Abstract
The recent discovery that the natriuretic peptide OvCNPb (Ornithorhynchus venom C-type natriuretic peptide B) from platypus (Ornithorynchus anatinus) venom contains a D-amino acid residue suggested that other D-amino-acid-containing peptides might be present in the venom. In the present study, we show that DLP-2 (defensin-like peptide-2), a 42-amino-acid residue polypeptide in the platypus venom, also contains a D-amino acid residue, D-methionine, at position 2, while DLP-4, which has an identical amino acid sequence, has all amino acids in the L-form. These findings were supported further by the detection of isomerase activity in the platypus gland venom extract that converts DLP-4 into DLP-2. In the light of this new information, the tertiary structure of DLP-2 was recalculated using a new structural template with D-Met2. The structure of DLP-4 was also determined in order to evaluate the effect of a D-amino acid at position 2 on the structure and possibly to explain the large retention time difference observed for the two molecules in reverse-phase HPLC. The solution structures of the DLP-2 and DLP-4 are very similar to each other and to the earlier reported structure of DLP-2, which assumed that all amino acids were in the L-form. Our results suggest that the incorporation of the D-amino acid at position 2 has minimal effect on the overall fold in solution.
Keywords: defensin-like peptide (DLP), nuclear Overhauser spectroscopy, peptide conformation, peptide isomerase, platypus venom peptide
Abbreviations: 2D, two-dimensional; CNS, crystallography and NMR system; DLP, defensin-like peptide; NOE, nuclear Overhauser effect; OvCNP, Ornithorhynchus venom C-type natriuretic peptide; RP-HPLC, reverse-phase HPLC; R.M.S.D., root mean square deviation
INTRODUCTION
The discovery of a biologically active peptide that contains D-amino acid residues in frog skin secretions more than 2 decades ago [1] refuted the longstanding notion that such types of unusual molecules only existed in lower forms of organisms, such as prokaryotes. This suggested the possibility that such peptides and proteins may also exist in higher organisms, possessing specific physiological functions. Accordingly, this led to a search in eukaryotes, and, to date, biologically active proteins containing D-amino acid residues have been discovered in frogs [1–3], molluscs [4–9], crustaceans [10–13], spiders [14,15] and, most recently, the platypus (Ornithorhynchus anatinus) [16], which is a mammal.
OvCNPa (Ornithorhynchus venom C-type natriuretic peptide A) and its D-amino-acid-containing isomer OvCNPb are the most biologically active polypeptides yet found in platypus venom [17–19]. These molecules cause rat uterine smooth muscle relaxation, oedema and mast cell histamine release. It has been shown previously that the different behaviour in HPLC, and structural folding in NMR spectroscopy, between the two forms of OvCNP, is due to the presence of the unusual D-leucine at the second position of the amino acid sequence of OvCNPb [16]. It is therefore possible that other D-amino-acid-containing peptides and a putative peptide-residue isomerase(s) responsible for their creation, are present in the platypus venom gland and possibly the venom itself.
The obvious candidate in the search for other D-amino-acid-containing peptides in the platypus is a family of polypeptides referred to as defensin-like peptides or DLPs [20,21]. These molecules are the most abundant peptides in the venom and have tertiary structures that resemble those of the mammalian anti-microbial β-defensins, but their role in the venom is currently unknown. Ever since the discovery of DLPs, many polypeptides of the same fold have been discovered in humans and in the toxins of various organisms, such as sea anemone and snakes [22].
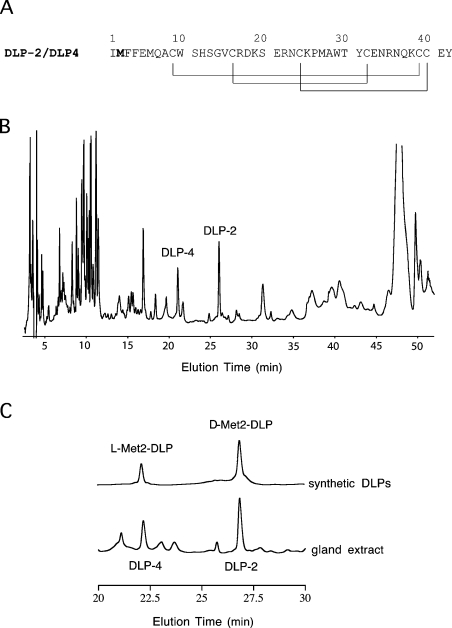
Two DLPs, namely DLP-4 and DLP-2, have identical amino acid sequences; however, they elute at significantly different times in RP-HPLC (reverse-phase HPLC) [16,21] (Figures 1A and 1B). Since the solution structure of native DLP-2 has been determined by NMR spectroscopy [21], it was speculated then that the difference between DLP-2 and DLP-4 may be due to the difference in the pattern of disulphide bond pairing, since there are six cysteine residues that may be paired in several possible combinations.
Figure 1. Primary structure and RP-HPLC of DLP-2 and DLP-4.
