Abstract
Tankyrase 1 is a PARP [poly(ADP-ribose) polymerase] that localizes to multiple subcellular sites, including telomeres and mitotic centrosomes. Previous studies demonstrated that cells deficient in tankyrase 1 suffered a block in resolution of sister telomeres and arrested in early anaphase [Dynek and Smith (2004) Science 304, 97–100]. This phenotype was dependent on the catalytic PARP activity of tankyrase 1. To identify critical acceptors of PARsylation [poly(ADP-ribosyl)ation] by tankyrase 1 in mitosis, tankyrase 1 immunoprecipitates were analysed for associated PARsylated proteins. We identified NuMA (nuclear mitotic apparatus protein) as a major acceptor of poly(ADP-ribose) from tankyrase 1 in mitosis. We showed by immunofluorescence and immunoprecipitation that association between tankyrase 1 and NuMA increases dramatically at the onset of mitosis, concomitant with PARsylation of NuMA. Knockdown of tankyrase 1 by siRNA (small interfering RNA) eliminates PARsylation of NuMA in mitosis, confirming tankyrase 1 as the PARP responsible for this modification. However, even in the absence of tankyrase 1 and PARsylation, NuMA localizes to spindle poles. By contrast, siRNA knockdown of NuMA results in complete loss of tankyrase 1 from spindle poles. We discuss our result in terms of a model where PARsylation of NuMA by tankyrase 1 in mitosis could play a role in sister telomere separation and/or mitotic progression.
Keywords: mitosis, nuclear mitotic apparatus protein (NuMA), poly(ADP-ribose) polymerase (PARP), spindle pole, tankyrase 1
Abbreviations: 3AB, 3-aminobenzamide; DAPI, 4,6-diamino-2-phenylindole; IRAP, insulin-responsive aminopeptidase; NuMA, nuclear mitotic apparatus protein; PAR, poly(ADP-ribose); PARP, poly(ADP-ribose) polymerase; PARsylation, poly(ADP-ribosyl)ation; siRNA, small interfering RNA; TAB182, 182 kDa tankyrase-binding protein; TRF, telomeric repeat binding factor
INTRODUCTION
Tankyrase 1 is a PARP [poly(ADP-ribose) polymerase] [1] that functions in telomere length regulation [2,3] and sister telomere cohesion [4] in human cells. Human telomeres consist of tandem arrays of TTAGGG repeats and associated proteins that function to protect and replicate telomeric DNA [5,6]. Telomere length maintenance depends upon telomerase, a reverse transcriptase that adds telomeric repeats to chromosome ends [7]. Tankyrase 1 was initially identified in a two-hybrid screen with the telomeric TTAGGG-repeat binding protein TRF1 (telomeric repeat binding factor 1) [8], a negative regulator of telomere length that acts in cis to block access of telomerase to telomeres [9,10]. Tankyrase 1 PARsylates [poly(ADP-ribosyl)ates] TRF1 in vitro, inhibiting its ability to bind telomeric DNA [1]. Upon overexpression of tankyrase 1 in the nucleus, TRF1 is removed from telomeres [2,3]. The DNA-unbound form of TRF1 is ubiquitinated and degraded by the proteasome [11]. Thus tankyrase 1 is thought to regulate telomere length by controlling TRF1 levels at telomeres.
Although studies indicate a role for tankyrase 1 at telomeres, unlike other telomeric proteins such as TRF1, which resides mainly at telomeres, only a small fraction of tankyrase 1 localizes to telomeres. Indeed, the majority of the protein resides elsewhere at multiple subcellular sites, including mitotic centrosomes [12], nuclear pore complexes [12] and the Golgi apparatus [13]. Consistent with these multiple localizations, two-hybrid screens have revealed a number of interacting proteins for tankyrase 1 (or its closely related human homologue tankyrase 2 [13–17]), including: a resident Golgi protein, IRAP (insulin-responsive aminopeptidase) [13]; an endosomal adaptor protein, Grb14 [17]; NuMA (nuclear mitotic apparatus protein) [18]; a heterochromatin and cortical actin staining protein, TAB182 (182 kDa tankyrase-binding protein) [19]; and the cytoplasmic, apoptosis-regulating Mcl-1 proteins [20].
It is perhaps not surprising that tankyrase 1 has multiple binding partners considering its primary structure, which comprises a number of protein–protein interaction motifs [1]. At its N-terminus tankyrase 1 contains an HPS region (homopolymeric tracts of histidine, proline and serine) of unknown function. The HPS region is followed by an ANK domain containing 24 ankyrin repeats [21]. This domain is responsible for interaction with most of the known partners of tankyrase 1 described above, including TRF1, for which it contains five distinct binding sites [19]. Next is a SAM (sterile alpha module) [22], which can promote homo- and hetero-multimerization between tankyrase 1 and tankyrase 2 [23–25]. Finally, the C-terminus contains the catalytic PARP domain. Tankyrase 1 is a member of the PARP superfamily of enzymes, which catalyse the formation of long chains of ADP-ribose polymers on to themselves and other protein acceptors [26,27]. These negatively charged polymers can drastically alter the properties of the protein acceptor, explaining for example how PARsylation of TRF1 by tankyrase 1 inhibits binding of TRF1 to telomeres [1–3]. In addition to TRF1, TAB182 and IRAP have been demonstrated as acceptors of PARsylation by tankyrase 1 in vitro [13,19].
To elucidate the function of tankyrase 1, we recently used siRNA (small interfering RNA) to knock down tankyrase 1 expression in human cells. We found, unexpectedly, that cells arrested in anaphase in the absence of tankyrase 1 [4]. Live cell imaging showed that, in tankyrase 1-deficient cells, chromosomes aligned normally on the metaphase plate, but sister chromatids were unable to segregate to daughter poles. Fluorescent in situ hybridization using chromosome-specific probes revealed that while sister chromatids were separated at their centromeres and along their arms, they remained associated at their telomeres, indicating that tankyrase 1 was required for separation of sister telomeres at mitosis. Finally, we showed that wild-type (but not PARP-dead) tankyrase 1 rescued the abnormal mitotic phenotype, indicating a requirement for PARsylation [4].
Here we identify NuMA as a major acceptor of PARsylation by tankyrase 1 in mitosis in human cells. NuMA is a large coiled-coil protein that shuttles between the nuclear matrix in interphase and the spindle poles in mitosis [28–31]. A number of functional studies indicate an essential role for NuMA in mitotic spindle assembly, where it is required to organize and stabilize a focused array of microtubules at spindle poles [30,32–35]. The role of NuMA at its interphase locale, the nuclear matrix, is less well understood. Our identification of NuMA as a major acceptor of PARsylation by tankyrase 1 in mitosis suggests the possibility that NuMA may play a critical role in tankyrase 1 function. We discuss our results in terms of a model for the regulation of sister telomere resolution and mitotic progression via PARsylation of NuMA by tankyrase 1.
EXPERIMENTAL
Cell cycle arrest and synchronization
To induce mitotic arrest, HeLaI.2.11 cells [36] were treated with 1.5 μg/ml nocodazole for 24 h.
