Abstract
Glucose acutely stimulates proinsulin synthesis in pancreatic β-cells through a poorly understood post-transcriptional mechanism. In the present study, we demonstrate in pancreatic β-cells that glucose stimulates the recruitment of ribosome-associated proinsulin mRNA, located in the cytoplasm, to the ER (endoplasmic reticulum), the site of proinsulin synthesis, and that this plays an important role in glucose-stimulated proinsulin synthesis. Interestingly, glucose has greater stimulatory effect on the recruitment of proinsulin mRNA to the ER compared with other mRNAs encoding secretory proteins. This, as far as we are aware, is the first example whereby mRNAs encoding secretory proteins are selectively recruited to the ER and provides a novel regulatory mechanism for secretory protein synthesis. Contrary to previous reports, and importantly in understanding the mechanism by which glucose stimulates proinsulin synthesis, we demonstrate that there is no large pool of ‘free’ proinsulin mRNA in the cytoplasm and that glucose does not increase the rate of de novo initiation on the proinsulin mRNA. However, we show that glucose does stimulate the rate of ribosome recruitment on to ribosome-associated proinsulin mRNA. In conclusion, our results provide evidence that the selective recruitment of proinsulin mRNA to the ER, together with increases in the rate of initiation are important mediators of glucose-stimulated proinsulin synthesis in pancreatic β-cells.
Keywords: β-cell, mRNA, polysome analysis, proinsulin, protein synthesis, signal recognition particle (SRP)
Abbreviations: CPH, carboxypeptidase-H; DTT, dithiothreitol; ER, endoplasmic reticulum; ERK, extracellular-signal-regulated kinase; KRB, Krebs–Ringer bicarbonate buffer; MIN6, mouse insulinoma cell line 6; mRNP, messenger ribonucleoprotein; PC2, prohormone convertase 2; poly(A)+, polyadenylated; RNC, ribosome nascent chain; RRL, rabbit reticulocyte lysate; SRP, signal recognition particle; SR, SRP receptor; UTR, untranslated region
INTRODUCTION
Nutrients such as glucose stimulate the rapid release of insulin from the pancreatic β-cell [1]. To ensure the immediate replenishment of insulin, there is a rapid and specific increase in proinsulin synthesis (10–20-fold within 40 min) [2,3]. This acute increase in proinsulin synthesis is mediated via a post-transcriptional mechanism as it occurs both in the absence of any change in proinsulin mRNA abundance [3] and in the presence of transcription inhibitors [4–6]. However, the mechanism by which glucose rapidly stimulates proinsulin synthesis is poorly understood.
Previous reports have indicated that de novo initiation (defined here as the recruitment of ribosomes on to ribosome-free mRNA) plays a key role in the regulation of proinsulin synthesis in response to glucose [3,7–9]. Indeed, based on these studies, it is thought that glucose stimulates proinsulin synthesis through the recruitment of ribosomes on to a large inert cytoplasmic pool of ‘free’ proinsulin mRNA (i.e. non-ribosome associated) [3,7–9]. The mechanism by which these reported increases in the rate of de novo initiation on the proinsulin mRNA occurs is unknown. The 5′-UTR (5′-untranslated region) of the proinsulin mRNA is predicted to form a stem–loop structure [10,11], which may act in a way analogous to that of the iron-responsive element found in the 5′-UTR of the ferritin mRNA [12]. Indeed, more recently, it has been demonstrated that the 5′-UTR of the proinsulin mRNA can promote glucose-stimulated proinsulin synthesis [13]. Other elements within the proinsulin mRNA have also been reported to be important for glucose-stimulated proinsulin synthesis [13–15]. The 3′-UTR has been shown to stabilize the proinsulin mRNA in response to glucose through binding to the polypyrimidine tract-binding protein [14,15].
It has also been reported that glucose may stimulate proinsulin synthesis by promoting the release of SRP (signal recognition particle) from the proinsulin mRNA RNC (ribosome nascent chain)–SRP complex through interaction with the SR (SRP receptor) [3,7–9] (see [16,17] for reviews on SRP/SR). This is primarily based on results showing that glucose stimulates the recruitment of approx. 15–38% of the proinsulin mRNA to the ER (endoplasmic reticulum), the site of proinsulin synthesis [3,7–9]. Additionally, it has been reported that proinsulin protein elongation rates are specifically up-regulated between 0 and 3.3 mM glucose [7]; however, these increases are fairly modest and occur only at low glucose concentrations and again are therefore unlikely to play a major role in glucose-stimulated proinsulin synthesis.
