Abstract
The Rho family of small GTPases are signalling molecules involved in cytoskeleton remodelling and gene transcription. Their activities are important for many cellular processes, including myogenesis. In particular, RhoA positively regulates skeletal-muscle differentiation. We report in the present study that the active form of RhoA increases the expression of utrophin, the autosomal homologue of dystrophin in the mouse C2C12 and rat L8 myoblastic cell lines. Even though this RhoA-dependent utrophin increase is higher in proliferating myoblasts, it is maintained during myogenic differentiation. This occurs via two mechanisms: (i) transcriptional activation of the utrophin promoter A and (ii) post-translational stabilization of utrophin. In addition, RhoA increases plasma-membrane localization of utrophin. Thus RhoA activation up-regulates utrophin levels and enhances its localization at the plasma membrane.
Keywords: dystrophin, plasma membrane, RhoA GTPase, skeletal muscle, utrophin stabilization, utrophin up-regulation
Abbreviations: CHX, cycloheximide; CMV, cytomegalovirus; DM, differentiation medium; DMD, Duchenne muscular dystrophy; DMEM, Dulbecco's modified Eagle's medium; NFAT, nuclear factor of activated T-cells
INTRODUCTION
DMD (Duchenne muscular dystrophy) is a severe X-linked muscle wasting disease caused by the absence of dystrophin, a cytoskeletal protein normally localized at the cytoplasmic surface of the sarcolemma, where it links the intracellular cytoskeleton network to the extracellular matrix and acts as a large protein scaffold referred to as the dystrophin–glycoprotein complex [1–3]. This disease affects 1 in 3500 males, and patients die from cardiac or respiratory complications in their twenties. Among the treatments proposed for DMD is compensation for dystrophin loss by replacement with utrophin [4,5]. Utrophin, the autosomal homologue of dystrophin, is associated with a complex of sarcolemmal proteins identical with that of dystrophin, and also binds F-actin [6,7]. Utrophin is expressed ubiquitously and, in adult muscle, this protein is found at the neuromuscular and myotendinous junctions [6,8]. In fetal skeletal muscle, utrophin is expressed throughout the sarcolemma from which it disappears and is replaced by dystrophin postnatally [9,10]. In addition, utrophin was found at the sarcolemma in mdx (dystrophin-deficient) skeletal muscle, which led to the hypothesis that utrophin might be capable, at least in part, of replacing dystrophin in adult tissues [11,12]. Overexpression of a utrophin transgene in the mdx mouse model of DMD rescued its myopathic phenotype, restoring muscle function and correct dystrophin-associated protein complex localization, suggesting that utrophin could functionally replace dystrophin in muscle [13–17]. Thus identifying the mechanisms that up-regulate utrophin expression and/or stabilization could have important implications in the treatment of DMD. Two promoters (A and B) are responsible for utrophin expression. Promoter A is responsible for synaptic and myogenic expression, whereas promoter B drives expression to the vascular endothelia [18]. Myogenic induction of promoter A is activated by the binding of the helix–loop–helix MRFs (myogenic regulatory factors) MyoD, myogenin and MRF4, which activate a number of genes in skeletal muscle [19]. These muscle-specific genes are controlled by different upstream pathways. The GTPase RhoA is crucial for myogenesis induction, since it controls both the expression and activity of serum response factor, a transcription factor that regulates the expression of the muscle-determining factor MyoD, as well as that of many muscle-specific genes [20–23].
In the present study, we have analysed the effect of an active form of RhoA (RhoAV14) on utrophin expression and subcellular distribution. Utrophin was analysed in cell lines from rat and mouse myoblasts stably expressing RhoAV14. We showed that utrophin expression levels are increased by approx. 2–7-fold by RhoAV14 through different pathways. Transcriptional activation is observed in RhoAV14-expressing myoblasts, as measured by mRNA levels and the activity of utrophin promoter A. In addition, we demonstrated that the utrophin protein is stabilized by RhoAV14 expression. Finally, plasma-membrane localization of utrophin is increased in RhoAV14-expressing myoblasts. These results suggest that RhoA expression or activation might offer a means to up-regulate utrophin expression.
MATERIALS AND METHODS
Establishment of stable cell lines
G418-resistant GP+E-86 clones expressing a constitutively activated form of RhoA (RhoAV14) were grown to collect retrovirus-containing cell-free supernatants. Infection of C2C12 and L8 myoblasts was performed as described in [21]. Cells were grown continuously in G418.
Cell culture
C2C12 mouse and L8 rat myoblasts were grown in DMEM (Dulbecco's modified Eagle's medium)/Ham's F-12 medium (ratio 1:1) supplemented with 10% fetal calf serum (Hyclone, Perbio Science, Tattenhall, Cheshire, U.K.). To induce differentiation, the growth medium was replaced with DM (differentiation medium) consisting of DMEM/Ham's F-12 medium supplemented with 2% fetal calf serum. A stable cell line was cultured under the same conditions in a medium supplemented with 1 mg/ml G418. Fresh CHX (cycloheximide) diluted in PBS was used at 10 μg/ml. Cytochalasin B was used at 2 μM and jasplakinolide at 0.1 μM.
