Abstract
Temperature is a primary environmental stress to which micro-organisms must be able to adapt and respond rapidly. Whereas some bacteria are restricted to specific niches and have limited abilities to survive changes in their environment, others, such as members of the Enterobacteriaceae, can withstand wide fluctuations in temperature. In addition to regulating cellular physiology, pathogenic bacteria use temperature as a cue for activating virulence gene expression. This work confirms that the nucleoid-associated protein H-NS (histone-like nucleoid structuring protein) is an essential component in thermoregulation of Salmonella. On increasing the temperature from 25 to 37 °C, more than 200 genes from Salmonella enterica serovar Typhimurium showed H-NS-dependent up-regulation. The thermal activation of gene expression is extremely rapid and change in temperature affects the DNA-binding properties of H-NS. The reduction in gene repression brought about by the increase in temperature is concomitant with a conformational change in the protein, resulting in the decrease in size of high-order oligomers and the appearance of increasing concentrations of discrete dimers of H-NS. The present study addresses one of the key complex mechanisms by which H-NS regulates gene expression.
Keywords: bacterial chromatin, conformational change, DNA microarray, histone-like nucleoid structuring protein (H-NS), oligomerization, temperature regulation
Abbreviations: AUC, analytical ultracentrifugation; H-NS, histone-like nucleoid structuring protein; ITC, isothermal titration calorimetry; LB, Luria–Bertani; RT, reverse transcriptase; SEC, size-exclusion chromatography; SPI-1, Salmonella pathogenicity island 1
INTRODUCTION
Pathogenic bacteria sense environmental stimuli by a range of mechanisms to ensure that virulence factors are only expressed in the correct location within the host [1]. The integrated response to diverse environmental stimuli is mediated via the bacterial nucleoid where chromosomal DNA is volumetrically constrained by interaction with a discrete set of bacterial proteins. The nucleoid is a highly dynamic structure and its topology responds to changes in the environment by means of alterations in the levels of DNA supercoiling and bending [1]. Temperature is a primary environmental factor determining the expression of many bacterial genes. Infection of the human host subjects enteric bacteria to a temperature shock, typically from ambient temperature to approx. 37 °C. Responses to this elevation of temperature include a significant increase in transcription of virulence genes [2] and in faster DNA replication, but does not include the classical heat-shock phenotype. These responses require a relaxation of DNA structure from the constraint of packaging proteins, allowing rapid access to the chromosome by key proteins involved in transcription [3].
The DNA of enteric bacteria is organized and compacted by nucleoid-associated proteins, of which H-NS (histone-like nucleoid structuring protein) is one of the major constituents [4–6]. In addition to its role in packaging DNA, H-NS exerts transcriptional control over a large number of unrelated genes in response to environmental stimuli [5,6]. Transcriptional profiling and proteomic studies of an Escherichia coli K-12 hns mutant indicated that approx. 5% of genes are under the control of this protein, many of which are linked to stress responses [5–8]. H-NS commonly behaves as a negative regulator of these genes [9–13]. This bifunctionality of H-NS requires a mechanism for efficient compaction of the bacterial DNA while simultaneously allowing for the rapid release of a defined set of genes from repression. Whether H-NS directly controls gene expression by binding to DNA and inhibiting the binding of RNA polymerase or by altering DNA topology remains the subject of debate [14,15].
The fundamental role of H-NS in the response of bacteria to temperature was first suggested for the human enteropathogen Shigella flexneri; transcription of the gene encoding the primary regulator of invasion functions, virF, is strictly temperature-dependent and is controlled by H-NS. Expression of the virF gene is prevented at low temperatures by H-NS-induced changes in the topology of the promoter region of DNA [16–19]. Increasing the temperature towards 37 °C decreases the ability of H-NS to bind to its target sequence at the virF promoter, enabling transcription to proceed [11]. Thermoregulation of the virF promoter has also been observed in the control of Yersinia enterocolitica virulence, which is also induced upon shifting to 37 °C [20].
H-NS is a small protein (136 amino acids) which consists of two functionally distinct domains: an N-terminal domain (contained within residues 1–89) involved in oligomerization and a C-terminal DNA-binding domain (approx. residues 91–136), linked via a short flexible region [6,21–23]. H-NS dimerizes through a coiled-coil interaction involving residues 22–49 of the N-terminal domain [24]. These dimers are then able to self-associate in a concentration-dependent manner [21,25–27], ultimately giving rise to high-order, polydisperse oligomeric states [23]. High-order self-association can be inhibited by truncation mutants lacking residues at the C-terminus [22,24,28,29] and the extreme N-terminus [24] of the oligomerization domain. This indicated that the oligomeric structure is based on a ‘head-to-tail’ interaction of the dimers [24]. H-NS1–89 self-associates in exactly the same way as the full-length protein to give high-order oligomers [23,30], but exhibits no DNA-binding capacity. Thus H-NS can be visualized as an extended protein core from which DNA-binding domains extend from flexible linkers. The ability of H-NS to dimerize, and then self-associate to form high-order oligomers, is essential for transcriptional regulation [22,30], as well as for chromosomal condensation [31].
This extended protein structure has multiple sites for oligonucleotide interaction, allowing H-NS to span extensive regions on bacterial chromosomal and plasmid DNA. At ambient temperatures, gel-retardation data reveal that the isolated C-terminal DNA-binding domain (H-NS91–136) binds to DNA with an affinity in the micromolar range [32]. However, these data have not been confirmed using other methods. The effects of binding of the C-terminal domain within the context of the full-length protein have been discussed, but not accurately quantified [33–35]. The structural effects of binding of H-NS oligomers to DNA have been demonstrated in scanning-force microscopy studies, which have revealed that, at sufficiently high protein concentrations, large regions of circular DNA (pUC19 plasmid) are constricted via lateral condensation [34]. The oligomeric state of H-NS at a given concentration is affected by temperature [27], which is likely to affect the formation of protein–DNA complexes. Force-extension measurements demonstrated that H-NS oligomerizes along extended tracts of DNA, resulting in a complex with an increased bending rigidity at higher protein concentrations [36,37]. Heating of the complex to 37 °C resulted in a loss of this rigidity and H-NS was found to be ineffective in protecting DNA from nuclease activity.
Two studies have reported the solution structure of the oligomerization domain of H-NS [24,38]. Although these two studies of domains with almost identical sequences report that the N-terminus exists as a homodimer which adopts a largely α-helical secondary structure consisting of two short helices (H1, residues 2–7; H2, residues 10–16) followed by a longer helix (H3, residues 22–49 [24]), there is a fundamental difference in the relative orientation of the helices. In one structure (NMR structure of residues 1–57 of H-NS from Salmonella typhimurium determined at 25 °C; PDB code 1LR1 [24]), H3 forms a parallel coiled-coil on to which the two N-terminal helices fold back. This arrangement is stabilized by a salt bridge between Arg14 and Glu23 and Glu26, and buries a significant hydrophobic surface area. The second structure (NMR structure of residues 1–46 of H-NS from E. coli at 30–37 °C; PDB code 1NI8 [38]) shows a novel fold in which residues of H2 and H3 form a U-shaped structure against which H1 lies perpendicularly. Residues 22–36 of H3 from each protomer form an antiparallel dimer interface from which the remaining ten C-terminal residues protrude. A similar structure was obtained from crystals of the H-NS homologue, VicH (residues 2–49), from Vibrio cholerae [39].
