H., Nakamoto T., Miyagawa K., Mitani K., Yazaki Y., Hirai H. c-Cbl is tyrosine-phosphorylated by interleukin-4 and enhances mitogenic and survival signals of interleukin-4 receptor by linking with the phosphatidylinositol 3′-kinase pathway. Blood. 1998;91:46–53. [PubMed] [Google Scholar]
- 56.Lazaar A. L., Krymskaya V. P., Das S. K. P. VCAM-1 activates phosphatidylinositol 3-kinase and induces p120Cbl phosphorylation in human airway smooth muscle cells. J. Immunol. 2001;166:155–161. doi: 10.4049/jimmunol.166.1.155. [DOI] [PubMed] [Google Scholar]
- 57.Arron J. R., Vologodskaia M., Wong B. R., Naramura M., Kim N., Gu H., Choi Y. A positive regulatory role for Cbl family proteins in tumor necrosis factor-related activation-induced cytokine (TRANCE) and CD40L-mediated Akt activation. J. Biol. Chem. 2001;276:30011–30017. doi: 10.1074/jbc.M100414200. [DOI] [PubMed] [Google Scholar]
- 58.Hofmann K., Bucher P. UBA domain: a sequence motif present in multiple enzyme classes of ubiquitin pathway. Trends Biochem. Sci. 1996;21:172–173. [PubMed] [Google Scholar]
- 59.Chen L., Shinde U., Ortolan T. G., Madura K. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep. 2001;2:933–938. doi: 10.1093/embo-reports/kve203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heessen S., Masucci M. G., Dantuma N. P. The UBA2 domain functions as an intrinsic stabilization signal that protects Rad23 from proteasomal degradation. Mol. Cell. 2005;18:225–235. doi: 10.1016/j.molcel.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 61.Davies G. C., Ettenberg S. A., Coats A. O., Mussante M., Ravicandran S., Collins J., Nau M. M., Lipkowitz S. Cbl-b interacts with ubiquitinated proteins; differential function of the UBA domains of c-Cbl and Cbl-b. Oncogene. 2004;23:7104–7115. doi: 10.1038/sj.onc.1207952. [DOI] [PubMed] [Google Scholar]
- 62.Thien C. B. F., Langdon W. Y. Cbl: many adaptations to regulate protein tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2001;2:294–305. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
- 63.Jongeward G. D., Clandinin T. R., Sternberg P. W. sli-1, a negative regulator of let-23-mediated signaling in C. elegans. Genetics. 1995;139:1553–1566. doi: 10.1093/genetics/139.4.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nau M. M., Lipkowitz S. Comparative genomic organization of the cbl genes. Gene. 2003;308:103–113. doi: 10.1016/s0378-1119(03)00471-2. [DOI] [PubMed] [Google Scholar]
- 65.Duan L., Reddi A. L., Ghosh A., Dimri M., Band H. The Cbl family and other ubiquitin ligases: destructive forces in control of antigen receptor signaling. Immunity. 2004;21:7–17. doi: 10.1016/j.immuni.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 66.Jang I.-K., Gu H. Negative regulation of TCR signaling and T cell activation by selective protein degradation. Curr. Opin. Immunol. 2003;15:315–320. doi: 10.1016/s0952-7915(03)00048-7. [DOI] [PubMed] [Google Scholar]
- 67.Mueller D. L. E3 ubiquitin ligases as T cell anergy factors. Nat. Immunol. 2004;5:883–890. doi: 10.1038/ni1106. [DOI] [PubMed] [Google Scholar]