Luciferase assays
Cells cultured in 35 mm dishes were transfected using the Lipofectamine™ method as described by the manufacturer (Life Technologies, Gaitherburg, MD, U.S.A.) with 1 μg of chimaeric construct containing the utrophin promoter A fused to the luciferase gene (plasmid 1,3 Fwt, kindly provided by Dr K. Davies, Department of Human Anatomy and Genetics, University of Oxford, Oxford, U.K.) and 40 ng of pRL CMV (cytomegalovirus) vector (Renilla luciferase CMV). The medium was replaced with growth medium 4 h after transfection. DM was added 4 h later. Cells were left for at least 2 days before extraction. Luciferase activity was measured using the Dual-Luciferase Reporter Assay system (Promega). All assays are performed in triplicate.
Gel electrophoresis and immunoblotting
Cell extracts were prepared as described in [24]. Protein concentration was determined with a BCA (bicinchoninic acid) protein assay kit (Pierce). Then, 20 μg of protein was resolved on a polyacrylamide gel (4–10%) and transferred on to Immobilon-P. Membranes were incubated with monoclonal antibodies against utrophin (NCL-DRP-2, Novocastra Laboratories, Newcastle upon Tyne, U.K.) or α-tubulin (1:100, hybridoma 356). Membranes were processed as described in [24].
Quantification of utrophin mRNA by real-time quantitative PCR
Total RNA was prepared from cell extracts collected at different times after DM addition using RNeasy minikit from Qiagen. cDNAs were synthesized from 5 mg of total RNA using SuperScript II Reverse Transcriptase (Invitrogen) and random hexamer primers. One fifth of the cDNA was directly used for the PCR set. The levels of the various cDNAs were determined by quanti-tative PCR using SYBR Green I from a Light cycler (Roche Diagnostics, Somerville, NJ, U.S.A.). The utrophin primers designed from mouse and rat sequences are 5′-GCACTGGCAGGTGAAGGATG-3′ and 5′-GTGGTGATGTTGAGGACGTTGAC-3′. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA was amplified as an internal control. The specificity of the primers was confirmed by DNA sequencing.
Immnocytochemistry
Cells grown on 35 mm dishes were fixed in 3.7% formaldehyde in PBS followed by a 5 min permeabilization in 0.1% Triton X-100 in PBS and incubated in PBS containing 0.1% BSA. Utrophin and β-catenin stainings were performed using NCL-DRP-2 (Novocastra Laboratories) or anti-β-catenin (Transduction Laboratories) revealed by an Alexa Fluor 546- or Alexa Fluor 488-conjugated goat anti-rabbit antibody (Molecular Probes, Interchim, Montlucon, France). Actin filaments were stained with rhodamine-conjugated phalloidin (0.5 unit/ml; Sigma). Images were captured with a MicroMax 1300 CCD camera (charge-coupled-device camera; RS-Princeton Instruments, Trenton, NJ, U.S.A.) driven by MetaMorph software (v.4.11; Universal Imaging Corp., Westchester, PA, U.S.A.). Images were processed using Adobe Photoshop and Illustrator.
Deconvolution and co-localization
Stacks of 16-bit files (Z step, 0.1 μm) were captured as described above, and epifluorescence images were first restored with Huygens (Scientific Volume Imaging b.v, Hilversum, The Netherlands). Huygens is an iterative program that can reassign light, after encoding as grey level, to its sources in the stack with a very high probability using a point spread function. This results in removing the fuzziness contained in the stack, while keeping the three-dimensional information. In the present study, the Maximum Likelihood Estimation algorithm was used throughout. Restored stacks were then further processed with Imaris (Bitplane, Zurich, Switzerland) for visualization and volume rendering. Respective co-localizations of utrophin and β-catenin fluorescence were analysed with the Imaris co-localization module.
RESULTS
Expression of active RhoA increases utrophin expression in cultured skeletal myoblasts
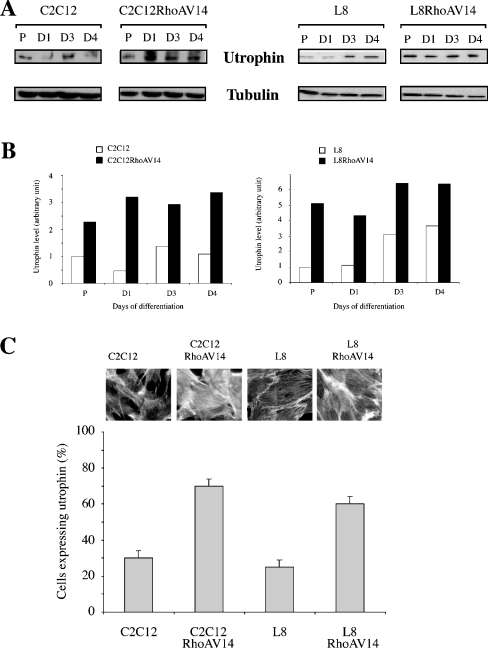
We have analysed utrophin expression in mouse C2C12 and rat L8 myoblast cell lines stably expressing a constitutively active form of RhoA GTPase (RhoAV14). Parental and RhoAV14-expressing cell lines were induced to differentiate, and utrophin expression was monitored at different times after the shift to DM. Figure 1(A) shows an immunoblot analysis of utrophin in parental and RhoAV14-expressing C2C12 and L8 myoblast during the course of differentiation. The histograms represent the amount of utrophin normalized to the amount of tubulin (Figure 1B). These results show that RhoAV14 increases the expression of utrophin in both C2C12 and L8 myoblasts (from 2- to 7-fold). This is observed in proliferative myoblasts and throughout the differentiation process. We also observed that utrophin expression is increased upon differentiation, as reported previously [19]. The amount of β-dystroglycan is similar in RhoAV14-expressing and control myoblasts (results not shown). Cell lines stably expressing RhoAV14 have been previously used and do not present defects in the differentiation process [21]. We also measured utrophin expression by immunocytochemistry in parental and RhoAV14-expressing myoblasts (Figure 1C). Quantification of several independent experiments also confirms the above results, namely that RhoAV14 increased utrophin expression in myogenic cell lines. Expression of activated RhoA protein was monitored by examining the modifications of F-actin-containing structures controlled by this protein, i.e. actin stress fibres [25]. Images of the F-actin staining show that RhoAV14-expressing cells had increased actin stress fibres (Figure 1C).