(A) Primary structure of DLP-2 and DLP-4. The methionine residue shown in bold is found to be in the L-form in DLP-4 and in the D-form in DLP-2. The disulphide-bonding pattern of DLP-2 as determined previously by NMR [21] is shown by lines below the sequences. It was found in the present study to be identical with that in DLP-4 based on chemical shift value comparison. (B) RP-HPLC of platypus venom gland extract. Homogenized gland (50 μl) was loaded on to a Synergi 4μ Hydro RP (250 mm×4.6 mm; Phenomenex) column in water containing 0.1% (v/v) trifluoroacetic acid. The components were eluted with a gradient of 20–45% acetonitrile containing 0.1% (v/v) trifluoroacetic acid at 1.0 ml·min−1, for 40 min at room temperature (21 °C). The absorbance was recorded at 215 nm. DLP-4 and DLP-2 eluted at ∼22 and ∼27 min respectively. (C) Sections of RP-HPLC of the venom-gland extract and solutions of synthetic DLPs. Both samples were loaded individually on to a Synergi 4μ Hydro RP (250 mm×4.6 mm; Phenomenex) column in water containing 0.1% (v/v) trifluoroacetic acid and eluted with a gradient as in (B).
With the availability of sufficient amounts of native DLP-4 for NMR experiments and knowing that other D-amino-acid-containing peptides may possibly exist in platypus venom, we show in the present paper that the difference between DLP-4 and DLP-2 is due to presence of a D-amino acid at position 2 in DLP-2. We also show that the platypus venom gland extract has isomerase activity that converts DLP-4 into DLP-2. In an effort to explain why the two molecules elute at significantly different times in RP-HPLC, we calculated the solution structures of DLP-2 and DLP-4 and compared their tertiary folds.
MATERIALS AND METHODS
Preparation of venom-gland extracts
The venom glands used in the present study were obtained from adult male platypuses that had been accidentally killed by motor vehicles. Each animal provided two glands that were dissected out after tying the ducts with ligatures. Glands were finely sliced, then the ligatures were removed, and the tissue was then placed in a container with 50 ml of cold 1% trehalose solution. The samples were homogenized with a Bamix cutter using short bursts to ensure that the temperature remained low. The resulting mixture was centrifuged at 3000 g for 5–10 min at 5 °C; the liquid in the extract was then decanted and passed through a 0.2 μm Sartorius Minisart filter (Göttingen, Germany). The filtrate was used as the starting material for the various experiments described below.
RP-HPLC of DLP samples
A GBC HPLC system (Gibco, Dandenong, Vic, Australia) with LC 1150 pump and LC 1210 UV/visible dual-wavelength detector was used in chromatography of the platypus venom gland extract and synthetic DLPs. A Phenomenex (Torrance, CA, U.S.A.) Synergi 4μ Hydro-RP analytical column (250 mm×4.60 mm) was used, and the solvent system consisted of 0.1% (v/v) trifluoroacetic acid in water (Buffer A) and 0.1% (v/v) trifluoroacetic acid in acetonitrile (Buffer B). Components were eluted at a flow rate of 1 ml·min−1 and were detected at either 215 nm or 280 nm. Two linear gradient profiles with different durations were implemented. For the 60 min acquisition run shown in Figure 1, the solvent gradient was 5–20% Buffer B for 5 min, 20–45% Buffer B for 40 min, 45–60% Buffer B for 5 min, 60% Buffer B for 5 min and 60–5% Buffer B for 5 min. For the 35 min acquisition run shown in Figure 3, the solvent gradient was 5–25% Buffer B for 5 min, 25–35% Buffer B for 16 min, 35–60% Buffer B for 2 min, 60% Buffer B for 2 min and 60–5% Buffer B for 2 min.
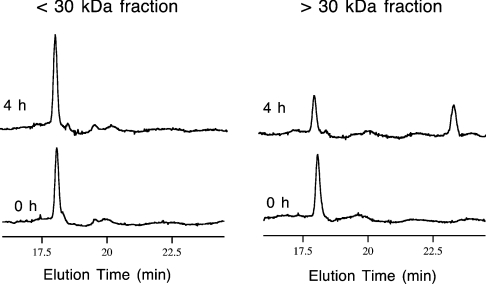
Figure 3. Isomerization of DLP-4 to DLP-2.
RP-HPLC traces of incubation mixtures obtained at the start (0 h) and after 4 h of incubation at 33 °C. The two venom-gland fractions were separated by centrifugal ultrafiltration with a 30 kDa nominal molecular mass cut-off filter. The incubation mixture contained the extract fraction, PBS, DLP-4 and EDTA in the amounts described in the Materials and methods section. The small increase in the DLP-4 HPLC peak after 4 h in the <30 kDa fraction was caused by inaccuracies in injection volume.