- 68.Langdon W. Y., Hyland C. D., Grumont R. J., Morse H. C., III The c-cbl proto-oncogene is preferentially expressed in thymus and testis tissue and encodes a nuclear protein. J. Virol. 1989;63:5420–5424. doi: 10.1128/jvi.63.12.5420-5424.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Donovan J. A., Wange R. L., Langdon W. Y., Samelson L. E. The protein product of the c-cbl protooncogene is the 120-kDa tyrosine-phosphorylated protein in Jurkat cells activated via the T cell antigen receptor. J. Biol. Chem. 1994;269:22921–22924. [PubMed] [Google Scholar]
- 70.Fukazawa T., Reedquist K. A., Trub T., Soltoff S., Panchamoorthy G., Druker B., Cantley L., Shoelson S. E., Band H. The SH3 domain-binding T cell tyrosyl phosphoprotein p120: demonstration of its identity with the c-cbl protooncogene and in vivo complexes with Fyn, Grb2, and phosphatidylinositol 3-kinase. J. Biol. Chem. 1995;270:19141–19150. doi: 10.1074/jbc.270.32.19141. [DOI] [PubMed] [Google Scholar]
- 71.Murphy M. A., Schnall R. G., Venter D. J., Barnett L., Bertoncello I., Thien C. B. F., Langdon W. Y., Bowtell D. D. L. Tissue hyperplasia and enhanced T cell signalling via ZAP-70 in c-Cbl deficient mice. Mol. Cell. Biol. 1998;18:4872–4882. doi: 10.1128/mcb.18.8.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Naramura M., Kole H. K., Hu R.-J., Gu H. Altered thymic positive selection and intracellular signals in Cbl-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 1998;95:15547–15552. doi: 10.1073/pnas.95.26.15547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bachmaier K., Krawczyk C., Kozieradzki I., Kong Y.-Y., Sasaki T., Oliveira-dos-Santos A., Mariathasan S., Bouchard D., Wakeham A., Itie A., et al. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature (London) 2000;403:211–216. doi: 10.1038/35003228. [DOI] [PubMed] [Google Scholar]
- 74.Chiang Y. J., Kole H. K., Brown K., Naramura M., Fukuhara S., Hu R.-J., Jang I. K., Gutkind J. S., Shevach E., Gu H. Cbl-b regulates the CD28 dependence of T-cell activation. Nature (London) 2000;403:216–220. doi: 10.1038/35003235. [DOI] [PubMed] [Google Scholar]
- 75.Krawczyk C., Bachmaier K., Sasaki T., Jones R. G., Snapper S. B., Bouchard D., Kozieradzki I., Ohashi P. S., Alt F. W., Penninger J. M. Cbl-b is a negative regulator of receptor clustering and raft aggregation in T cells. Immunity. 2000;13:463–473. doi: 10.1016/s1074-7613(00)00046-7. [DOI] [PubMed] [Google Scholar]
- 76.Naramura M., Jang I. K., Kole H., Huang F., Haines D., Gu H. c-Cbl and Cbl-b regulate T cell responsiveness by promoting ligand-induced TCR down-modulation. Nat. Immunol. 2002;3:1192–1199. doi: 10.1038/ni855. [DOI] [PubMed] [Google Scholar]
- 77.Griffiths E. K., Sanchez O., Mill P., Krawczyk C., Hojilla C. V., Rubin E., Nau M. M., Khokha R., Lipkowitz S., Hui C.-C., Penninger J. M. Cbl-3-deficient mice exhibit normal epithelial development. Mol. Cell. Biol. 2003;23:7708–7718. doi: 10.1128/MCB.23.21.7708-7718.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thien C. B. F., Bowtell D. D. L., Langdon W. Y. Perturbed regulation of ZAP-70 and sustained tyrosine phosphorylation of LAT and SLP-76 in c-Cbl-deficient thymocytes. J. Immunol. 1999;162:7133–7139. [PubMed] [Google Scholar]
