Abstract
Little is known about fungal biofilms, which may cause infection and antibiotic resistance. In this study, biofilm formation by different Candida species, particularly Candida albicans and C. parapsilosis, was evaluated by using a clinically relevant model of Candida biofilm on medical devices. Candida biofilms were allowed to form on silicone elastomer and were quantified by tetrazolium (XTT) and dry weight (DW) assays. Formed biofilm was visualized by using fluorescence microscopy and confocal scanning laser microscopy with Calcofluor White (Sigma Chemical Co., St. Louis, Mo.), concanavalin A-Alexafluor 488 (Molecular Probes, Eugene, Oreg.), and FUN-1 (Molecular Probes) dyes. Although minimal variations in biofilm production among invasive C. albicans isolates were seen, significant differences between invasive and noninvasive isolates (P < 0.001) were noted. C. albicans isolates produced more biofilm than C. parapsilosis, C. glabrata, and C. tropicalis isolates, as determined by DW assays (P was <0.001 for all comparisons) and microscopy. Interestingly, noninvasive isolates demonstrated a higher level of XTT activity than invasive isolates. On microscopy, C. albicans biofilms had a morphology different from that of other species, consisting of a basal blastospore layer with a dense overlying matrix composed of exopolysaccharides and hyphae. In contrast, C. parapsilosis biofilms had less volume than C. albicans biofilms and were comprised exclusively of clumped blastospores. Unlike planktonically grown cells, Candida biofilms rapidly (within 6 h) developed fluconazole resistance (MIC, >128 μg/ml). Importantly, XTT and FUN-1 activity showed biofilm cells to be metabolically active. In conclusion, our data show that C. albicans produces quantitatively larger and qualitatively more complex biofilms than other species, in particular, C. parapsilosis.
Biofilms represent the most prevalent type of microbial growth in nature and are crucial to the development of clinical infections (14, 34). In the latter setting, they serve as a nidus for disease and are associated with high-level antibiotic resistance of the associated organisms (29). Although bacterial biofilms involving organisms such as Pseudomonas have been well characterized (14, 34), the study of fungal biofilms is still in its infancy.
Candida is the fourth most common cause of bloodstream infections in hospitalized patients (5). Up to 40% of patients with Candida strains isolated from intravenous catheters have underlying fungemia (1, 32), and the mortality rate of patients with catheter-related candidemia approaches 40% (32). While Candida albicans is the most commonly isolated fungal species, other species are being isolated with increasing frequency (26, 36). In several studies, C. parapsilosis has become the second most commonly isolated fungal organism (25, 35). This species is of special concern in critically ill neonates, in whom it is known to be associated with the use of central lines and parenteral nutrition (42, 43, 50).
Candidiasis associated with intravenous lines and bioprosthetic devices is problematic, since these devices can act as substrates for biofilm growth. Antifungal therapy alone is insufficient for cure; affected devices generally need to be removed (30, 40). Removal of these devices has serious implications in the setting of heart valves, joint prostheses, and central nervous system shunts. Until recently, the reason for the need for device removal has been a mystery. However, our laboratories and others have demonstrated nearly total resistance of biofilm-associated organisms to antifungal agents (2, 10, 11, 21).
Previous Candida biofilm model systems have had numerous limitations. In some studies, common pathogenic yeast isolates or clinically relevant materials were not used (41). In other studies, experiments were performed with sections of catheter material, which are difficult to work with, quantify, or image in detail (20). Thus, to study Candida biofilms representative of those formed on biomedical materials, it was necessary to develop a new, more physiologically relevant model of biofilm formation on intravascular catheters and indwelling bioprosthetic devices.
Our goal was to design a model enabling detailed quantitation and confocal microscopic analysis of biofilms. In this study, we developed such a model and used it to compare biofilms from clinical C. albicans strains obtained from both different body sites and different types of infections. We also compared biofilm production by various Candida species. In particular, we compared C. albicans with C. parapsilosis because of the increasing prevalence, importance as an invasive pathogen (especially in neonatal disease), and association with intravenous lines of the latter (42, 43, 50). Given the preeminence of C. albicans in clinical infections, we hypothesized that strains of this species isolated from sites of invasive disease (i.e., obtained from normally sterile sites) would be better biofilm formers than those obtained from nonsterile sites. We also postulated that C. albicans would form more biofilms than non-C albicans species. Finally, to determine if the model had functional relevance, we examined the fluconazole susceptibility of C. albicans biofilms.
MATERIALS AND METHODS
Organisms.
The various Candida isolates (C. albicans, C. parapsilosis, C. glabrata, and C. tropicalis) used in this study were obtained by subculturing clinical specimens from the microbiology laboratory at the University Hospitals of Cleveland. Species identification was performed by using routine germ tube and API testing methods. C. albicans strain M61 was obtained at University Hospitals of Cleveland from an intravascular line culture, and C. parapsilosis strain A71 was obtained from a sputum culture. C. albicans strain GDH2346 was obtained from a patient with documented denture stomatitis (obtained from L. Julia Douglas, University of Glasgow, Glasgow, United Kingdom) and was previously shown to produce biofilms (11).
Medium and growth conditions.
All Candida strains were grown in yeast nitrogen base (YNB) medium (Difco Laboratories, Detroit, Mich.) supplemented with 50 mM glucose (11). Fifty milliliters of medium (in 250-ml Erlenmeyer flasks) was inoculated with Candida from fresh Sabouraud dextrose agar plates (Difco) and incubated for 24 h at 37°C in an orbital water bath shaker at 60 rpm. Cells were harvested and washed twice with 0.15 M phosphate-buffered saline (PBS; pH 7.2, Ca2+- and Mg2+-free). Cells were resuspended in 10 ml of PBS, counted after serial dilution, standardized, and used immediately.
Substrate material.
Silicone elastomer (SE) sheets were obtained from Cardiovascular Instrument Corp., Wakefield, Mass. This material was chosen due to its similarity to material used in indwelling devices, its availability as flat medical-grade sheeting (which is not the case for materials such as catheter-grade polyvinyl chloride), and the documented ability of SE to promote Candida biofilm formation (20). A flat profile facilitates quantitation and imaging of biofilms. In accordance with the manufacturer's instructions, the material was cleaned by washing in hand soap and water, rinsed with distilled water, and autoclaved. Flat circular disks, 1.5 cm in diameter, were obtained by cutting with a cork borer.
Biofilm formation.
