Abstract
Dopachrome tautomerase (Dct) is a type I membrane protein and an important regulatory enzyme that plays a pivotal role in the biosynthesis of melanin and in the rapid metabolism of its toxic intermediates. Dct-mutant melanocytes carrying the slaty or slaty light mutations were derived from the skin of newborn congenic C57BL/6J non-agouti black mice and were used to study the effect(s) of these mutations on the intracellular trafficking of Dct and on the pigmentation of the cells. Dct activity is 3-fold lower in slaty cells compared with non-agouti black melanocytes, whereas slaty light melanocytes have a surprisingly 28-fold lower Dct activity. Homology modelling of the active site of Dct suggests that the slaty mutation [R194Q (Arg194→Gln)] is located in the active site and may alter the ability of the enzyme to transform the substrate. Transmembrane prediction methods indicate that the slaty light mutation [G486R (Gly486→Arg)] may result in the sliding of the transmembrane domain towards the N-terminus, thus interfering with Dct function. Chemical analysis showed that both Dct mutations increase pheomelanin and reduce eumelanin produced by melanocytes in culture. Thus the enzymatic activity of Dct may play a role in determining whether the eumelanin or pheomelanin pathway is preferred for pigment biosynthesis.
Keywords: dopachrome tautomerase (Dct), eumelanin, melanocyte, pheomelanin, pigmentation, slaty
Abbreviations: 4-AHP, 4-amino-3-hydroxyphenylalanine; BiP, immunoglobulin heavy-chain binding protein; DAPI, 4,6-diamidino-2-phenylindole; Dct, dopachrome tautomerase; DHICA, 5,6-dihydroxyindole-2-carboxylic acid; endo H, endoglycosidase H; ER, endoplasmic reticulum; HRP, horseradish peroxidase; NP40, Nonidet P40; OCA, oculocutaneous albinism; PNGase F, peptide N-glycosidase F; PTCA, pyrrole-2,3,5-tricarboxylic acid; RT, reverse transcriptase
INTRODUCTION
Dopachrome tautomerase (Dct; EC 5.3.3.12) is an enzyme in the biosynthetic pathway of melanin that isomerizes the orange-coloured melanogenic intermediate dopachrome to DHICA (5,6-dihydroxyindole-2-carboxylic acid) [1], a precursor of DHICA-rich eumelanins. The gene encoding Dct maps to mouse chromosome 14, at the coat colour locus slaty. Two recessive mutant alleles at this locus, slaty (Dctslt) and slaty2J (Dct2J) [2,3], and a third, semi-dominant allele, namely slaty light (Dctslt–lt) [2], result in the dilution of coat colour from black to varying shades of grey. Dct is a member of the tyrosinase-related protein family, and the two other members of this family, tyrosinase (Tyr) and Tyrp1, are also melanogenic enzymes with distinct functions [4,5]. Interestingly, mutations in the genes encoding tyrosinase or Tyrp1 also lead to hypopigmentation and, in humans, have been associated with two different pigmentary diseases, OCA (oculocutaneous albinism) types I and III respectively [6,7]. In melanosomes, which are the membrane-bound organelles wherein melanin is produced, Tyr, Tyrp1 and Dct form a complex [8], and mutations in Tyr or Tyrp1 affect the processing, stability and function of each other, but neither seems to affect Dct [6,7]. Studies employing transfection of COS-7 cells with mutant tyrosinase-related proteins showed that Dct increases tyrosinase activity by stabilizing the protein [9]. To date, Dct has not been associated with a human pigmentary disease, but might be expected to be associated with a mild form of OCA. However, the mechanism(s) whereby mutations in Dct affect pigmentation is unknown at this point.
Mouse Dct is a type I membrane protein that plays a pivotal role in the biosynthesis of melanin and in the rapid metabolism of its toxic intermediates [10]. All three tyrosinase-related proteins have two metal-binding motifs that are critical to their enzymatic function, tyrosinase requiring copper, Tyrp1 requiring iron and Dct requiring zinc [11,12]. In mice, mutations in the Dct gene result in premature melanocyte death, probably from cytotoxic intermediates generated in its absence [13], and therefore Dct-mutant melanocytes are extremely fragile and have been impossible to culture until now. However, we recently developed a tissue culture system that allows primary melanocytes derived from the skin of newborn Dct-mutant mice to grow and eventually to be immortalized [14]. Therefore, in the present study, we focused on characterizing the effects of the slaty and slaty light mutations using those immortalized melanocytes. Mutant Dct produced by slaty mice has a single amino acid difference compared with wild-type Dct, namely an R194Q (Arg194→Gln) substitution in the first metal-binding domain. A point mutation in exon 8 was identified in the Dct gene of slaty light mice [2], which results in a G486R (Gly486→Arg) substitution in the transmembrane domain. In this study, we attempted to define the effects of those two mutations of the Dct gene on the catalytic functions of the mutant proteins, their processing and sorting to melanosomes, and on the melanins produced. Our results show that both mutations in the Dct gene not only dramatically decreased the enzymatic activities of the mutant proteins and decreased eumelanin production as expected, but surprisingly had no effect on the post-translational processing and trafficking of the mutant proteins, and even more unexpectedly, significantly increased the production of pheomelanin.
EXPERIMENTAL
Materials
αPEP1, αPEP7 and αPEP8 are rabbit antibodies raised in our laboratory against the C-terminal peptide of Tyrp1, Tyr and Dct respectively as described previously [15,16]. Anti-rabbit IgG HRP (horseradish peroxidase)-linked antibody and anti-mouse IgG HRP-linked antibody were obtained from Amersham Biosciences (Piscataway, NJ, U.S.A.). Monoclonal antibody directed to Vti1B was from B&D (Palo Alto, CA, U.S.A.) and HMB-45 monoclonal antibody [17] was purchased from Dako (Carpinteria, CA, U.S.A.). The antibody directed to BiP (immunoglobulin heavy-chain binding protein) was obtained from BD Transduction Laboratories (San Jose, CA, U.S.A.) and the anti-mouse IgG HRP-linked (whole antibody) was from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.). Normal horse serum, normal goat serum, Texas Red anti-rabbit IgG (H+L) and FITC anti-mouse IgG (H+L) were all from Vector Laboratories (Burlingame, CA, U.S.A.). Endo H (endoglycosidase H) and PNGase F (peptide N-glycosidase F) were from New England Biolabs (Beverly, MA, U.S.A.). The glycoprotein deglycosylation kit was from Chemicon (Temecula, CA, U.S.A.).
Cell culture
Primary black, slaty and slaty light melanocytes were derived from the dorsal skins of 1-day-old C57BL/6J congenic non-agouti black (a/a Tyrp1+ Dct+), slaty (a/a Tyrp1+ Dctslt) and slaty light (a/a Tyrp1+ Dctslt–lt) mice respectively and were immortalized using a method described previously [14]. To remove fibroblasts, melanocytes were cultured with 100 μg/ml Geneticin (G418 sulphate; Invitrogen) for 2–3 days (repeated if necessary as long as fibroblasts persisted).
Cellular fractionation
Melanocytes were harvested with trypsin/EDTA, washed three times with cold PBS without CaCl2 and MgCl2 and were solubilized in extraction buffer [1% NP40 (Nonidet P40) in PBS containing a protease inhibitor cocktail; Boehringer Mannheim, Roche, Mannheim, Germany] as described previously [18]. Protein concentrations were determined with a BCA (bicinchoninic acid) assay kit (Pierce, Rockford, IL, U.S.A.) using BSA as a standard. To purify melanosomes, we used sucrose density-gradient ultracentrifugation, as described previously [5]. Melanosomes that localized at various layers of the gradient were recovered by pipette and were further analysed by Western immunoblotting. To investigate the presence of Dct in the melanosomal membrane, a sodium carbonate extraction was performed as reported previously [19]. Briefly, melanosomal fractions separated as noted above were incubated with 100 mM sodium carbonate (pH 11.5) for 1 h at 4 °C and were then centrifuged at 235000 g in a Ti45 rotor for 1 h. The pellet and the supernatant fractions were saved and tested for the presence of Dct and BiP (used as a control) by Western blotting.