To generate staged cell extracts, exponentially growing HeLaI.2.11 cells were treated with 2 mM thymidine for 14 h, released into fresh medium for 11 h, treated again with 2 mM thymidine for 14 h, and released into fresh medium containing 30 ng/ml nocodazole for 12 h. Cells were harvested for analysis at intervals from 0 to 12 h during the nocodazole incubation. Following 12 h in nocodazole, cells were collected by mitotic shake-off, replated in fresh medium and harvested for analysis at intervals from 0 to 3 h.
To collect mitotic cells without using nocodazole, cells were synchronized by double thymidine block as described above, and rounded mitotic cells were collected by shake-off between 8 and 9 h after release into fresh medium.
The cell cycle was verified by FACS analysis. Cells were collected by trypsinization, resuspended in PBS containing 2 mM EDTA, and fixed with cold 70% (v/v) ethanol. Cells were stained with propidium iodide (50 μg/ml) and analysed with a Becton-Dickinson FACScan and Modfit 3.0 software to determine relative DNA content.
Cell extracts
HeLaI.2.11 cells were resuspended in 4 vol. of buffer C [20 mM Hepes/KOH, pH 7.9, 420 mM KCl, 25% glycerol, 0.1 mM EDTA, 5 mM MgCl2, 0.2% Nonidet P40, 1 mM dithiothreitol and 2.5% protease inhibitor cocktail (Sigma)] or TNE buffer (10 mM Tris, pH 7.8, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P40 and 2.5% protease inhibitor cocktail) for 1 h on ice. Suspensions were pelleted at 8000 g for 10 min. Aliquots of 25 μg (determined by Bio-Rad protein assay) of supernatant proteins were fractionated by SDS/PAGE and analysed by immunoblotting.
Immunoprecipitation, phosphatase treatment and in vitro PARP assays
For immunoprecipitations, HeLaI.2.11 cell extracts were generated in TNE buffer or buffer C (generally one or two 15 cm dishes in 0.5–1.0 ml of buffer) as indicated in the Figure legends. Supernatants were pre-cleared with rabbit IgG or normal mouse serum and Protein G–Sepharose, rotating at 4 °C for 30 min. Non-specific antibody complexes and protein aggregates were removed by centrifugation at 800 g for 3 min. The supernatant was then incubated with rabbit IgG (1.8 μg/ml), rabbit anti-tankyrase 1 antibody 465 (1.8 μg/ml) [1], mouse anti-NuMA (0.5 μg/ml; Oncogene) or normal mouse serum (1 μl/ml) at 4 °C with rocking for 1.5 h. Antigen–antibody complexes were then bound to Protein G–Sepharose at 4 °C with rocking for 1.5 h. After binding, beads from TNE extracts were washed in TNE buffer, and beads from buffer C extracts were washed in buffer D (20 mM Hepes, 100 mM KCl, 20% glycerol, 0.2 mM EDTA, 0.2 mM EGTA, 0.1% Triton X-100 and 0.1% Nonidet P40).
For phosphatase treatment, immunocomplexes bound to beads were washed three times with HBS buffer (20 mM Tris, pH 5.5, 0.5% Nonidet P40, 0.5 M NaCl and protease inhibitor cocktail) containing 1 mM 3AB (3-aminobenzamide), incubated in a 12.5 μl of a buffered reaction containing 1500 units of λ-phosphatase (New England Biolabs) for 30 min at 37 °C, and washed with HBS buffer.
For in vitro PARP assays, immunocomplexes bound to beads were washed three times with HBS containing 1 mM 3AB, washed once in 100 μl of PARP reaction buffer (50 mM Tris, pH 8.0, 4 mM MgCl2, 0.2 mM dithiothreitol), and incubated in 100 μl of PARP reaction buffer containing 50 μCi of [32P]NAD+ (1000 Ci/mmol; Amersham) for 30 min at 25 °C. Where indicated, 2.5 μg of recombinant tankyrase 1 [1] or PARP-1 (Biomol Research Laboratories), or various concentrations (0–1 mM) of NAD+, were added. After the PARP reaction, beads were washed with HBS buffer.
Bound proteins were resuspended in 2×sample buffer, fractionated by SDS/PAGE and transferred electrophoretically to nitrocellulose for immunoblot analysis, or gels were dried for detection by autoradiography.
Immunoblotting
Immunoblots were incubated with the following primary antibodies: rabbit anti-tankyrase 1 antibody 465 (1.8 μg/ml) [1], mouse anti-α-tubulin (1:500000; Sigma), rabbit anti-cyclin A (1 μg/ml; Upstate), rabbit anti-cyclin B1 (0.2 μg/ml; Santa Cruz), mouse anti-NuMA (0.1 μg/ml; Oncogene), rabbit anti-β-actin (0.2 ug/ml; Santa Cruz) or rabbit anti-PAR [poly(ADP-ribose)] (1:1000; Alexis 96-10, or a gift from Guy Poirier, Faculty of Medicine, Laval University, Ste-Foy, Quebec G1V 4G2, Canada), followed by horseradish peroxidase-conjugated donkey anti-rabbit or anti-mouse IgG (Amersham; 1:2500). Bound antibody was detected by SuperSignal West Pico or Dura kits (Pierce).
siRNA
HeLaI.2.11 cells were transfected without (mock) or with siRNA oligonucleotides (synthesized by Dharmacon Research Inc.) directed against tankyrase 1 (5′-AACAAUUCACCGUCGUCCUCU-3′) [4] or NuMA (5′-GGCGUGGCAGGAGAAGUUC-3′) [37] using Oligofectamine (Invitrogen) for 48 h, as described by the manufacturer.
Indirect immunofluorescence analysis
Methanol-fixed HeLaI.2.11 cells were incubated with mouse anti-NuMA (0.05 μg/ml; Oncogene), mouse anti-NuMA antibody 1F1 (ascites 1:100) [38] (kindly provided by Andreas Merdes, Wellcome Trust Centre for Cell Biology, ICMB, University of Edinburgh, U.K., and Don Cleveland, Ludwig Institute for Cancer Research and Department of Cellular and Molecular Medicine, University of California at San Diego, La Jolla, CA 92093, U.S.A.), or rabbit anti-tankyrase 1 antibody 609 (1 μg/ml) [2]. Primary antibodies were detected with FITC- or TRITC (tetramethylrhodamine isothiocyanate)-conjugated donkey anti-rabbit or anti-mouse antibodies (Jackson Laboratories) respectively at 1:100. DNA was stained with DAPI (4,6-diamino-2-phenylindole; 0.2 μg/ml). Images were acquired on a Zeiss Axioplan 2 microscope with a Photometrix SenSyn camera. Photographs were processed and merged using IPLab software. Images in Figures 6(A) and 6(C) were collected at the same exposure times and processed identically.