SE disks were placed in 12-well tissue culture plates (Becton Dickinson, Franklin Lakes, N.J.) and incubated in fetal bovine serum (FBS) for 24 h at 37°C on a rocker table (Bellco Glass Inc., Vineland, N.J.) (pretreatment phase) (10, 11). The rocker table was used to provide quasi-linear medium flow over the surface of the disks. The disks were then moved to new plates and washed with PBS to remove residual FBS. To ensure uniform biofilm formation on disks, we immersed them in a Candida cell suspension. Three milliliters of standardized cell suspension, containing 107 blastospores/ml, was added to the wells, and the disks were incubated for 90 min at 37°C on a rocker table (adhesion phase). The disks were gently agitated and transferred to new plates to ensure the removal of nonadherent cells. The disks were then immersed in YNB medium with 50 mM glucose and incubated for 48 h at 37°C on a rocker table (biofilm formation phase). For controls, disks were processed in identical fashion, except that no Candida cells were added. All assays were carried out in quadruplicate and on different days.
Quantitation of biofilms.
Quantitation of Candida biofilms was performed as described previously (11) by using both a biochemical assay, i.e., the 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT; Sigma Chemical Co., St. Louis, Mo.) reduction assay (20), and dry weight (DW) measurements. XTT is reduced by mitochondrial dehydrogenase into a water-soluble formazan product that is measured spectrophotometrically. Following the biofilm formation phase, SE disks containing C. albicans biofilms were transferred to new 12-well tissue culture plates containing 3 ml of PBS per well. Fifty microliters of XTT salt solution (1 mg/ml in PBS) and 4 μl of menadione solution (1 mM in acetone; Sigma) were added to each well. The plates were incubated at 37°C for 5 h, and then the medium was removed and centrifuged for 5 min at 6,000 × g to pellet any suspended cells or debris. XTT formazan in the supernatant was measured at 492 nm by using a spectrophotometer (Genesys 5; Spectronic Instruments, Rochester, N.Y.).
DW measurements represent total biofilm mass, including fungal cells and extracellular matrix (for details of this technique, see references 11 and 20). Briefly, biofilms were scraped off the surface of the disks by using a cell scraper (Becton Dickinson), and both disks and scrapers were rinsed with PBS to remove residual biofilms. The material was filtered by using a 0.45-μm-pore-size Millipore filter, dried in an incubator at 37°C for 48 h, and weighed.
Wet weight (WW) measurements represent the entire, hydrated mass of biofilm. After biofilm formation, preweighed SE disks with attached biofilm were removed from culture plates, carefully side blotted to remove excess medium without disrupting the biofilm, and weighed.
FM.
Evaluation of gross biofilm morphology was carried out by using fluorescence microscopy (FM). Intact biofilms on SE disks were transferred to microscope slides and stained with 1 drop of Calcofluor White (0.05% [vol/vol]; Sigma). The samples were examined on a ZVS-47E Axioskop (Carl Zeiss Inc., Thornwood, N.Y.) platform (11) at a magnification of ×10 to ×20 by using a Zeiss short-arc mercury lamp with excitation at 395 to 440 nm, beam splitting at 460 nm, and emission at 470 nm. Images were captured by using Zeiss Axiovision v3.0.6 software.
Confocal microscopy.
Biofilm staining and confocal scanning laser microscopy (CSLM) were performed as described previously (10). FUN-1 (Molecular Probes, Eugene, Oreg.) is a fluorescent dye taken up by fungal cells; in the presence of metabolic viability, it is converted from a diffuse yellow cytoplasmic stain to red, rod-like collections. Concanavalin A-Alexafluor 488 conjugate (CAAF; Molecular Probes) selectively binds to polysaccharides including α-mannopyranosyl and α-glucopyranosyl residues and gives green fluorescence. Following biofilm formation, disks were removed and transferred to new 12-well plates. FUN-1 (10 μM) from a 10 mM stock and 25 μg of CAAF/ml from a 5-mg/ml stock were mixed in 4 ml of PBS. This mixture was added to wells containing biofilm disks. The plates were then incubated for 45 min at 37°C. The disks were removed from the wells, placed in 35-mm glass-bottom microwell dishes (MatTek Corp., Ashland, Mass.), and observed by using a Zeiss Axiovert 100 M confocal scanning laser microscope (with a rhodamine-fluorescein isothiocyanate protocol and with excitation at 543 [HeNe laser] and 488 nm [argon laser], beam splitting at 488 and 543 nm, and emission at 560 and 505 nm for FUN-1 [red] and CAAF [green], respectively). The lenses used included Zeiss Achroplan 20x/0.5 and C-Apochromat 40x/1.2 water immersion objectives. Images were captured and processed by using Zeiss LSM510 v2.8 and Photoshop v5.5 (Adobe Systems, Inc., San Jose, Calif.) software.
Antifungal susceptibility.
Fluconazole was obtained from Pfizer Pharmaceuticals Group (New York, N.Y.). We used a published method to determine the antifungal susceptibility of biofilm-grown Candida (11, 21). Briefly, following biofilm formation, disks were gently agitated and transferred to new culture plates to remove non-biofilm-associated cells. YNB medium (3 ml) containing different concentrations of fluconazole was added to each well to produce final concentrations of 0.5 to 128 μg/ml. Biofilm activity at 48 h was determined by using the XTT assay as described above. The antifungal concentration which caused a 50% reduction in the metabolic activity of the biofilm compared with the activity of the control (incubated in the absence of drug) was then determined by using the XTT colorimetric results (21). As shown previously, when this assay is used, the 50% reduction in metabolic activity is equivalent to the MIC at which 50% of tested isolates are inhibited (MIC50), as determined by the NCCLS M27-A method (11, 23, 31). The antifungal susceptibility of planktonically grown cells to fluconazole was determined by using both the established XTT assay (23) and the NCCLS M27-A method (31) to ensure validity. Fluconazole resistance was defined as an MIC of ≥64 (41).
Statistical analysis.
Each experiment was performed in quadruplicate on at least two separate days; data shown in the figures are from one representative experiment. Comparative results for different isolates were normalized to C. albicans strain M61 which, by definition, was considered to have 100% activity. Statistical analysis, including analysis of variance post hoc analysis with the Bonferroni-Dunn calculation, was performed by using StatView v5.0.1 software (SAS Institute, Cary, N.C.). Significance levels for P values are given in the figure legends.
RESULTS
Optimization of biofilm growth on SE disks.