Western blotting and glycan analysis by glycosidase digestion
For Western blotting, samples were separated by electrophoresis under reducing conditions, as described previously [18]. For limited PNGase F digestion, the samples were digested with 0, 0.4, 1, 10, 100 or 1000 units of enzyme overnight at 37 °C and were further analysed by PAGE. Neuraminidase and O-glycan deglycosylations were performed according to the manufacturer's instructions.
Metabolic labelling
Metabolic labelling and immunoprecipitation experiments were performed as reported previously [19]. Melanocytes were cultured in six-well tissue culture plates for 48 h before labelling. Then the cells were preincubated in methionine- and cysteine-free Dulbecco's modified Eagle's medium (Gibco BRL, Grand Island, NY, U.S.A.) for 30 min at 37 °C in a humidified incubator with 5% CO2 and were then labelled for 30 min with 0.5 mCi of [35S]-methionine and cysteine mixture (Redivue Promix; Amersham Biosciences). Following the 30 min labelling period, the isotope-supplemented medium was removed and a complete medium containing 1 mM unlabelled methionine was added. Cells were harvested by scraping at 0 min, 1, 2 and 4 h of chase. The cells were washed three times with PBS at 4 °C and then solubilized for 1 h at 4 °C in lysis buffer (50 mM Tris/HCl, pH 7.4, containing 1% NP40, 0.01% SDS and protease inhibitor cocktail). 35S-labelled cell lysates were precleared with 20 μl of Protein G–Sepharose 4 Fast Flow (Amersham Biosciences) for 2 h at 4 °C with mixing. The supernatants were collected following centrifugation at 5000 g for 1 min at 4 °C and then incubated with αPEP8 or αPEP7 for 2 h at 4 °C with mixing. The immunocomplexes were separated by incubation with 20 μl of Protein G–Sepharose 4 Fast Flow for 2 h at 4 °C and were further washed three times by centrifugation with the lysis buffer. The final pellets were resuspended in sample buffer (1% NP40 in PBS mixed with protease inhibitors), heated at 100 °C for 5 min and centrifuged. The samples were separated on 8–16% Tris/glycine gels (Invitrogen). The dried gels were exposed in a storage phosphor screen, scanned with a STORM PhosphorImager (Molecular Dynamics, Sunnyvale, CA, U.S.A.) and were then analysed with the Image Quant program.
Production of cDNA, cloning and sequence analysis
We confirmed the presence of the slaty and slaty light mutations in the two immortalized cell lines using RT (reverse transcriptase)–PCR. Total cytoplasmic RNA was isolated from cultured melanocytes using the RNeasy Mini kit (Qiagen, Valencia, CA, U.S.A.) according to the manufacturer's instructions. The SuperScript™ II RNase H− RT (Invitrogen) was used to synthesize first-strand cDNA with oligo-dT12–18 primer (Invitrogen) from 1–5 μg of total RNA at 42 °C for 50 min. The cDNA was used in initial PCR amplification with the primers designed against the regions carrying the specific mutations. The sequences of the primers are: 5′-cctggccaagaagagtatcc-3′ (slaty1) and 5′-cacgtcacactcgttcttcc-3′ (slaty2); 5′-gtggtcctccactcttttacagacgc-3′ (slaty light1) and 5′-tgaagagttccacctgtctcaagatgagcg-3′ (slaty light2). Amplifications were performed in a total volume of 50 μl containing 5 μl of 10× PCR reaction buffer (Roche, Indianapolis, IN, U.S.A.), 1 μl of dNTP mix (10 mM each), 1 μl of forward and reverse primers, 1 μl of FastStartTaq DNA polymerase (50 units/μl; Roche), 36 μl of diethyl pyrocarbonate-treated water (KD Medical, Columbia, MD, U.S.A.) and 5 μl of cDNA template. The cycling reaction was performed in a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, U.S.A.) for 1 cycle of 95 °C for 4 min, 36 cycles of 95 °C for 30 s, 58 °C for 30 s and 72 °C for 40 s, followed by 1 cycle of 72 °C for 7 min. PCR products were separated on 1.2% agarose gels (Invitrogen) and visualized under UV light. PCR products were purified from gel using the QIAquick gel extraction kit (Qiagen) and cloned into the pGEM-T Easy Vector (Promega, Madison, WI, U.S.A.). Recombinants were identified through blue–white colour selection in LB AMP/X-GAL/IPTG (ampicillin/5-bromo-4-chloroindol-3-yl β-D-galactopyranoside/isopropyl β-D-thiogalactoside) plates (KD Medical). Plasmid DNA was isolated by the QIA Prep Spin Miniprep procedure (Qiagen). Cloned fragments were analysed by automatic sequencing using the ABI PRISM Big Dye Terminator reaction kit (Applied Biosystems) and an automatic sequencer (ABI PRISM Cycle sequencing; PerkinElmer, Norwalk, CT, U.S.A.) and sequencing files were analysed with Sequencher 4.0.5 software. Nucleotidic sequences were compared using the BLAST (NCBI, Bethesda, MD, U.S.A.) program.
Northern blotting
Northern blotting was performed following the NorthernMax procedure (Ambion, Austin, TX, U.S.A.). Briefly, 10 μg of total RNA was denatured in formaldehyde buffer for 15 min at 65 °C and then separated by electrophoresis on 1% agarose gels. After transfer to a BrightStar-Plus membrane (Ambion), the RNA was cross-linked using UV irradiation (Stratalinker; Stratagene, La Jolla, CA, U.S.A.). A 444 nt fragment of the mouse Dct exonic region (PCR-amplified on cDNA using Dct1: 5′-cagacaccagaccctggagtggc-3′ and Dct2: 5′-gtacctgtgccacgtgacaaaggcag-3′ and sequence-verified) was radiolabelled with deoxycytidine 5′-[α-32P]triphosphate by random priming using DECAprime II labelling kit (Ambion). Northern blots were hybridized for 16 h at 42 °C, then washed twice for 5 min in 2×SSC (1×SSC is 0.15 M NaCl/0.015 M sodium citrate) with 0.1% SDS and once for 15 min in 0.1×SSC with 0.1% SDS at 42 °C. Membranes were exposed on a Molecular Dynamics phosphor screen for 1 day before analysis with a STORM 860 PhosphorImager (Molecular Dynamics).
Quantitative real-time PCR
Comparative determinations of Dct transcript levels were performed by quantitative RT–PCR using specific primers and SyBr Green detection (Light Cycler; Roche). We quantified the transcripts relative to the gapdh housekeeping gene by determining the difference between the crossing point (Cp) of amplification of the target RNA and the gapdh RNA (ΔCp gapdh); Cp is defined as the point when the amplification starts the exponential phase. Comparison of transcript levels then relies on differences between the ΔCps (ΔΔCp, ddcp). The numbers of fold activation were calculated as follows: given the relation nb fold=[nb copies Dct (mutant)]/[nb copies Dct (black)], nb fold can be expressed as 10x, where x=log 10{log[nbcopiesDct(mutant)]}−{log[nbcopiesDct(black)]}.
Melanogenic activity assays
Dct activity was measured by HPLC as the disappearance of dopachrome substrate and the production of DHICA rather than 5,6-dihydroxyindole, as reported previously [20]. Measurement of peaks resolved by HPLC and comparison with known standards permitted the conversion of data into pmol of product. The results are expressed as pmol of product·(μg of protein)−1·h−1 at 37 °C. Tyrosine hydroxylase activity was measured using the [3H]tyrosine assay [21]. The 3H2O produced during the hydroxylation of tyrosine to L-dopa was measured. The amount of product (pmol) is calculated following measurements of radioactivity by liquid-scintillation counting. Melanin production was measured using the [14C]tyrosine assay as described previously [22].