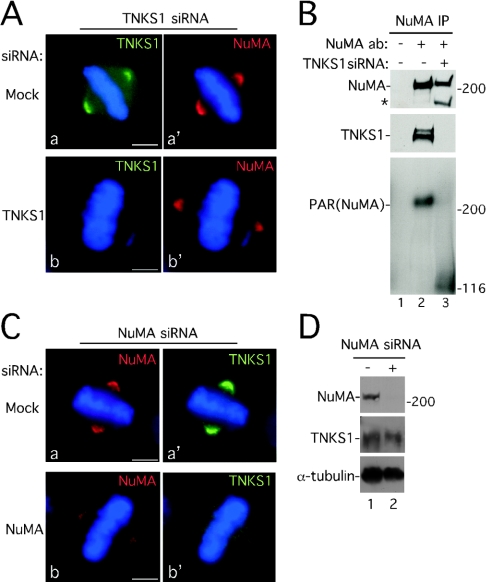
Figure 6. Tankyrase 1 is required for PARsylation of NuMA, but not for localization of NuMA to spindle poles.
(A) NuMA localizes to spindle poles in the absence of tankyrase 1. Immunofluorescence analysis of methanol-fixed HeLaI.2.11 cells is shown after 48 h of transfection with tankyrase 1 siRNA. Cells were stained with anti-tankyrase 1 antibody 609 (TNKS1; green) (a and b) or anti-NuMA 1F1 (red) (a′ and b′). DAPI staining of the DNA is in blue. Bar=5 μm. (B) siRNA of tankyrase 1 eliminates PARsylation of NuMA in mitosis. Cells were incubated with (+) or without (−) tankyrase 1 siRNA for 48 h and treated with nocodazole for 24 h prior to harvesting. Extracts generated in buffer C were immunoprecipitated (IP) with control serum (−) or anti-NuMA antibodies (ab) (+). Products were detected by immunoblotting with anti-NuMA (top panel), anti-tankyrase 1 antibody 465 (TNKS1; middle panel), or anti-PAR (bottom panel). The asterisk (*) indicates a NuMA breakdown product described previously [55,56]. (C) Knockdown of NuMA results in loss of tankyrase 1 at spindle poles. Immunofluorescence analysis of methanol-fixed HeLaI.2.11 cells is shown after 48 h of transfection with NuMA siRNA. Cells were stained with anti-NuMA antibody 1F1 (red) (a and b) or anti-tankyrase 1 antibody 609 (TNKS1; green) (a′ and b′). DAPI staining of the DNA is in blue. Bar=5 μm. (D) Immunoblot analysis of HeLa.I.2.11 cells mock transfected (−) or transfected with NuMA siRNA (+) for 48 h. Cell extracts were generated in buffer C and proteins were detected by immunoblotting with anti-NuMA, anti-tankyrase 1 465 (TNKS1) or anti-α-tubulin antibodies.
RESULTS
Tankyrase 1 is phosphorylated in mitosis
In order to investigate a role for tankyrase 1 specifically in mitosis, we characterized the state of the protein across the cell cycle. To generate staged extracts across mitosis, HeLaI.2.11 cells were released from an S phase double thymidine block into nocodazole for 12 h. At this time, mitotic cells were collected by shake-off and released out of nocodazole for 3 h. The cell cycle stage was determined by FACS analysis (Figure 1A) and cell extracts were examined by immunoblotting (Figure 1B). Upon entry into mitosis, tankyrase 1 shifted to a slower-migrating form (Figure 1B, lane 5); this mobility shift was lost upon entry into G1 (Figure 1B, lane 10). The G2/M phase timing of the tankyrase 1 mobility shift was confirmed by comparison with the mitotic cyclins; tankyrase 1 mobility shifted just prior to cyclin A degradation and persisted until cyclin B degradation (Figure 1B). Together, these data indicate a cell-cycle-regulated change in tankyrase 1 in mitosis.
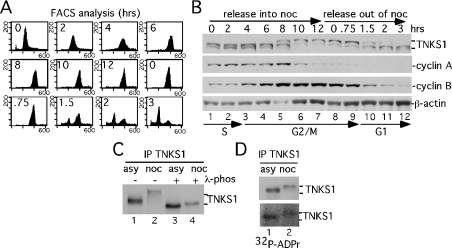
Figure 1. Tankyrase 1 is modified in mitosis.
(A, B) Analysis of staged cell cycle extracts shows a mitosis-specific mobility shift in tankyrase 1. Following arrest in S phase by a double thymidine block, HeLaI.2.11 cells were released into nocodazole for 12 h, collected by shake-off and released out of nocodazole for 3 h. (A) FACS analysis of staged extracts: y axis, cell numbers; x axis, relative DNA content based on propidium iodide staining. (B) Immunoblot analysis of staged cell extracts generated in buffer C. Products were detected with anti-tankyrase 1 antibody 465 (TNKS1), anti-cyclin A, anti-cyclin B or anti-β-actin antibodies. Cell cycle stages S, G2/M and G1 as determined by FACS (A) are indicated. (C) The tankyrase 1 mobility shift is due to phosphorylation. Extracts generated in TNE buffer from asynchronous (asy) or nocodazole (noc)-arrested cells were immunoprecipitated (IP) with anti-tankyrase 1 antibody 465, incubated with (+) or without (−) λ-phosphatase and products detected by immunoblotting with anti-tankyrase 1 465 (TNKS1). (D) Mitotic tankyrase 1 does not show an increase in autoPARsylation. Tankyrase 1 was immunoprecipitated as in (C) and incubated in a PARP assay with [32P]NAD+. Products were detected by immunoblotting with anti-tankyrase 1 antibody 465 (TNKS1; upper panel) or by autoradiography (32P-ADPr; lower panel).
To determine if the observed mobility shift reflected phosphorylation, tankyrase 1 was immunoprecipitated from cell extracts generated from asynchronous or nocodazole-arrested (mitotic) cells and incubated with λ-phosphatase. As shown in Figure 1(C), phosphatase treatment of mitotic tankyrase 1 eliminated the mobility shift (compare Figure 1C, lanes 2 and 4), indicating that tankyrase 1 was phosphorylated in mitosis. Interestingly, phosphatase treatment of asynchronous tankyrase 1 also resulted in a slight increase in tankyrase 1 mobility (compare Figure 1C, lanes 1 and 3), indicating that interphase tankyrase 1 was also phosphorylated to some extent.
The observation of a novel form of tankyrase 1 in mitosis raised the possibility that phosphorylation might influence its catalytic activity. To address this question, the ability of asynchronous compared with mitotic tankyrase 1 to undergo auto-ADP-ribosylation was determined. Tankyrase 1 was immunoprecipitated from asynchronous or mitotic cells and subjected to a PARP assay in vitro using [32P]NAD+ as a substrate. As shown in Figure 1(D), no significant difference in tankyrase 1 automodification was observed, suggesting that phosphorylation at mitosis did not dramatically alter the catalytic activity of tankyrase 1 in vitro. Nonetheless, phosphorylation of tankyrase 1 could influence its binding partners and potential acceptors of ADP-ribosylation.