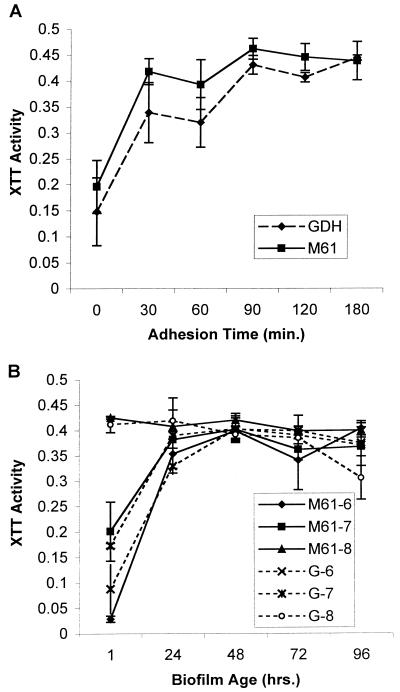
Preliminary experiments showed that preincubation of SE disks with FBS consistently produced more abundant biofilm than incubation of disks with PBS (data not shown). In previous studies, microliter amounts of cell suspensions were used for the adhesion phase of biofilm formation on catheter material (11, 20). We found it difficult to obtain consistent results with such a method, likely because of the highly hydrophobic surface of the SE disks. As a result, we immersed disks in 3 ml of cell suspension during the adhesion phase. We measured biofilm growth by using the XTT assay. When confined to a single species or strain, this method correlates with the more laborious biofilm DW measurement method (11). Figure 1 illustrates biofilm growth obtained with C. albicans strains M61 and GDH2346 to determine optimal adhesion time, inoculum concentration, and duration of growth. The latter strain was used to provide a reference to an established dental biofilm model system (10, 11). As shown in Fig. 1A, adhesion under static conditions for 90 min produced maximal subsequent biofilm formation at 48 h. Figure 1B shows that biofilm formation by both M61 and GDH2346 on SE appeared to reach a plateau by 48 h, regardless of inoculum size. The XTT, DW, and FM analyses revealed that the inoculum size of 107 blastospores produced the most abundant and stable mature biofilms. As measured by the DW method at 48 h and under optimal conditions, 3 to 4 mg of biofilm was produced per disk.
FIG. 1.
Optimization of Candida biofilm growth on SE disks. (A) Effect of adhesion time on biofilm formation by C. albicans (strains M61 and GDH2346 [GDH]). XTT activity was measured at 492 nm. (B) Effect of C. albicans inoculum concentration on subsequent biofilm formation. The scheme for strain names is as follows: M61-7, C. albicans M61 at 107; G-8, GDH2346 at 108; and so on. Assays were performed in quadruplicate. Each result is representative of at least two experiments. All values are means and standard deviations.
Invasive C. albicans isolates form more biofilm than noninvasive isolates.
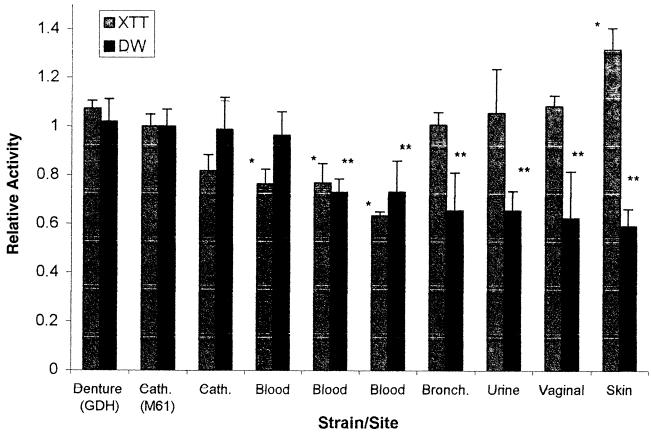
Figure 2 shows a comparison of biofilm formation by C. albicans isolates obtained from different sites. None of the isolates obtained from locations other than catheters or blood came from patients with known systemic infections. The results were normalized to the activity of strain M61. C. albicans GDH2346 is a documented biofilm producer from a patient with denture stomatitis (11). Our results show that catheter and blood isolates had consistently higher DWs (representing actual biofilm formation) than commensal isolates (the P value was <0.001 when M61 was compared to bronchial, urine, vaginal, and skin isolates). An apparent bimodal distribution was also observed among C. albicans blood isolates, with some forming larger amounts of biofilm than others, although the differences were not statistically significant.
FIG. 2.
Comparison of biofilms formed by different C. albicans isolates. The graph shows XTT and DW values for isolates of C. albicans obtained from different clinical sites. Cath., catheter; Bronch., bronchial site. Results were normalized to those for C. albicans strain M61. Assays were performed in quadruplicate. Each result is representative of at least two experiments. All values are means and standard deviations. Comparisons are significant for P values of <0.0011. A single asterisk indicates a P value of <0.001 for an XTT value of an isolate compared to M61; a double asterisk indicates a P value of <0.001 for a DW value of an isolate compared to M61.
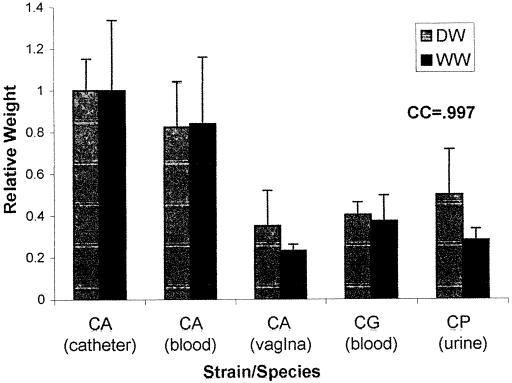
In contrast, the level of XTT activity of biofilms formed by noninvasive isolates was as high as or higher than that seen with invasive isolates (i.e., those found in normally sterile sites, such as catheters or blood; the P value was <0.001 for skin specimens only) and exhibited a marked discrepancy from relative biofilm DWs which, as shown above, were higher in invasive isolates. In fact, there was often discordance between biofilm formation measured by XTT activity and that determined by DW measurements. This result was presumably due to a distinction between the presence of the blastospore layer, different metabolic rates of individual isolates, and the amount of matrix produced. The basal cell layer appeared responsible for nearly all XTT activity, since XTT conversion was noted to be unchanged in disks from which the biofilm matrix was manually removed. To ensure that DW measurements were not misrepresenting biofilm comparisons by overlooking the aqueous component of the matrix (52), we compared DW and WW and found them to be well correlated (Fig. 3), with a correlation coefficient of 0.997.
FIG. 3.
Correlation of DW and WW measurements. The graph shows DW and WW measurements for different Candida isolates. CA, CG, and CP, C. albicans, C. glabrata, and C. parapsilosis, respectively. Results were normalized to those for C. albicans strain M61. Assays were performed in quadruplicate. Each result is representative of at least two experiments. All values are means and standard deviations. CC, correlation coefficient for the comparison of DW and WW measurements for all five isolates.