Analytical methods
The eumelanin and pheomelanin contents of samples were quantified as described previously [23]. For the determination of eumelanin, the samples were oxidized with permanganate to give PTCA (pyrrole-2,3,5-tricarboxylic acid), which was quantified by HPLC using UV detection [24]. Each determination was performed in duplicate. PTCA is almost exclusively produced from DHICA-derived units in eumelanin [25]. For the HPLC determination of pheomelanin, the samples were hydrolysed with hydriodic acid to give 4-AHP (4-amino-3-hydroxyphenylalanine), which was quantified using electrochemical detection as follows: 1 ng of PTCA and 1 ng of 4-AHP correspond to 45 ng of eumelanin and 9 ng of pheomelanin respectively [26]. For the spectrophotometric determination of total melanin, samples were heated with 900 μl of Soluene-350 (Packard, Meriden, CT, U.S.A.) and analysed for absorbance (A) at 500 nm [26].
The chromatographic determination of thiols (cysteine and the reduced form of glutathione) in extracts of non-agouti black and Dct-mutant melanocytes was performed using a method described previously [27]. Briefly, 1.5×106 cells for each sample were harvested by trypsinization, washed three times with cold PBS and then extracted with 1 ml of cold 0.4 M HClO4. The mixture was sonicated on ice for 30 s, centrifuged at 14000 g for 30 min at 4 °C, and the supernatants were then used for the determination of thiols.
Immunohistochemical staining
Dual labelling using immunofluorescence and laser scanning confocal fluorescence microscopy was used to evaluate the localization of melanogenic proteins in non-agouti black and in Dct-mutant melanocytes, as detailed previously [5]. The following antibody dilutions were used: αPEP7, 1:40; αPEP8, 1:40; Vti1b, 1:10; HMB-45, 1:20. Nuclei were counterstained with DAPI (4,6-diamidino-2-phenylindole, blue fluorescence; Vector Laboratories) for 15 min at room temperature (23 °C). Immunoreactive cells were classified into three categories according to whether they showed green, red or yellow fluorescence (the latter colour indicating co-localization of the red and green signals). All preparations were examined with a Model TCS4D DMIRBE confocal microscope (Leica, Heidelberg, Germany) equipped with argon and argon–krypton laser sources. Controls included sections stained as detailed above, but omitting the first antibody.
Electron microscopy
Confluent flasks of non-agouti black and of Dct-mutant melanocytes were fixed with 2% (v/v) glutaraldehyde for 24 h at 4 °C, collected by centrifugation and then washed twice with cold PBS. All samples were post-fixed in 1% osmium tetroxide in 0.1 M phosphate buffer (pH 7.2) for 1 h, dehydrated through a graded ethanol series and embedded in PolyBed 812 at 60 °C for 48 h. Ultrathin sections were cut and stained with uranyl acetate and examined in a JEOL JEM-1200 EX electron microscope.
Homology modelling
Homology modelling was used to build a three-dimensional model of Dct in the region of the active site of the enzyme. The identification of a suitable template was performed by fold recognition [28]. The target and template sequences were then aligned using MULTALIN [29], and the alignment was further optimized manually in several steps, and tuned to fit information on secondary structure and on residues involved in metal binding. Prediction of the target secondary structure was performed using several methods: PHD [30], PsiPred [31], SAM_T99 [32] and SSPRO [33]. The optimized sequence alignment in the metal-binding domains was used to build a three-dimensional model. This was further subjected to simulated annealing in the region of non-conserved loops and, finally, to repeated rounds of energy minimization in order to relieve steric conflicts. Structure modelling, refinement and analysis were performed using the programs Insight II, Discover and Homology (Accelrys, San Diego, CA, U.S.A.), on a Silicon Graphics Octane 2 station. Modelling of the C-terminal region was based on transmembrane domain prediction using DAS [34], TOPPRED [35], TMHMM [36] and TMPRED [37] and hydrophobicity analysis based on the Kyte and Doolittle scale.
RESULTS
Dilution of pigmentation resulting from slaty and slaty light mutations
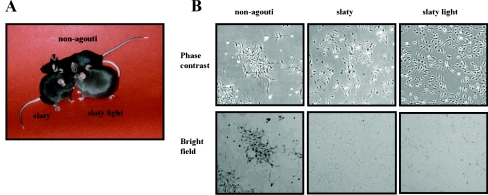
Figure 1(A) shows the dilution of coat colour in Dct-mutant mice compared with non-agouti black mice, as reported previously [3,38,39]. Young slaty mice exhibit a darker coat colour and tend to get depigmented as they get older, showing slightly greyer hair compared with non-agouti black mice of the same age. They also tend to lose their hair earlier in their lifetime compared with non-agouti black mice [40,41], and this may be due to the production of toxic intermediates in the hair bulb, due to the Dct gene mutations. The slaty light phenotype is more severe than the slaty phenotype, i.e. the mice have even lighter colour coats.
Figure 1. Comparison of non-agouti black and Dct-mutant mice.
(A) Note the dilution of coat colour in Dct-mutants. (B) Phase-contrast and bright field photomicrographs of non-agouti black, slaty and slaty light melanocytes. All photomicrographs are shown at the same magnification (×10).
Dct-mutant melanocytes derived from Dct-mutant mice grew as adherent monolayers and were dendritic (Figure 1B). Bright-field microscopy shows the highly pigmented population of non-agouti black melanocytes and the more diluted pigmentation of Dct-mutant cells, which is in good agreement with the phenotypes of the mice (Figure 1A).
Melanin biosynthesis in Dct-mutant melanocytes
Chemical analysis of the melanins revealed striking differences between the three cell lines tested, showing reductions of eumelanin and increases in pheomelanin produced by melanocytes carrying mutations in the Dct gene (Table 1). Eumelanin content of slaty melanocytes was only 4% and that of slaty light melanocytes was 24% of that of non-agouti black melanocytes. Surprisingly, the pheomelanin content is 5-fold higher in slaty and 26-fold higher in slaty light cells compared with non-agouti black melanocytes. The PTCA/total melanin ratio is an indicator of DHICA-derived units. In this regard, the decreased ratio for slaty melanocytes is consistent with the low level of Dct, and similar results were obtained for hair extracts as reported previously [42]. However, we are unable to explain the relatively high ratio for slaty light at this time.
Table 1. Enzymatic activity of Dct and tyrosinase, content of melanin and thiol in non-agouti black versus Dct-mutant melanocytes in culture.
| Melanin content* | Enzymatic activity‡ | Thiol content§ | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Phenotype | PTCA-eumelanin | 4-AHP-pheomelanin | Total melanin† | PTCA/total melanin† | Dct | Tyrosinase | Melanin formation | Cysteine | Glutathione |
| Non-agouti black | 1280±16 | 26±0.5 | 599 | 2.14 | 23.58±0.82 | 26.20±8.55 | 53.30±3.29 | 0.27 | 2.78 |
| Slaty | 53±3 | 130±10 | 81 | 0.66 | 8.36±0.16 | 25.81±1.42 | 61.46±4.77 | 0.27 | 3.07 |
| Slaty light | 302±39 | 662±21 | 198 | 1.53 | 0.88±0.02 | 23.45±3.66 | 60.56±2.77 | 0.60 | 0.29 |
* Expressed in ng/106 cells.
† Expressed in terms of A500×1000/106 cells.
‡ Expressed in terms of pmol·(μg of protein)−1·h−1.
§ Expressed in terms of nmol/106 cells.