In the present study, we demonstrate, in pancreatic β-cells, that glucose stimulates the selective recruitment of the proinsulin mRNA from a cytoplasmic pool of ribosome-associated proinsulin mRNA to the ER. In vitro translation of proinsulin mRNA isolated from membranes or translation of proinsulin mRNA in situ (i.e. on the ER) demonstrates that the recruitment of the proinsulin mRNA to the ER plays a significant role in glucose-stimulated proinsulin synthesis. Contrary to previous reports [1,3–7], we show that: (i) there is no detectable inert pool of ‘free’ proinsulin mRNA and (ii) glucose does not stimulate de novo initiation on the proinsulin mRNA. However, we demonstrate that glucose does stimulate an increase in ribosome recruitment on to ribosome-associated proinsulin mRNA.
EXPERIMENTAL
Chemicals and materials
Analytical grade biochemicals were purchased from Fisher Scientific (Loughborough, Leics., U.K.) and Sigma, unless otherwise specified. RRLs (rabbit reticulocyte lysates; nuclease-treated), mRNA selection poly(A)+ (polyadenylated) tract mRNA isolation system II, Klenow fragment and dNTPs were obtained from Promega (Chilworth, Southampton, U.K.). Hybond-N membrane, [α-32P]dCTP redivue tips, RNA Guard™ and Probequant™ G50 columns were obtained from Amersham Biosciences. Collagenase, foetal calf serum, [35S]methionine and digitonin were obtained from Serva (Heidelberg, Germany), Invitrogen, MP Biomedicals (Irvino, CA, U.S.A.) and Merck Biosciences (Beeston, Nottingham, U.K.) respectively.
Cell culture and treatment
MIN6 cells (mouse insulinoma cell line 6; kindly provided by Professor J.-I. Miyazaki, Department of Surgery and Surgical Basic Science, Graduate School of Medicine, Kyoto University, Kyoto, Japan) were used at approx. 80% confluence between passages 16 and 28. MIN6 cells were grown in Dulbecco's modified Eagle's medium containing 25 mM glucose supplemented with 15% (v/v) heat-inactivated foetal calf serum, 100 μg/ml streptomycin, 100 units/ml penicillin sulphate and 75 μM 2-mercaptoethanol, equilibrated with 5% CO2 and 95% air at 37 °C. Before treatment, the medium was removed and the cells were washed twice in Hepes-balanced KRB (Krebs–Ringer bicarbonate buffer; 115 mM NaCl, 5 mM KCl, 10 mM NaHCO3, 2.5 mM MgCl2, 2.5 mM CaCl2 and 20 mM Hepes, pH 7.4) containing 0.5% BSA. Cells were then preincubated for 1 h at 37 °C in KRB containing 2 mM glucose (unless otherwise stated in the Figure legends) before incubation in KRB containing 2 or 20 mM glucose for a further 1 h (unless otherwise stated in the Figure legends). During the final 10 min, cycloheximide was added to the cells at a concentration of 100 μg/ml to prevent ribosomal run-off (unless otherwise stated).
Polysome analysis
After treatment, cells were lysed in polysome buffer (20 mM Hepes, pH 7.6, 15 mM MgCl2, 300 mM KCl, 1 mg/ml heparin, 0.1 mg/ml cycloheximide, 1 mM DTT (dithiothreitol), 1 μl/ml RNA Guard™, 1 mM benzamidine/HCl, 0.1 mM PMSF, 1 μg/ml leupeptin and 1 μg/ml pepstatin) supplemented with 1% Triton. Cell lysates were centrifuged at 13000 g for 10 min at 4 °C to remove nuclei and cell debris. The supernatants were then layered on to either 7–47% or 20–50% sucrose gradients (made in polysome buffer) and centrifuged at 39000 rev./min for 2 h at 4 °C in a Sorvall TH64.I rotor (radius 7.19–15.32 cm). The gradients were fractionated using an ISCO gradient fractionator that continuously measured absorbance A at 254 nm. RNA, for Northern-blot analysis, was precipitated overnight at −80 °C from each fraction by the addition of 3 vol. of 8 M guanidinium chloride and 4 vol. of ethanol.