C. albicans produces quantitatively more biofilm than other Candida species.
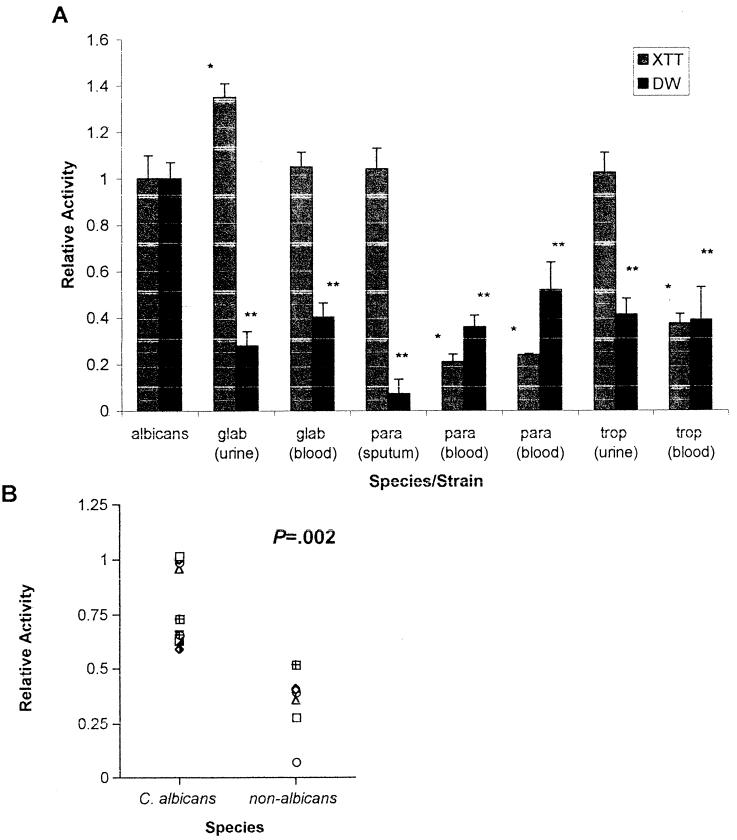
Figure 4A shows biofilm formation by different Candida species, as measured by the XTT and DW methods. C. albicans consistently formed more biofilm than non-C. albicans species, as shown by DW analysis (the P value was <0.001 for all isolates in a comparison with C. albicans M61) and as supported by subsequent microscopy (see below). In particular, biofilm DW measurements for C. parapsilosis were consistently smaller than those for C. albicans. To ensure that the much smaller amount of biofilm formed by C. parapsilosis was not due to unfavorable growth parameters (since clinically, C. parapsilosis favors sites exposed to higher glucose concentrations provided by parenteral feeding), we repeated the experiments with 275 mM glucose (equivalent to a 5% dextrose solution) and obtained similar results (data not shown). Figure 4B shows a comparison of all C. albicans (n = 10) and non-C. albicans (n = 7) isolates. C. albicans isolates consistently produced more biofilm, as determined by DW measurements, than non-C. albicans isolates (the P value was 0.002 when the means of the two groups were compared).
FIG. 4.
Comparison of biofilms formed by different Candida species. (A) XTT and DW measurements for different species of Candida. albicans, glab, para, and trop, C. albicans, C. glabrata, C. parapsilosis, and C. tropicalis, respectively. Results were normalized to those for C. albicans strain M61. Assays were performed in quadruplicate. Each result is representative of at least two experiments. All values are means and standard deviations. Comparisons are significant for P values of <0.0011. A single asterisk indicates a P value of <0.001 for an XTT value of an isolate compared to M61; a double asterisk indicates a P value of <0.001 for a DW value of an isolate compared to M61. (B) DW values for C. albicans versus non-C. albicans species shown in Fig. 2 and 4A, respectively.
Interestingly, examination of XTT activity across species showed that, in line with the data above, noninvasive isolates exhibited a higher level of XTT activity than invasive isolates (Fig. 4) (the P value was <0.001 for all intraspecies comparisons, except for that with C. glabrata). The noninvasive isolates also had a consistently high relative XTT/relative DW ratio, like that of the noninvasive C. albicans isolates in Fig. 2 (the single exception was the high XTT/DW ratio of the C. glabrata blood isolate). A DW comparison of non-C. albicans invasive and noninvasive isolates revealed that, in general, invasive isolates produced more biofilm than noninvasive isolates (although the differences were not statistically significant), except for C. tropicalis. To resolve the differences between XTT and DW, we examined the formed biofilms by using FM and found that, despite their high level of XTT activity, noninvasive isolates produced much less biofilm (data not shown), parallel to the DW results.
C. albicans biofilms grown on SE form a complex structure.
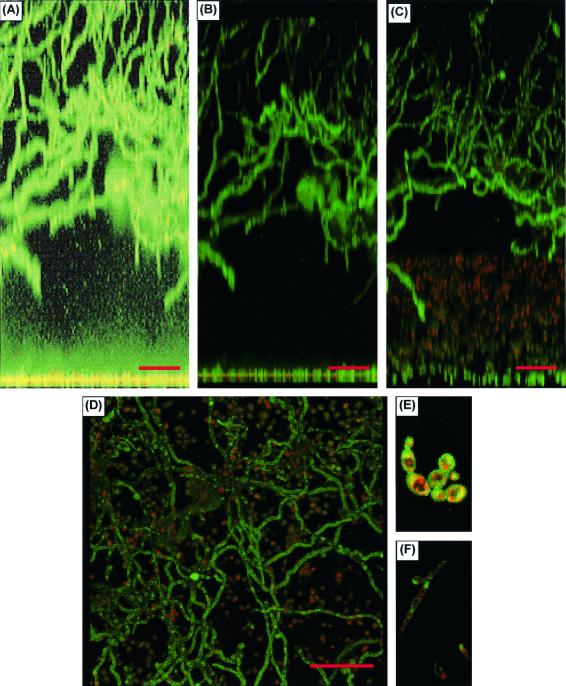
As shown in Figures 5 and 6, the biofilm formed by pathogenic C. albicans was a complex phenomenon. There was a basal blastospore layer, which was one to two cells deep (Fig. 5A and 6A). FUN-1 staining showed this layer to be metabolically active, as indicated by the red fluorescence in Fig. 5B to E. The basal layer was covered by a thick (450- to 550-μm) biphasic matrix, consisting of a dense extracellular component comprised of cell wall-like compounds (as shown in Fig. 5A by diffuse CAAF staining and in Fig. 6B by diffuse Calcofluor White staining) and abundant hyphal elements (Fig. 5A to D). CAAF staining throughout the matrix indicated that it was composed of polysaccharide elements. The matrix was differentiated into a lower, largely cell-free element and an upper layer interwoven with dense hyphal forms (Fig. 5A to D and 6B). On CSLM, the matrix was stable and nonmobile and exhibited uniform CAAF staining (10). Like the basal blastospore layer, the hyphal elements were metabolically active, as indicated by FUN-1 conversion (Fig. 5F). No gaps in CAAF staining, which would have been indicative of water channels, were discerned. Biofilm morphology, as depicted by FM (Fig. 6B) and CSLM (top-down reconstruction in Fig. 5D), showed similar features.
FIG. 5.