- 79.Thien C. B. F., Scaife R. M., Papadimitriou J. M., Murphy M. A., Bowtell D. D. L., Langdon W. Y. A mouse with a loss-of-function mutation in the c-Cbl TKB domain shows perturbed thymocyte signaling without enhancing the activity of the ZAP-70 tyrosine kinase. J. Exp. Med. 2003;197:503–513. doi: 10.1084/jem.20021498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang H. Y., Altman Y., Fang D., Elly C., Dai Y., Shao Y., Liu Y. C. Cbl promotes ubiquitination of the T cell receptor zeta through an adaptor function of Zap-70. J. Biol. Chem. 2001;276:26004–26011. doi: 10.1074/jbc.M010738200. [DOI] [PubMed] [Google Scholar]
- 81.Niederberger N., Holmberg K., Alam S. M., Sakati W., Naramura M., Gu H., Gascoigne N. R. J. Allelic exclusion of the TCR α-chain is an active process requiring TCR-mediated signaling and c-Cbl. J. Immunol. 2003;170:4557–4563. doi: 10.4049/jimmunol.170.9.4557. [DOI] [PubMed] [Google Scholar]
- 82.Krawczyk C. M., Jones R. G., Atfield A., Bachmaier K., Arya S., Odermatt B., Ohashi P. S., Penninger J. M. Differential control of CD28-regulated in vivo immunity by the E3 ligase Cbl-b. J. Immunol. 2005;174:1472–1478. doi: 10.4049/jimmunol.174.3.1472. [DOI] [PubMed] [Google Scholar]
- 83.Schwartz R. H. T cell anergy. Annu. Rev. Immunol. 2003;21:305–334. doi: 10.1146/annurev.immunol.21.120601.141110. [DOI] [PubMed] [Google Scholar]
- 84.Gronski M. A., Boulter J. M., Moskophidis D., Nguyen L. T., Holmberg K., Elford A. R., Deenick E. K., Kim H. O., Penninger J. M., Odermatt B., et al. TCR affinity and negative regulation limit autoimmunity. Nat. Med. 2004;10:1234–1239. doi: 10.1038/nm1114. [DOI] [PubMed] [Google Scholar]
- 85.Yokoi N., Komeda K., Wang H.-Y., Yano H., Kitada K., Saitoh Y., Seino Y., Yasuda K., Serikawa T., Seino S. Cblb is a major susceptibility gene for rat type 1 diabetes mellitus. Nat. Genet. 2002;31:391–394. doi: 10.1038/ng927. [DOI] [PubMed] [Google Scholar]
- 86.Bergholdt R., Taxvig C., Eising S., Nerup J., Pociot F. CBLB variants in type 1 diabetes and their genetic interaction with CTLA4. J. Leukocyte Biol. 2005;77:579–585. doi: 10.1189/jlb.0904524. [DOI] [PubMed] [Google Scholar]
- 87.Zhou Q. L., Park J. G., Jiang Z. Y., Holik J. J., Mitra P., Semiz S., Guilherme A., Powelka A. M., Tang X., Virbasius J., Czech M. P. Analysis of insulin signalling by RNAi-based gene silencing. Biochem. Soc. Trans. 2004;32:817–821. doi: 10.1042/BST0320817. [DOI] [PubMed] [Google Scholar]
- 88.Mitra P., Zheng X., Czech M. P. RNAi-based analysis of CAP, Cbl, and CrkII function in the regulation of GLUT4 by insulin. J. Biol. Chem. 2004;279:37431–37435. doi: 10.1074/jbc.C400180200. [DOI] [PubMed] [Google Scholar]
- 89.Baumann C. A., Ribon V., Kanzaki M., Thurmond D. C., Mora S., Shigematsu S., Bickel P. E., Pessin J. E., Saltiel A. R. CAP defines a second signalling pathway required for insulin-stimulated glucose transport. Nature (London) 2000;407:202–207. doi: 10.1038/35025089. [DOI] [PubMed] [Google Scholar]
- 90.Wohlfert E. A., Callahan M. K., Clark R. B. Resistance to CD4+CD25+ regulatory T cells and TGF-β in Cbl-b−/− mice. J. Immunol. 2004;173:1059–1065. doi: 10.4049/jimmunol.173.2.1059. [DOI] [PubMed] [Google Scholar]
- 91.Thornton A. M., Shevach E. M. CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J. Exp. Med. 1998;188:287–296. doi: 10.1084/jem.188.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shevach E. M., McHugh R. S., Piccirillo C. A., Thornton A. M. Control of T-cell activation by CD4+CD25+ suppressor T cells. Immunol. Rev. 2001;182:58–67. doi: 10.1034/j.1600-065x.2001.1820104.x. [DOI] [PubMed] [Google Scholar]