A novel acceptor of PARsylation by tankyrase 1 in mitosis
The observation of a novel form of tankyrase 1 in mitosis raised the possibility that phosphorylated tankyrase 1 might have unique binding partners that could be acceptors of ADP-ribosylation. To identify acceptors of tankyrase 1-mediated ADP-ribosylation in mitosis, extracts derived from nocodazole-arrested cells were immunoprecipitated with anti-tankyrase 1 or control antibody and then subjected to a PARP assay in vitro using [32P]NAD+ as a substrate. In addition to tankyrase 1, a 32P-labelled protein, which migrated with an apparent molecular mass greater than 200 kDa, specifically co-immunoprecipitated with anti-tankyrase 1, but not control IgG (Figure 2A, lane 2, lower panel). In addition, a 32P-labelled protein of approx. 116 kDa was co-immunoprecipitated non-specifically (i.e. with either control or anti-tankyrase 1 antibody; Figure 2B, lanes 1 and 2, lower panel). This non-specific signal probably resulted from contaminating automodified PARP1 [39], which is a very abundant PARP that migrates at 116 kDa. However, as detailed below, modification of the novel acceptor is not due to PARP1.
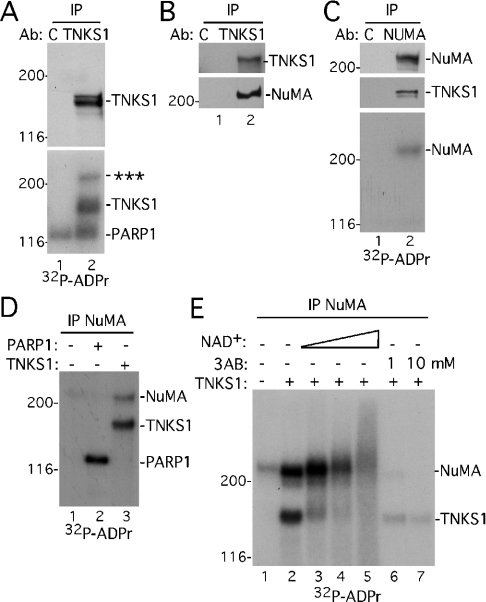
Figure 2. A novel acceptor of PAR from tankyrase 1 in mitosis.
(A) A novel acceptor of PAR co-immunoprecipitates with tankyrase 1. Extracts generated in TNE buffer from nocodazole-arrested HeLa.I.2.11 cells were immunoprecipitated (IP) with control IgG (C) or anti-tankyrase 1 antibody (Ab) 465 (TNKS1), and incubated in a PARP assay with [32P]NAD+. Products were detected by immunoblotting with anti-tankyrase 1 antibody 465 (TNKS1; upper panel) or by autoradiography (32P-ADPr; lower panel); *** indicates the novel acceptor. (B) NuMA co-immunoprecipitates with tankyrase 1. Extracts were immunoprecipitated as in (A) and products detected by immunoblotting with anti-tankyrase 1 antibody 465 or anti-NuMA. (C) NuMA is PARsylated. Extracts generated in TNE buffer from nocodazole-arrested cells were immunoprecipitated with control serum (C) or anti-NuMA and incubated in a PARP assay with [32P]NAD+. Products were detected by immunoblotting with anti-NuMA (top panel) or anti-tankyrase 1 antibody 465 (TNKS1; middle panel) or by autoradiography (32P-ADPr; bottom panel). (D) NuMA is PARsylated by recombinant tankyrase 1, not PARP1. NuMA was immunoprecipitated as in (C) and incubated in a PARP assay with [32P]NAD+ in the absence (−) or presence (+) of 2.5 μg of recombinant PARP1 or TNKS1. Products were detected by autoradiography. (E) PARsylation of NuMA is stimulated by NAD+ and inhibited by 3AB. NuMA was immunoprecipitated as in (C) and incubated in a PARP assay with [32P]NAD+ in the absence (−) or presence (+) of 2.5 μg of recombinant TNKS1 and increasing amounts of unlabelled NAD+ (0.04, 0.2 or 1 mM) or 3AB (1 or 10 mM). Products were detected by autoradiography.
We sought to pursue the novel high-molecular-mass ADP-ribosylated protein. One likely candidate for a tankyrase 1-interacting protein of >200 kDa was NuMA (∼240 kDa). Previous studies indicated that a C-terminal fragment of NuMA could interact with tankyrase 1 by co-immunoprecipitation of transfected proteins [18]. To determine if NuMA was the acceptor protein, tankyrase 1 immunoprecipitates from nocodazole-arrested cells were analysed by immunoblotting with anti-NuMA antibody. As shown in Figure 2(B), NuMA specifically co-immunoprecipitated with tankyrase 1. Conversely, immunoprecipitation of NuMA resulted in tankyrase 1 co-immunoprecipitation (Figure 2C). Moreover, when NuMA immunoprecipitates were subjected to a PARP assay in vitro, 32P-labelled NuMA was detected (Figure 2C, lane 2, bottom panel), indicating that NuMA was ADP-ribosylated. In this case 32P-labelled tankyrase was not detected (Figure 2C, lane 2), probably due to reduced amounts of tankyrase 1 in the co-immunoprecipitate compared with direct immunoprecipitation of tankyrase 1 (Figure 2A, lane 2).
Recombinant proteins were used to confirm that it was tankyrase 1, and not the contaminating PARP1, that was responsible for ADP-ribosylation of NuMA. NuMA was immunoprecipitated from nocodazole-arrested cells and subjected to an in vitro PARP assay in the absence or presence of recombinant tankyrase 1 or PARP-1. As shown in Figure 2(D), addition of recombinant tankyrase 1 stimulated ADP-ribosylation of NuMA (compare lanes 1 and 3), whereas addition of recombinant PARP1 had no effect (compare lanes 1 and 2). To verify that tankyrase 1-mediated modification of NuMA was PARsylation, and not mono(ADP-ribosyl)ation, the in vitro PARP assay was performed in the presence of increasing amounts of the substrate, NAD+. In the presence of increasing concentrations of NAD+, both 32P-labelled tankyrase 1 and NuMA were converted into slower migrating diffuse bands, indicative of PARsylation (Figure 2E, lanes 3–5). Modification of tankyrase 1 and NuMA was inhibited by the PARP inhibitor 3AB (Figure 2E, lanes 6 and 7). Together, these data indicate that NuMA is PARsylated by tankyrase 1 in vitro.