Figure 1. Effect of RhoAV14 on utrophin expression.
(A) Protein extracts (20 μg/well) from parental and RhoAV14-expressing C2C12 and L8 myoblasts collected at the indicated periods were immunoblotted for utrophin and α-tubulin expression. Results are representative of at least three independent experiments. P, proliferative myoblasts; D, days in DM. (B) The histogram represents the quantification of utrophin expression normalized for the amount of α-tubulin. These results are representative for three independent experiments. (C) Utrophin expression analysed by indirect immunofluorescence in parental and RhoAV14-expressing C2C12 and L8 myoblasts. The histogram represents the number of cells expressing utrophin under each condition. Results are representative of three independent experiments (∼100 cells were counted in each experiment). Images show parental and RhoAV14-expressing myoblasts stained for filamentous actin distribution with rhodamine-conjugated phalloidin.
Expression of active RhoA increases utrophin mRNA level and activates utrophin promoter A
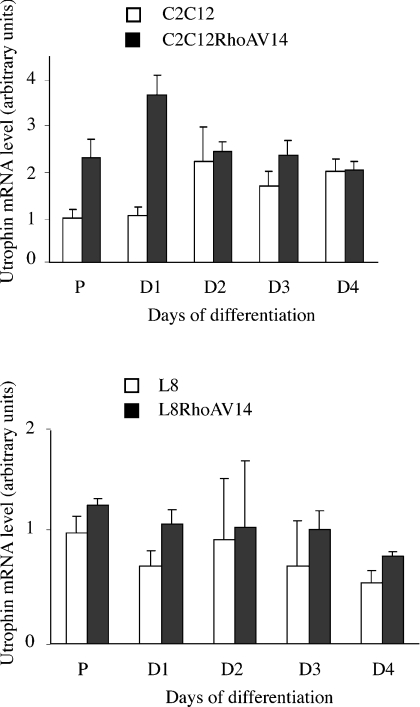
In order to analyse the mechanisms that lead to increased utrophin expression, we first measured utrophin mRNA expression using real-time quantitative PCR (Figure 2). RNA was extracted from parental or RhoAV14-expressing myoblasts under normal proliferating conditions or after the shift to DM. RhoAV14 induced a 2–3.5-fold increase in utrophin mRNA in proliferative C2C12 myoblasts and after 1 day in DM, but not later during differentiation. In addition, this effect is not found in L8 myoblasts at either time point of the differentiation process.
Figure 2. Utrophin mRNA level is increased in C2C12 myoblasts.
The Figure shows real-time quantitative mRNA expression analysis in parental and RhoAV14-expressing C2C12 and L8 myoblasts. mRNA extraction was performed in proliferative myoblasts or at the indicated days after the addition of DM. Results are means±S.E.M. for three independent experiments.
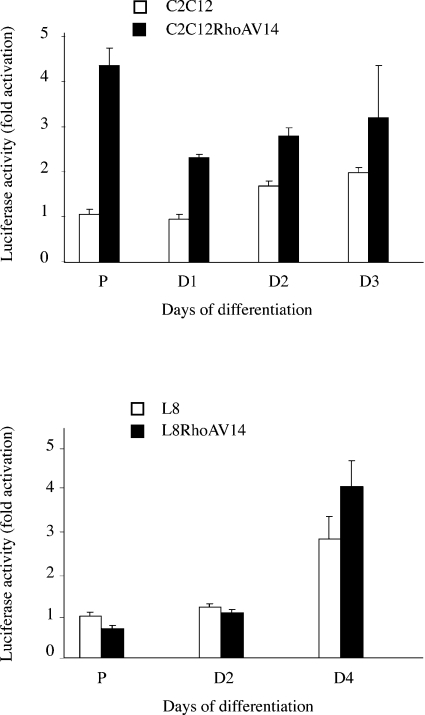
To determine whether RhoAV14 can induce the utrophin promoter, we analysed the activity of a reporter construct in which the expression of luciferase is driven by the utrophin promoter A [19]. RhoAV14 increases the activity of the utrophin reporter gene in C2C12 myoblasts but not in L8 cells (Figure 3), supporting our previous results on mRNA expression.