- 93.Ohashi P. S. T-cell signalling and autoimmunity: molecular mechanisms of disease. Nat. Rev. Immunol. 2002;2:427–438. doi: 10.1038/nri822. [DOI] [PubMed] [Google Scholar]
- 94.von Boehmer H. Mechanisms of suppression by suppressor T cells. Nat. Immunol. 2005;6:338–344. doi: 10.1038/ni1180. [DOI] [PubMed] [Google Scholar]
- 95.Baecher-Allan C., Viglietta V., Hafler D. A. Inhibition of human CD4+CD25+high regulatory T cell function. J. Immunol. 2002;169:6210–6217. doi: 10.4049/jimmunol.169.11.6210. [DOI] [PubMed] [Google Scholar]
- 96.Heissmeyer V., Macián F., Im S.-H., Varma R., Feske S., Venuprasad K., Gu H., Liu Y. C., Dustin M. L., Rao A. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat. Immunol. 2004;5:255–265. doi: 10.1038/ni1047. [DOI] [PubMed] [Google Scholar]
- 97.Boussiotis V. A., Freeman G. J., Berezovskaya A., Barber D. L., Nadler L. M. Maintenance of human T cell anergy: blocking of IL-2 gene transcription by activated Rap1. Science. 1997;278:124–128. doi: 10.1126/science.278.5335.124. [DOI] [PubMed] [Google Scholar]
- 98.Elly C., Witte S., Zhang Z., Rosnet O., Lipkowitz S., Altman A., Liu Y.-C. Tyrosine phosphorylation and complex formation of Cbl-b upon T cell receptor stimulation. Oncogene. 1999;18:1147–1156. doi: 10.1038/sj.onc.1202411. [DOI] [PubMed] [Google Scholar]
- 99.Shao Y., Elly C., Liu Y.-C. Negative regulation of Rap1 activation by the Cbl E3 ubiquitin ligase. EMBO Rep. 2003;4:425–431. doi: 10.1038/sj.embor.embor813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang W., Shao Y., Fang D., Huang J., Jeon M.-S., Liu Y.-C. Negative regulation of T cell antigen receptor-mediated Crkl-L-C3G signaling and cell adhesion by Cbl-b. J. Biol. Chem. 2003;278:23978–23983. doi: 10.1074/jbc.M212671200. [DOI] [PubMed] [Google Scholar]
- 101.Sebzda E., Bracke M., Tugal T., Hogg N., Cantrell D. A. Rap1A positively regulates T cells via integrin activation rather than inhibiting lymphocyte signaling. Nat. Immunol. 2002;3:251–258. doi: 10.1038/ni765. [DOI] [PubMed] [Google Scholar]
- 102.Krawczyk C., Penninger J. M. Molecular controls of antigen receptor clustering and autoimmunity. Trends Cell Biol. 2001;11:212–220. doi: 10.1016/s0962-8924(01)01981-x. [DOI] [PubMed] [Google Scholar]
- 103.Kong G., Dalton M., Wardenburg J. B., Straus D., Kurosaki T., Chan A. C. Distinct tyrosine phosphorylation sites in ZAP-70 mediate activation and negative regulation of antigen receptor function. Mol. Cell. Biol. 1996;16:5026–5035. doi: 10.1128/mcb.16.9.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhao Q., Weiss A. Enhancement of lymphocyte responsiveness by a gain-of-function mutation of ZAP-70. Mol. Cell. Biol. 1996;16:6765–6774. doi: 10.1128/mcb.16.12.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Carpino N., Turner S., Mekala D., Takahashi Y., Zang H., Geiger T. L., Doherty P., Ihle J. N. Regulation of ZAP-70 activation and TCR signaling by two related proteins, Sts-1 and Sts-2. Immunity. 2004;20:37–46. doi: 10.1016/s1074-7613(03)00351-0. [DOI] [PubMed] [Google Scholar]
- 106.Feshchenko E. A., Smirnova E. V., Swaminathan G., Teckchandani A. M., Agrawal R., Band H., Zhang X., Annan R. S., Carr S. A., Tsygankov A. Y. TULA: an SH3- and UBA-containing protein that binds to c-Cbl and ubiquitin. Oncogene. 2004;23:4690–4706. doi: 10.1038/sj.onc.1207627. [DOI] [PubMed] [Google Scholar]