NuMA is PARsylated in vivo in mitosis
Our studies thus far indicated that NuMA and tankyrase 1 form a complex in mitotic cells. To determine if this association also occurred in interphase, we compared nocodazole-arrested mitotic with asynchronous cells. As shown in Figure 3(A), immunoblot analysis indicated similar levels of tankyrase 1 and NuMA in asynchronous (lane 1) and mitotic (lane 2) cell extracts. Immunoprecipitation with NuMA followed by immunoblot analysis with anti-tankyrase 1 indicated that the proteins were complexed in extracts from asynchronous (lane 4) as well as mitotic (lane 6) cells. We next probed the blots with antibodies against PAR. This type of analysis differs from the in vitro PARP assay with [32P]NAD+ in that it can detect PARsylations that occur in vivo. Analysis of whole-cell extracts with anti-PAR revealed a protein of >200 kDa (similar to NuMA) in mitotic (Figure 3A, lane 2, bottom panel) but not asynchronous (lane 1) extracts, as well as a 116 kDa protein (probably PARP1). Immunoblot analysis of the NuMA immunoprecipitates with anti-PAR indicated that the high-molecular-mass protein was indeed PAR–NuMA (Figure 3A, lane 6, bottom panel). Interestingly, despite the presence of similar amounts of NuMA and tankyrase 1 in the immunoprecipitates from asynchronous or mitotic cells (Figure 3A, lanes 4 and 6, top panel), PAR–NuMA was significantly increased in mitotic cells (compare lanes 4 and 6, bottom panel).
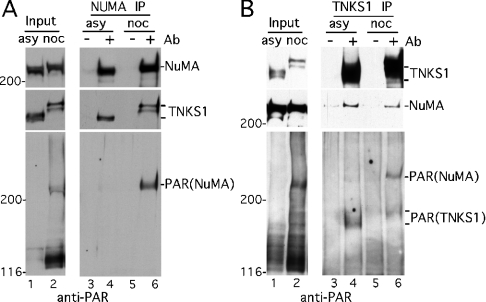
Figure 3. NuMA is PARsylated in mitotic cells.
(A) NuMA immunoprecipitated from mitotic cells is PARsylated. Extracts generated in buffer C from asynchronous (asy) or nocodazole (noc)-arrested HeLaI.2.11 cells were analysed directly (input) or immunoprecipitated (IP) with control serum (−) or anti-NuMA antibodies (Ab) (+). Products were detected by immunoblotting with anti-NuMA, anti-tankyrase 1 465 (TNKS1) or anti-PAR antibodies. (B) Tankyrase 1 immunoprecipitates from mitotic cells contain PARsylated NuMA. Extracts generated as in (A) were analysed directly (Input) or immunoprecipitated with control IgG (−) or anti-tankyrase 1 antibody 465 (+). Products were detected by immunoblotting with anti-tankyrase 1 465 (TNKS1), anti-NuMA or anti-PAR.
When cell extracts were analysed by immunoprecipitation with anti-tankyrase 1 antibodies followed by immunoblotting with anti-PAR, PAR–tankyrase 1 was detected in immunoprecipitates from either asynchronous or mitotic extracts (Figure 3B, lanes 4 and 6, bottom panel). In contrast, although NuMA was present in similar amounts in these immunoprecipitates, PAR–NuMA was (again) considerably increased in mitotic cells (Figure 3B, compare lanes 4 and 6). Together these studies suggest that NuMA is a major acceptor of PARsylation in mitosis.
Tankyrase 1 associates with NuMA at the onset of mitosis
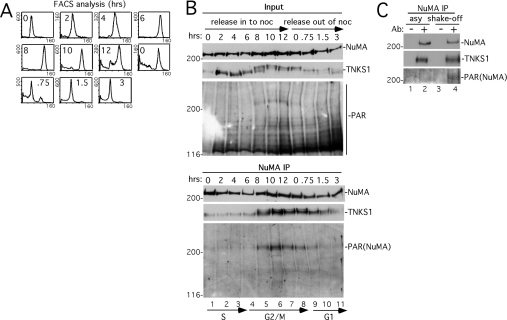
To delineate further precisely when tankyrase 1 and NuMA interact, their association was analysed across the cell cycle. HeLaI.2.11 cells were synchronized as in Figure 1(A) and analysed by FACS (Figure 4A). Immunoblot analysis of whole-cell extracts (input) indicated that NuMA and tankyrase 1 levels were constant through S and G2/M phases (Figure 4B). In contrast, when the blot was probed with anti-PAR, a band that migrated at the same molecular mass as NuMA (>200 kDa) was restricted to the G2/M phase of the cell cycle (Figure 4B, lanes 5–7). Cell extracts were immunoprecipitated with anti-NuMA antibodies followed by immunoblotting with antibodies against NuMA, tankyrase 1 or PAR. As shown in Figure 4(B), while the NuMA immunoprecipitates contained similar amounts of NuMA across the cell cycle, tankyrase 1 and PAR–NuMA were dramatically increased in G2/M (lanes 5–8). These findings suggested that NuMA was PARsylated in a cell cycle-dependent manner by tankyrase 1 at G2/M.
Figure 4. Tankyrase 1 associates with and PARsylates NuMA at the onset of mitosis.
(A, B) Analysis of staged cell cycle extracts. Following arrest in S phase by a double thymidine block, HeLaI.2.11 cells were released into nocodazole for 12 h, collected by shake-off and released out of nocodazole for 3 h. (A) FACS analysis of staged extracts: y axis, cell numbers; x axis, relative DNA content based on propidium iodide staining. (B) Staged cell extracts generated in buffer C were analysed directly (Input) or immunoprecipitated (IP) with anti-NuMA. Products were detected by immunoblotting with anti-NuMA, anti-tankyrase 1 antibody 465 (TNKS1) or anti-PAR. Cell cycle stages S, G2/M and G1 as determined by FACS as in (A) are indicated. (C) PARsylation of NuMA does not depend on nocodazole. Extracts generated in buffer C from asynchronous (asy) or from mitotic shake-off 8–9 h after release from a double thymidine block (shake-off) were immunoprecipitated with control serum (−) or anti-NuMA antibodies (Ab) (+). Products were detected by immunoblotting with anti-NuMA, anti-tankyrase 1 antibody 465 (TNKS1) or anti-PAR.
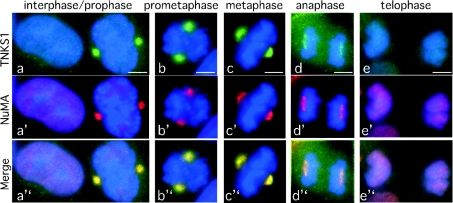
In the experiments described thus far, nocodazole was used to increase the number of cells that accumulated in mitosis. To ensure that the observed modification was not dependent on nocodazole-induced arrest, cells were isolated in mitosis by mitotic shake-off. Thus cells were released synchronously in S phase from a double thymidine block, and then rounded cells were isolated by mitotic shake-off after 8–9 h in culture. PAR–NuMA was significantly increased in the mitotic cells (Figure 4C, lane 4), indicating that the modification did not depend on nocodazole. Finally, immunofluorescence analysis was used to determine the timing of the association between NuMA and tankyrase 1 in an asynchronously growing population of cells. The earliest time that co-localization between NuMA and tankyrase 1 was observed was in prophase (Figure 5a). This co-localization persisted during prometaphase (Figure 5b), metaphase (Figure 5c) and anaphase (Figure 5d), but was then lost in telophase (Figure 5e) as both proteins were no longer detected at centrosomes.
Figure 5. Immunofluorescence analysis indicates co-localization of NuMA and tankyrase 1 from prophase through to anaphase.