Preparation of DLP samples
Native DLP-2 and DLP-4 were isolated from the venom-gland extracts by RP-HPLC as described above or as described by Torres et al. [21]. Chemical syntheses of L-Met2-DLP and D-Met2-DLP were performed manually using HBTU [2-(1-H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate] activation of Boc (t-butoxycarbonyl)–amino acids with in situ neutralization chemistry, as described previously [23]. Details of the procedure were similar to the synthesis of OvCNPa and D-OvCNPb as described by Torres et al. [16]. NMR samples were prepared by dissolving 1–2 mg of the freeze-dried DLPs in 0.350 ml of H2O/2H2O (9:1, v/v) in a 5-mm magnetic-susceptibility matched Shigemi (Allison Park, PA, U.S.A.) NMR tube.
Assay for L-to-D-amino acid residue isomerase activity
A platypus gland extract that contains no DLP-2 and DLP-4 components was fractionated into two samples by centrifugal ultrafiltration using Amicon Ultrafree-MC with 30 kDa nominal molecular mass cut-off. Gland extract (400 μl) was added to the filter cup and this was spun at 4500 g for ∼15–30 min. The filtrate obtained at this stage contained components with nominal molecular mass <30 kDa and was subsequently used in the isomerase assay. The retentate which contained components with nominal molecular mass >30 kDa was washed twice to remove traces of components with molecular mass <30 kDa by reconstituting it with two 350 μl volumes of 50 mM PBS containing 0.9% (w/v) NaCl at pH 7.4 and repeating the filtration. Finally, the retentate was reconstituted with 350 μl of 50 mM PBS and was used in the isomerase assay.
In the assay, 20 μl of retentate and filtrate was mixed with 20 μl of 50 mM PBS, 10 μl of 0.3 mM synthetic DLP-4 and 6 μl of 40 mM EDTA. This mixture was incubated at 33 °C (close to platypus average body temperature) in a water bath, for 24 h. The conversion of DLP-4 into DLP-2 was monitored by RP-HPLC with a total elution time of 35 min.
NMR spectroscopy
All NMR experiments were performed on a Bruker (Karlsruhe, Germany) AVANCE-600 DRX spectrometer using a 5-mm 1H-inverse probe operating at a sample temperature of 25 °C. The 2D (two-dimensional) NMR experiments that were performed were TOCSY [24], with a spin-lock period of 60 ms, and NOESY [25], with a mixing time of 200 ms. The spectra were acquired in phase-sensitive mode using time-proportional phase detection [26]. Solvent-signal suppression was achieved either by presaturation or by using the WATERGATE [27] pulse sequence. All spectra were processed using XWIN-NMR software (Bruker) and were analysed using the program XEASY [28].
Structure calculations
The DLP-2 structure was recalculated using all the constraint data obtained earlier [21], and a modified CNS (crystallography and NMR system) [29] structural parameter set (protein-allhdg.param, protein-allhdg.top and protein.link) that incorporates a D-methionine residue at position 2, as suggested in the default CNS protein-allhdg.param file. The standard dynamic annealing protocol anneal.inp in CNS was used to calculate 500 structures starting from a DLP-2 structure in an extended conformation. The ‘best’ 20 structures, with lowest overall energy, were considered to be representative structures of DLP-2.
An initial structure calculation strategy to that used earlier for DLP-2 [21] was adopted for DLP-4. The NOESY spectrum recorded at 25 °C with a mixing time of 200 ms provided 432 non-redundant NOE (nuclear Overhauser effect)-derived distances used for the structure calculations. The program INFIT [30] was implemented to yield 3JNHα coupling constants from 2D NOESY spectra, which then provided seven Φ dihedral angle constraints which were restrained similarly to those used in the DLP-2 structure calculation [21]. These data, together with the four disulphide bonds that were inferred in the present study and 12 hydrogen bonds, were used as structural constraints (see Table 1). Hydrogen-bonding pairs were deduced from 1H/2H exchange experiments, and the preliminary calculated structures that were obtained using the NOAH protocol [31,32] in the torsion-angle dynamics program DYANA [33]. Hydrogen-bonding constraints were only introduced in the intermediate stages of the structure calculation that implemented the standard annealing protocol in DYANA; these constraints were assigned upper distance-limits of 2.2 Å (1 Å=0.1 nm) for NHi to Oj and 3.2 Å for Ni to Oj. The final stage in DLP-4 calculation was performed in CNS using a protocol that was identical with that described above for DLP-2. Secondary structures and the locations of well-defined regions in DLP-2 and DLP-4 were determined using MOLMOL [34].
Table 1. Structural statistics for the ensembles of 20 DLP-2 and 20 DLP-4 structures.