Figure 3. Utrophin promoter A is activated in C2C12 myoblasts.
C2C12 and L8 myoblasts were co-transfected with plasmids containing the luciferase reporter gene driven by the utrophin promoter A and a Renilla luciferase reporter gene driven by the CMV promoter. After the different treatments indicated on the graph, luciferase activity was measured and corrected with respect to Renilla luciferase activity. Results are means±S.E.M. for three independent experiments.
Expression of active RhoA stabilizes utrophin
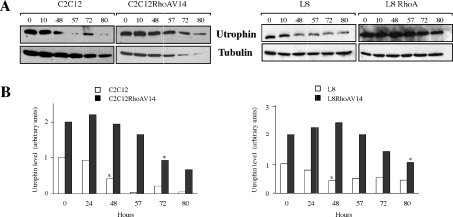
We next assessed whether RhoAV14 might also influence the half-life of utrophin protein. Control and RhoAV14-expressing C2C12 and L8 myoblasts were treated with CHX, and the amount of utrophin was analysed by Western blotting at different times thereafter (Figure 4). The half-life of utrophin is strongly increased in RhoA-expressing cells, going from 48 h in control to 70–80 h in RhoAV14-expressing myoblasts. These results show that active RhoA induces the stabilization of the utrophin protein in both C2C12 and L8 myoblasts.
Figure 4. Effect of RhoAV14 expression on utrophin half-life.
(A) Parental and RhoAV14-expressing C2C12 and L8 myoblasts were treated with CHX (10 μg/ml) for 24, 48, 57, 72 and 80 h, and the level of utrophin protein was analysed by Western blotting. (B) The histogram represents the quantification of utrophin protein level after CHX treatment normalized for the amount of α-tubulin. These results are representative of three independent experiments. Asterisks (*) indicate the time required to have half of the protein left.
Expression of active RhoA triggers plasma-membrane localization of utrophin
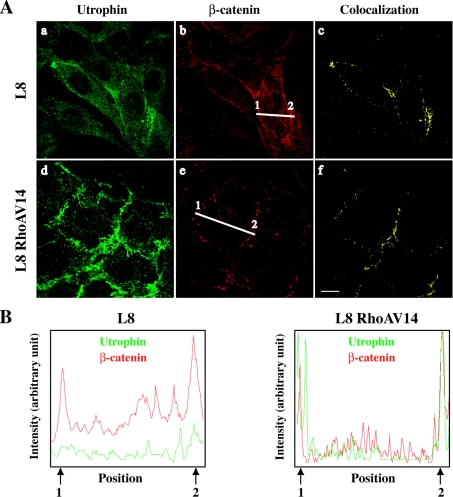
Utrophin localization was analysed by immunocytochemistry in control and RhoAV14-expressing C2C12 and L8 myoblasts. As shown in Figure 5(A), utrophin weakly accumulates at the plasma membrane in parental L8 myoblasts, and the accumulation is increased in RhoAV14-expressing L8 myoblasts [Figures 5A(a) and 5A(d) respectively]. To further confirm utrophin association with the plasma membrane, we performed a co-localization analysis with β-catenin [Figures 5A(b) and 5A(e)], a membrane-associated protein that accumulates at cell–cell contact sites [24]. Image stacks were deconvoluted using the Huygens System image restoration software. The co-localization of utrophin and β-catenin fluorescence was studied with the Imaris co-localization module [Figures 5A(c) and 5A(f)]. As shown in the line scan analysis (Figure 5B), utrophin co-localized with β-catenin at the plasma membrane (arrows 1 and 2). This co-localization is strongly increased in RhoAV14-expressing cells. A similar but weaker increase of utrophin localization at the plasma membrane was also observed in C2C12 myoblasts (results not shown). Taken together, these results show that active RhoA increases utrophin localization at the plasma membrane.
Figure 5. Effect of RhoAV14 on utrophin localization.
(A) Utrophin and β-catenin localizations were analysed by indirect immunofluorescence in control L8 (a, b) and L8 RhoAV14-expressing myoblasts (d, e). Deconvoluted stacks of images were visualized using Imaris software. The respective voxel co-localizations of utrophin with β-catenin were obtained using the Imaris co-localization module and are shown in (c, f). Scale bar, 10 μm. (B) Co-localization of the utrophin and β-catenin signals was analysed along the line indicated on the images in (A) by line scan (MetaMorph software).