Cycling cells were methanol-fixed and stained with anti-tankyrase 1 antibody 609 (green) and anti-NuMA antibody 1F1 (red). DAPI staining of the DNA is blue. Co-localization is indicated as yellow in the merged images. Cell cycle stage is indicated at the top. Bar=5 μm.
Knockdown of tankyrase 1 eliminates PARsylation of NuMA, but not its localization to spindle poles
To determine if NuMA depends on tankyrase 1 for its localization to spindle poles, tankyrase 1 expression was knocked down using siRNA and the cells were subjected to immunofluorescence analysis. In the absence of tankyrase 1, NuMA localized to spindle poles (Figure 6A, panels b and b′). To confirm that NuMA was no longer PARsylated in the absence of tankyrase 1, nocodazole-arrested tankyrase 1 siRNA cells were immunoprecipitated with anti-NuMA antibody and analysed by immunoblotting with anti-PAR. In the absence of tankyrase 1, NuMA was no longer PARsylated (Figure 6B, compare lanes 2 and 3). These data confirm tankyrase 1 as the PARP responsible for PARsylation of NuMA in vivo and, furthermore, indicate that PARsylation of NuMA is not required for its localization to spindle poles. To determine if tankyrase 1 depends on NuMA for its localization to spindle poles, NuMA expression was knocked down by siRNA and the cells were subjected to immunofluorescence analysis. Knockdown of NuMA resulted in loss of tankyrase 1 from spindle poles (Figure 6C, panels b and b′). Note that whereas NuMA siRNA resulted in only a partial knockdown of NuMA at spindle poles, it induced a complete loss of tankyrase 1 at the poles. Indeed 80 out of 100 metaphases from NuMA siRNA-treated cells lacked tankyrase 1 at spindle poles. By contrast, 0 out of 100 metaphases from mock siRNA-treated cells showed loss of tankyrase 1 at spindle poles. Immunoblot analysis of NuMA siRNA cells showed that levels of NuMA were decreased, whereas tankyrase 1 was unaffected (Figure 6D). These data indicate that NuMA is required for localization of tankyrase 1 to spindle poles in mitosis.
DISCUSSION
Previous studies indicated that PARP-active tankyrase 1 was required for the separation of sister telomeres and mitotic progression [4]. Our finding that NuMA is a major acceptor of PARsylation by tankyrase 1 specifically in mitosis raises the possibility that this modification could play a role in sister telomere resolution and/or mitotic progression.
Tankyrase 1 in mitosis
We demonstrate that tankyrase 1 is quantitatively phosphorylated in mitosis. A previous study indicated that insulin-stimulated phosphorylation of tankyrase 1 increased its PARP activity [13]. We did not detect a reproducible significant increase in the PARP activity of tankyrase 1 at mitosis, as measured by automodification in vitro (Figure 1D) or by detection with anti-PAR antibodies (Figure 3B). Thus the phosphorylated mitotic tankyrase 1 observed here may differ from the insulin-stimulated form. Phosphorylation at mitosis could influence the subcellular localization and/or binding partners of tankyrase 1. For example, phosphorylation could be required to release tankyrase 1 from its interphase binding partners (which may reside in the Golgi or nuclear pore complex) and allow its association with mitotic binding partners at telomeres or spindle poles.
Phosphorylation does not appear to be required for the association of tankyrase 1 with NuMA, since NuMA can be co-immunoprecipitated with unmodified tankyrase 1 from either asynchronous or mitotic cell extracts (Figure 3). Interestingly, however, anti-PAR detection of NuMA immunoisolated from asynchronous cells was greatly reduced compared with that from mitotic cells, despite the presence of tankyrase 1 in the complex (Figure 3). One possibility is that the tankyrase 1–NuMA complex detected in asynchronous cell extracts in Figure 3 may not actually be formed in vivo, but rather only after cell lysis. Indeed, analysis of staged cell cycle extracts indicated a dramatic increase at G2/M in the association of tankyrase 1 with NuMA and a concomitant increase in the PARsylation of NuMA (Figure 4B).
Tankyrase 1 PARsylates NuMA in mitosis and co-localizes with NuMA to spindle poles. However, in the absence of tankyrase 1 and NuMA PARsylation, NuMA still localizes to spindle poles (Figures 6A and 6B), probably due to its association with tubulin and other components of the mitotic spindle. Thus PARsylation per se is not required for localization of NuMA to spindle poles. Nonetheless, this modification may influence NuMA's association with other proteins and its function during mitosis. By contrast, siRNA knockdown of NuMA resulted in loss of tankyrase 1 at spindle poles (Figure 6C), indicating that localization of tankyrase 1 to spindle poles depends on NuMA.
PARsylation of NuMA in the nuclear matrix
During interphase, NuMA resides in the nucleus, as a filamentous component of the nuclear matrix [33,40–44]. The nuclear matrix is the insoluble non-chromatin structure that remains after detergent extraction with DNase and high salt. The matrix may serve as a scaffold to organize nuclear components and to compartmentalize nuclear functions. Interestingly, biochemical studies indicate that telomeric DNA and the telomeric DNA binding protein TRF1 associate with the nuclear matrix [45,46]. Ultrasructural analysis indicates that in human cells telomeres exist as matrix-associated higher-order complexes dispersed throughout the nucleus [46]. One interesting possibility is that PARsylation of NuMA by tankyrase 1 could influence the association of telomeres with the nuclear matrix. For example, in late G2 or early prophase, it may be necessary to release telomeres from a nuclear scaffold to allow resolution of sister telomere cohesion.
Tankyrase 1, NuMA, sister telomere cohesion, and mitotic progression
In the absence of tankyrase 1, sister telomeres fail to separate and cells arrest in anaphase. Rescue of this phenotype required a PARP-active tankyrase 1 [4], prompting a search for acceptors of PAR by tankyrase 1 in mitosis. We show here that NuMA is the main acceptor of PARsylation by tankyrase 1 in mitosis, thereby begging the question of whether PARsylation of NuMA is required for sister telomere resolution and/or mitotic progression. Here it is worth mentioning that while it is clear that sister telomeres do not separate in the absence of tankyrase 1, what is less clear is why tankyrase 1 siRNA cells arrest specifically in anaphase. Unseparated telomeres may serve as a physical block to chromosome segregation or act as a signal to a checkpoint for anaphase arrest. Alternatively, tankyrase 1 may act at two stages: first, in late G2/early prophase to separate telomeres, and secondly, in anaphase to promote exit from mitosis. NuMA could play a role at either or both of these stages. Thus PARsylation of NuMA may (as described above) release telomeres from the nuclear matrix in G2/early prophase to allow dissociation of sister telomeres. Alternatively, or in addition, TRF1 could be the critical acceptor of PARsylation in this first step. Regardless, at the second step PARsylation of NuMA may be required for anaphase exit. A recent study showed that prolonged stability of spindle poles in anaphase correlated with NuMA binding to the poles [47]. Although highly speculative, perhaps PARsylation of NuMA is necessary for dissociation of NuMA from the poles and spindle disassembly. Interestingly, recent studies in Saccharomyces cerevisiae indicate that completion of the cell cycle requires co-ordination of chromosome segregation with mitotic spindle disassembly, and that this is achieved though a single protein, the CDC14 phosphatase [48,49]. Tankyrase 1 could play a similar role in mammalian cells, co-ordinating sister telomere separation with mitotic exit.