In the present study we demonstrate that H-NS is involved in the bacterial response to temperature shift. Using gene-expression profiling, we demonstrate that culturing S. typhimurium at 37 °C results in the transcription of a large number of genes that are repressed by H-NS in ambient conditions (25 °C). We propose a mechanism involving a conformational change of the H-NS dimer which enables the protein to respond rapidly to temperature. This effect results in a reduction in the oligomeric state of the protein. The resultant loss in co-operativity of binding of oligomeric H-NS facilitates dissociation from DNA at 37 °C, thus enabling transcription of temperature-regulated genes.
EXPERIMENTAL
Bacterial strains
The wild-type strain used in this study was S. typhimurium LT2 (designated LT2a, originally provided by Dr Bruce Ames, Molecular Microbiology Group, Institute of Food Research, Norwich, U.K.). Construction of the hns deletion mutant was performed as follows. PCR was used to amplify 415 bp and 620 bp DNA fragments, corresponding to the regions up- and down-stream of hns. The upstream fragment was amplified using primers sthns-F (5′-CACATTCCTTCACAAAGCATGCCG-3′) and stΔhns-R (5′-TAAGGCCTGTACGAATTCTGTAGTAATCTCAAACT-3′), and the downstream region with primers stΔhns-F (5′-CAGAATTCGTACAGGCCTTAATTTACTTCCTGGAT-3′) and sthns-R (5′-ACGAATATGGATCCGACCTGTCTC-3′). Primers stΔhns-R and stΔhns-F contained overlapping, complementary sequences that included a StuI restriction site. Following amplification, these two PCR products were linked together in a recombinant PCR reaction using primers sthns-F and sthns-R, and cloned into pUC18 [40] to form pMDG100. An 800 bp chloramphenicol-resistance cassette from Campylobacter coli (Genbank accession no. M35190) was cloned into the StuI site of pMDG100. The insert from pMDG100 was subcloned into the suicide vector pCVD442 to form pMDG101 and integrated into the chromosome of a spontaneously arising streptomycin-resistant derivative of LT2 by allelic replacement to form a merodiploid [41]. Prior to resolving-out the suicide plasmid, it was necessary to transform the merodiploid with a low-copy-number H-NS-expressing plasmid pLG703, based upon pLG339 [42]. pLG703 was constructed by cloning a 736 bp EcoRI–StuI fragment encoding hns from E. coli into the EcoRI–HincII sites of pLG339. After successfully removing the chromosomal copy of hns, pLG703 was subsequently cured by incubating the strain overnight in LB (Luria–Bertani) medium supplemented with 50 μg/ml novobiocin. The resulting hns deletion mutant was designated JH4000 (genotype, LT2 rpsL Δhns::cam). All bacterial cultures were grown in LB medium.
Microarray analysis
Duplicate 250 ml flasks containing 50-ml volumes of LB broth were inoculated with 1:100 vols of cultures grown overnight of LT2 or JH4000, and were shaken at 25 or 37 °C in a New Brunswick Innova 3100 water bath until they reached a D600 of 0.600. Aliquots (3.3 ml) of each culture were transferred into 50 ml polypropylene centrifuge tubes containing one-fifth vol. of 5% (v/v) phenol (pH 4.0) (Sigma–Aldrich)/95% (v/v) ethanol (BDH) and incubated on ice for ≥30 min. The RNA from each culture was isolated and purified as described by the Microarray Facility, Institute of Food Research, Norwich, U.K. (see http://www.ifr.ac.uk/safety/microarrays/protocols.html). The RNA was reverse-transcribed, labelled with Cy5-dCTP and co-hybridized with LT2 genomic DNA labelled with Cy3-dCTP to microarray slides containing 4418 open reading frames, representing 97% of the S. typhimurium LT2 genes (see http://www.ifr.ac.uk/safety/microarrays/protocols.html). Duplicate RNA samples from each culture and condition were hybridized to two arrays (i.e. four arrays/strain per condition). After hybridization, slides were washed twice for 10 min in each of the following successive washes: 2× SSC (where 1×SSC is 0.15 M NaCl/0.015 M sodium citrate), 0.1% SDS at 65 °C; 1×SSC at room temperature and 0.2× SSC at room temperature. The slides were dried by centrifugation and scanned on a GenePix 4000A scanner (Axon Instruments). The fluorescent intensities for each cDNA/genomic DNA spot were measured using GenePix Pro 3.0 software (Axon Instruments). All spots with a fluorescent intensity of less than 2 S.D. above background were excluded from further analysis. Inequalities in dye incorporation or template concentration were compensated by data centring, bringing the natural logarithm of the ratios for each group of spots printed by the same pin to zero. The complete data set is available as Supplementary Table S1 (http://www.BiochemJ.org/bj/391/bj3910203add.htm). Data passing the quality controls was analysed using GeneSpring 6.2 software (Silicon Genetics). Significance of the data at P<0.01 was measured using a parametric-based test, adjusting the individual P value with the Benjamini and Hochberg false-discovery rate multiple-test correction. Hierarchical clustering of gene expression profiles was undertaken for the 531 gene data set in GeneSpring 6.2, using the Pearson correlation.
Temperature-induction experiment
An overnight culture of LT2 was grown as described above to D600 of 0.600, and a 3.3 ml sample was taken for RNA extraction. The 25 °C culture was immediately transferred into a 37 °C water bath and samples taken at 1, 3, 6, 10, 15, 20, 30 and 60 min post-temperature up-shift for RNA isolation and attenuance measurements. The actual temperature transition of the cultures was measured using a temperature-logging device (Hanna Instruments). RNA was purified from each of the samples as described above. Quantitative real-time PCR was performed on the temperature-shift RNA samples and on a sample of RNA from LT2 grown at 37 °C and harvested at D600 of 0.600, and levels of hilA, hilC and hilD mRNA measured. The primers and probes used for the assays were designed using the Assays by Design service (Applied Biosystems). RT (reverse transcriptase)-PCR reactions were performed with the TaqMan One-Step RT-PCR kit (Applied Biosystems), according to the manufacturer's instructions. The primers and the FAM/TAMRA (carboxyfluorescein/carboxytetramethylrhodamine)-labelled probes for each assay were as follows: hilC-F, 5′-GGATGCTCGTATGAATCAGGCTAT-3′; hilC-R, 5′-GCGGTGTAATCTTAAAATGCCGTTTA-3′; hilC probe, 5′-GCGGTGTAATCTTAAAATGCCGTTTA-3′; hilD-F, 5′-GGCGGTACCCACAGAGAAAG-3′; hilD-R, 5′-AAATACCTCTCTTCTGGCAGGAAAG-3′; hilD probe, 5′-TCAGGCGTATAGAAGATC-3′; hilA-F, 5′-TGCTGCCGGTGACCATTA-3′; hilA-R, 5′-GCGTAATTGATCCATGAGCTCAAGA-3′; hilA probe, 5′-CTGCGGCAGTTCTT-3′. Reactions were performed on a TaqMan 7700 sequence detection system, incubating the samples at 48 °C for 30 min followed by a 10 min incubation period at 95 °C and 40 cycles of 15 s at 95 °C and 60 s at 60 °C. The threshold intensities for the temperature-shift samples were compared with a standard curve generated using doubling concentrations of RNA ranging from 0.5 to 8 ng, from LT2 grown at 37 °C and harvested at a culture at D600 of 0.600.