As shown in Table 1, Dct activity in slaty melanocytes is reduced to approx. 36% of the enzyme level in non-agouti black cells and, more surprisingly, slaty light melanocytes have only approx. 4% of the activity of non-agouti black melanocytes. Tyrosinase activity, measured by the 3H and 14C melanogenic assays, showed no differences between the three cell lines, suggesting that altered levels of tyrosinase are not responsible for the reduced pigmentation of Dct-mutant cells. The thiol content of Dct-mutant cells revealed a 2-fold higher concentration of cysteine and a very low concentration of glutathione in slaty light melanocytes compared with non-agouti black melanocytes; the glutathione level in slaty cells is slightly increased compared with non-agouti black melanocytes, and there was no difference in the cysteine levels between the two cell lines.
Intracellular trafficking and targeting of Dct to melanosomes in Dct-mutant melanocytes
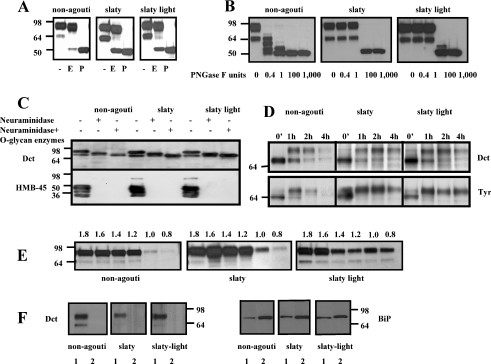
We then investigated the intracellular trafficking and targeting of Dct to melanosomes in non-agouti black and in Dct-mutant melanocytes. Western-blot analysis of crude extracts of non-agouti black and Dct-mutant melanocytes (Figure 2A) show that Dct is detectable as two distinct bands (at ∼69 and 85 kDa) in all three cell lines studied. PNGase F digestion of the lysates, which removes the attached N-glycans from the polypeptide chains, caused Dct to migrate as a single band at approx. 55 kDa, which indicates that the two bands are glycoforms of the same Dct polypeptide. Untreated extracts from slaty and from slaty light melanocytes show a more predominant 69 kDa band compared with non-agouti black cells, which may indicate a prolonged glycosylation and/or a longer retention in the ER (endoplasmic reticulum)/trans-Golgi network. These results show that the slaty and the slaty light mutations do not affect the synthesis of Dct protein. Western blots of tyrosinase and tyrosinase-related protein 1 also showed a similar profile in all three cell lines, suggesting that there was no influence of mutant Dct on the stability or trafficking of those other melanogenic proteins (results not shown).
Figure 2. Western-blot analysis of Dct glycosylation in extracts of melanocytes.
(A) Equal amounts of protein (1 μg) were electrophoretically separated by SDS/PAGE (8% polyacrylamide). Glycosylation of Dct was determined after digestion with buffer (−), endo H (E) or PNGase F (P), as indicated; the blots were incubated with αPEP8 (1:1000) and with the HRP-linked secondary antibody (1:1500). (B) Limited digestion of Dct using different concentrations of PNGase F as noted. (C) Neuraminidase digestion and O-glycan deglycosylation of Dct in non-agouti, slaty and slaty light melanocytes. Blots were also probed with HMB-45 antibody as a positive control to show the reactivity of neuraminidase. (D) Metabolic pulse–chase labelling of non-agouti black and Dct-mutant melanocytes; melanocytes were labelled for 30 min with 35S, harvested at the chase times indicated, solubilized and immunoprecipitated using specific antibodies for Dct and tyrosinase. (E) Distribution of Dct in melanosomal fractions of non-agouti black and Dct-mutant melanocytes: 0.8 and 1.0 M, stage I/II; 1.2 and 1.4 M, stage II; 1.6 and 1.8 M, stage III/VI. (F) Sodium carbonate extraction of membrane proteins; 1, pellet (membrane fraction of melanosomes purified by sucrose gradient, not extracted by sodium carbonate); 2, supernatant (soluble) fraction of purified melanosomes (extracted by sodium carbonate). BiP was used as a control for the sodium carbonate extraction.
Limited digestion with PNGase F showed a differential resistance to this enzyme of the Dct produced by the three types of melanocytes (Figure 2B). The presence of the major 85 kDa band in all samples suggests that all glycosylation sites are occupied. Wild-type Dct can be hydrolysed at all N-glycosylation sites one by one, even with a small amount of PNGase F (0.4 unit), suggesting that all sites are equally exposed. However, Dct produced in slaty and in slaty light melanocytes is more resistant (there is no hydrolysis until 100 units of PNGase F is used), suggesting that not all sites are equally exposed. Thus the folding of Dct may be different in these mutant cells, which suggests that the Dct produced by Dct-mutant melanocytes has one key N-glycosylation site allowing the access to all others and that the difficulty to access these sites may delay the intracellular processing of the mutant protein.
Pulse–chase metabolic labelling analysis shows that the mutations of the Dct gene do not affect the glycosylation, maturation or intracellular trafficking of the mutant proteins (Figures 2C and 2D). Neuraminidase digestion (Figure 2C) shows no difference in Dct between the three cell lines and also proves that the majority of Dct undergoes complex glycosylation in the late stages of the Golgi apparatus. The digestion of Dct in the presence of neuraminidase and O-glycan enzymes reduces the molecular mass of the protein even further, suggesting that Dct may also carry O-glycan residues (including sialic acid) that are added to the protein during late stages of the intracellular trafficking. The blots were probed with HMB-45 antibody as a positive control for neuraminidase activity, since that antibody does not recognize the Pmel17 epitope if sialic acid residues are removed [5]. The metabolic labelling experiments (Figure 2D) show no disruptions in the biosynthesis or processing of Dct or tyrosinase in the Dct mutant cell lines compared with non-agouti black melanocytes. Dct and tyrosinase are both synthesized as 55 kDa polypeptides that almost instantly undergo early glycosylation in the ER and are then seen as bands of 64 kDa. No significant changes were noted in the stability of Dct in the mutant melanocytes compared with control non-agouti melanocytes.
Melanosomes in various stages of maturation can be separated by sucrose-density-gradient ultracentrifugation. Figure 2(E) shows the distribution of Dct in melanosomes at different stages of maturation (i.e. in various parts of the sucrose density gradient). The fully mature form of Dct reaches melanosomes in all three types of melanocytes and is found to some extent in early stage I and II melanosomes, but is mainly distributed in late stage III and IV melanosomes. These data confirm that the slaty light mutation does not cause a defect in the transport of the protein to melanosomes, and that the hypopigmentation of these mice must be due to other factors that decrease the enzymatic activity of mutant Dct.
To determine if either mutation of Dct affects the transmembrane location of the mutant protein, we used sodium carbonate extraction [19]. The presence of wild-type and mutant Dcts in the melanosomal membrane (including the slaty light Dct, which has a mutation in the transmembrane domain) is shown in Figure 2(F). Following the sodium carbonate extraction, Dct was found in the insoluble membrane fraction in all three cell lines, suggesting that even the mutant proteins remain membrane-integrated. BiP was used as a control [43] to show that the sodium carbonate extraction removed the bulk of peripheral membrane proteins in all three cell lines (Figure 2F, right panel).