- 107.Kowanetz K., Crosetto N., Haglund K., Schmidt M. H. H., Heldin C.-H., Dikic I. Suppressors of T-cell receptor signaling Sts-1 and Sts-2 bind to Cbl and inhibit endocytosis of receptor tyrosine kinases. J. Biol. Chem. 2004;279:32786–32795. doi: 10.1074/jbc.M403759200. [DOI] [PubMed] [Google Scholar]
- 108.Starr T. K., Jameson S. C., Hogquist K. A. Positive and negative selection of T cells. Annu. Rev. Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 109.Palmer E. Negative selection – clearing out the bad apples from the T-cell repertoire. Nat. Rev. Immunol. 2003;3:383–391. doi: 10.1038/nri1085. [DOI] [PubMed] [Google Scholar]
- 110.Ohashi P. S. Negative selection and autoimmunity. Curr. Opin. Immunol. 2003;15:668–676. doi: 10.1016/j.coi.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 111.Chiang Y. J., Sommer C. L., Jordan M. S., Gu H., Samelson L. E., Koretzky G. A., Hodes R. J. Inactivation of c-Cbl reverses neonatal lethality and T cell developmental arrest of SLP-76-deficient mice. J. Exp. Med. 2004;200:25–34. doi: 10.1084/jem.20040262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Richie L. I., Ebert P. J. R., Wu L. C., Krummel M. F., Owen J. J. T., Davis M. M. Imaging synapse formation during thymocyte selection: inability of CD3ζ to form a stable central accumulation during negative selection. Immunity. 2002;16:595–606. doi: 10.1016/s1074-7613(02)00299-6. [DOI] [PubMed] [Google Scholar]
- 113.Minter L. M., Osborne B. A. Cell death in the thymus – it's all a matter of contacts. Semin. Immunol. 2003;15:135–144. doi: 10.1016/s1044-5323(03)00029-0. [DOI] [PubMed] [Google Scholar]
- 114.Wiedemann A., Müller S., Favier B., Penna D., Guiraud M., Delmas C., Champagne E., Valitutti S. T-cell activation is accompanied by an ubiquitination process occurring at the immunological synapse. Immunol. Lett. 2005;98:57–61. doi: 10.1016/j.imlet.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 115.Garcia G. G., Miller R. A. Single-cell analyses reveal two defects in peptide-specific activation of naive T cells from aged mice. J. Immunol. 2001;166:3151–3157. doi: 10.4049/jimmunol.166.5.3151. [DOI] [PubMed] [Google Scholar]
- 116.Hawash I. Y., Kesavan K. P., Magee A. I., Geahlen R. L., Harrison M. L. The Lck SH3 domain negatively regulates localization to lipid rafts through an interaction with c-Cbl. J. Biol. Chem. 2002;277:5683–5691. doi: 10.1074/jbc.M110002200. [DOI] [PubMed] [Google Scholar]
- 117.Muñoz P., Navarro M.-d.-C., Pavón E. J., Salmerón J., Malavasi F., Sancho J., Zubiaur M. CD38 signaling in T cells is initiated within a subset of membrane rafts containing Lck and the CD3-ζ subunit of the T cell antigen receptor. J. Biol. Chem. 2003;278:50791–50802. doi: 10.1074/jbc.M308034200. [DOI] [PubMed] [Google Scholar]
- 118.Dykstra M., Cherukuri A., Sohn H. W., Tzeng S.-J., Pierce S. K. Location is everything: lipid rafts and immune cell signaling. Annu. Rev. Immunol. 2003;21:457–481. doi: 10.1146/annurev.immunol.21.120601.141021. [DOI] [PubMed] [Google Scholar]
- 119.Xavier R., Brennan T., Li Q., McCormack C., Seed B. Membrane compartmentation is required for efficient T cell activation. Immunity. 1998;8:723–732. doi: 10.1016/s1074-7613(00)80577-4. [DOI] [PubMed] [Google Scholar]