Utrophin localization is dependent on the actin cytoskeleton
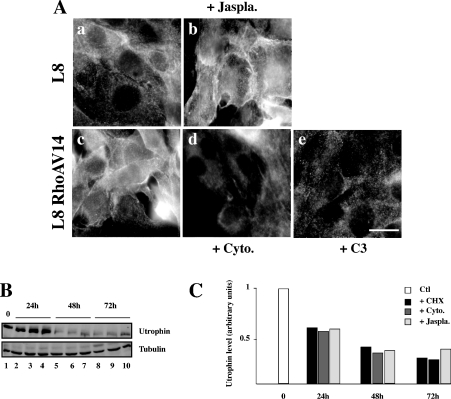
We next investigated whether this increased utrophin localization at the plasma membrane results from change in actin polymerization, since RhoA GTPase controls actin stress fibre assembly [25]. Utrophin localization was analysed by immunocytochemistry in control L8 myoblasts [Figure 6A(a)] and in L8 myoblasts treated with jasplakinolide [Figure 6A(b)], a peptide inducing actin polymerization [26]. This treatment increases utrophin localization at the plasma membrane. We also analysed utrophin localization in RhoAV14-expressing L8 myoblasts [Figure 6A(c)] and in RhoAV14-expressing L8 myoblasts treated with cytochalasin B [Figure 6A(d)], which disrupts the F-actin cytoskeleton, or with exoenzyme C3 [Figure 6A(e)], which inactivates the GTPases RhoA, RhoB and RhoC and induces the loss of stress fibres [27]. These two treatments translocated utrophin from the plasma membrane. We thus examined whether this correlates with utrophin stabilization. L8 myoblasts were treated with CHX alone or with either cytochalasin B or jasplakinolide, and the amount of utrophin was analysed at different times thereafter (Figures 6B and 6C). The half-life of utrophin is not modified by these drugs. These results show that utrophin localization is dependent on the assembly of the F-actin cytoskeleton, whereas its stability is not.
Figure 6. Effect of actin cytoskeleton modifications on utrophin localization.
(A) Utrophin localization was analysed by indirect immunofluorescence in control L8 myoblasts (a), L8 myoblasts treated with jasplakinolide (0.1 μM) (b), L8 RhoAV14-expressing myoblasts (c), L8 RhoAV14-expressing myoblasts treated with cytochalasin B (2 μM) (d) or with exoenzyme C3 (25 μg/ml) (e). Scale bar, 10 μm. (B) Myoblasts were treated with CHX (+CHX, 10 μg/ml), CHX+cytochalasin B (+cyto) or CHX+jasplakinolide (+jaspla) and the amount of utrophin was analysed by Western blotting at different times thereafter. (C) The histogram represents the quantification of utrophin protein level normalized for the amount of α-tubulin.
DISCUSSION
We demonstrate in the present study that expression of an active form of RhoA induces increased expression and plasma-membrane association of utrophin. These results add to our understanding of the mechanism of utrophin expression that could be of therapeutic benefit in DMD. Furthermore, the observation that RhoA induces significant up-regulation of utrophin along the plasma membrane places RhoA on the list of potential ‘booster genes’, once again suggesting the potential for the development of pharmacological strategies for treating various forms of muscular dystrophy [28]. Indeed, humans with DMD and mdx mice carry X-linked recessive mutations in the gene encoding dystrophin, a large membrane-associated cytoskeletal protein located just beneath the sarcolemma membrane. The lack of dystrophin also results in a secondary loss or reduced expression of dystrophin-associated glycoproteins, including dystroglycans and sarcoglycans [29]. Transgenic overexpression of utrophin can compensate for the lack of dystrophin in mdx mice and relieves the symptoms at both the structural and functional level [13,30]. Thus utrophin can replace dystrophin, restore the expression of dystrophin-associated glycoproteins in the mdx mouse and thereby directly prevent or delay the onset of pathology [31].
We have identified at least two mechanisms to explain the up-regulation of utrophin in active RhoA-expressing cells. First, RhoAV14 induces utrophin promoter activity and mRNA accumulation in mouse C2C12 myoblasts. However, this effect was not found in rat L8 myoblasts. Why this mechanism does not function in this cell line remains unclear. Our results confirm previous observations of a 2-fold increase in A-utrophin promoter activity and mRNA level during differentiation of C2C12 myoblasts [19,32]. In addition, previous studies have identified glucocorticoid, heregulin (a polypeptide growth factor homologue) and the phosphatase inhibitor okadaic acid as activators of utrophin promoter A [33–35]. In all cases, the effect is in the same range as the one we observe with RhoA. Studies of the utrophin promoter A show the existence of different regulatory elements, such as E-box, N-box, Sp1 (specificity protein 1) and NFAT (nuclear factor of activated T-cells)-binding sites, which co-ordinately regulate its transcription [19,36,37]. The RhoA-dependent signalling targets remain to be determined, although it might regulate the activity of the E-box, Sp1- and NFAT-binding sites. Indeed, RhoA has been shown to control the expression of the muscle-determining factor MyoD, which, similar to other myogenic factors, binds to the utrophin E-box [19–21,23,38]. In addition, RhoA might modulate the activity of Sp family and NFAT transcription factors [39,40].