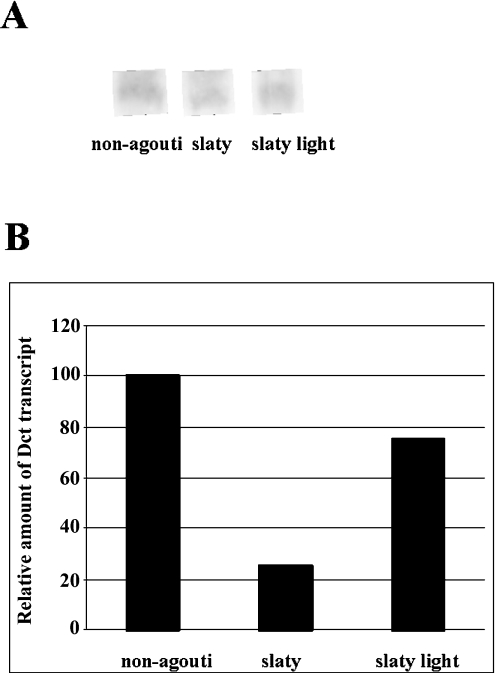
Northern blotting was used to detect Dct mRNA transcripts in black, slaty and slaty light melanocytes (Figure 3A), and semiquantitative real-time RT–PCR was also performed to compare the levels of Dct mRNAs in these cell lines (Figure 3B). Decreases in Dct transcripts were observed in Dct-mutant melanocytes using both techniques, especially in slaty melanocytes. We quantified the transcripts by determining the difference between the crossing point of amplification of Dct RNA and the gapdh housekeeping gene RNA (ΔCp). Thus the comparison of transcript levels relies on the difference between ΔCps (ΔΔCp) in each sample.
Figure 3. Northern blotting of Dct transcripts.
(A) Northern blotting shows one isoform of approx. 444 nt. (B) Real-time PCR analysis of Dct transcripts. Relative amounts of Dct transcripts in slaty and slaty light melanocytes are compared with non-agouti black melanocytes. Results presented were obtained from two independent experiments with two points of measurements each and represent means±S.D. (S.D. values are too small to be seen).
Subcellular distribution of Dct in Dct-mutant melanocytes and ultrastructural characterization of their melanosomes
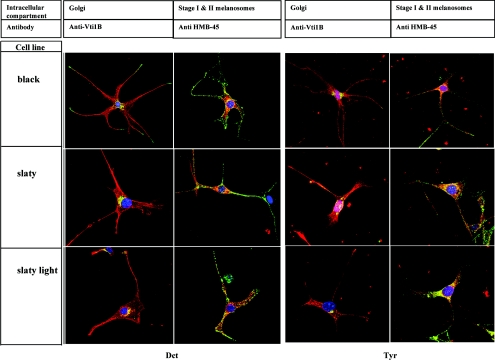
To investigate the subcellular localization of Dct in non-agouti black and in Dct-mutant melanocytes, we used immunohistochemical staining to compare its distribution with Vti1B (a Golgi marker) and HMB-45 (a marker for early melanosomes) (Figure 4, left panels). Dct was detected by red fluorescence and HMB-45 and Vti1B were detected by green fluorescence; in the merged images, yellow indicates co-localization of the two signals. The distribution of Dct in non-agouti black melanocytes was mainly granular throughout the cytoplasm and in dendrites, as well as in the perinuclear area where the Golgi apparatus and stage I melanosomes are found. These results confirm that Dct is processed in the ER and Golgi and is then distributed primarily to melanosomes, as expected from the results presented above. The distribution of Dct in the Dct-mutant cells shows that the mutant Dct is also transported to the Golgi compartment and to the melanosomes, suggesting again that there is no significant trafficking defect responsible for the hypopigmented phenotype of Dct-mutant melanocytes. Figure 4 (right panels) shows that there is no disruption in the intracellular distribution of tyrosinase, suggesting that the mutations in Dct gene do not affect tyrosinase biosynthesis, maturation or trafficking; similar results were obtained for Tyrp1 (results not shown).
Figure 4. Subcellular distribution of Dct (left) and tyrosinase (right) in non-agouti black, slaty and slaty light melanocytes.
Cells were stained with antibodies and fluorescent probes as noted in the Experimental section, and localization of Dct or tyrosinase (red) with Vti1B or HMB-45 (green) was analysed by confocal microscopy. Co-localization of antibodies is indicated by the yellow colour. Nuclei are stained blue with DAPI. Scale bar, 5 μm.
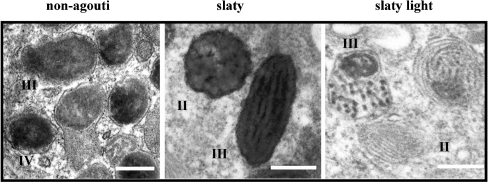
Electron microscopy revealed that melanocytes derived from non-agouti black mice contain numerous highly melanized melanosomes, mainly in stages III and IV, with very well-organized structures (Figure 5). In contrast, melanosomes in Dct-mutant cells had different characteristics: slaty melanocytes contained dark, flocculent melanosomes, showing internal lamellae not fully covered by pigment (which resembled immature stage II–III eumelanin-like organelles). On the other hand, slaty light melanocytes contained melanosomes with mixed pigment distribution including deposits consistent with black eumelanin and aggregations of pheomelanin-like pigments.
Figure 5. Ultrastructure of melanosomes from non-agouti black and Dct-mutant melanocytes.
Figures show different melanosome stages and arrangements of their internal matrices. Scale bar, 0.2 μm.
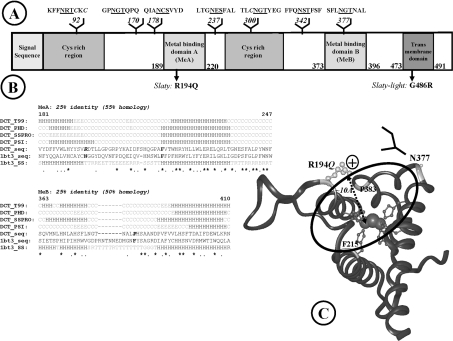
Remote homology modelling of metal-binding domains, containing the slaty mutation
Dct is a highly N-linked glycosylated type I membrane protein containing an N-terminal signal sequence, an epidermal growth factor repeat and other conserved cysteine residues that may be involved in protein–protein interactions, two metal-binding domains (MeA and MeB) that serve as the catalytic site, and a C-terminal transmembrane domain with a short cytoplasmic tail (Figure 6A). Starting with its known primary structure, we used in silico methods to model the structural changes induced in Dct by the slaty and slaty light mutations. Template identification based on fold recognition [28] for the overall Dct sequence resulted in one putative template: catechol oxidase from Ipomoea batatas (PDB code 1bt3), which has been resolved at 2.5 Å resolution (1 Å=10−10 m). Although the e-value is low (0.0156), indicating a 95% confidence in the fold assignment, the level of sequence identity is 15%, which is lower than the limit generally accepted for homology modelling and makes even remote homology techniques uncertain.
Figure 6. Modelling the active site containing the slaty mutation.
(A) Domain organization of proteins in the tyrosinase family. (B) Refined sequence alignment for metal-binding domains. Conserved histidine residues involved in metal binding are highlighted in grey; identical residues are marked with an asterisk, whereas similar residues are marked with a dot; the deleted loop from the template and the replacing potential N-glycosylation site (located on the edge of the binding site) are dot-underlined B, residue in isolated β bridge; E, extended β-strand; G, 310 helix; H, helix; S, bend; T, hydrogen-bonded turn; C, coil (other than helix and extended β in secondary structure prediction); R, random (not assigned secondary structure in the template). (C) Three-dimensional model of the active site of Dct. Histidine residues co-ordinating the metal ions are represented by ball-and-stick, coloured dark grey. Arg194 affected by the slaty mutation is light grey, the N-glycan replacing the loop in the template is indicated schematically by a branched structure. Arg194 is on the edge of the active-site cavity, at the appropriate distance for substrate binding.
We therefore focused on modelling only the active-site region, where the identity level is 25% (Figure 6B). This contains two non-contiguous metal-binding domains (MeA and MeB), brought in contact during the folding process, which form a typical four-α-helix bundle conserved in a large number of proteins from the phenol-oxidase class. Each metal-binding domain consists of two anti-parallel α-helices that bring together three histidine residues that bind the metal ion. Within the two metal-binding domains, the alignment was manually refined to best preserve secondary-structure elements and to conserve the three histidine residues involved in metal binding. In MeB, this imposes an eight-amino- acid deletion in an exposed hairpin loop of the template. By this procedure, we obtained a significant increase in the identity levels, 25% identity (55% homology) for MeA and 25% identity (50% homology) for MeB, which is satisfactory for remote homology techniques.