Competitive gel-retardation assays
H-NS protein from S. typhimurium was purified as described previously [23] and was used throughout the present study. A 2127 bp DNA fragment encoding hilC and the upstream gene STM2686 was amplified by PCR using primers hilC-F (5′-GGCGTCATTAAGCATGCTCTTGATG-3′) and hilC-R (5′-GCGACTACTGCGCAAGTAGATAAC-3′). The DNA was digested with DraI and purified using a Qiaquick PCR purification kit (Qiagen). In each assay, 0.5 μg of digested DNA was incubated for 20 min with 1× binding buffer [10 mM Tris/HCl, pH 7.5, 1 mM EDTA, 80 mM NaCl, 10 mM β-mercaptoethanol, 4% (v/v) glycerol and 0.01% (w/v) Bromophenol Blue] and purified H-NS in a 10 μl reaction volume to achieve final concentrations of 6.5, 13, 19.5, 26, 39 and 52 μM. The assays were incubated at 25 °C or 37 °C for 20 min and loaded on to 1% (w/v) TAE (Tris/acetate/EDTA)/agarose gels and electrophoresed overnight at 1 V/cm at 25 °C or 37 °C. The gels were subsequently stained with ethidium bromide to reveal the DNA–protein complexes.
SEC (size-exclusion chromatography)
The H-NS1–89 polypeptide was injected on to a Superdex 75 Hiload XK16/60 column at a concentration of 168 μM in 20 mM sodium phosphate (pH 7.0) and 300 mM NaCl. The flow rate was set at 1.5 ml/min. The column temperature was controlled by an external water bath set at temperature 17.5 to 45 °C. Protein elution was detected at A280. Molecular masses were calculated from a curve based on globular protein standards. The molecular mass was determined from the linear fit of the standard curve [i.e. log mass=−0.026(elution volume)+6.28]. In this, and in previously reported data, these standards have been shown to over-estimate the size of the H-NS oligomeric state. This is likely to reflect the non-globular and ellipsoid nature of H-NS and oligomers thereof [24,29].
ITC (isothermal titration calorimetry)
ITC experiments were conducted on a VP ITC (Microcal, Northampton, MA, U.S.A.), as described previously [43,44]. Experiments were performed at a temperature range of 15 to 45 °C. Oligonucleotides were derived by suspending calf thymus DNA in 20 mM sodium phosphate buffer, pH 7.0, 300 mM NaCl and sonicating at 4 °C for 180 pulses of 30 s until average molecular mass was decreased to approx. 300 bp. All titrations were performed in 20 mM sodium phosphate buffer (pH 7.0)/300 mM NaCl. DNA (20 mM bp) were titrated into 40 μM of H-NS. At all temperatures, the heats of dilution for the protein were determined by titration of H-NS into the ITC cell containing only buffer solution. The heats per injection from these experiments were subtracted from those of the raw-binding data. All binding data were analysed by fitting the binding isotherm to a simple independent binding-site model using Origin software provided with the ITC (MicroCal Inc.). Binding data were additionally fitted to a co-operative model [45] in which full-length H-NS was initially assumed to bind to DNA as a dimeric unit to a single oligonucleotide (i.e only one H-NS binding site was occupied in the dimer), representing the homogeneous lattice. This model, although not satisfying the expected interaction of two oligomers (both protein and DNA), provided some evalution of co-operative binding. Dissociation constants based on this model were in the μM range (results not shown).
RESULTS
Most temperature-induced genes are under the control of H-NS
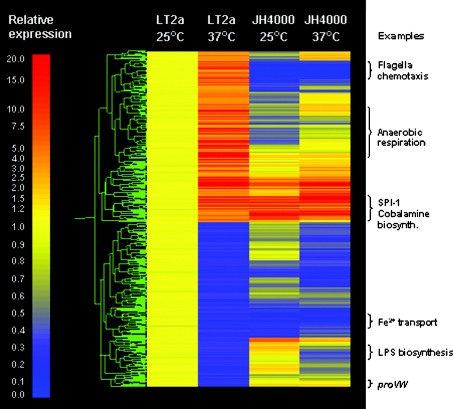
To demonstrate the role of H-NS in direct or indirect regulation, expression profiling was used to identify thermoregulated genes in S. typhimurium. This was performed using duplicate mid-exponential phase cultures of LT2 (wild-type) and JH4000 (hns null mutant), grown in LB medium at 25 °C or 37 °C. We defined thermoregulated genes as those showing a difference of ≥3-fold in expression between growth at 25 and 37 °C in LT2 (P<0.01) (Supplementary Table S1, http://www.BiochemJ.org/bj/391/bj3910203add.htm). Genes that responded to temperature in LT2, but showed no significant temperature-related change (less than 3-fold) in JH4000, were considered to show H-NS-dependent thermoregulation. Genes that showed a difference in temperature responsiveness of less than 1-fold between the two strains were excluded. A total of 531 out of 4451 genes responded to temperature in LT2 (Supplementary Tables S2a and S2b, http://www.BiochemJ.org/bj/391/bj3910203add.htm). Of these, the temperature regulation of 408 genes (77%) was found to be either directly or indirectly dependent on H-NS; 210 of these genes were up-regulated during growth at 37 °C (Supplementary Table S2c, http://www.BiochemJ.org/bj/391/bj3910203add.htm). Figure 1 shows a gene tree of expression profiles of the 531 temperature-responsive genes in S. typhimurium. The data have been normalized to LT2 grown at 25 °C and illustrate how the majority of the LT2-temperature responsive genes are not significantly thermoregulated in JH4000.
Figure 1. H-NS controls the expression of 77% of the thermoregulated genes of S. typhimurium.
The cluster diagram shows the expression profile of S. typhimurium LT2 and the hns null mutant JH4000 grown at 25 and 37 °C, relative to the expression in LT2 at 25 °C. Out of 4451 genes, 531 showed an expression differential of ≥3-fold between incubation temperatures of 25 and 37 °C in LT2 and were therefore defined as temperature-responsive. The temperature response of 408 of these genes was found to be H-NS-regulated, as demonstrated by the similar expression levels observed at the two temperatures in JH4000. Each horizontal line represents one gene; red indicates an increase in expression, yellow indicates no change, and blue indicates a decrease in expression (relative expressions levels are indicated on the left-hand side).
A closer examination of the 210 genes which show H-NS-dependent up-regulation at 37 °C revealed that many are located within the SPI-1 (Salmonella pathogenicity island 1) or belong to the flagellar/chemotaxis regulon. In addition, there are a large number of genes involved with anaerobic/aerobic respiration and the cbi operon encoding synthesis of the vitamin B12 adenosyl cobalamide precursor (Supplementary Table S2c, http://www.BiochemJ.org/bj/391/bj3910203add.htm). It is important to note that these data do not necessarily reflect a direct involvement of H-NS in gene regulation. The effects observed can potentially include contributions from other mechanisms including other transcriptional regulators whose expression is temperature-dependent, or other global mechanisms that are connected to nucleoid compaction, such as supercoiling. Nonetheless, all of these additional factors are indirectly affected by H-NS. In the present work we focus solely on the mechanism of temperature-induced up-regulation of genes. However, since H-NS normally functions as a repressor of transcription, it is surprising to note that H-NS controls the expression of similar numbers of genes that are activated and down-regulated by growth at 37 °C. The down-regulation of 198 genes during growth at 37 °C could be an indirect effect, resulting from changes in expression of another regulator that is controlled by H-NS, for example, a regulatory RNA, as several of these are known to be repressed by H-NS (these genes were not represented on our microarray).