1 kcal=4.184 kJ. EvdW, van der Waals energy.
| Quantity | DLP-2 | DLP-4 |
|---|---|---|
| Distance restraints | ||
| Intra-residue (i−j=0) | 242 | 197 |
| Sequential (|i−j|=1) | 190 | 116 |
| Medium-range (|i−j|≤5) | 95 | 36 |
| Long-range (|i−j|>5) | 172 | 83 |
| Hydrogen-bonds | 16 | 12 |
| Total | 715 | 444 |
| Dihedral angle restraints | ||
| Φ | 24 | 7 |
| Mean energies (kcal·mol−1) | ||
| ENOE | 6.79±0.63 | 7.97±1.48 |
| Econstraineddihedralangle | 0.04±0.02 | 0.01±0.01 |
| EvdW | 8.34±0.98 | 7.94±1.77 |
| Ebond | 2.14±0.12 | 1.58±0.43 |
| Eimproper | 0.89±0.13 | 0.81±0.16 |
| Eangle | 20.71±0.45 | 19.91±0.94 |
| Etotal | 38.91±1.14 | 38.22±2.25 |
| Atomic R.M.S.D. with the mean (Å) | ||
| Backbone atoms (1–42) | 1.48±0.30 | 1.71±0.46 |
| Heavy atoms (1–42) | 2.26±0.26 | 2.39±0.43 |
| Backbone atoms (7–42) | 0.38±0.09 | 0.67±0.22 |
| Heavy atoms (7–42) | 1.08±0.14 | 1.33±0.35 |
The co-ordinates for the ensemble of 20 structures of DLP-2 and DLP-4 have been deposited in the RSCB Protein Data Bank under accession codes 1ZUE and 1ZUF respectively.
RESULTS AND DISCUSSION
Discovery of a D-amino acid in DLP-2
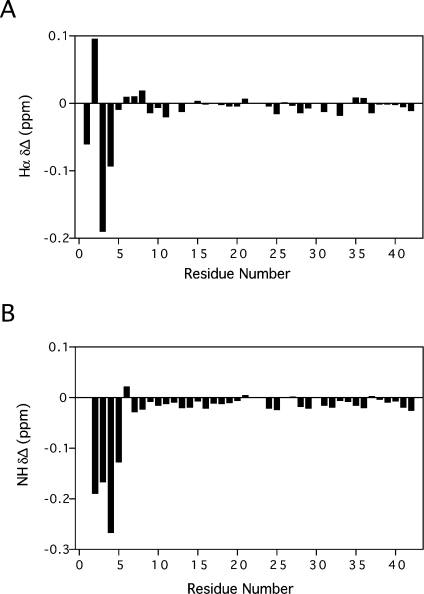
Comparison of 2D TOCSY and 2D NOESY NMR data of native DLP-4 obtained in the present study and those of DLP-2 obtained from the previous study [21] revealed that there were significant differences in chemical shifts of 1Hs in residues near the N-terminus. Specifically, the backbone proton NH and Hα NMR resonances of the first five amino acid residues in DLP-2 and DLP-4 differed significantly in chemical shift (Figure 2). This extent of the differences was similar to those observed between OvCNPa and OvCNPb [16], and is what led to the discovery of D-leucine at position 2 in OvCNPb. This outcome suggested that a D-amino acid may also be present in either DLP-2 or DLP-4, and probably at position 2 of the sequence.
Figure 2. 1H NMR spectral differences between DLP-2 and DLP-4.
Hα (A) and NH (B) 1H NMR chemical-shift differences for all amino acid residues of the two DLPs. Values were obtained by subtracting DLP-2 chemical shifts from those of DLP-4.
Besides this, the chemical shifts of the six cysteine protons of DLP-4 and DLP-2 were found to be very similar, suggesting that their cysteine-pairing configurations were identical. This contradicts the earlier suggestion that the difference between the two isomers could be due to a difference in disulphide-bonding pattern [21]. In short, all these NMR data implied that the structural difference between DLP-2 and DLP-4 was localized to the N-terminal region of the peptide.
To settle conclusively the basis of this difference, two defensin-like polypeptide isomers were chemically synthesized: the first had the same amino acid sequence as DLP-2 (and DLP-4) with all amino acids in the L-form, while the second had the same amino acid sequence except that L-methionine at position 2 was replaced by D-methionine. Comparison of RP-HPLC traces of the two chemically synthesized peptides, native DLP-2 and native DLP-4 resolved the difference between DLP-2 and DLP-4 (Figure 1C). The retention times of DLP-4 and DLP-2 were the same as those of the first and second chemically synthesized peptides respectively. This finding was supported further by 1D (one-dimensional) and 2D NMR experiments performed on native DLP-2, native DLP-4 and the two chemically synthesized peptides. The observed 1H chemical shifts and the 2D TOCSY peak patterns of synthetic L-Met2-DLP and D-Met2-DLP matched exactly those of native DLP-4 and native DLP-2 respectively. Thus it can be stated beyond doubt that the difference between DLP-2 and DLP-4 is the unusual presence of D-methionine at position 2 in DLP-2.
L-to-D-amino acid residue isomerase activity
Preliminary assay experiments by straight addition of synthetic DLP-4 to the whole gland extract with no endogenous DLPs already showed conversion into DLP-2, but the activity was observed to be very low. It was suspected that some components in the venom might be interfering with the isomerization by competitive inhibition, so that fractionation by centrifugal ultrafiltration was performed. In the assay experiment, two fractionated venom-gland extracts were used to which synthetic DLP-4 were added: one containing proteins with nominal molecular mass <30 kDa and the other containing proteins with nominal molecular mass >30 kDa. After 4 h of incubation at 33 °C, the HPLC trace of the incubation mixture with >30 kDa components showed a substantial decrease of the peak corresponding to DLP-4, while a new peak corresponding to DLP-2 emerged (Figure 3). The incubation mixture with <30 kDa components, on the other hand, did not show any DLP-2 peak. These results showed the conversion of DLP-4 into DLP-2 in the incubation mixture with >30 kDa components and confirmed the presence of the L-to-D-amino acid residue isomerase activity in that fraction. Efforts are now underway to characterize and isolate the enzyme responsible for this isomerase activity in the platypus venom gland.