The active-site model (Figure 6C) indicates that the α-helices of the two metal-binding domains are almost orthogonal, forming a cavity containing the two Zn ions. In this model, the R194Q slaty mutation (represented in light grey in Figure 6C) is located at the end of the first α-helix in MeA, on the edge of this cavity; the loop that has been deleted to optimize the target-template alignment is also located on the edge of the binding-site cavity, and does not affect the structure packing within the model.
Analysis of the transmembrane domain containing the slaty light mutation
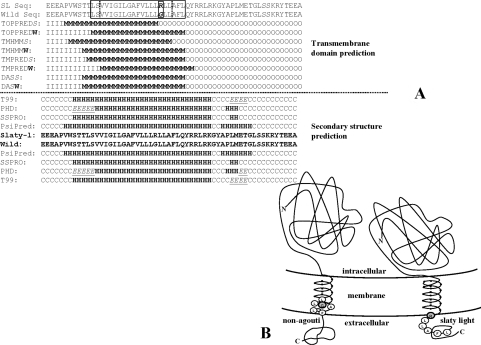
A Kyte and Doolittle hydrophobicity profile of the C-terminal domains of wild-type and slaty light Dct indicates a slight decrease in hydrophobicity due to the presence of an arginine residue. However, this does not affect the ability of the protein to set inserted into the membrane, as suggested by the transmembrane domain and secondary-structure predictions (Figure 7A). Three out of four transmembrane prediction methods indicate that rather than disrupting the transmembrane structure, the mutation would simply slide the transmembrane domain by approx. four amino acids towards the N-terminus (Figure 7B).
Figure 7. Analysis of the C-terminal transmembrane domains of wild-type and slaty light Dct.
(A) G486R mutation does not disrupt the insertion of the protein within the membrane, but it slides the transmembrane region as a whole by approximately four amino acids upstream of the sequence. C, coil (other than helix and extended β in secondary-structure prediction); E, extended β-strand; H, helix; I, intracellular; M, membrane; O, outside (extramelanosomal). (B) Schematic representation of the transmembrane domain sliding in slaty light (right) compared with wild-type Dct (left). Sliding of the transmembrane domain could alter either the properties of the cytosolic domain or the proper position of the globular domain within the melanosome, with respect to the other melanosomal proteins.
DISCUSSION
Mutant Dct produced in slaty mice has a single amino acid substitution from wild-type Dct (R194Q) in the first metal-binding domain. That mutation results in a 3–4-fold decrease in the catalytic function of Dct in vivo, which results in a decrease in the pigmentation of mutant mice to a dark grey/brown colour rather than black as produced in mice wild-type at that locus [44]. In the present study, we show that the enzymatic activity of Dct is 3-fold lower in vitro in slaty melanocytes derived from newborn Dct-mutant mice compared with non-agouti black melanocytes. In slaty light mice, a single amino acid substitution (G486R) was identified [2]. Here, we show that the enzymatic activity of Dct in slaty light melanocytes is even more dramatically lower (28-fold) compared with non-agouti black melanocytes (Table 1), which elicits an even more diluted coat colour in mice carrying this mutation (Figure 1A). When melanins in hair are analysed, the slaty mutation leads to a reduction of approx. 40% of total melanin [42], which is significantly less than what is found in cells in culture (Table 1). In that same report (and in [39]), no effect on pheomelanin synthesis was seen in the hair of slaty-mutant mice. Such differences in values of pheomelanin and total melanins are no doubt due in part to differences in tissue culture (melanocytes only) and in situ (where transfer of melanosomes to the hair shaft is a key event). In addition, the melanocyte population in situ may be significantly compromised in Dct-mutant mice compared with black mice whereas, in tissue culture, only viable melanocytes are co-cultured, passaged and studied. Furthermore, such differences may also reflect the influence of physiological factors in the skin that affect melanocyte function.
In order to explain the link between mutations in the Dct gene and the enzymatic activity/pigmentation of Dct-mutant mice/melanocytes, we followed the processing and intracellular trafficking of Dct in non-agouti black and in Dct-mutant melanocytes. Our results demonstrate clearly that mutant Dct is correctly processed and transported to melanosomes in both types of Dct-mutant melanocytes. Therefore we conclude that there are no dramatic disruptions of the trafficking pathway of Dct resulting from the slaty or slaty light mutations, despite the fact that those mutations severely disrupt the enzymatic activities of the mutant proteins. Furthermore, we also conclude that there is no influence of mutant Dct on the processing, stability or trafficking of other melanogenic proteins (tyrosinase and Tyrp1). These results are consistent with recent findings showing that in mouse melanoma cells, Dct is sorted and trafficked beyond the ER independent of Tyrp1 [45] and that mutations in Tyr or Tyrp1 had no effect on Dct localization [6,7]. Dct mutations in mice dramatically decrease eumelanin content in Dct-mutant melanocytes and unexpectedly increase pheomelanin content (Table 1). One possible hypothesis for the mechanism underlying this increase in pheomelanin would be that the low enzymatic activity of Dct in these Dct-mutant cell lines results in increased levels of its substrate dopachrome and the precursor of dopachrome (dopaquinone), and that those compounds could then be trapped more readily by thiols, thus raising the pheomelanin content. Table 1 shows that there is a significant reduction in the level of glutathione in slaty light cells, indicating a modification of thiol metabolism that might be related to the high level of pheomelanin in these cells. Guyonneau et al. [46] recently generated Dct-knockout mice, and the primary melanocytes derived from them showed a normal distribution of tyrosinase and Tyrp1. Pigment production and coat colour of the Dct knockout mice were affected but were not as severe as the slaty light mutation, suggesting that the presence of mutant protein has a further impact on melanocytes. It remains possible that other unknown melanogenic enzymes may be influenced by Dct function. Since Dct is dramatically down-regulated during the switch to pheomelanogenesis [47], it is tempting to speculate that Dct plays a role in the switching over to pheomelanin production that has not been previously suspected.
We used in silico methods to model the structural changes induced in Dct by the slaty and slaty light mutations. The Dct active site is placed in a cavity formed by four α-helices, containing two Zn ions each co-ordinated by three histidine residues (Figure 6C). Only one of the two ligand-binding amino acids of catechol oxidase (Phe114 and Phe261) is conserved in Dct (Phe215), the other being replaced with a proline residue (Pro383), which is consistent with the fact that Dct and catechol oxidase bind different substrates and perform different functions. In a catalytic mechanism proposed for Dct [48], the phenolic and ketonic oxygens of the dopachrome substrate interact with the Zn ions, and this induces the rearrangement of double bonds in the indolequinone ring. The carboxylic oxygen, on the other hand, is also crucial for guiding the dopachrome in the right stereochemistry into the active site, by interacting with a putative electron acceptor group on the enzyme. In the model that we present here (Figure 6C), the Arg194 involved in the slaty mutation is placed on the edge of the active-site cavity at approx. 10 Å mean distance from the Zn ion co-ordinated by MeA. Considering that, in dopachrome, the distance from the carboxylic oxygen to the ketonic oxygen is approx. 8.5 Å, this may suggest that Arg194 is the electron acceptor group required for substrate binding. The loop of the template that was deleted during the alignment optimization is situated above the metal-binding site, thus suggesting a potential protective role. Interestingly, in the tyrosinase-related protein family, the missing loop is replaced by an N-glycosylation. Occupation of the Asn371 site (sequon Asn-Gly-Thr) located exactly in place of the missing loop, has been shown to be important for tyrosinase activity [49] and there is a high probability that this site is also occupied in Dct, as suggested by a local composition favouring occupancy [50] (Gly and Thr in the sequon and aromatic amino acids upstream of the site).