Thermoregulated genes react rapidly to temperature transition
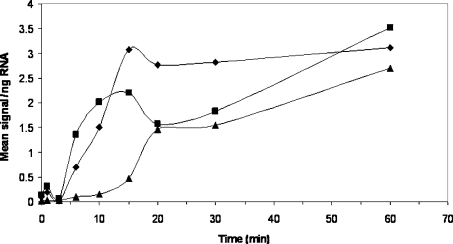
To corroborate the microarray data for a single set of genes and to determine the rate at which H-NS-regulated genes react to changes in temperature, a temperature-shift experiment was performed. A mid-exponential phase culture of LT2 was transferred from one shaking waterbath at 25 °C to another at 37 °C. The culture was monitored with a temperature logging device and took 2 min to equilibrate from 25 to 37 °C (results not shown). The expression of three SPI-1 genes which showed an H-NS-dependent thermo-induction (Supplementary Table S2c, http://www.BiochemJ.org/bj/391/bj3910203add.htm), hilA, hilC and hilD, were measured by quantitative RT-PCR. Prior to transferring the culture from 25 to 37 °C, expression of these genes was very low compared with LT2 grown at 37 °C from the outset (Figure 2). However, within 4 to 7 min of transferring the culture to 37 °C, all three genes showed significant induction which continued to rise throughout the experiment. Levels of hilA, hilC and hilD transcripts were found to be induced by 64-, 17- and 10-fold respectively, within 1 h of transfer to 37 °C. The detection of induction at relatively early time points suggests a rapid response to temperature change mediated through H-NS.
Figure 2. Kinetics of temperature induction.
A culture of LT2a was incubated at 25 °C in LB broth to a D600 of 0.60 and transferred to 37 °C. Samples were harvested just prior to temperature up-shift and at intervals thereafter for RNA extraction. Expression of hilA (▲), hilC (◆) and hilD (■) was measured by quantitative RT-PCR and normalized to an LT2 culture grown at 37 °C throughout, and harvested at D600 of 0.60.
H-NS binds to the hilC promoter and structural gene in a temperature-dependent manner
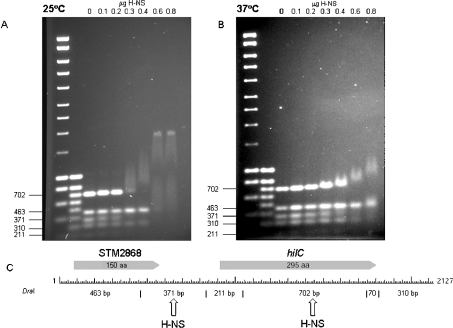
To demonstrate the direct involvement of H-NS in the thermo-regulation of hilC, we studied the binding of H-NS by competitive gel-retardation assays at 25 and 37 °C. A 2128 bp DNA fragment, encoding hilC and the up-stream gene STM2868, was digested with DraI and incubated with increasing concentrations of H-NS to test: (i) whether the promoter of this gene binds H-NS, and (ii) whether this binding is temperature-dependent. The range of H-NS concentrations tested was based upon amounts that have been previously shown to retard the proU promoter and its downstream regulatory element [21]. Figure 3 shows that H-NS preferentially binds to two DNA fragments. The first 371 bp region contains the hilC promoter (the 3′ end of this fragment is located 48 bp upstream of the hilC transcriptional start site [46]). The second 702 bp fragment encodes much of the hilC structural gene. The binding of H-NS to these two fragments is temperature-dependent. At 25 °C, strong retardation of the two DNA fragments was observed. In contrast, at 37 °C, only slight retardation of the two fragments was observed. It is apparent that the 463, 211 and 310 bp fragments encoding STM2868, the 5′-end of the hilC structural gene and region downstream of hilC respectively, were retarded at 25 °C only at the highest H-NS concentrations used. These results agree well with those for the proU operon when H-NS binds within the proV operon [21].
Figure 3. Competitive gel-shift assays with H-NS at 25 and 37 °C.
A 2128 bp DNA fragment encoding hilC and the up-stream gene STM2868 were digested with DraI. Digested DNA (0.5 μg) was incubated with a range of concentrations of H-NS at 25 °C (A) or 37 °C (B) and electrophoresis was carried out at the respective temperatures. A restriction map (C) shows the location of the genes and DraI restriction sites. Arrows indicate the fragments displaying a high affinity for H-NS.
Structural response of H-NS to increased temperature
To substantiate the observation of the interaction of H-NS being directly affected by temperature, a biophysical investigation of the thermal behaviour of the protein in vitro was performed. The high thermal stability of H-NS has been reported previously; the melting temperature TM of the full-length protein at 0.22 mM is 58 °C and the monomeric C-terminal domain (residues 89–136) has a Tm of 56.7–61 °C [23]. CD spectroscopy shows that full-length H-NS and the C-terminal deleted H-NS1–89 are essentially fully folded at the temperature range 25–35 °C, showing no net change in α-helix content as measured at 222 nm (results not shown). Therefore, the mechanism of H-NS-dependent thermo-induction of gene expression observed above is not due to the thermal denaturation of H-NS. Consequently, the protein must experience a temperature-dependent structural change which reduces the affinity of H-NS for DNA and increases access to promoter sites by RNA polymerase. This could occur by modulation of the oligomeric state of H-NS which would affect the co-operativity of binding to DNA.
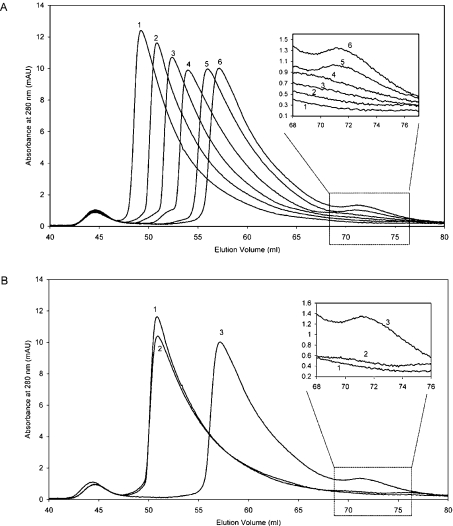
To investigate this effect, the oligomeric state of H-NS was determined by SEC over a range of temperatures. Since we are only interested in the oligomerization process, the experiment was performed on the truncated N-terminal region of H-NS (H-NS1–89). This polypeptide has identical oligomerization properties as the full-length protein [23]. Increasing the temperature resulted in a significant reduction of the average molecular mass of the high-order polydisperse H-NS1–89, as seen in the increasing elution volume of the highest molecular mass species (Figure 4A). Thus elevation of temperature impedes the ability of H-NS to form high-order states. The reduction in size of the oligomers is accompanied by the appearance of a low molecular mass species which is eluted at 71.5 ml, a volume corresponding to a molecular mass of approx. 26.3 kDa (see the Experimental section) which is close to that of a dimer of the 1–89 polypeptide (21.0 kDa; Figure 4A, inset). At higher temperatures, the concentration of dimeric H-NS increases. This reduction of high-order oligomerization and the concomitant appearance of dimer with elevated temperature is fully reversible (Figure 4B). AUC (analytical ultracentrifugation) performed at 25 °C and 37 °C under equilibrium conditions revealed a similar reduction in the molecular mass of the high-order oligomeric species (results not shown).
Figure 4. Change in H-NS oligomerization with increasing temperature over the range 17.5–45 °C (A) and reversibility of H-NS oligomerization between 25 to 45 °C (B).
(A) SEC graphs for H-NS1–89 at a range of temperatures (lines: 1, 17.5 °C; 2, 25 °C; 3, 30 °C; 4, 35 °C; 5, 40 °C; 6, 45 °C). The inset shows an expansion of the graph demonstrating the increase in the peak size at an elution volume of approx. 71.5 ml corresponding to dimeric H-NS1–89. (B) SEC graphs for H-NS1–89. H-NS1–89 was initially run on the column at 25 °C (line 1). The sample was then kept at 45 °C for 15 h and re-run on the column at that temperature (line 3). It was then subsequently cooled to 4 °C and incubated at this temperature for 6 h before being heated to 25 °C and injected on to the chromatographic column (line 2). The sample shows almost complete reversibility over the temperature range studied. The inset shows that the formation of dimer (elution volume 71.5 ml) is also highly reversible.