We also demonstrated that RhoA affects utrophin protein levels. This was particularly obvious in rat L8 myoblasts, where RhoA has no transcriptional effect. This post-transcriptional effect is also observed in C2C12 myoblasts. Such a regulation has also been observed in vivo, since, in mdx mice, utrophin mRNA is not up-regulated [41]. Since utrophin binds only to F-actin and not to G-actin and links costameric actin filaments to the sarcolemma [7,17], we then tested whether plasma-membrane localization might protect utrophin from degradation in the cytoplasm. Whereas RhoA-induced utrophin recruitment to the membrane might be linked to an increase in actin polymerization, this is not true for the stability of the protein. Thus the most likely mechanism is protection from protein degradation. Indeed, utrophin can be cleaved by calpains [42], and we can imagine that RhoA modifies their activity. RhoA GTPase activity is mediated by downstream effectors, such as the Rho kinase family of serine/threonine kinases and the proline-rich formin homology domain-containing proteins mDia1 and mDia2, which regulate actin stress fibre organization and alignment of actin bundles and microtubules [43]. Whether these RhoA effectors participate in the regulation of utrophin localization through signalling towards actin filaments remains to be determined. Recently, the modulation of the tyrosine phosphorylation status of β-dystroglycan has been shown to regulate its association with utrophin [44]. The potential role of RhoA in β-dystroglycan phosphorylation and utrophin binding will be interesting to analyse. Finally, perturbation of the RhoA signalling pathway has been observed in the dy mice [dystrophic mice, in which the laminin-alpha(2) component of the dystrophin-associated complex is mutated], suggesting a major role for this molecule in various forms of muscle dystrophies [45].
RhoA activities are thus potential targets for pharmacological intervention that would up-regulate the expression of utrophin along the extrasynaptic sarcolemma of the dystrophic muscle.
Acknowledgments
We thank P. Travo for constant support (http://www.mri.cnrs.fr), Y. Boublik (CNRS, Centre National de la Recherche Scientifique for C3 exoenzyme and B. Hipskind and P. Fort (CNRS) for a critical reading of this paper. This work was supported by the Association Francaise contre les Myopathies.
References
- 1.Koenig M., Hoffman E. P., Bertelson C. J., Monaco A. P., Feener C., Kunkel L. M. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell (Cambridge, Mass.) 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 2.Matsumura K., Campbell K. P. Dystrophin-glycoprotein complex: its role in the molecular pathogenesis of muscular dystrophies. Muscle Nerve. 1994;17:2–15. doi: 10.1002/mus.880170103. [DOI] [PubMed] [Google Scholar]
- 3.Campbell K. P. Three muscular dystrophies: loss of cytoskeleton-extracellular matrix linkage. Cell (Cambridge, Mass.) 1995;80:675–679. doi: 10.1016/0092-8674(95)90344-5. [DOI] [PubMed] [Google Scholar]
- 4.Kapsa R., Kornberg A. J., Byrne E. Novel therapies for Duchenne muscular dystrophy. Lancet Neurol. 2003;2:299–310. doi: 10.1016/s1474-4422(03)00382-x. [DOI] [PubMed] [Google Scholar]
- 5.Nowak K. J., Davies K. E. Duchenne muscular dystrophy and dystrophin: pathogenesis and opportunities for treatment. EMBO Rep. 2004;5:872–876. doi: 10.1038/sj.embor.7400221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohlendieck K., Ervasti J. M., Matsumura K., Kahl S. D., Leveille C. J., Campbell K. P. Dystrophin-related protein is localized to neuromuscular junctions of adult skeletal muscle. Neuron. 1991;7:499–508. doi: 10.1016/0896-6273(91)90301-f. [DOI] [PubMed] [Google Scholar]
- 7.Winder S. J., Hemmings L., Maciver S. K., Bolton S. J., Tinsley J. M., Davies K. E., Critchley D. R., Kendrick-Jones J. Utrophin actin binding domain: analysis of actin binding and cellular targeting. J. Cell Sci. 1995;108:63–71. doi: 10.1242/jcs.108.1.63. [DOI] [PubMed] [Google Scholar]
- 8.Law D. J., Allen D. L., Tidball J. G. Talin, vinculin and DRP (utrophin) concentrations are increased at mdx myotendinous junctions following onset of necrosis. J. Cell Sci. 1994;107:1477–1483. doi: 10.1242/jcs.107.6.1477. [DOI] [PubMed] [Google Scholar]
- 9.Clerk A., Morris G. E., Dubowitz V., Davies K. E., Sewry C. A. Dystrophin-related protein, utrophin, in normal and dystrophic human fetal skeletal muscle. Histochem. J. 1993;25:554–561. [PubMed] [Google Scholar]
- 10.Rigoletto C., Prelle A., Ciscato P., Moggio M., Comi G., Fortunato F., Scarlato G. Utrophin expression during human fetal development. Int. J. Dev. Neurosci. 1995;13:585–593. doi: 10.1016/0736-5748(95)00039-j. [DOI] [PubMed] [Google Scholar]