Transmembrane domain prediction indicates that the G486R mutation would slide the transmembrane domain as a whole by approximately four amino acids upstream (Figure 7B) rather than disrupting the insertion of the protein into the membrane. This represents one turn of the α-helix, equivalent to one pitch (∼5.4 Å) along the z-axis, perpendicular to the membrane plane. Therefore the mechanism by which this mutation may affect the enzymatic activity is more subtle than simply disrupting the membrane insertion of Dct (which we confirmed by the sodium carbonate extraction experiment). Sliding could interfere with Dct function either by altering the properties of the cytosolic domain or by altering the proper positioning of the globular domain within the melanosome, perhaps interfering with its interactions with other tyrosinase-related proteins in the complex.
In conclusion, we show in the present study that the hypopigmented coat colour of mice carrying mutations in the Dct gene is due to reduced enzymatic functions of Dct, which results either from interference with the active site (slaty) or from its improper insertion into the melanosomal membrane (slaty light). The post-translational processing and trafficking of Dct to melanosomes is not affected by either mutation. Homology modelling of the active site of Dct suggests that the slaty mutation occurs in a critical part of the active site of the enzyme and may alter its ability to transform the substrate. Transmembrane prediction methods indicate that the slaty light mutation may determine the sliding of the transmembrane domain towards the N-terminus by approximately four amino acids, thus interfering with Dct function, perhaps by disrupting the ability of Dct to interact properly with other melanosomal proteins. Despite intensive searching by many groups, no pigmentary disease has been associated with mutations in the DCT gene in humans, which makes it one of the few pigmentation genes without an associated human pigmentary disease. This could be because of the expected mild phenotype that results from mutations in Dct. Further characterization of melanocytes carrying mutations in Dct will contribute to a better understanding of the importance of Dct in melanocyte function and its implications in the metabolism of melanin toxic intermediates and in cellular survival mechanisms.
Acknowledgments
We thank M. Yamaguchi and M. Spencer from the Pathology Core, National Heart, Lung and Blood Institute (Bethesda, MD, U.S.A.), for their help with the electron microscopy and electron micrographs. We thank Dr H. Watabe (National Cancer Institute, NIH), Dr G. Negroiu (Institute of Biochemistry, Bucharest, Romania), Dr L. Tabak (National Institute of Dental and Carniofacial Research, NIH) and Dr S. Koyota (National Institute of Diabetes and Digestive and Kidney Diseases, NIH) for useful discussions regarding technical details of the methods used in this study. A.L.M. and A.-J.P. acknowledge the Wellcome Trust support from grant CRIG 067361.
References
- 1.Körner A. M., Pawelek J. M. DOPAchrome conversion: a possible control point in melanin biosynthesis. J. Invest. Dermatol. 1980;75:192–195. doi: 10.1111/1523-1747.ep12522650. [DOI] [PubMed] [Google Scholar]
- 2.Budd P. S., Jackson I. J. Structure of the mouse tyrosinase-related protein-2/dopachrome tautomerase (Tyrp2/Dct) gene and sequence of two novel slaty alleles. Genomics. 1995;29:35–43. doi: 10.1006/geno.1995.1212. [DOI] [PubMed] [Google Scholar]
- 3.Silvers W. K. Basel: Springer-Verlag; 1979. The Coat Colors of Mice: A Model for Mammalian Gene Action and Interaction. [Google Scholar]
- 4.Furumura M., Sakai C., Potterf S. B., Vieira W., Barsh G. S., Hearing V. J. Characterization of genes modulated during pheomelanogenesis using differential display. Proc. Natl. Acad. Sci. U.S.A. 1998;95:7374–7378. doi: 10.1073/pnas.95.13.7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kushimoto T., Basrur V., Matsunaga J., Vieira W. D., Muller J., Appella E., Hearing V. J. A new model for melanosome biogenesis based on the purification and mapping of early melanosomes. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10698–10703. doi: 10.1073/pnas.191184798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toyofuku K., Wada I., Spritz R. A., Hearing V. J. The molecular basis of oculocutaneous albinism type 1 (OCA1): sorting failure and degradation of mutant tyrosinase results in a lack of pigmentation. Biochem. J. 2001;355:259–269. doi: 10.1042/0264-6021:3550259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toyofuku K., Wada I., Valencia J. C., Kushimoto T., Ferrans V. J., Hearing V. J. Oculocutaneous albinism (OCA) types 1 and 3 are ER retention diseases: mutations in tyrosinase and/or Tyrp1 influence the maturation, degradation of calnexin association of the other. FASEB J. 2001;15:2149–2161. doi: 10.1096/fj.01-0216com. [DOI] [PubMed] [Google Scholar]
- 8.Orlow S. J., Zhou B. K., Chakraborty A. K., Drucker M., Pifko-Hirst S., Pawelek J. M. High-molecular-weight forms of tyrosinase and the tyrosinase-related proteins: evidence for a melanogenic complex. J. Invest. Dermatol. 1994;103:196–201. doi: 10.1111/1523-1747.ep12392743. [DOI] [PubMed] [Google Scholar]
- 9.Manga P., Sato K., Ye L., Beermann F., Lamoreux M. L., Orlow S. J. Mutational analysis of the modulation of tyrosinase by tyrosinase-related proteins 1 and 2 in vitro. Pigment Cell Res. 2000;13:364–374. doi: 10.1034/j.1600-0749.2000.130510.x. [DOI] [PubMed] [Google Scholar]
- 10.Jackson I. J., Chambers D. M., Tsukamoto K., Copeland N. G., Gilbert D. J., Jenkins N. A., Hearing V. J. A second tyrosinase-related protein, TRP2, maps to and is mutated at the mouse slaty locus. EMBO J. 1992;11:527–535. doi: 10.1002/j.1460-2075.1992.tb05083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furumura M., Solano F., Matsunaga N., Sakai C., Spritz R. A., Hearing V. J. Metal ligand binding specificities of the tyrosinase related proteins. Biochem. Biophys. Res. Commun. 1998;242:579–585. doi: 10.1006/bbrc.1997.8007. [DOI] [PubMed] [Google Scholar]
- 12.Spritz R. A., Ho L., Furumura M., Hearing V. J. Mutational analysis of copper-binding by human tyrosinase. J. Invest. Dermatol. 1997;109:207–212. doi: 10.1111/1523-1747.ep12319351. [DOI] [PubMed] [Google Scholar]
- 13.Urabe K., Aroca P., Tsukamoto K., Mascagna D., Palumbo A., Prota G., Hearing V. J. The inherent cytotoxicity of melanogenic intermediates: a revision. Biochim. Biophys. Acta. 1994;1221:272–278. doi: 10.1016/0167-4889(94)90250-x. [DOI] [PubMed] [Google Scholar]
- 14.Costin E. G., Vieira W. D., Valencia J. C., Rouzaud F., Lamoreux M. L., Hearing V. J. Immortalization of mouse melanocytes carrying mutations in various pigmentation genes. Anal. Biochem. 2004;335:171–174. doi: 10.1016/j.ab.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 15.Jiménez M., Maloy W. L., Hearing V. J. Specific identification of an authentic clone for mammalian tyrosinase. J. Biol. Chem. 1989;264:3397–3403. [PubMed] [Google Scholar]