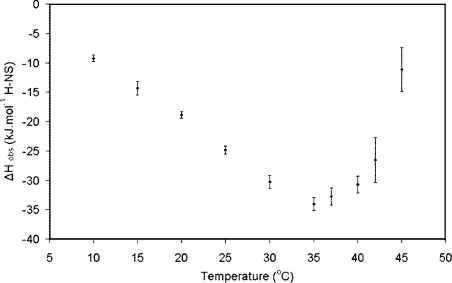
The thermodynamic parameters associated with the interaction of H-NS with DNA were determined using ITC. This technique was adopted because ITC experiments accurately measure the observed change in enthalpy, ΔH, which occurs on forming a complex. This term includes heat associated with binding, as well as any conformational changes which may occur in the interacting moieties on forming the complex. In the case of a simple interaction of ‘rigid bodies’ in which no additional equilibria are involved, the temperature-dependence of this term (otherwise known as the change in heat capacity; ΔCp=dΔH/dT) is negative and linear. ΔCp correlates with the change of surface area exposed to bulk solvent on forming a biomolecular complex. In a case where an additional equilibrium occurs involving events whose ΔCp is different to that of binding alone, a deviation from the linear relationship in the temperature dependence of ΔH will be observed [47]. To measure the binding to oligomers formed by the full-length protein, and to avoid possible complications associated with an oligonucleotide to which putative specific and non-specific interactions could occur simultaneously, H-NS was titrated with a 300 bp non-specific calf thymus oligonucleotide at 300 mM NaCl. The data were fitted to a simple model based on multiple independent binding sites. The ΔCp for the interaction remains negative up to approx. 30 °C (Figure 5). This is a general signature of the burial of predominantly hydrophobic surface area away from contact with bulk solvent. This is likely to be derived from the binding of the C-terminal domain to DNA and is common for protein–DNA interactions [48]. The value of the ΔCp for the binding of full-length H-NS for the interaction over the temperature range 10–25 °C is low for a protein–DNA interaction (approx. 1 kJ·mol−1·K−1) and is consistent with a non-specific interaction [48,49]. As the temperature was increased through 30 °C to where the population of dimer is clearly apparent in the SEC studies (see above), the ΔCp value gradually changes to a positive value. This unusual response upon complex formation between H-NS and the oligonucleotide at elevated temperatures is consistent with an event in which hydrophobic surface is exposed to solvent. Clearly this does not emanate from the formation of the complex between the C-terminal domain of H-NS and DNA. This could, however, be caused by the exposure of surface area on dissociation from oligomeric to dimeric states, or from a conformational change of the protein. Importantly, the appearance of a positive slope provides strong evidence that the isolated protein is not unfolding at the elevated temperature. This is because H-NS binds DNA in its folded form [32] and therefore any unfolded protein would have to refold to bind to the DNA. This refolding would involve the burial of hydrophobic surface area, resulting in a more negative slope in Figure 5 [41], rather than the opposite effect that we observed.
Figure 5. Change in observed enthalpy with temperature.
Plot of the change in observed enthalpy, ΔH, against temperature for the interaction of full-length H-NS with an approx. 300 bp oligonucleotide derived from sonicated calf thymus.
The affinity of the interaction was determined based on fitting data to the independent binding-sites model (i.e. neglecting any effect of co-operativity). Using this model, the KD for DNA binding to H-NS increases from approx. 0.2 mM to 0.7 mM over the temperature range 10–45 °C (Table 1). The KD values derived by ITC for this non-specific interaction are lower than those previously obtained using gel-shift analysis [32], but are of the same magnitude as values obtained for binding studies between the isolated C-terminal domain of H-NS (residues 89–136) and a 20 bp non-specific oligonucleotide (S. Ono and J. E. Ladbury, unpublished work). These data show that, without incorporating a term in the fitting algorithm to accommodate the effect of co-operativity of binding between the oligomeric H-NS and the oligonucleotide, the affinity of the C-terminal domain to DNA does not change dramatically over the temperature range investigated. This suggests that the effects of temperature on gene expression observed above are not the result of the temperature dependence of the direct interaction between protein and DNA. This implicates the effect of co-operativity derived from the oligomerization as the source of differential binding of H-NS to the bacterial DNA. We attempted to model the effect of the potential co-operative effect of the binding of H-NS to DNA using various methods, but these were largely inadequate in accommodating the binding of one oligomeric species with another (see the Experimental section). The fit from the ITC data gives a value for the number of bp (N) which are occupied per H-NS molecule, N=14.9 (see Table 1). This value agrees well with the number of bp in the binding site as determined by different methods [32,36].
Table 1. Binding data for the interaction of H-NS1–136 with DNA at a range of temperatures.
Data determined from ITC results using a single independent binding-site model (see the Experimental section). N is the number of bp occupied per binding site on the protein.
| Temperature ( °C) | ΔH (kJ·mol−1) | KD (μM) | ΔG (kJ·mol−1) | N |
|---|---|---|---|---|
| 10 | −9.23±0.52 | 167 | −20.45 | 16.6 |
| 15 | −14.31±1.14 | 236 | −19.96 | 15.9 |
| 20 | −18.87±0.57 | 171 | −21.09 | 15.5 |
| 25 | −24.87±0.68 | 260 | −20.42 | 15.7 |
| 30 | −30.30±1.11 | 314 | −20.29 | 16.2 |
| 35 | −34.05±1.10 | 375 | −20.17 | 15.5 |
| 37 | −32.77±1.47 | 442 | −19.87 | 14.4 |
| 40 | −30.68±1.45 | 637 | −19.12 | 11.7 |
| 42 | −26.55±3.82 | 735 | −18.86 | 12.5 |
| 45 | −11.16±3.76 | 685 | −19.23 | 14.9 |
DISCUSSION
The rapidity with which microbial pathogens respond to the host environment is remarkable. However, the need to allow rapid access of transcriptional activator proteins to large tracts of DNA in response to changes in temperature cannot be allowed to compromise the packaging of the bacterial chromosome. Any shift in temperature represents a challenge for the proteins that maintain nucleoid structure. H-NS, as one of the most abundant nucleoid-associated proteins, controls most of the genes which function either to combat stress or to ensure successful infection and are thermoregulated [7]. Microarray analysis of S. typhimurium LT2 and the hns null mutant JH4000 strains identified those genes that are differentially expressed in response to temperature and showed that the thermoregulation of a majority of genes (77%) are H-NS dependent. Interestingly, and in common with other pathogens, the SPI-1 pathogenicity island is only expressed at 37 °C in wild-type S. typhimurium, but is expressed at high levels at both 25 °C and 37 °C in the absence of H-NS. Selected genes from the SPI-1 pathogenicity island show rapid induction. Furthermore, the binding of H-NS has a direct effect on the temperature regulation of these genes consistent with the concept of H-NS acting as a direct transcriptional repressor at the non-permissive temperature of 25 °C, yet allowing expression at 37 °C. These data are also consistent with idea that the reduced ability of H-NS to oligomerize at 37 °C impedes its binding to DNA [36].