- 120.Avota E., Müller N., Klett M., Schneider-Schaulies S. Measles virus interacts with and alters signal transduction in T-cell lipid rafts. J. Virol. 2004;78:9552–9559. doi: 10.1128/JVI.78.17.9552-9559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lafont F., Simons K. Raft-partitioning of the ubiquitin ligases Cbl and Nedd4 upon IgE-triggered cell signaling. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3180–3184. doi: 10.1073/pnas.051003498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kimura A., Baumann C. A., Chiang S.-H., Saltiel A. R. The sorbin homology domain: a motif for the targeting of proteins to lipid rafts. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9098–9103. doi: 10.1073/pnas.151252898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ebert P. J. R., Baker J. F., Punt J. A. Immature CD4+CD8+ thymocytes do not polarize lipid rafts in response to TCR-mediated signals. J. Immunol. 2000;165:5435–5442. doi: 10.4049/jimmunol.165.10.5435. [DOI] [PubMed] [Google Scholar]
- 124.Fang D., Liu Y. C. Proteolysis-independent regulation of PI3K by Cbl-b-mediated ubiquitination in T cells. Nat. Immunol. 2001;2:870–875. doi: 10.1038/ni0901-870. [DOI] [PubMed] [Google Scholar]
- 125.Liu Y. C., Gu H. Cbl and Cbl-b in T-cell regulation. Trends Immunol. 2002;23:140–143. doi: 10.1016/s1471-4906(01)02157-3. [DOI] [PubMed] [Google Scholar]
- 126.Fang D., Wang H.-Y., Fang N., Altman Y., Elly C., Liu Y.-C. Cbl-b, a RING-type E3 ubiquitin ligase, targets phosphatidylinositol 3-kinase for ubiquitination in T cells. J. Biol. Chem. 2001;276:4872–4878. doi: 10.1074/jbc.M008901200. [DOI] [PubMed] [Google Scholar]
- 127.Cohen S., Dadi H., Shaoul E., Sharfe N., Roifman C. M. Cloning and characterization of a lymphoid-specific, inducible human protein tyrosine phosphatase, Lyp. Blood. 1999;93:2013–2024. [PubMed] [Google Scholar]
- 128.Zhang J., Chiang Y. J., Hodes R. J., Siraganian R. P. Inactivation of c-Cbl or Cbl-b differentially affects signaling from the high affinity IgE receptor. J. Immunol. 2004;173:1811–1818. doi: 10.4049/jimmunol.173.3.1811. [DOI] [PubMed] [Google Scholar]
- 129.Qu X., Sada K., Kyo S., Maeno K., Miah S. M. S., Yamamura H. Negative regulation of FcεRI-mediated mast cell activation by ubiquitin-protein ligase Cbl-b. Blood. 2004;103:1779–1786. doi: 10.1182/blood-2003-07-2260. [DOI] [PubMed] [Google Scholar]
- 130.Rao N., Ghosh A. K., Ota S., Zhou P., Reddi A. L., Hakezi K., Druker B. K., Wu J., Band H. The non-receptor tyrosine kinase Syk is a target of Cbl-mediated ubiquitylation upon B-cell receptor stimulation. EMBO J. 2001;20:7085–7095. doi: 10.1093/emboj/20.24.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Paolini R., Molfetta R., Beitz L. O., Zhang J., Scharenberg A. M., Piccoli M., Frati L., Siraganian R., Santoni A. Activation of Syk tyrosine kinase is required for c-Cbl-mediated ubiquitination of FcεRI and Syk in RBL cells. J. Biol. Chem. 2002;277:36940–36947. doi: 10.1074/jbc.M204948200. [DOI] [PubMed] [Google Scholar]
- 132.Yasuda T., Tezuka T., Maeda A., Inazu T., Yamanashi Y., Gu H., Kurosaki T. Cbl-b positively regulates Btk-mediated activation of phospholipase C-γ2 in B cells. J. Exp. Med. 2002;196:51–63. doi: 10.1084/jem.20020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang Z., Elly C., Altman A., Liu Y.-C. Dual regulation of T cell receptor-mediated signaling by oncogenic Cbl mutant 70Z. J. Biol. Chem. 1999;274:4883–4889. doi: 10.1074/jbc.274.8.4883. [DOI] [PubMed] [Google Scholar]