PAR and spindle function
NuMA is a large multifaceted protein. It has an intrinsic ability to self assemble into fibrous structures [50] and it interacts with a number of essential mitotic components, including microtubules [51,52], dynein/dynactin [34] and the mammalian Pins homologue, LGN [53]. Thus NuMA is likely to have multiple functions in mitosis. PARsylation of NuMA may influence the structural integrity of the poles and/or essential protein interactions required for spindle function. Interestingly, a recent study demonstrated a requirement for PAR in mitotic spindle assembly and structure [54]. Our identification of NuMA as an acceptor of PARsylation in mitosis provides a candidate for a critical acceptor of PAR in the mitotic spindle.
Acknowledgments
We thank Guy Poirier for anti-PAR antibodies, and Tom Meier and members of the Smith lab for comments on the manuscript. J. N. D. was supported by NIH Predoctoral Training Program in Cell and Molecular Biology (GM07238). This work was supported by a grant from the NIH (RO1 CA95099).
References
- 1.Smith S., Giriat I., Schmitt A., de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 2.Cook B. D., Dynek J. N., Chang W., Shostak G., Smith S. Role for the related poly(ADP-ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol. Cell. Biol. 2002;22:332–342. doi: 10.1128/MCB.22.1.332-342.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith S., de Lange T. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 2000;10:1299–1302. doi: 10.1016/s0960-9822(00)00752-1. [DOI] [PubMed] [Google Scholar]
- 4.Dynek J. N., Smith S. Resolution of sister telomere association is required for progression through mitosis. Science. 2004;304:97–100. doi: 10.1126/science.1094754. [DOI] [PubMed] [Google Scholar]
- 5.de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 6.Smorgorzewska A., de Lange T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 7.Greider C. W., Blackburn E. H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 8.Chong L., van Steensel B., Broccoli D., Erdjument-Bromage H., Hanish J., Tempst P., de Lange T. A human telomeric protein. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 9.Ancelin K., Brunori M., Bauwens S., Koering C. E., Brun C., Ricoul M., Pommier J. P., Sabatier L., Gilson E. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol. Cell. Biol. 2002;22:3474–3487. doi: 10.1128/MCB.22.10.3474-3487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Steensel B., de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature (London) 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 11.Chang W., Dynek J. N., Smith S. TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev. 2003;17:1328–1333. doi: 10.1101/gad.1077103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith S., de Lange T. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J. Cell Sci. 1999;112:3649–3656. doi: 10.1242/jcs.112.21.3649. [DOI] [PubMed] [Google Scholar]
- 13.Chi N. W., Lodish H. F. Tankyrase is a Golgi-associated MAP kinase substrate that interacts with IRAP in GLUT4 vesicles. J. Biol. Chem. 2000;275:38437–38444. doi: 10.1074/jbc.M007635200. [DOI] [PubMed] [Google Scholar]
- 14.Kaminker P. G., Kim S. H., Taylor R. D., Zebarjadian Y., Funk W. D., Morin G. B., Yaswen P., Campisi J. TANK2, a new TRF1-associated poly(ADP-ribose) polymerase, causes rapid induction of cell death upon overexpression. J. Biol. Chem. 2001;276:35891–35899. doi: 10.1074/jbc.M105968200. [DOI] [PubMed] [Google Scholar]
- 15.Kuimov A. N., Kuprash D. V., Petrov V. N., Vdovichenko K. K., Scanlan M. J., Jongeneel C. V., Lagarkova M. A., Nedospasov S. A. Cloning and characterization of TNKL, a member of tankyrase gene family. Genes Immun. 2001;2:52–55. doi: 10.1038/sj.gene.6363722. [DOI] [PubMed] [Google Scholar]
- 16.Monz D., Munnia A., Comtesse N., Fischer U., Steudel W. I., Feiden W., Glass B., Meese E. U. Novel tankyrase-related gene detected with meningioma-specific sera. Clin. Cancer Res. 2001;7:113–119. [PubMed] [Google Scholar]
- 17.Lyons R. J., Deane R., Lynch D. K., Ye Z. S., Sanderson G. M., Eyre H. J., Sutherland G. R., Daly R. J. Identification of a novel human tankyrase through its interaction with the adapter protein Grb14. J. Biol. Chem. 2001;276:17172–17180. doi: 10.1074/jbc.M009756200. [DOI] [PubMed] [Google Scholar]
- 18.Sbodio J. I., Chi N. W. Identification of a tankyrase-binding motif shared by IRAP, TAB182, and human TRF1 but not mouse TRF1. NuMA contains this RXXPDG motif and is a novel tankyrase partner. J. Biol. Chem. 2002;277:31887–31892. doi: 10.1074/jbc.M203916200. [DOI] [PubMed] [Google Scholar]
- 19.Seimiya H., Smith S. The telomeric poly(ADP-ribose) polymerase, tankyrase 1, contains multiple binding sites for telomeric repeat binding factor 1 (TRF1) and a novel acceptor, 182-kDa tankyrase-binding protein (TAB182) J. Biol. Chem. 2002;277:14116–14126. doi: 10.1074/jbc.M112266200. [DOI] [PubMed] [Google Scholar]
- 20.Bae J., Donigian J. R., Hsueh A. J. Tankyrase 1 interacts with Mcl-1 proteins and inhibits their regulation of apoptosis. J. Biol. Chem. 2003;278:5195–5204. doi: 10.1074/jbc.M201988200. [DOI] [PubMed] [Google Scholar]
- 21.Mosavi L. K., Cammett T. J., Desrosiers D. C., Peng Z. Y. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim C. A., Bowie J. U. SAM domains: uniform structure, diversity of function. Trends Biochem. Sci. 2003;28:625–628. doi: 10.1016/j.tibs.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 23.De Rycker M., Price C. M. Tankyrase polymerization is controlled by its sterile alpha motif and poly(ADP-ribose) polymerase domains. Mol. Cell. Biol. 2004;24:9802–9812. doi: 10.1128/MCB.24.22.9802-9812.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Rycker M., Venkatesan R. N., Wei C., Price C. M. Vertebrate tankyrase domain structure and sterile alpha motif (SAM)-mediated multimerization. Biochem. J. 2003;372:87–96. doi: 10.1042/BJ20021450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sbodio J. I., Lodish H. F., Chi N. W. Tankyrase-2 oligomerizes with tankyrase-1 and binds to both TRF1 (telomere-repeat-binding factor 1) and IRAP (insulin-responsive aminopeptidase) Biochem. J. 2002;361:451–459. doi: 10.1042/0264-6021:3610451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ame J. C., Spenlehauer C., de Murcia G. The PARP superfamily. BioEssays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 27.Smith S. The world according to PARP. Trends Biochem. Sci. 2001;26:174–179. doi: 10.1016/s0968-0004(00)01780-1. [DOI] [PubMed] [Google Scholar]