Another interesting observation from these experiments is that H-NS binds to the hilC structural gene in addition to the promoter. This resembles the situation observed for the H-NS-regulated proU operon where H-NS binds to the DRE (downstream-regulatory element) within the proV structural gene [50]. It could be argued that the temperature-dependent changes in retardation of the 371 and 702 bp fragments encoding the hilC promoter and structural gene could be the result of alterations in DNA conformation with temperature, rather than the oligomerization status of H-NS. It is therefore important to note that even those DNA fragments for which H-NS has a poor affinity and are only weakly retarded by the highest concentrations of H-NS at 25 °C also show reduced levels of retardation at 37 °C. This supports the view that H-NS undergoes a structural transition resulting in reduced DNA-binding affinity. In addition, it is important to point out that the concentrations of protein used in the gel retardation experiment are lower than those used in the SEC experiments above. Through the Law of Mass Action the high-order oligomer is more stable than the dimer. Under the experimental temperatures adopted with these lower H-NS concentrations, the relative population of dimeric compared with oligomeric state is higher than in the SEC.
Our data suggest that H-NS can perform the dual roles of facilitating gene expression while maintaining the DNA in a condensed state by acting as a subtle temperature-dependent conformational switch. The mechanism by which this is achieved as the temperature is raised is based on a concentration-dependent number of H-NS molecules from the high-order polydisperse protein scaffold undergoing a change in structure, resulting in the appearance of dimers (the lowest functional state for the protein) and a reduction in the average molecular mass of these oligomers.
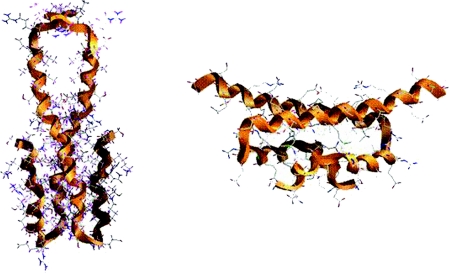
We speculate that the previously reported structures of the oligomerization domain of H-NS might reflect the hypothesized conformational change, since they show two distinct juxtapositions of the α-helices forming the dimer (Figure 6). The two solution structures of the N-terminal of H-NS were determined at two different temperatures, 25 °C [24] (Figure 6A) and approx. 37 °C [38] (Figure 6B). Although these structures are essentially identical in terms of secondary structure, they exhibit radically different alignment of the primary helix (H3) involved in the coiled-coil tertiary structure. The structure determined at 25 °C is a dimer formed via a parallel coiled-coil interaction of H3 of each protomer. In the structure determined at higher temperature, the H3 helices are flipped from being aligned in a parallel coiled-coil to adopt an obtuse angle to each other. Although these structures only represent the N-termini of the oligomerization domain, at higher temperature the head-to-tail mechanism for oligomerization [24], in which the N-terminal residues of one H-NS dimeric domain interact with the C-terminal residues of another, could be compromised as the C-terminal residues of each protomer become orientated away from each other [38]. The conformational change of the protein at higher temperature correlates with the appearance of a population of a dimeric species (as seen in the data from SEC experiments, Figure 4), but results in no net change of the secondary structure and negligible change in sedimentation and hydrodynamic properties (confirmed by both equilibrium and sedimentation velocity AUC experiments; results not shown).
Figure 6. Ribbon representation of the structures of H-NS1–57 (left-hand panel; PDB code, 1LR1 [22]) and H-NS1–46 (right-hand panel; PDB code, 1NI8 [38]).
Helix 3 (H3) is the longest helix in both structures.
The herein proposed model suggests that H-NS can bind to DNA in either of the conformations described above (the isolated C-terminal DNA-binding domain is known to be stable at much higher temperatures [23] and is not affected by the conformational change which is proposed for the N-terminus). However, the loss in co-operative binding when dimerization is induced results in the higher temperature form of the protein being easier to remove from DNA promoter sites, allowing greater access for the initiation of transcription following temperature up-shift (Figure 7). The low ΔCp for the binding to DNA determined at the lower temperatures (up to 30 °C) is consistent with the binding interface being small, non-specific and/or water-mediated in nature [48,49,51]. The small, but significant, increase of ΔH with temperature (resulting in the apparent positive ΔCp) for this interaction above 30 °C, suggests the increase in influence of an enthalpic contribution from the formation of dimers. This positive ΔCp correlates with an increase in exposure of apolar surface area either from the change in dimer conformation itself or from the resulting separation of dimers from the oligomeric structure. This is consistent with the finding that 345 Å2 of additional hydrophobic surface area is exposed in the high-temperature structure (2482 Å2 for residues 1–46 for the structure with PDB code 1LR1 compared with 2837 Å2 for 1NI8 [52]).
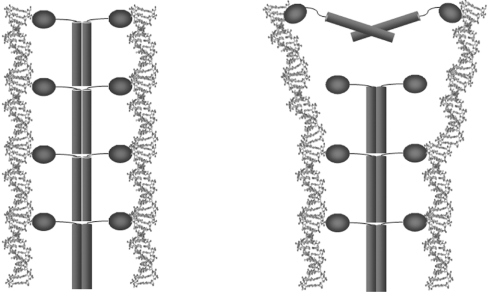
Figure 7. Representation of the proposed mechanism for H-NS responding to a temperature rise from 25 to 37 °C.
In the left-hand panel (25 °C) full-length H-NS is able to bind to strands of DNA in a co-operative manner based on the high-order oligomeric structure. The right-hand panel (37 °C) shows the effect of the conformational change on one of the H-NS dimers due to increased temperature. The dimer is no longer able to interact with the high-order protein oligomer and therefore no longer binds to DNA in a co-operative manner. The resulting loss in affinity affects the DNA topology and makes it more accessible to RNA polymerase, leading to gene transcription.
Importantly, the SEC studies demonstrate that appearance of dimer is not an ‘all-or-nothing’ response to thermal increase under the temperature conditions experienced by enteric bacteria during infection. The retention of a level of co-operative binding is likely to be crucial to maintaining control of the condensation of the nucleoid, and to avoid cellular disfunction. Furthermore, growth at 37 °C does not result in complete derepression, as demonstrated for the H-NS-dependent repression of the hly operon controlling the expression of the toxin α-haemolysin produced by many uropathogenic E. coli strains [53]. This H-NS-dependent temperature-induced phenotype resembles the effect observed in mutations of residues in the first 20 amino acids of the protein [21,22,30,38]. This suggests that mutations in the N-terminal region are likely to disrupt the high-order oligomerization capacity of the protein and achieve a similar effect to that observed on thermal elevation.
The mechanism by which H-NS acts as a thermal switch does not preclude the involvement of other proteins in the thermo-regulation of gene expression. Indeed, it is highly likely that subsidiary activator proteins are required to achieve specificity in this process, by recognizing appropriate genes for the initiation of transcription [54,55]. Equally, the H-NS-dependent mechanism proposed here does not rule out the role of previously reported temperature-dependent DNA structural effects in dictating thermoregulation of gene expression. The thermally controlled changes in the high-order structure and binding to DNA by H-NS may well act in concert with DNA structural change to achieve a fine-tuned response [56]. These findings contribute to the exciting paradigm of gene regulation being mediated by the oligomeric state of a protein, resulting in a bacterium using this protein as the primary thermometer to translate a sensory event into an effective response at the level of global gene expression.
Online data
Acknowledgments
J.E.L. is a Wellcome Trust Senior Research Fellow. This work was funded by a BBSRC (Biotechnology and Biological Sciences Research Council) studentship to S.O., and by the BBSRC Core Strategic Grant to J.C.D.H. T.O. was funded by a Wellcome Trust Vacation Scholarship. We gratefully acknowledge data on AUC of H-NS and discussions with Arsen Petrovic, John Eccleston, Julian Eaton and Hannah Gilbert. M.D.G. appreciated the support of Charles Penn in preparing the manuscript. This study benefited from the contribution of several members of the Hinton laboratory, particularly Vittoria Danino, Sacha Lucchini, Matt Rolfe and Arthur Thompson. We also are grateful to Ray Dixon for critical reading of the manuscript.