Structure of DLP-2 and DLP-4
The large difference in retention time observed between DLP-2 and DLP-4 in RP-HPLC is intriguing and required further investigation because it implies that there could be a significant conformational difference between the two isomers. The overall fold of DLP-4 was thus calculated using the available 2D NOESY and 2D TOCSY data. It is important to note that, for the reported DLP-2 structure calculations, we assumed that all the amino acids were in the L-form, which, according to the present study, is no longer valid. The 3D (three-dimensional) structure of DLP-2 was therefore recalculated using a new structural template that incorporates a D-Met2 plus the atom-to-atom distance data used previously [21].
Table 1 presents a summary of the characteristics of the calculated DLP-2 and DLP-4 structures. The number of distance and dihedral angle restraints for DLP-2 is much greater than that for DLP-4. This is basically because of the more comprehensive experiments performed for DLP-2 than for DLP-4. This resulted in a lower R.M.S.D. for DLP-2 on the well-defined region of the structure.
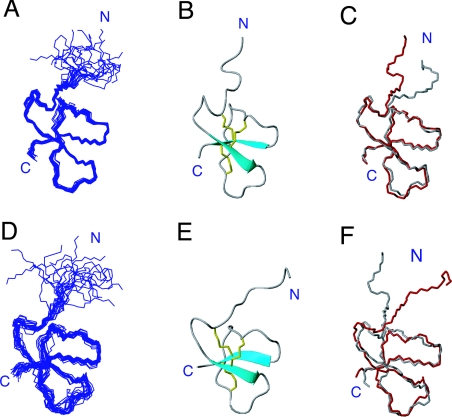
The overall fold of the recalculated DLP-2 with D-Met2 is very similar to the old DLP-2 structure with L-Met2 (Figures 4A–4C). The R.M.S.D. (root mean square deviation) is 0.66 Å when the backbone atoms of residues 7–42 (the well-defined region) of one of the new DLP-2 structures is superimposed over one of the old DLP-2 structures. This is expected, given that the same structural constraints were used in the calculation and Met2 is located in the disordered region of the molecule and had no medium- or long-range NOEs with other residues. The small difference observed is perhaps due to the absence of the short helix at residues 9–12, but this is not a real difference, since the helix is not consistently present in the ensemble of old DLP-2 structures, and two of the structures in the ensemble of the new DLP-2 structures do have it. The anti-parallel β-sheets defined by residues 15–18 and 37–40 were consistently present in both sets of calculated structures.
Figure 4. Structures of DLP-2 and DLP-4.
(A) Ensemble of the 20 ‘best’ recalculated DLP-2 structures aligned by superimposing the backbone atoms of residues 7–42. (B) Ribbon diagram of the recalculated DLP-2 structure showing the secondary-structural elements and disulphide connectivities. (C) Superposition of the recalculated (red) and previously reported DLP-2 structures (grey; Protein Data Bank code 1D6B) over the backbone of residues 7–42. Only the polypeptide backbone chains are shown. (D) Ensemble of the 20 ‘best’ DLP-4 structures aligned by superimposing the backbone atom of residues 7–42. (E) Ribbon diagram showing the secondary-structural elements and disulphide connectivities. (F) Superposition of the recalculated DLP-2 (grey) and DLP-4 (red) structures over the backbone of residues 7–42.
The structure of DLP-4 as shown in Figures 4(D) and 4(E) has very strong similarity to the DLP-2 shown in Figures 4(A) and 4(B), but its resolution is lower (see also Table 1). This can be explained by the fact that, although the amount of data used in calculating the DLP-4 structure was significantly less than that used for DLP-2, comparison of their NOESY data showed that the medium- and long-range NOE patterns were very similar. Hence, this does not suggest any significant difference in their overall folds and secondary structures. The N-terminal region up to residue 7 of DLP-4, which contained the important L-Met2 residue, was found to be similarly disordered owing to a lack of medium and long-range NOEs in this section of the molecule. Superposition of one of the structures of DLP-4 and DLP-2 on the backbone of the well-defined region, as defined by residues 7–42, resulted in an R.M.S.D. of 1.46 Å; this showed clearly that the tertiary structures of both molecules were very similar (Figure 4F). It is not relevant to compare the local structures near residue 2 in detail because these parts are disordered in both molecules; however, the calculated structures obtained here imply that the substitution of D-Met2 for L-Met2 had negligible effect on the overall fold of the DLP.