- 16.Tsukamoto K., Jackson I. J., Urabe K., Montague P. M., Hearing V. J. A second tyrosinase-related protein, TRP2, is a melanogenic enzyme termed DOPAchrome tautomerase. EMBO J. 1992;11:519–526. doi: 10.1002/j.1460-2075.1992.tb05082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaumburg-Lever G., Metzler G., Kaiserling E. Ultrastructural localization of HMB-45 binding sites. J. Cutan. Pathol. 1991;18:432–435. doi: 10.1111/j.1600-0560.1991.tb01380.x. [DOI] [PubMed] [Google Scholar]
- 18.Costin G. E., Valencia J. C., Vieira W. D., Lamoreux M. L., Hearing V. J. Tyrosinase processing and intracellular trafficking is disrupted in mouse primary melanocytes carrying the uw mutation: a model for oculocutaneous albinism (OCA) type 4. J. Cell Sci. 2003;116:3203–3212. doi: 10.1242/jcs.00598. [DOI] [PubMed] [Google Scholar]
- 19.Fujiki Y., Hubbard A. L., Fowler S., Lazarow P. B. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palumbo A., d'Ischia M., Misuraca G., Prota G. Effect of metal ions on the rearrangement of DOPAchrome. Biochim. Biophys. Acta. 1987;925:203–209. doi: 10.1016/0304-4165(87)90110-3. [DOI] [PubMed] [Google Scholar]
- 21.Pomerantz S. H. L-tyrosine-3,5-3H assay for tyrosinase development in skin of newborn hamsters. Science. 1969;164:838–839. doi: 10.1126/science.164.3881.838. [DOI] [PubMed] [Google Scholar]
- 22.Hearing V. J., Ekel T. M. Mammalian tyrosinase: a comparison of tyrosine hydroxylation and melanin formation. Biochem. J. 1976;157:549–557. doi: 10.1042/bj1570549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wakamatsu K., Ito S. Advanced chemical methods in melanin determination. Pigment Cell Res. 2002;15:174–183. doi: 10.1034/j.1600-0749.2002.02017.x. [DOI] [PubMed] [Google Scholar]
- 24.Ito S., Wakamatsu K. An improved modification of permanganate oxidation of eumelanin that gives a constant yield of pyrrole-2,3,5-tricarboxylic acid. Pigment Cell Res. 1994;7:141–144. doi: 10.1111/j.1600-0749.1994.tb00041.x. [DOI] [PubMed] [Google Scholar]
- 25.Ito S., Wakamatsu K., Ozeki H. Chemical analysis of melanins and its application to the study of the regulation of melanogenesis. Pigment Cell Res. 2000;13:103–109. doi: 10.1034/j.1600-0749.13.s8.19.x. [DOI] [PubMed] [Google Scholar]
- 26.Ozeki H., Ito S., Wakamatsu K., Thody A. J. Spectrophotometric characterization of eumelanin and pheomelanin in hair. Pigment Cell Res. 1996;9:265–270. doi: 10.1111/j.1600-0749.1996.tb00116.x. [DOI] [PubMed] [Google Scholar]
- 27.Imai Y., Ito S., Fujita K. Determination of natural thiols by liquid chromatography after derivatization with 3,5-di-tert-butyl-1,2-benzoquinone. J. Chromatogr. 1987;420:404–410. doi: 10.1016/0378-4347(87)80197-4. [DOI] [PubMed] [Google Scholar]
- 28.Kelley L. A., MacCallum R. M., Sternberg M. J. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 29.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rost B., Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 31.McGuffin L. J., Bryson K., Jones D. T. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 32.Karplus K., Barrett C., Hughey R. Hidden Markov models for detecting remote protein homologies. Bioinformatics. 1998;14:846–856. doi: 10.1093/bioinformatics/14.10.846. [DOI] [PubMed] [Google Scholar]
- 33.Pollastri G., Przybylski D., Rost B., Baldi P. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins. 2002;47:228–235. doi: 10.1002/prot.10082. [DOI] [PubMed] [Google Scholar]
- 34.Cserzo M., Wallin E., Simon I., von Heijne G., Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 35.Claros M. G., von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 1994;10:685–686. [Google Scholar]
- 36.Krogh A., Larsson B., von Heijne G., Sonnhammer E. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2002;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 37.Hofmann K., Stoffel W. TMbase – a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler. 1993;374:166. [Google Scholar]
- 38.Bennett D. C., Lamoreux M. L. The color loci of mice – a genetic century. Pigment Cell Res. 2003;16:333–344. doi: 10.1034/j.1600-0749.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- 39.Ozeki H., Ito S., Wakamatsu K., Hirobe T. Chemical characterization of hair melanins in various coat-color mutants of mice. J. Invest. Dermatol. 1995;105:361–366. doi: 10.1111/1523-1747.ep12320792. [DOI] [PubMed] [Google Scholar]
- 40.Pierro L. J., Chase H. B. Slate – a new coat color mutant in the mouse. J. Hered. 1963;54:47–50. doi: 10.1093/oxfordjournals.jhered.a107219. [DOI] [PubMed] [Google Scholar]
- 41.Pierro L. J., Chase H. B. Temporary hair loss associated with the slate mutation of coat color in the mouse. Nature (London) 1965;205:579–580. [Google Scholar]
- 42.Lamoreux M. L., Wakamatsu K., Ito S. Interaction of major coat color gene functions in mice as studied by chemical analysis of eumelanin and pheomelanin. Pigment Cell Res. 2001;14:23–31. doi: 10.1034/j.1600-0749.2001.140105.x. [DOI] [PubMed] [Google Scholar]
- 43.Semenza J. C., Hardwick K. G., Dean N., Pelham H. R. ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell (Cambridge, Mass.) 1990;61:1349–1357. doi: 10.1016/0092-8674(90)90698-e. [DOI] [PubMed] [Google Scholar]
- 44.Kroumpouzos G., Urabe K., Kobayashi T., Sakai C., Hearing V. J. Functional analysis of the slaty gene product (TRP2) as DOPAchrome tautomerase, and the effect of a point mutation on its catalytic function. Biochem. Biophys. Res. Commun. 1994;202:1060–1068. doi: 10.1006/bbrc.1994.2036. [DOI] [PubMed] [Google Scholar]
- 45.Negroiu G., Dwek R. A., Petrescu S. M. Tyrosinase-related protein-2 and -1 are trafficked on distinct routes in B16 melanoma cells. Biochem. Biophys. Res. Commun. 2005;328:914–921. doi: 10.1016/j.bbrc.2005.01.040. [DOI] [PubMed] [Google Scholar]
- 46.Guyonneau L., Murisier F., Rossier A., Moulin A., Beermann F. Melanocytes and pigmentation are affected in dopachrome tautomerase knockout mice. Mol. Cell. Biol. 2004;24:3396–3403. doi: 10.1128/MCB.24.8.3396-3403.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakai C., Ollmann M., Kobayashi T., Abdel-Malek Z. A., Muller J., Vieira W. D., Imokawa G., Barsh G. S., Hearing V. J. Modulation of murine melanocyte function in vitro by agouti signal protein. EMBO J. 1997;16:3544–3552. doi: 10.1093/emboj/16.12.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Solano F., Martinez-Liarte J. H., Jiménez-Cervantes C., García-Borrón J. C., Jara J. R., Lozano J. A. Molecular mechanism for catalysis by a new zinc-enzyme: DOPAchrome tautomerase. Biochem. J. 1996;313:447–453. doi: 10.1042/bj3130447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Branza-Nichita N., Negroiu G., Petrescu A. J., Garman E. F., Platt F. M., Wormald M. R., Dwek R. A., Petrescu S. M. Mutations at critical N-glycosylation sites reduce tyrosinase activity by altering folding and quality control. J. Biol. Chem. 2000;275:8169–8175. doi: 10.1074/jbc.275.11.8169. [DOI] [PubMed] [Google Scholar]
- 50.Petrescu A. J., Milac A. L., Petrescu S. M., Dwek R. A., Wormald M. R. Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure, and folding. Glycobiology. 2004;14:103–114. doi: 10.1093/glycob/cwh008. [DOI] [PubMed] [Google Scholar]