- 11.Karpati G., Carpenter S., Morris G. E., Davies K. E., Guerin C., Holland P. Localization and quantitation of the chromosome 6-encoded dystrophin-related protein in normal and pathological human muscle. J. Neuropathol. Exp. Neurol. 1993;52:119–128. doi: 10.1097/00005072-199303000-00004. [DOI] [PubMed] [Google Scholar]
- 12.Weir A. P., Morgan J. E., Davies K. E. A-utrophin up-regulation in mdx skeletal muscle is independent of regeneration. Neuromuscul. Disord. 2004;14:19–23. doi: 10.1016/j.nmd.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Tinsley J., Deconinck N., Fisher R., Kahn D., Phelps S., Gillis J. M., Davies K. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat. Med. 1998;4:1441–1444. doi: 10.1038/4033. [DOI] [PubMed] [Google Scholar]
- 14.Rafael J. A., Tinsley J. M., Potter A. C., Deconinck A. E., Davies K. E. Skeletal muscle-specific expression of a utrophin transgene rescues utrophin-dystrophin deficient mice. Nat. Genet. 1998;19:79–82. doi: 10.1038/ng0598-79. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert R., Nalbanoglu J., Tinsley J. M., Massie B., Davies K. E., Karpati G. Efficient utrophin expression following adenovirus gene transfer in dystrophic muscle. Biochem. Biophys. Res. Commun. 1998;242:244–247. doi: 10.1006/bbrc.1997.7936. [DOI] [PubMed] [Google Scholar]
- 16.Squire S., Raymackers J. M., Vandebrouck C., Potter A., Tinsley J., Fisher R., Gillis J. M., Davies K. E. Prevention of pathology in mdx mice by expression of utrophin: analysis using an inducible transgenic expression system. Hum. Mol. Genet. 2002;11:3333–3344. doi: 10.1093/hmg/11.26.3333. [DOI] [PubMed] [Google Scholar]
- 17.Rybakova I. N., Patel J. R., Davies K. E., Yurchenco P. D., Ervasti J. M. Utrophin binds laterally along actin filaments and can couple costameric actin with sarcolemma when overexpressed in dystrophin-deficient muscle. Mol. Biol. Cell. 2002;13:1512–1521. doi: 10.1091/mbc.01-09-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weir A. P., Burton E. A., Harrod G., Davies K. E. A- and B-utrophin have different expression patterns and are differentially up-regulated in mdx muscle. J. Biol. Chem. 2002;277:45285–45290. doi: 10.1074/jbc.M205177200. [DOI] [PubMed] [Google Scholar]
- 19.Perkins K. J., Burton E. A., Davies K. E. The role of basal and myogenic factors in the transcriptional activation of utrophin promoter A: implications for therapeutic up-regulation in Duchenne muscular dystrophy. Nucleic Acids Res. 2001;29:4843–4850. doi: 10.1093/nar/29.23.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carnac G., Primig M., Kitzmann M., Chafey P., Tuil D., Lamb N., Fernandez A. RhoA GTPase and serum response factor control selectively the expression of MyoD without affecting Myf5 in mouse myoblasts. Mol. Biol. Cell. 1998;9:1891–1902. doi: 10.1091/mbc.9.7.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meriane M., Roux P., Primig M., Fort P., Gauthier-Rouviere C. Critical activities of Rac1 and Cdc42Hs in skeletal myogenesis: antagonistic effects of JNK and p38 pathways. Mol. Biol. Cell. 2000;11:2513–2528. doi: 10.1091/mbc.11.8.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charrasse S., Causeret M., Comunale F., Bonet-Kerrache A., Gauthier-Rouviere C. Rho GTPases and cadherin-based cell adhesion in skeletal muscle development. J. Muscle Res. Cell Motil. 2003;24:309–313. [PubMed] [Google Scholar]
- 23.Wei L., Zhou W., Croissant J. D., Johansen F. E., Prywes R., Balasubramanyam A., Schwartz R. J. RhoA signaling via serum response factor plays an obligatory role in myogenic differentiation. J. Biol. Chem. 1998;273:30287–30294. doi: 10.1074/jbc.273.46.30287. [DOI] [PubMed] [Google Scholar]
- 24.Charrasse S., Meriane M., Comunale F., Blangy A., Gauthier-Rouviere C. N-cadherin-dependent cell-cell contact regulates Rho GTPases and beta-catenin localization in mouse C2C12 myoblasts. J. Cell Biol. 2002;158:953–965. doi: 10.1083/jcb.200202034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paterson H. F., Self A. J., Garrett M. D., Just I., Aktories K., Hall A. Microinjection of recombinant p21rho induces rapid changes in cell morphology. J. Cell Biol. 1990;111:1001–1007. doi: 10.1083/jcb.111.3.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bubb M. R., Senderowicz A. M., Sausville E. A., Duncan K. L., Korn E. D. Jasplakinolide, a cytotoxic natural product, induces actin polymerization and competitively inhibits the binding of phalloidin to F-actin. J. Biol. Chem. 1994;269:14869–14871. [PubMed] [Google Scholar]
- 27.Barbieri J. T., Riese M. J., Aktories K. Bacterial toxins that modify the actin cytoskeleton. Annu. Rev. Cell Dev. Biol. 2002;18:315–344. doi: 10.1146/annurev.cellbio.18.012502.134748. [DOI] [PubMed] [Google Scholar]
- 28.Engvall E., Wewer U. M. The new frontier in muscular dystrophy research: booster genes. FASEB J. 2003;17:1579–1584. doi: 10.1096/fj.02-1215rev. [DOI] [PubMed] [Google Scholar]