CSLM examination of C. albicans biofilms. (A) CSLM image obtained with CAAF and FUN-1 staining and processed by a transparent projection technique to show a lateral view of the biofilm, which is ≈450 μm deep. A ×20 water immersion objective was used. Images were enhanced to highlight CAAF. The green CAAF staining highlights cell walls of the basal blastospore layer (bottom) and the upper hyphal layer. The diffuse staining of the extracellular space reflects CAAF binding matrix polysaccharides throughout the biofilm (the mild basilar enhancement is due to an out-of-focus artifact enhanced by the processing protocol). (B) Same lateral view but processed to show both CAAF and FUN-1 staining. (C) Tilt view (≈30°) illustrating red emission by converted FUN-1 in blastospores. (D) Top-down reconstructed view of the same image stack. (E and F) Representative red-enhanced image of the basal blastospore layer (E) and a similar image of hyphae in the upper matrix (F). Both panels E and F show FUN-1 conversion, as indicated by red dots within the cells.
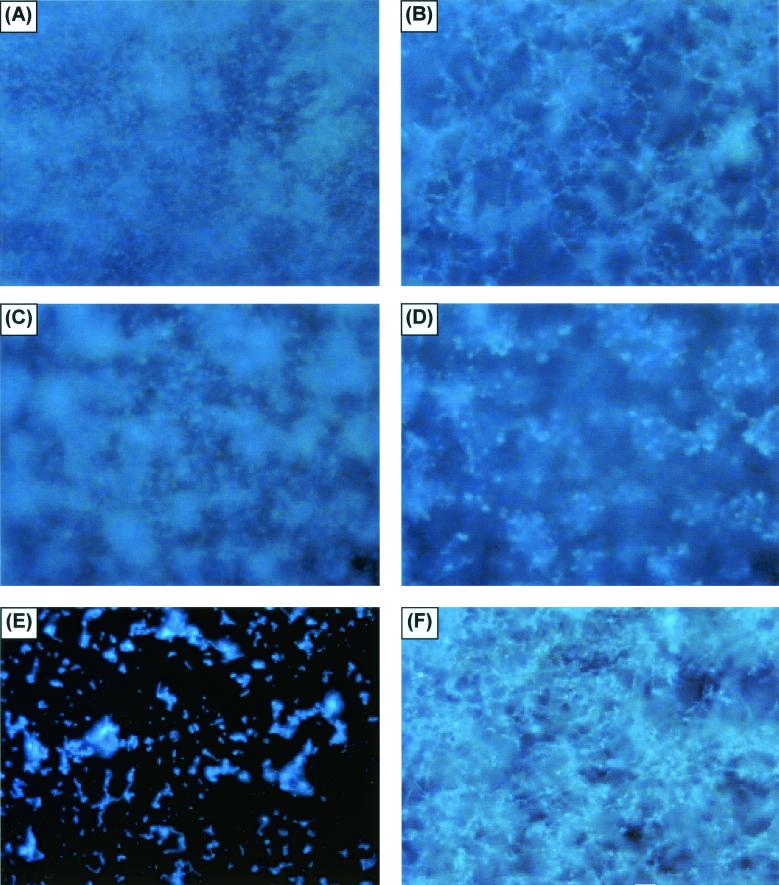
FIG. 6.
FM examination of fungal biofilms formed by different Candida species. (A and B) FM examination of the basal and upper layers of a C. albicans biofilm, respectively. The biofilm was stained with Calcofluor White and viewed at ×10. (C and D) Similar views of C. parapsilosis, although the view of the upper layer is at a lower altitude than that in panel B, due to the limited thickness of the matrix. (E and F) Views of C. glabrata and C. tropicalis, respectively. Other samples of C. tropicalis and C. parapsilosis had the same appearance as that in panel E.
Microscopy revealed that C. albicans produces biofilms qualitatively different from those produced by non-C. albicans species, in particular, C. parapsilosis.
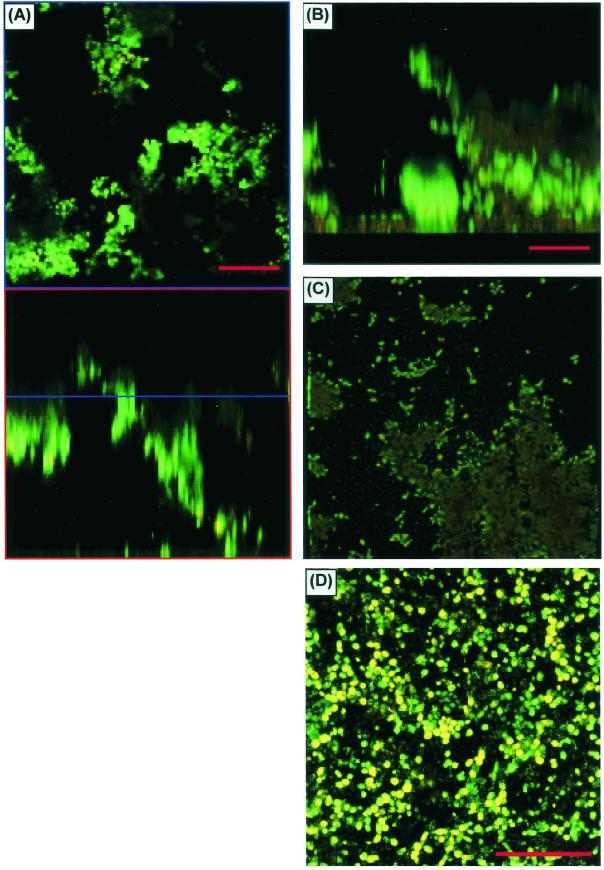
Figure 6 shows an FM comparison of biofilms formed by different Candida species. To provide correlation with the XTT and DW results and to ensure that the marked microscopy differences seen in biofilm formation between C. albicans and C. parapsilosis were not simply due to inadequate species sampling, we examined biofilm formation by other species as well. On FM, C. parapsilosis, C. glabrata, and C. tropicalis produced much less biofilm than C. albicans or no biofilm. The biofilm produced by the isolate of C. parapsilosis (obtained from a blood specimen) was considerably less thick and showed distinct clumping (Fig. 6D). CSLM imaging of the same C. parapsilosis biofilm is shown in Fig. 7A to C. C. parapsilosis formed a much shallower biofilm than C. albicans (75 to 125 μm for the former and 450 to 550 μm for the latter). This biofilm consisted of irregular groupings of blastospores, consistent with the FM images and reminiscent of structures seen in bacterial biofilms (14, 27, 34, 51). Also in contrast to the C. albicans biofilm, the C. parapsilosis biofilm exhibited minimal, if any, visible matrix (Fig. 7A to C). Other isolates of C. parapsilosis produced only a basal blastospore layer (Fig. 7D). All of the C. glabrata isolates examined produced a scant population of blastospores on the SE surface (Fig. 6E), and a similar pattern was seen with several other isolates of C. parapsilosis and C. tropicalis. One isolate of C. tropicalis produced a thin (≈30-μm) layer of matrix-encased hyphae (Fig. 6F); interestingly, however, there was no visible basal blastospore layer.