The strong similarity between the solution structures of DLP-2 and DLP-4 obtained in the present study does not explain the significant RP-HPLC retention time difference between the two molecules. It is easy to accept that for small peptides of fewer than ten residues, the conversion of an L- into a D-amino acid will result in a substantial change in structure that leads to a significant difference in chromatographic behaviour [7]. For larger polypeptides, such as the DLPs, the effect of D-amino acid substitution on structure and hence chromatographic behaviour should be less prominent. It is clear from the different elution times that the hydrophobicities of the two molecules in RP-HPLC are considerably different. Therefore a possible explanation that accounts for the results is that the structures of the two polypeptides when placed in a more hydrophobic environment are different. Note that the N-termini of DLP-2 and DLP-4 are highly hydrophobic, as four of first five residues are hydrophobic (see Figure 1A). It is possible that, in the RP-HPLC environment, the flexible N-terminus that incorporates Met2 interacts with the surface of the compact invariant part of the molecule to form a ‘denatured’ state whose structure and hydrophobicity are strongly dependent on the chirality of Met2. For example, the N-terminal hydrophobic side chain might be more buried in DLP-4, making the whole molecule more hydrophilic, while, in DLP-2, this side chain might be more exposed to the solvent, making the whole molecule more hydrophobic.
Biological implications
The discovery of a D-amino acid residue in DLP-2 means that there are now known to be two polypeptides, DLP-2 and OvCNPb, in the platypus venom that contain a D-amino acid at position 2. Both these molecules are larger (DLP-2 and OvCNPb consist of 42 and 39 amino acid residues respectively) than many of the D-amino-acid-containing peptides discovered elsewhere. The importance of this post-translational modification in the platypus is yet to be established, but, in other studies with other organisms, it has been shown that the D-amino-acid-containing peptide is not the only potent form, or it is at least more potent than the isomer that contains all L-forms of the residues [7]. The present study suggests the presence of at least one peptide-residue isomerase that catalyses the conversion of the L-form of the second amino acid residue in a peptide into its D-form. Since the second amino acid residues in OvCNPb and in DLP-2 are different (D-Leu2 in OvCNPb and D-Met2 in DLP-2), it is possible that there are two enzymes that are specific for each substrate. However, we cannot discount the possibility that there is only one isomerase that has broad specificity, acting on both OvCNPa and DLP-4. In the present study, we have successfully shown that there is an isomerase activity in the venom that converts DLP-4 into DLP-2. It will be important to characterize this enzyme comprehensively to see whether it is related to other common proteins and whether it can act on other molecules besides DLP-4.
Note that the presence of a D-amino acid residue at the second position in a peptide is not unique to the platypus, since it has also been observed in secreted peptides from frogs and cone-snails [1–3,5,6], but our findings are the first for a mammal. The new information obtained in the present study therefore suggests the presence of a common or related type of isomerase in these organisms. Recently, Jilek et al. [35] discovered an isomerase in frog skin secretions that converts the L-form of the second amino acid residue into the D-form in a model peptide patterned after bombinin H. The enzyme is a glycoprotein with an apparent size of 52 kDa and has similarity to the N-terminal H-domain of the human IgG Fc-binding protein [36]. It has been speculated that a similar enzyme and D-amino-acid-containing peptides, such as hormones, antibacterial and neuropeptides, might be present in other vertebrate species. Our present work supports this idea, since DLPs are structurally related to mammalian β-defensins. Thus it will be worthwhile to search for similar proteins in the mammalian genome to explore the possibility that D-amino-acid-containing biologically active proteins or peptides exist in mammals, including humans.
The occurrence of the D-amino acid residue is not limited to only the second position in the peptide sequences: some peptides in cone snail venoms [4,8,9] and crustacean eyestalks [11,13], for example, have D-amino acid residues at positions 4 and 3, while in ω-Aga IVB (ω-Aga-TK) in funnelweb spider venom [14,15] and in another cone snail venom [7], the D-amino acid residue is situated at position 3 from the C-terminus. These modifications most certainly require a different type of enzyme, as has already been found for the funnelweb spider venom [14]. Since detecting D-amino acids in peptides and proteins is not straightforward, it is quite possible that many molecules of these sorts and their respective enzymes are more prevalent in Nature than previously anticipated and are awaiting discovery.
Acknowledgments
This work was supported by an Australian Research Council Discovery Grant to P.W.K. and to Dr J.I. Vandenberg, who is also thanked for valuable discussions. We thank Mr Nicholas Mok for conducting initial experiments in this work, Dr Bill Bubb for help with the NMR experiments, and Mr Bill Lowe for expert technical assistance.