- 29.Di Blasi C., Morandi L., Barresi R., Blasevich F., Cornelio F., Mora M. Dystrophin-associated protein abnormalities in dystrophin-deficient muscle fibers from symptomatic and asymptomatic Duchenne/Becker muscular dystrophy carriers. Acta Neuropathol. (Berlin) 1996;92:369–377. doi: 10.1007/s004010050532. [DOI] [PubMed] [Google Scholar]
- 30.Deconinck N., Tinsley J., De Backer F., Fisher R., Kahn D., Phelps S., Davies K., Gillis J. M. Expression of truncated utrophin leads to major functional improvements in dystrophin-deficient muscles of mice. Nat. Med. 1997;3:1216–1221. doi: 10.1038/nm1197-1216. [DOI] [PubMed] [Google Scholar]
- 31.Perkins K. J., Davies K. E. The role of utrophin in the potential therapy of Duchenne muscular dystrophy. Neuromuscul. Disord. 2002;12(Suppl. 1):S78–S89. doi: 10.1016/s0960-8966(02)00087-1. [DOI] [PubMed] [Google Scholar]
- 32.Gramolini A. O., Jasmin B. J. Expression of the utrophin gene during myogenic differentiation. Nucleic Acids Res. 1999;27:3603–3609. doi: 10.1093/nar/27.17.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Courdier-Fruh I., Barman L., Briguet A., Meier T. Glucocorticoid-mediated regulation of utrophin levels in human muscle fibers. Neuromuscul. Disord. 2002;12(Suppl. 1):S95–S104. doi: 10.1016/s0960-8966(02)00089-5. [DOI] [PubMed] [Google Scholar]
- 34.Khurana T. S., Rosmarin A. G., Shang J., Krag T. O., Das S., Gammeltoft S. Activation of utrophin promoter by heregulin via the ets-related transcription factor complex GA-binding protein alpha/beta. Mol. Biol. Cell. 1999;10:2075–2086. doi: 10.1091/mbc.10.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodova M., Brownback K., Werle M. J. Okadaic acid augments utrophin in myogenic cells. Neurosci. Lett. 2004;363:163–167. doi: 10.1016/j.neulet.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 36.Dennis C. L., Tinsley J. M., Deconinck A. E., Davies K. E. Molecular and functional analysis of the utrophin promoter. Nucleic Acids Res. 1996;24:1646–1652. doi: 10.1093/nar/24.9.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chakkalakal J. V., Stocksley M. A., Harrison M. A., Angus L. M., Deschenes-Furry J., St-Pierre S., Megeney L. A., Chin E. R., Michel R. N., Jasmin B. J. Expression of utrophin A mRNA correlates with the oxidative capacity of skeletal muscle fiber types and is regulated by calcineurin/NFAT signalling. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7791–7796. doi: 10.1073/pnas.0932671100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei L., Zhou W., Wang L., Schwartz R. J. beta(1)-integrin and PI 3-kinase regulate RhoA-dependent activation of skeletal alpha-actin promoter in myoblasts. Am. J. Physiol. Heart Circ. Physiol. 2000;278:H1736–H1743. doi: 10.1152/ajpheart.2000.278.6.H1736. [DOI] [PubMed] [Google Scholar]
- 39.Ashcroft F. J., Varro A., Dimaline R., Dockray G. J. Control of expression of the lectin-like protein Reg-1 by gastrin: role of the Rho family GTPase RhoA and a C-rich promoter element. Biochem. J. 2004;381:397–403. doi: 10.1042/BJ20031793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foucault I., Le Bras S., Charvet C., Moon C., Altman A., Deckert M. The adaptor protein 3BP2 associates VAV guanine nucleotide exchange factors to regulate NFAT activation by the B cell antigen receptor. Blood. 2004;105:1106–1113. doi: 10.1182/blood-2003-08-2965. [DOI] [PubMed] [Google Scholar]
- 41.Roma J., Munell F., Fargas A., Roig M. Evolution of pathological changes in the gastrocnemius of the mdx mice correlate with utrophin and beta-dystroglycan expression. Acta Neuropathol. (Berlin) 2004;108:443–452. doi: 10.1007/s00401-004-0908-1. [DOI] [PubMed] [Google Scholar]
- 42.Earnest J. P., Santos G. F., Zuerbig S., Fox J. E. Dystrophin-related protein in the platelet membrane skeleton. Integrin-induced change in detergent-insolubility and cleavage by calpain in aggregating platelets. J. Biol. Chem. 1995;270:27259–27265. doi: 10.1074/jbc.270.45.27259. [DOI] [PubMed] [Google Scholar]
- 43.Bishop A. L., Hall A. Rho GTPases and their effector proteins. Biochem. J. 2000;348:241–255. [PMC free article] [PubMed] [Google Scholar]
- 44.James M., Nuttall A., Ilsley J. L., Ottersbach K., Tinsley J. M., Sudol M., Winder S. J. Adhesion-dependent tyrosine phosphorylation of (beta)-dystroglycan regulates its interaction with utrophin. J. Cell Sci. 2000;113:1717–1726. doi: 10.1242/jcs.113.10.1717. [DOI] [PubMed] [Google Scholar]
- 45.Sakuma K., Nakao R., Inashima S., Hirata M., Kubo T., Yasuhara M. Marked reduction of focal adhesion kinase, serum response factor and myocyte enhancer factor 2C, but increase in RhoA and myostatin in the hindlimb dy mouse muscles. Acta Neuropathol. (Berlin) 2004;108:241–249. doi: 10.1007/s00401-004-0884-5. [DOI] [PubMed] [Google Scholar]