FIG. 7.
CSLM characterization of C. parapsilosis biofilms. (A) Horizontal (top) and vertical (bottom) slice reconstructions of a C. parapsilosis biofilm examined by CSLM with CAAF and FUN-1. Images were obtained with a ×20 water immersion objective. Green haze is an out-of-focus artifact, not extracellular matrix. (B) Image reconstructed by using transparent projection to show the lateral aspect, which is ≈117 μm deep. (C) Top-down view of the same image stack. (D) View of a different C. parapsilosis isolate, which failed to form appreciable matrix.
C. albicans biofilms rapidly develop fluconazole resistance.
Bacterial and fungal biofilms exhibit antimicrobial drug resistance (2, 7, 10, 11). We selected two representative C. albicans strains (M61 and GDH2346) known to form mature biofilm in our model system and tested their susceptibility to fluconazole. As shown in Table 1, both GDH2346 and M61 biofilms developed fluconazole resistance (MIC50, >128 μg/ml) in a rapid temporal manner, and the process was complete by 6 h. The same organisms grown under planktonic conditions retained their fluconazole susceptibility; the MIC50 was ≤1 μg/ml.
TABLE 1.
Fluconazole susceptibility of planktonically grown and biofilm-grown C. albicans, as measured by MIC50
| Strain | MIC50 (μg/ml) in indicated cell typea:
|
|||||
|---|---|---|---|---|---|---|
| Planktonic
|
Biofilm (XTT) at age (h):
|
|||||
| NCCLS M27-A | XTT | 0 | 6 | 24 | ||
| M61 | 1 | 1 | 4 | >128 | >128 | |
| GDH2346 | 0.25 | 0.25 | 4 | >128 | >128 | |
For details of the methods used, see the text.
Metabolic quiescence has been proposed as a mechanism of biofilm antimicrobial resistance in bacteria (28) and could account for the phenomenon in Candida biofilms. While our experiments were not focused on resistance mechanisms, it is important to note that C. albicans cells grown in biofilm actively metabolized XTT. Furthermore, Fig. 5B to F show that C. albicans cells in biofilms metabolized FUN-1, a process which occurred by a biochemical pathway different from that of XTT, confirming that these cells were metabolically active.
DISCUSSION
We developed a novel system for modeling the behavior of biofilms produced by pathogenic fungi and used it to compare the abilities of different C. albicans strains and non-C. albicans species to form biofilms. The advantages of this system are multiple. The growth system is simple and does not require elaborate flow chamber devices, which are costly, complicated, and labor-intensive. Medium circulation provided by rocker table motion induces quasi-linear flow stimulating biofilm growth, similar to the Calgary Biofilm Device (9). The enhancement of biofilm formation by FBS preincubation is similar to the effects of saliva shown in a previously described denture biofilm model (11) and thereby mimics physiologic conditions. The use of flat sheeting of a medically relevant substrate is new, allows easy quantitation and, for the first time, produces reproducible imaging of intact, living, and nondehydrated specimens by various microscopic techniques, including CSLM. Moreover, this system allows for the high-volume production of biofilms (gram amounts), which will facilitate future genetic and proteomic studies aimed at understanding the biology and drug resistance of biofilms.
The choice of substrate is not trivial, since prior studies indicated that chemical composition, degree of hydrophobicity, and the presence of sera, protein, or amino acids affect Candida binding to surfaces (11, 17, 22, 33, 49). Other studies have shown that SE (the material used in the current study) allows better biofilm formation than other substrates (20), and our own unpublished observations support this finding. Finally, it is well documented that different substrates induce separate sets of structural genes critical for attachment in bacterial systems (39, 47). Similar work needs to be conducted for Candida species.
Our quantitative results are in contrast to those of most prior studies, which have reported few consistent differences in biofilm activity between strains or species (20), and indicate that more invasive isolates (i.e., those obtained from normally sterile sites, such as catheters or blood) of C. albicans have an increased ability to form biofilms. Only one other study has suggested a similar phenomenon for C. parapsilosis (37). Whether this finding reflects innate virulence or up-regulation of virulence following invasion remains to be determined. Distinguishing blood and catheter isolates as invasive compared to those obtained from other sites is clinically justifiable (40). C. albicans also appears more prone to biofilm formation than other potentially invasive but less commonly pathogenic species, such as C. parapsilosis, C. glabrata, and C. tropicalis. The marked differences in biofilm production and morphology between pathogenic and less pathogenic strains have implications for future work and suggest that studies of biochemical and genetic mechanisms in biofilms need to be performed by using pathogenic species from clinical specimens and nearly physiologic conditions (10).
The observed differences between the XTT and DW results have important implications for biofilm studies, since to date it was assumed that metabolic measurements based on XTT activity were sufficient to quantitate biofilms (11, 20). While XTT measurements alone may be used to monitor biofilm formation or progression in a particular strain, DW and microscopic analyses are critical for strain and species comparisons. One hypothesis is that given the abundance of biofilm production, such cells may be shifting metabolism away from routine functions (38, 46). Thus, strains producing low levels of biofilm or no biofilm might in fact be expected to show a higher level of expression of routine markers of metabolic activity, such as XTT. Importantly, this hypothesis was supported by the higher levels of XTT metabolism exhibited by noninvasive C. albicans strains as well as less commonly pathogenic Candida species than by C. albicans catheter and blood strains, although further study is needed to prove the significance of these findings. This article does not address issues of cell division (46) or quorum sensing (15, 16), known to be important in bacterial biofilms and a subject of our current investigations.
Our microscopy studies showed that C. albicans forms complex biofilms consisting of an essentially confluent basal blastospore layer covered by a thick matrix composed of extracellular material and hyphal elements. This biphasic structure has support from earlier, preliminary microscopy (3), and it has been shown elsewhere that this structure is similar to that seen on infected intravenous catheters obtained from patients (10). The matrix is composed of cell wall-like polysaccharides, as indicated by diffuse CAAF staining as well as Calcofluor White (which binds to β-linked polysaccharides; see reference 10) staining, and is supported by an initial biochemical analysis (19). The hyphal elements are metabolically active. The thickness of the specimens, on the order of 500 μm, indicates that the cells are producing huge quantities of extracellular material. This behavior is similar to that of Pseudomonas biofilm systems that produce copious extracellular alginate (18) as well as other bacteria (13, 48) in which exopolysaccharides comprise up to 90% of biofilm organic matter (52). In our C. albicans biofilm model, there is an apparent absence of the water channels present in some bacterial systems (8, 51), but this finding needs further investigation.