References
- 1.Montecucchi P. C., de Castiglione R., Piani S., Gozzini L., Erspamer V. Amino acid composition and sequence of dermorphin, a novel opiate-like peptide from the skin of Phyllomedusa sauvagei. Int. J. Pept. Protein Res. 1981;17:275–283. doi: 10.1111/j.1399-3011.1981.tb01993.x. [DOI] [PubMed] [Google Scholar]
- 2.Kreil G. D-Amino acids in animal peptides. Annu. Rev. Biochem. 1997;66:337–345. doi: 10.1146/annurev.biochem.66.1.337. [DOI] [PubMed] [Google Scholar]
- 3.Richter K., Egger R., Kreil G. D-Alanine in the frog-skin peptide dermorphin is derived from L-alanine in the precursor. Science. 1987;238:200–202. doi: 10.1126/science.3659910. [DOI] [PubMed] [Google Scholar]
- 4.Jimenez E. C., Olivera B. M., Gray W. R., Cruz L. J. Contryphan is a D-tryptophan-containing Conus peptide. J. Biol. Chem. 1996;271:28002–28005. doi: 10.1074/jbc.271.45.28002. [DOI] [PubMed] [Google Scholar]
- 5.Kamatani Y., Minakata H., Kenny P. T., Iwashita T., Watanabe K., Funase K., Sun X. P., Yongsiri A., Kim K. H., Novales-Li P. Achatin-I, an endogenous neuroexcitatory tetrapeptide from Achatina fulica Ferussac containing a D-amino acid residue. Biochem. Biophys. Res. Commun. 1989;160:1015–1020. doi: 10.1016/s0006-291x(89)80103-2. [DOI] [PubMed] [Google Scholar]
- 6.Ohta N., Kubota I., Takao T., Shimonishi Y., Yasudakamatani Y., Minakata H., Nomoto K., Muneoka Y., Kobayashi M. Fulicin, a novel neuropeptide containing a D-amino-acid residue isolated from the ganglia of Achatina fulica. Biochem. Biophys. Res. Commun. 1991;178:486–493. doi: 10.1016/0006-291x(91)90133-r. [DOI] [PubMed] [Google Scholar]
- 7.Buczek O., Yoshikami D., Bulaj G., Jimenez E. C., Olivera B. M. Post-translational amino acid isomerization: a functionally important D-amino acid in an excitatory peptide. J. Biol. Chem. 2005;280:4247–4253. doi: 10.1074/jbc.M405835200. [DOI] [PubMed] [Google Scholar]
- 8.Jacobsen R., Jimenez E. C., Grilley M., Watkins M., Hillyard D., Cruz L. J., Olivera B. M. The contryphans, a D-tryptophan-containing family of Conus peptides: interconversion between conformers. J. Pept. Res. 1998;51:173–179. doi: 10.1111/j.1399-3011.1998.tb01213.x. [DOI] [PubMed] [Google Scholar]
- 9.Jacobsen R. B., Jimenez E. C., De la Cruz R. G. C., Gray W. R., Cruz L. J., Olivera B. M. A novel D-leucine-containing Conus peptide: diverse conformational dynamics in the contryphan family. J. Pept. Res. 1999;54:93–99. doi: 10.1034/j.1399-3011.1999.00093.x. [DOI] [PubMed] [Google Scholar]
- 10.Soyez D., Toullec J. Y., Ollivaux C., Geraud G. L to D amino acid isomerization in a peptide hormone is a late post-translational event occurring in specialized neurosecretory cells. J. Biol. Chem. 2000;275:37870–37875. doi: 10.1074/jbc.M007302200. [DOI] [PubMed] [Google Scholar]
- 11.Soyez D., Van Herp F., Rossier J., Le Caer J. P., Tensen C. P., Lafont R. Evidence for a conformational polymorphism of invertebrate neurohormones: D-amino acid residue in crustacean hyperglycemic peptides. J. Biol. Chem. 1994;269:18295–18298. [PubMed] [Google Scholar]
- 12.Gallois D., Brisorgueil M. J., Conrath M., Mailly P., Soyez D. Posttranslational isomerization of a neuropeptide in crustacean neurosecretory cells studied by ultrastructural immunocytochemistry. Eur. J. Cell Biol. 2003;82:431–440. doi: 10.1078/0171-9335-00329. [DOI] [PubMed] [Google Scholar]
- 13.Yasuda A., Yasuda Y., Fujita T., Naya Y. Characterization of crustacean hyperglycemic hormone from the crayfish (Procambarus clarkii): multiplicity of molecular forms by stereoinversion and diverse functions. Gen. Comp. Endocrinol. 1994;95:387–398. doi: 10.1006/gcen.1994.1138. [DOI] [PubMed] [Google Scholar]
- 14.Heck S. D., Siok C. J., Krapcho K. J., Kelbaugh P. R., Thadeio P. F., Welch M. J., Williams R. D., Ganong A. H., Kelly M. E., Lanzetti A. J. Functional consequences of posttranslational isomerization of Ser46 in a calcium channel toxin. Science. 1994;266:1065–1068. doi: 10.1126/science.7973665. [DOI] [PubMed] [Google Scholar]
- 15.Shikata Y., Watanabe T., Teramoto T., Inoue A., Kawakami Y., Nishizawa Y., Katayama K., Kuwada M. Isolation and characterization of a peptide isomerase from funnel web spider venom. J. Biol. Chem. 1995;270:16719–16723. doi: 10.1074/jbc.270.28.16719. [DOI] [PubMed] [Google Scholar]
- 16.Torres A. M., Menz I., Alewood P. F., Bansal P., Lahnstein J., Gallagher C. H., Kuchel P. W. D-Amino acid residue in the C-type natriuretic peptide from the venom of the mammal, Ornithorhynchus anatinus, the Australian platypus. FEBS Lett. 2002;524:172–176. doi: 10.1016/s0014-5793(02)03050-8. [DOI] [PubMed] [Google Scholar]