References
- 1.Dorman C. J. H-NS: A universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 2.Eriksson S., Hurme R., Rhen M. Low-temperature sensors in bacteria. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2002;357:887–893. doi: 10.1098/rstb.2002.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marshall D. G., Bowe F., Hale C., Dougan G., Dorman C. DNA topology and adaptation of Salmonella typhimurium to an intracellular environment. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000;355:565–574. doi: 10.1098/rstb.2000.0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drlica K., Rouviere-Yaniv J. Histone-like proteins of bacteria. Microbiol. Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams R. M., Rimsky S. Escherichia coli nucleoid-associated protein H-NS: a central controller of gene regulatory networks. FEMS Microbiol. Lett. 1997;156:175–185. doi: 10.1111/j.1574-6968.1997.tb12724.x. [DOI] [PubMed] [Google Scholar]
- 6.Dorman C. J., Hinton J. C. D., Free A. Domain organisation and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol. 1999;7:124–128. doi: 10.1016/s0966-842x(99)01455-9. [DOI] [PubMed] [Google Scholar]
- 7.Hommais F., Krin E., Laurent-Winter C., Soutourina O., Malpertuy A., LeCaer J.-P., Danchin A., Bertin P. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 2001;40:20–36. doi: 10.1046/j.1365-2958.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- 8.Laurent-Winter C., Ngo S., Danchin A., Bertin P. Role of Escherichia coli histone-like nucleoid structuring protein in bacterial metabolism and stress response. Eur. J. Biochem. 1997;244:767–773. doi: 10.1111/j.1432-1033.1997.00767.x. [DOI] [PubMed] [Google Scholar]
- 9.Spassky A., Rimsky S., Garreau H., Buc H. H1a, an E. coli DNA-binding protein which accumulates in stationary phase, strongly compacts DNA in vitro. Nucleic Acids Res. 1984;12:5321–5340. doi: 10.1093/nar/12.13.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falconi M., Higgins N. P., Spurio R., Pon C. L., Gualerzi C. O. Expression of the gene encoding the major bacterial nucleoid protein H-NS is subject to transcriptional auto-repression. Mol. Microbiol. 1993;10:273–282. doi: 10.1111/j.1365-2958.1993.tb01953.x. [DOI] [PubMed] [Google Scholar]
- 11.Falconi M., Colonna B., Prosseda G., Micheli G., Gualerzi C. O. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 1998;17:7033–7043. doi: 10.1093/emboj/17.23.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ueguchi C., Mizuno T. The Escherichia coli nucleoid protein H-NS functions directly as a transcriptional repressor. EMBO J. 1993;12:1039–1046. doi: 10.1002/j.1460-2075.1993.tb05745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Afflerbach H., Schroder O., Wagner R. Conformational changes of the upstream DNA mediated by H-NS and FIS regulate E. coli RrnB P1 promoter activity. J. Mol. Biol. 1999;286:339–353. doi: 10.1006/jmbi.1998.2494. [DOI] [PubMed] [Google Scholar]
- 14.Göransson M., Sonden B., Nilsson P., Dagberg B., Forsman K., Emanuelsson K., Uhlin B. E. Transcriptional silencing and thermoregulation of gene expression in Escherichia coli. Nature (London) 1990;344:682–685. doi: 10.1038/344682a0. [DOI] [PubMed] [Google Scholar]
- 15.Tupper A. E., Owen-Hughes T. A., Ussery D. W., Santos D. S., Ferguson D. J. P., Sidebotham J. M., Hinton J. C. D., Higgins C. F. The chromatin-associated protein H-NS alters DNA topology in vitro. EMBO J. 1994;13:258–268. doi: 10.1002/j.1460-2075.1994.tb06256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tobe T., Yoshokawa M., Mizuno T., Sasakawa C. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by VirF and repression by H-NS. J. Bacteriol. 1993;175:6142–6149. doi: 10.1128/jb.175.19.6142-6149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colonna B., Casalino M., Fradiani P. A., Zagaglia C., Naitza S., Leoni L., Prosseda G., Coppo A., Gherlardini P., Nicoletti M. H-NS regulation of virulence gene expression in enteroinvasive Escherichia coli harbouring the virulence plasmid integrated into the host chromosome. J. Bacteriol. 1995;177:4703–4712. doi: 10.1128/jb.177.16.4703-4712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorman C. J., Porter M. E. The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol. Microbiol. 1998;29:677–684. doi: 10.1046/j.1365-2958.1998.00902.x. [DOI] [PubMed] [Google Scholar]
- 19.Prosseda G., Falconi M., Giangrossi M., Gualerzi C. O., Gioacchino M., Colonna B. The virF promoter in Shigella: more than just a curved DNA stretch. Mol. Microbiol. 2004;51:523–537. doi: 10.1046/j.1365-2958.2003.03848.x. [DOI] [PubMed] [Google Scholar]
- 20.Rohde J. R., Fox J. M., Minnich T. Thermoregulation in Yersinia enterocolitica is coincident with changes in DNA supercoiling. Mol. Microbiol. 1994;12:187–199. doi: 10.1111/j.1365-2958.1994.tb01008.x. [DOI] [PubMed] [Google Scholar]
- 21.Ueguchi C., Suzuki T., Yoshida T., Tanaka K., Mizuno T. Systematic mutational analysis revealing the functional domain organization of the Escherichia coli nucleoid protein H-NS. J. Mol. Biol. 1996;263:149–162. doi: 10.1006/jmbi.1996.0566. [DOI] [PubMed] [Google Scholar]
- 22.Ueguchi C., Seto C., Suzuki T., Mizuno T. Clarification of the dimerization domain and its functional significance for the Escherichia coli nucleoid protein H-NS. J. Mol. Biol. 1997;274:145–151. doi: 10.1006/jmbi.1997.1381. [DOI] [PubMed] [Google Scholar]
- 23.Smyth C. P., Lundbäck T., Renzoni D., Siligardi G., Beavil R., Layton M., Sidebotham J., Hinton J. C. D., Driscoll P. C., Higgins C. F., Ladbury J. E. Oligomerization of the chromatin-structuring protein H-NS. Mol. Microbiol. 2000;36:962–972. doi: 10.1046/j.1365-2958.2000.01917.x. [DOI] [PubMed] [Google Scholar]
- 24.Esposito D., Petrovic A., Harris R., Ono S., Eccleston J. F., Mbabaali A., Haq I., Higgins C. F., Hinton J. C. D., Driscoll P. C., Ladbury J. E. H-NS oligomerization domain structure reveals the mechanism for high order self-association of the intact protein. J. Mol. Biol. 2002;324:841–850. doi: 10.1016/s0022-2836(02)01141-5. [DOI] [PubMed] [Google Scholar]
- 25.Falconi M., Gualtieri M. T., LaTeana A., Losso M. A., Pon C. L. Proteins from the prokaryotic nucleoid: primary and quarternary structure of the 15-kDa Escherichia coli DNA binding protein H-NS. Mol. Microbiol. 1998;2:323–329. doi: 10.1111/j.1365-2958.1988.tb00035.x. [DOI] [PubMed] [Google Scholar]
- 26.Spurio R., Falconi M., Brandi A., Pon C. L., Gualerzi C. O. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for bending. EMBO J. 1997;16:1795–1805. doi: 10.1093/emboj/16.7.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ceschini S., Lupidi G., Coletta M., Pon C. L., Fioretti E., Angeletti M. Multimeric self-assembly equilibrium involving the histone-like protein H-NS. J. Biol. Chem. 2000;275:729–734. doi: 10.1074/jbc.275.2.729. [DOI] [PubMed] [Google Scholar]