Microscopy findings correlated with the quantitative results, confirming that C. albicans produces more biofilm than less commonly pathogenic species, including C. parapsilosis, C. glabrata, and C. tropicalis. Most of the latter species appear to form only basal blastospore layers, similar to those of Saccharomyces cerevisiae (10, 41). Compared to C. albicans strains, C. parapsilosis strains which formed biofilms produced both a different morphology, with the biofilms being apparently devoid of extracellular matrix material and hyphae, and much less thickness. Notably, the morphology of C. parapsilosis biofilms may at times approximate that seen in bacteria (14, 34, 46, 51), perhaps reflective of the established tendency of the former organism to flocculate in cultures. The heterogeneity in the ability of C. parapsilosis strains to form biofilms is similar to that noted in earlier work on “slime” formation by this species (6, 37). The morphology of the C. tropicalis strain which produced a biofilm was striking, as no basal blastospore layer was in evidence (data not shown), in marked contrast to biofilms formed by C. albicans and C. parapsilosis.
Biofilm drug resistance is a phenomenon consistently expressed across model systems (2, 10, 11) and likely of great clinical relevance (12). The fluconazole resistance displayed by our model biofilms emerges rapidly, similar to results obtained earlier with a model of dental Candida biofilm (10, 11). Multiple mechanisms have been proposed for the biofilm resistance phenomenon (12, 28, 29). One of the most favored has been metabolic quiescence, which appears to be relevant in bacterial systems (29). However, our results show that fungal biofilms metabolize both XTT and FUN-1, the latter of which occurs via a separate pathway. Therefore, it appears that profound metabolic quiescence is not likely to be a mechanism of drug resistance. Studies are needed to address other mechanisms, including mitotic inhibition (29); expression of drug resistance genes, such as CDR1, CDR2, and MDR (27, 44); and drug absorption by a biofilm matrix (4, 24, 45).
In conclusion, we developed a simple, inexpensive, and reproducible system for the study of catheter and bioprosthetic material-associated Candida biofilms. Using this model, we have shown that invasive Candida strains and species appear to be superior biofilm formers relative to noninvasive ones; however, further work involving more clinical samples is needed to determine the validity of this observation. Finally, C. albicans exhibits unique biofilm morphology compared to C. parapsilosis.
Acknowledgments
We thank A. Niemenen's Core CSLM Facility (NCI P30 CA43703-12); S. Leidich, G. Reyes, N. Isham, and A. Morrisey for advice; and Kathleen Smith for secretarial assistance.
Funding was provided by the NIH (Infectious Disease/Geographic Medicine training grant AI07024 to D.M.K.; grant AI35097 to M.A.G.), the Steris Foundation, and Pfizer Inc.
Editor: T. R. Kozel
REFERENCES
- 1.Anaissie, E. J., J. H. Rex, O. Uzon, and S. Vartivarian. 1998. Predictors of adverse outcome in cancer patients with candidemia. Am. J. Med. 104:238-245. [DOI] [PubMed] [Google Scholar]
- 2.Baillie, G. S., and L. J. Douglas. 1999. Candida biofilms and their susceptibility to antifungal agents. Methods Enzymol. 310:644-656. [DOI] [PubMed] [Google Scholar]
- 3.Baillie, G. S., and L. J. Douglas. 1999. Role of dimorphism in the development of Candida albicans biofilms. J. Med. Microbiol. 48:671-679. [DOI] [PubMed] [Google Scholar]
- 4.Baillie, G. S., and L. J. Douglas. 2000. Matrix polymers of Candida biofilms and their possible role in biofilm resistance to antifungal agents. J. Antimicrob. Chemother. 46:397-403. [DOI] [PubMed] [Google Scholar]
- 5.Banejee, S. N., G. T. Emori, D. H. Culver, R. P. Gaynes, W. R. Jarvis, T. Horan, J. R. Edwards, et al. 1991. Secular trends in nosocomial primary bloodstream infections in the United States, 1980-1989. Am. J. Med. 91(Suppl. 3B):86-89. [DOI] [PubMed] [Google Scholar]
- 6.Branchini, M. L., M. A. Pfaller, J. Rhine-Chalberg, T. Frempong, and H. D. Isenberg. 1994. Genotypic variation and slime production among blood and catheter isolates of Candida parapsilosis. J. Clin. Microbiol. 32:452-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, M. R., and P. Gilbert. 1993. Sensitivity of biofilms to antimicrobial agents. J. Appl. Bacteriol. 74(Suppl.):87S-97S. [DOI] [PubMed]
- 8.Bryers, J. D., and F. Drummond. 1998. Local macromolecule diffusion coefficients in structurally non-uniform bacterial biofilms using fluorescence recovery after photobleaching (FRAP). Biotechnol. Bioeng. 60:462-473. [PubMed] [Google Scholar]
- 9.Ceri, H., M. E. Olson, C. Stremick, R. R. Read, D. Morck, and A. Buret. 1999. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities in bacterial biofilms. J. Clin. Microbiol. 37:1771-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandra, J., D. M. Kuhn, P. K. Mukherjee, L. L. Hoyer, and M. A. Ghannoum. 2001. Biofilm formation by the pathogen C. albicans--development, architecture, and drug resistance. J. Bacteriol. 183:5385-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandra, J., P. K. Mukherjee, S. D. Leidich, F. F. Faddoul, L. L. Hoyer, L. J. Douglas, and M. A. Ghannoum. 2001. Antifungal resistance of Candida biofilms formed on denture acrylic in vitro. J. Dent. Res. 80:903-908. [DOI] [PubMed] [Google Scholar]
- 12.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 13.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 16.De Kievit, T. R., R. Gillis, S. Marx, C. Brown, and B. H. Iglewski. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol. 67:1865-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Azizi, M., and N. Khardori. 1999. Factors influencing adherence of Candida spp. to host tissues and plastic surfaces. Indian J. Exp. Biol. 37:941-951. [PubMed] [Google Scholar]
- 18.Gacesa, P. 1998. Bacterial alginate biosynthesis--recent progress and future prospects. Microbiology 144:1133-1143. [DOI] [PubMed] [Google Scholar]
- 19.Hawser, S. P., G. S. Baillie, and L. J. Douglas. 1998. Production of extracellular matrix by Candida albicans biofilms. J. Med. Microbiol. 47:253-256. [DOI] [PubMed] [Google Scholar]