- 17.de Plater G. M., Martin R. L., Milburn P. J. A C-type natriuretic peptide from the venom of the platypus (Ornithorhynchus anatinus): structure and pharmacology. Comp. Biochem. Physiol. Part C Pharmacol. Toxicol. Endocrinol. 1998;120:99–110. doi: 10.1016/s0742-8413(98)00030-9. [DOI] [PubMed] [Google Scholar]
- 18.de Plater G. M., Martin R. L., Milburn P. J. The natriuretic peptide (ovCNP-39) from platypus (Ornithorhynchus anatinus) venom relaxes the isolated rat uterus and promotes oedema and mast cell histamine release. Toxicon. 1998;36:847–857. doi: 10.1016/s0041-0101(97)00176-1. [DOI] [PubMed] [Google Scholar]
- 19.Torres A. M., Alewood D., Alewood P. F., Gallagher C. H., Kuchel P. W. Conformations of platypus venom C-type natriuretic peptide in aqueous solution and sodium dodecyl sulfate. Toxicon. 2002;40:711–719. doi: 10.1016/s0041-0101(01)00266-5. [DOI] [PubMed] [Google Scholar]
- 20.Torres A. M., Wang X., Fletcher J. I., Alewood D., Alewood P. F., Smith R., Simpson R. J., Nicholson G. M., Sutherland S. K., Gallagher C. H., et al. Solution structure of a defensin-like peptide from platypus venom. Biochem. J. 1999;341:785–794. [PMC free article] [PubMed] [Google Scholar]
- 21.Torres A. M., de Plater G. M., Doverskog M., Birinyi-Strachan L. C., Nicholson G. M., Gallagher C. H., Kuchel P. W. Defensin-like peptide-2 from platypus venom: member of a class of peptides with a distinct structural fold. Biochem. J. 2000;348:649–656. [PMC free article] [PubMed] [Google Scholar]
- 22.Torres A. M., Kuchel P. W. The β-defensin-fold family of polypeptides. Toxicon. 2004;44:581–588. doi: 10.1016/j.toxicon.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 23.Schnolzer M., Alewood P., Jones A., Alewood D., Kent S. B. In situ neutralization in Boc-chemistry solid phase peptide synthesis: rapid, high yield assembly of difficult sequences. Int. J. Pept. Protein Res. 1992;40:180–193. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 24.Bax A., Davis D. G. Mlev-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985;65:355–360. [Google Scholar]
- 25.Kumar A., Ernst R. R., Wüthrich K. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton–proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 1980;95:1–6. doi: 10.1016/0006-291x(80)90695-6. [DOI] [PubMed] [Google Scholar]
- 26.Marion D., Wüthrich K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H–1H spin–spin coupling constants in proteins. Biochem. Biophys. Res. Commun. 1983;113:967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- 27.Piotto M., Saudek V., Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 28.Bartels C., Xia T. H., Billeter M., Guntert P., Wüthrich K. The program Xeasy for computer-supported NMR spectral-analysis of biological macromolecules. J. Biomol. NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 29.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Szyperski T., Guntert P., Otting G., Wüthrich K. Determination of scalar coupling-constants by inverse Fourier transformation of in-phase multiplets. J. Magn. Reson. 1992;99:552–560. [Google Scholar]
- 31.Mumenthaler C., Braun W. Automated assignment of simulated and experimental NOESY spectra of proteins by feedback filtering and self-correcting distance geometry. J. Mol. Biol. 1995;254:465–480. doi: 10.1006/jmbi.1995.0631. [DOI] [PubMed] [Google Scholar]
- 32.Mumenthaler C., Guntert P., Braun W., Wüthrich K. Automated combined assignment of NOESY spectra and three-dimensional protein structure determination. J. Biomol. NMR. 1997;10:351–362. doi: 10.1023/a:1018383106236. [DOI] [PubMed] [Google Scholar]
- 33.Guntert P., Mumenthaler C., Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 34.Koradi R., Billeter M., Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:51–55–29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 35.Jilek A., Mollay C., Tippelt C., Grassi J., Mignogna G., Mullegger J., Sander V., Fehrer C., Barra D., Kreil G. Biosynthesis of a D-amino acid in peptide linkage by an enzyme from frog skin secretions. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4235–4239. doi: 10.1073/pnas.0500789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harada N., Iijima S., Kobayashi K., Yoshida T., Brown W. R., Hibi T., Oshima A., Morikawa M. Human IgGFc binding protein (FcγBP) in colonic epithelial cells exhibits mucin-like structure. J. Biol. Chem. 1997;272:15232–15241. doi: 10.1074/jbc.272.24.15232. [DOI] [PubMed] [Google Scholar]