- 28.Cleveland D. W. NuMA: a protein involved in nuclear structure, spindle assembly, and nuclear re-formation. Trends Cell Biol. 1995;5:60–64. doi: 10.1016/s0962-8924(00)88947-3. [DOI] [PubMed] [Google Scholar]
- 29.Compton D. A., Cleveland D. W. NuMA, a nuclear protein involved in mitosis and nuclear reformation. Curr. Opin. Cell Biol. 1994;6:343–346. doi: 10.1016/0955-0674(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 30.Fant X., Merdes A., Haren L. Cell and molecular biology of spindle poles and NuMA. Int. Rev. Cytol. 2004;238:1–57. doi: 10.1016/S0074-7696(04)38001-0. [DOI] [PubMed] [Google Scholar]
- 31.Lydersen B. K., Pettijohn D. E. Human-specific nuclear protein that associates with the polar region of the mitotic apparatus: distribution in a human/hamster hybrid cell. Cell. 1980;22:489–499. doi: 10.1016/0092-8674(80)90359-1. [DOI] [PubMed] [Google Scholar]
- 32.Gaglio T., Saredi A., Compton D. A. NuMA is required for the organization of microtubules into aster-like mitotic arrays. J. Cell Biol. 1995;131:693–708. doi: 10.1083/jcb.131.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kallajoki M., Weber K., Osborn M. A 210 kDa nuclear matrix protein is a functional part of the mitotic spindle; a microinjection study using SPN monoclonal antibodies. EMBO J. 1991;10:3351–3362. doi: 10.1002/j.1460-2075.1991.tb04899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merdes A., Ramyar K., Vechio J. D., Cleveland D. W. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- 35.Yang C. H., Snyder M. The nuclear-mitotic apparatus protein is important in the establishment and maintenance of the bipolar mitotic spindle apparatus. Mol. Biol. Cell. 1992;3:1259–1267. doi: 10.1091/mbc.3.11.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Steensel B., Smogorzewska A., de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 37.Elbashir S. M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature (London) 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 38.Compton D. A., Yen T. J., Cleveland D. W. Identification of novel centromere/kinetochore-associated proteins using monoclonal antibodies generated against human mitotic chromosome scaffolds. J. Cell Biol. 1991;112:1083–1097. doi: 10.1083/jcb.112.6.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D'Amours D., Desnoyers S., D'silva I., Poirier G. G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 40.Barboro P., D'Arrigo C., Diaspro A., Mormino M., Alberti I., Parodi S., Patrone E., Balbi C. Unraveling the organization of the internal nuclear matrix: RNA-dependent anchoring of NuMA to a lamin scaffold. Exp. Cell Res. 2002;279:202–218. doi: 10.1006/excr.2002.5605. [DOI] [PubMed] [Google Scholar]
- 41.Gueth-Hallonet C., Wang J., Harborth J., Weber K., Osborn M. Induction of a regular nuclear lattice by overexpression of NuMA. Exp. Cell Res. 1998;243:434–452. doi: 10.1006/excr.1998.4178. [DOI] [PubMed] [Google Scholar]
- 42.Harborth J., Weber K., Osborn M. Epitope mapping and direct visualization of the parallel, in-register arrangement of the double-stranded coiled-coil in the NuMA protein. EMBO J. 1995;14:2447–2460. doi: 10.1002/j.1460-2075.1995.tb07242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luderus M. E., den Blaauwen J. L., de Smit O. J., Compton D. A., van Driel R. Binding of matrix attachment regions to lamin polymers involves single-stranded regions and the minor groove. Mol. Cell. Biol. 1994;14:6297–6305. doi: 10.1128/mcb.14.9.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng C., He D., Brinkley B. R. Localization of NuMA protein isoforms in the nuclear matrix of mammalian cells. Cell Motil. Cytoskeleton. 1994;29:167–176. doi: 10.1002/cm.970290208. [DOI] [PubMed] [Google Scholar]
- 45.de Lange T. Human telomeres are attached to the nuclear matrix. EMBO J. 1992;11:717–724. doi: 10.1002/j.1460-2075.1992.tb05104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luderus M. E., van Steensel B., Chong L., Sibon O. C., Cremers F. F., de Lange T. Structure, subnuclear distribution, and nuclear matrix association of the mammalian telomeric complex. J. Cell Biol. 1996;135:867–881. doi: 10.1083/jcb.135.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gehmlich K., Haren L., Merdes A. Cyclin B degradation leads to NuMA release from dynein/dynactin and from spindle poles. EMBO Rep. 2004;5:97–103. doi: 10.1038/sj.embor.7400046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.D'Amours D., Stegmeier F., Amon A. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell. 2004;117:455–469. doi: 10.1016/s0092-8674(04)00413-1. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan M., Higuchi T., Katis V. L., Uhlmann F. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell. 2004;117:471–482. doi: 10.1016/s0092-8674(04)00415-5. [DOI] [PubMed] [Google Scholar]
- 50.Harborth J., Wang J., Gueth-Hallonet C., Weber K., Osborn M. Self assembly of NuMA: multiarm oligomers as structural units of a nuclear lattice. EMBO J. 1999;18:1689–1700. doi: 10.1093/emboj/18.6.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du Q., Taylor L., Compton D. A., Macara I. G. LGN blocks the ability of NuMA to bind and stabilize microtubules. A mechanism for mitotic spindle assembly regulation. Curr. Biol. 2002;12:1928–1933. doi: 10.1016/s0960-9822(02)01298-8. [DOI] [PubMed] [Google Scholar]
- 52.Haren L., Merdes A. Direct binding of NuMA to tubulin is mediated by a novel sequence motif in the tail domain that bundles and stabilizes microtubules. J. Cell Sci. 2002;115:1815–1824. doi: 10.1242/jcs.115.9.1815. [DOI] [PubMed] [Google Scholar]
- 53.Du Q., Stukenberg P. T., Macara I. G. A mammalian Partner of inscuteable binds NuMA and regulates mitotic spindle organization. Nat. Cell Biol. 2001;3:1069–1075. doi: 10.1038/ncb1201-1069. [DOI] [PubMed] [Google Scholar]
- 54.Chang P., Jacobson M. K., Mitchison T. J. Poly(ADP-ribose) is required for spindle assembly and structure. Nature (London) 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- 55.Gueth-Hallonet C., Weber K., Osborn M. Cleavage of the nuclear matrix protein NuMA during apoptosis. Exp. Cell Res. 1997;233:21–24. doi: 10.1006/excr.1997.3557. [DOI] [PubMed] [Google Scholar]
- 56.Hsu H. L., Yeh N. H. Dynamic changes of NuMA during the cell cycle and possible appearance of a truncated form of NuMA during apoptosis. J. Cell Sci. 1996;109:277–288. doi: 10.1242/jcs.109.2.277. [DOI] [PubMed] [Google Scholar]