- 20.Hawser, S. P., and L. J. Douglas. 1994. Biofilm formation by Candida species on the surface of catheter materials in vitro. Infect. Immun. 62:915-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawser, S. P., and L. J. Douglas. 1995. Resistance of Candida albicans biofilms to antifungal agents in vitro. Antimicrob. Agents Chemother. 39:2128-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hawser, S. P., and K. Islam. 1998. Binding of C. albicans to immobilized amino acids and bovine serum albumen. Infect. Immun. 66:140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawser, S. P., H. Norris, C. J. Jessup, and M. A. Ghannoum. 1998. Comparison of a 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT) colorimetric method with the standardized National Committee for Clinical Laboratory Standards method of testing clinical yeast isolates for susceptibility to antifungal agents. J. Clin. Microbiol. 36:1450-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoyle, B. D., J. Jass, and J. W. Costerton. 1990. The biofilm glycocalyx as a resistance factor. J. Antimicrob. Chemother. 26:1-5. [DOI] [PubMed] [Google Scholar]
- 25.Kremery, V., and G. Kovacicova, Jr. 2000. Longitudinal 10-year prospective survey of fungemia in the Slovak Republic: trends in etiology in 310 episodes. Slovak Fungaemic Group. Diagn. Microbiol. Infect. Dis. 36:7-11. [DOI] [PubMed] [Google Scholar]
- 26.Kremery, V., Jr., M. Mrazova, A. Kunora, E. Grey, J. Mardiak, L. Jurga, A. Sabo, et al. 1999. Nosocomial candidemias due to species other than Candida albicans in cancer patients. Aetiology, risk factors, and outcome of 45 episodes within 10 years in a single cancer institution. Support Care Cancer 7:428-431. [DOI] [PubMed] [Google Scholar]
- 27.Kuchma, S. L., and G. A. O'Toole. 2000. Surface-induced and biofilm-induced changes in gene expression. Curr. Opin. Biotechnol. 11:429-433. [DOI] [PubMed] [Google Scholar]
- 28.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahmoud, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 30.Mermel, L. A., B. M. Farr, R. J. Sherertz, I. I. Raad, N. O'Grady, J. S. Harris, and O. E. Craven. 2001. Guidelines for the management of intravascular catheter-related infections. Clin. Infect. Dis. 32:1249-1272. [DOI] [PubMed] [Google Scholar]
- 31.National Committee for Clinical Laboratory Standards. 1997. Reference method for broth dilution antifungal susceptibility testing of yeasts. M27-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 32.Nguyen, M. H., J. E. Peacock Jr., D. C. Tanner, A. J. Morris, M. L. Nguyen, D. R. Snydman, M. M. Wagener, et al. 1995. Therapeutic approaches in patients with candidemia: evaluation in a multicenter, prospective, observational study. Arch. Intern. Med. 155:2429-2435. [PubMed] [Google Scholar]
- 33.Nikawa, H., H. Nishimura, S. Makihara, T. Hamada, S. Sadamori, and L. P. Samaranayake. 2000. Effect of serum concentration on Candida biofilm formation on acrylic surfaces. Mycoses 43:139-143. [DOI] [PubMed] [Google Scholar]
- 34.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 35.Pagano, L., A. Antinori, A. Ammassari, L. Mele, A. Nosari, L. Melillo, B. Martino, et al. 1999. Retrospective study of candidemia in patients with hematological malignancies. Clinical features, risk factors, and outcome of 76 episodes. Eur. J. Haematol. 63:77-85. [DOI] [PubMed] [Google Scholar]
- 36.Pfaller, M. A., R. N. Jones, G. W. Doern, H. S. Sader, S. A. Messer, A. Houston, S. Coffman, et al. 2000. Bloodstream infections due to Candida species: SENTRY antimicrobial surveillance program in North America and Latin America, 1997-1998. Antimicrob. Agents Chemother. 44:747-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfaller, M. A., S. A. Messer, and R. J. Hollis. 1995. Variations in DNA subtype, antifungal susceptibility, and slime production among clinical isolates of Candida parapsilosis. Diagn. Microbiol. Infect. Dis. 21:9-14. [DOI] [PubMed] [Google Scholar]
- 38.Prigent-Combaret, C., C. O. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pruzzo, C., A. Crippa, S. Bertone, L. Pane, and A. Carli. 1996. Attachment of Vibrio alginolyticus to chitin mediated by chitin-binding proteins. Microbiology 142:2181-2186. [DOI] [PubMed] [Google Scholar]
- 40.Rex, J. H., T. J. Walsh, J. D. Sobel, S. G. Filler, P. G. Pappas, W. E. Dismukes, and J. E. Edwards. 2000. Practice guidelines for the treatment of candidiasis. Clin. Infect. Dis. 30:662-668. [DOI] [PubMed] [Google Scholar]
- 41.Reynolds, T. B., and G. R. Fink. 2001. Bakers' yeast, a model for fungal biofilm formation. Science 291:878-881. [DOI] [PubMed] [Google Scholar]
- 42.Saiman, L., E. Ludington, M. Pfaller, S. Rangel-Frausto, R. T. Wiblin, J. Dawson, H. M. Blumberg, et al. 2000. Risk factors for candidemia in neonatal intensive care unit patients. The National Epidemiology of Mycosis Survey Study Group. Pediatr. Infect. Dis. J. 19:319-324. [DOI] [PubMed] [Google Scholar]
- 43.Sanchez, V., J. A. Vasquez, D. Barth-Jones, L. Dembry, J. D. Sobel, and M. J. Zervus. 1993. Nosocomial acquisition of Candida parapsilosis: an epidemiologic study. Am. J. Med. 94:577-582. [DOI] [PubMed] [Google Scholar]
- 44.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143:405-416. [DOI] [PubMed] [Google Scholar]
- 45.Shigeta, M., G. Tanaka, H. Komatsuzawa, M. Sugai, H. Suginaka, and T. Usui. 1997. Permeation of antimicrobial agents Pseudomonas aeruginosa biofilms: a simple method. Chemotherapy (Tokyo) 43:340-345. [DOI] [PubMed] [Google Scholar]
- 46.Watnick, P., and R. Kolter. 2000. Biofilm, city of microbes. J. Bacteriol. 182:2675-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watnick, P. I., K. J. Fullner, and R. Kolter. 1999. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J. Bacteriol. 181:3606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watnick, P. I., and Kolter, R. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Webb, J. S., H. C. Van der Mei, M. Nixon, I. M. Eastwood, M. Greenhalgh, S. J. Read, G. D. Robson, et al. 1999. Plasticizers increase adhesion of the deteriogenic fungus Aureobasidium pullulans to polyvinyl chloride. Appl. Environ. Microbiol. 65:3575-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weems, J. J., Jr., M. E. Cumberland, J. Ward, M. Willy, A. A. Padhye, and S. L. Solomon. 1987. Candida parapsilosis fungemia associated with parenteral nutrition and contaminated blood pressure transducers. J. Clin. Microbiol. 25:1029-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wimpenny, J., W. Manz, and U. Szewzyk. 2000. Heterogeneity in biofilms. FEMS Microbiol. Rev. 24:661-671. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, X. Q., P. L. Bishop, and M. J. Kupferle. 1998. Measurement of polysaccharides and proteins in biofilm extracellular polymers. Water Sci. Technol. 37:345-348. [Google Scholar]