- 134.Li D., Gál I., Vermes C., Alegre M.-L., Chong A. S. F., Chen L., Shao Q., Adarichev V., Xu X., Koreny T., et al. Cbl-b: one of the key molecules tuning CD28- and CTLA-4-mediated T cell costimulation. J. Immunol. 2004;173:7135–7139. doi: 10.4049/jimmunol.173.12.7135. [DOI] [PubMed] [Google Scholar]
- 135.Zhang J., Bárdos T., Li D., Gál I., Vermes C., Xu J., Mikecz K., Finnegan A., Lipkowitz S., Glant T. T. Regulation of T cell activation threshold by CD28 costimulation through targeting Cbl-b for ubiquitination. J. Immunol. 2002;169:2236–2240. doi: 10.4049/jimmunol.169.5.2236. [DOI] [PubMed] [Google Scholar]
- 136.McLean L. A., Gathmann I., Capdeville R., Polymeropoulos M. H., Dressman M. Pharmacogenomic analysis of cytogenetic response in chronic myeloid leukemia patients treated with imatinib. Clin. Cancer Res. 2004;10:155–165. doi: 10.1158/1078-0432.ccr-0784-3. [DOI] [PubMed] [Google Scholar]
- 137.Kaneta Y., Kagami Y., Katagiri T., Tsunoda T., Jin-nai I., Taguchi H., Hirai H., Ohnishi K., Ueda T., Emi N., et al. Prediction of sensitivity to STI571 among chronic myeloid leukemia patients by genome-wide cDNA microarray analysis. Jpn. J. Cancer Res. 2002;93:849–856. doi: 10.1111/j.1349-7006.2002.tb01328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sattler M., Pride Y. B., Quinnan L. R., Verma S., Malouf N. A., Husson H., Salgia R., Lipkowitz S., Griffin J. D. Differential expression and signaling of CBL and CBL-B in BCR/ABL transformed cells. Oncogene. 2002;21:1423–1433. doi: 10.1038/sj.onc.1205202. [DOI] [PubMed] [Google Scholar]
- 139.Leng Q., Borkow G., Bentwich Z. Attenuated signaling associated with immune activation in HIV-1-infected individuals. Biochem. Biophys. Res. Commun. 2002;298:464–467. doi: 10.1016/s0006-291x(02)02460-9. [DOI] [PubMed] [Google Scholar]
- 140.Trebak M., Lambert C. A., Rahmouni S., Debrus S., Defresne M.-P., Regnier D. C., Boniver J., Moutschen M. Decreased protein levels of the c-Cbl protooncogene in murine AIDS. Cell. Immunol. 1998;188:151–157. doi: 10.1006/cimm.1998.1348. [DOI] [PubMed] [Google Scholar]
- 141.Papin J. A., Hunter T., Palsson B. O., Subramaniam S. Reconstruction of cellular signalling networks and analysis of their properties. Nat. Rev. Mol. Cell Biol. 2005;6:99–111. doi: 10.1038/nrm1570. [DOI] [PubMed] [Google Scholar]
- 142.Tsutsui S., Noorbakhsh F., Sullivan A., Henderson A. J., Warren K., Toney-Earley K., Waltz S. E., Power C. RON-regulated innate immunity is protective in an animal model of multiple sclerosis. Ann. Neurol. 2005;57:883–895. doi: 10.1002/ana.20502. [DOI] [PubMed] [Google Scholar]
- 143.Penengo L., Rubin C., Yarden Y., Gaudino G. c-Cbl is a critical modulator of the Ron tyrosine kinase receptor. Oncogene. 2003;22:3669–3679. doi: 10.1038/sj.onc.1206585. [DOI] [PubMed] [Google Scholar]
- 144.Huang F., Sorkin A. Growth factor receptor binding protein 2-mediated recruitment of the RING domain of Cbl to the epidermal growth factor receptor is essential and sufficient to support receptor endocytosis. Mol. Biol. Cell. 2005;16:1268–1281. doi: 10.1091/mbc.E04-09-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]