- 28.Williams R. M., Rimsky S., Buc H. Probing the structure, function and interactions of the Escherichia coli H-NS and StpA proteins using dominant negative derivatives. J. Bacteriol. 1996;178:4335–4343. doi: 10.1128/jb.178.15.4335-4343.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renzoni D., Esposito D., Pfuhl M., Hinton J. C. D., Higgins C. F., Driscoll P. C., Ladbury J. E. Structural characterization of the N-terminal oligomerization domain of the bacterial chromatin-structuring protein, H-NS. J. Mol. Biol. 2001;306:1127–1137. doi: 10.1006/jmbi.2001.4471. [DOI] [PubMed] [Google Scholar]
- 30.Nye M. B., Taylor R. K. Vibrio cholerae H-NS domain structure and function with respect to transcriptional repression of ToxR regulon genes reveals differences among H-NS family members. Mol. Microbiol. 2003;50:427–444. doi: 10.1046/j.1365-2958.2003.03701.x. [DOI] [PubMed] [Google Scholar]
- 31.Badaut C., Williams R., Arluison V., Bouffartigues E., Robert B., Buc H., Rimsky S. The degree of oligomerization of the H-NS nucleoid structuring protein is related to specific binding to DNA. J. Biol. Chem. 2002;277:41657–41666. doi: 10.1074/jbc.M206037200. [DOI] [PubMed] [Google Scholar]
- 32.Shindo H., Ohnuki A., Ginba H., Katoh E., Ueguchi C., Mizuno T., Yamazaki T. Identification of the DNA binding surface of H-NS protein from Escherichia coli by heteronuclear NMR spectroscopy. FEBS Microbiol. Lett. 1999;455:63–69. doi: 10.1016/s0014-5793(99)00862-5. [DOI] [PubMed] [Google Scholar]
- 33.Caramel A., Schnetz K. Lac and Lambda repressors relieve silencing of the Escherichia coli bgl promoter. Activation by alteration of a repressing nucleoprotein complex. J. Mol. Biol. 1998;284:875–883. doi: 10.1006/jmbi.1998.2191. [DOI] [PubMed] [Google Scholar]
- 34.Dame R. T., Wyman C., Goosen N. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acid Res. 2000;28:3504–3510. doi: 10.1093/nar/28.18.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rimsky S., Zuber F., Buckle M., Buc H. A molecular mechanism for the repression of transcription by the H-NS protein. Mol. Microbiol. 2001;42:1311–1323. doi: 10.1046/j.1365-2958.2001.02706.x. [DOI] [PubMed] [Google Scholar]
- 36.Amit R., Oppenheim A. B., Stavans J. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys. J. 2003;84:2467–2473. doi: 10.1016/S0006-3495(03)75051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dame R. T., Wuite G. J. On the role of H-NS in the organization of bacterial chromatin: from the bulk to the single molecule. Biophys. J. 2003;85:4146–4148. doi: 10.1016/S0006-3495(03)74826-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bloch V., Yang Y., Margeat E., Chavanieu A., Auge M. T., Robert B., Arold S., Rimsky S., Kochoyan M. The H-NS dimerization domain defines a new fold contributing to DNA recognition. Nat. Struct. Biol. 2003;10:212–218. doi: 10.1038/nsb904. [DOI] [PubMed] [Google Scholar]
- 39.Cerdan R., Bloch V., Yang Y., Bertin P., Dumas C., Rimsky S., Kochoyon M., Arold S. T. Crystal structure of the N-terminal dimerisation domain of VicH, the H-NS-like protein from Vibrio cholerae. J. Mol. Biol. 2003;334:179–185. doi: 10.1016/j.jmb.2003.09.051. [DOI] [PubMed] [Google Scholar]
- 40.Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 41.Donnenberg M. S., Kaper J. B. Construction of an EAE deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoker N. G., Fairweather N. F., Spratt B. G. Versatile low-copy number plasmid vectors for cloning in Escherichia coli. Gene. 1982;18:335–341. doi: 10.1016/0378-1119(82)90172-x. [DOI] [PubMed] [Google Scholar]
- 43.Wiseman T., Williston S., Brandts J. F., Lin N.-L. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 44.Ladbury J. E., Chowdhry B. Z. Sensing the heat: the application of isothermal titration calorimetry to thermodynamic studies of biomolecular interactions. Chem. Biol. 1996;3:791–801. doi: 10.1016/s1074-5521(96)90063-0. [DOI] [PubMed] [Google Scholar]
- 45.McGhee J. D., von Hippel P. H. Theoretical aspects of DNA–protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 46.Olekhnovich I. N., Kadner R. J. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 2002;184:4148–4160. doi: 10.1128/JB.184.15.4148-4160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams M. A., Ladbury J. E. The extended interface: measuring non-local effects in biomolecular interactions. Curr. Opin. Struct. Biol. 2004;14:562–569. doi: 10.1016/j.sbi.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 48.Ladbury J. E. Counting the calories to stay in the groove. Structure. 1995;3:635–639. doi: 10.1016/s0969-2126(01)00197-6. [DOI] [PubMed] [Google Scholar]
- 49.Morton C. J., Ladbury J. E. Water mediated protein–DNA interactions: the relationship of thermodynamics to structural detail. Protein Sci. 1996;5:2115–2118. doi: 10.1002/pro.5560051018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fletcher S. A., Csonka L. N. Fine-structure deletion analysis of the transcriptional silencer of the proU operon of Salmonella typhimurium. J. Bacteriol. 1995;177:4508–4513. doi: 10.1128/jb.177.15.4508-4513.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ladbury J. E., Wright J. G., Sturtevant J. M., Sigler P. B. A thermodynamic study of the trp repressor-operator interaction. J. Mol. Biol. 1994;238:669–681. doi: 10.1006/jmbi.1994.1328. [DOI] [PubMed] [Google Scholar]
- 52.Hubbard S. J., Thornton J. M. London, UK: Computer program, Department of Biochemistry and Molecular Biology, University College; 1993. “NACCESS”. [Google Scholar]
- 53.Madrid C., Neito J. M., Paytubi S., Falconi M., Gualerzi C. O., Juarez A. Temperature and H-NS dependent regulation of a plasmid-encoded virulence operon expressing Escherichia coli hemolysin. J. Bacteriol. 2002;184:5058–5066. doi: 10.1128/JB.184.18.5058-5066.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Motin V. L., Georgescu A. M., Fitch J. P., Gu P. P., Nelson D. O., Mabery S. L., Garnham J. B., Sokhansanj B. A., Ott L. L., Coleman M. A., et al. Temporal global changes in gene expression during temperature transition in Yersinia pestis. J. Bacteriol. 2004;186:6298–6305. doi: 10.1128/JB.186.18.6298-6305.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garcia J., Cordeiro T. N., Nieto J. M., Pons I., Juarez A., Pons M. Interaction between the bacterial nucleoid associated protein Hha and H-NS involves a conformational change of Hha. Biochem. J. 2005;388:755–762. doi: 10.1042/BJ20050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rimsky S. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr. Opin. Microbiol. 2004;7:1–6. doi: 10.1016/j.mib.2004.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.