Subcellular fractionation: consecutive centrifugation method
After treatment, as described above and in the Figure legends, cells were disrupted in homogenization buffer (250 mM sucrose, 250 mM KCl, 10 mM MgCl2, 10 mM Tris/HCl, pH 7.5, 1 mM EDTA, 2 mM DTT, 1 μl/ml RNA Guard™, 0.1 mg/ml cycloheximide, 1 mM benzamidine/HCl, 0.1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin and 1 mg/ml heparin) and homogenized with 12 strokes in a Dounce homogenizer (20 strokes for islets). The homogenate was centrifuged at 1200 g for 2 min at 4 °C to pellet unlysed cells and nuclei. The supernatant was then centrifuged at 6000 g for 20 min at 4 °C to pellet membranes (P6 fraction). The supernatant was removed and centrifuged at 18000 g for 20 min at 4 °C to pellet any residual membranes (P18 fraction). The supernatant was finally centrifuged at 200000 g for 20 min at 4 °C resulting in a pellet containing ribosomes and mRNPs (messenger ribonucleoproteins; P200) and the supernatant containing ‘free’ mRNA (S200). The pellets were resuspended in Triton lysis buffer (1% Triton, 20 mM Hepes, pH 7.6, 300 mM KCl, 15 mM MgCl2, 2 mM DTT, 1 μl/ml RNA Guard™, 0.1 mg/ml cycloheximide, 1 mg/ml heparin, 1 mM benzamidine/HCl, 0.2 mM PMSF, 1 μg/ml leupeptin and 1 μg/ml pepstatin). Laemmli sample buffer [31] was added to an aliquot of each fraction for Western-blot analysis and RNA was precipitated from the remaining samples for Northern-blot analysis. This consecutive centrifugation method of fractionation is an adaptation of a method by Frey et al. [18].
Subcellular fractionation: digitonin cell permeabilization method
After treatment, as described above and in the Figure legends, the cells were washed with PBS containing 0.1 mg/ml cycloheximide. The cells were then detached from the plate using trypsin/EDTA solution (Invitrogen) supplemented with 0.1 mg/ml cycloheximide. Cells were pelleted at 250 g for 2 min at 4 °C and the supernatant was removed. The cells were then resuspended in polysome buffer containing 80 μg/ml digitonin (Merck Biosciences) and incubated on ice for 5 min. Digitonin-permeabilized cells were pelleted at 250 g for 3 min at 4 °C. The supernatant containing the cytosol was called the cytosolic fraction and the digitonin-permeabilized cell pellet was called the membrane fraction. Laemmli sample buffer was added to an aliquot of each fraction for Western-blot analysis and RNA was isolated from the remaining samples using TRI reagent (Sigma) for Northern-blot analysis.
SDS/PAGE, Western blotting and immunoprecipitation
Anti-ERK2 (where ERK2 stands for extracellular-signal-regulated kinase 2) antiserum was purchased from New England Biolabs (Hitchin, Herts., U.K.). Anti-bovine proinsulin antibody was purchased from Sigma. Anti-SRP54 antibody was purchased from Becton Dickinson (Cowley, Oxford, U.K.). Western blotting, SDS/PAGE and immunoprecipitations were performed as described previously [19].
In vitro and in situ translations
RRLs were used either as provided by the manufacturer or depleted of ribosomes (ribosomes were pelleted by two consecutive centrifugations at 200000 g for 20 min at 4 °C). For in situ and in vitro translations, membranes were isolated using the consecutive centrifugation method of subcellular fractionation. For in situ translation, the membrane fraction was washed twice in KHM buffer (25 mM K-Hepes, pH 7.2, 50 mM KOAc, 5 mM MgOAc, 1 mM DTT and 1 μl/ml RNA Guard™) and resuspended in KHM buffer. The membranes were then added to an RRL in vitro translation system and associated mRNAs were translated in the presence of [35S]methionine for 1 h at 30 °C according to the manufacturer's instructions. After the in situ translation reaction, the membranes, containing proteins translocated into the ER, were pelleted at 6000 g for 20 min at 4 °C and resuspended in Laemmli sample buffer. For in vitro translations, total RNA was isolated from the membrane fractions using TRI reagent. mRNA was then isolated from total RNA using the poly(A)+ tract mRNA isolation system II. mRNA was then translated using the RRL translation system in the presence of [35S]methionine for 1 h at 30 °C according to the manufacturer's instructions. Proinsulin was immunoprecipitated from the reaction and Laemmli sample buffer was added. All translated products were electrophoresed through SDS/polyacrylamide gels and visualized by autoradiography.
Northern blotting
RNA samples were heated at 65 °C for 10 min in RNA sample buffer (Sigma). RNA samples were cooled on ice and run on 1% agarose formaldehyde gels. RNA was transferred by capillary action on to Hybond-N membrane (Amersham Biosciences) and fixed. [α-32P]dCTP radiolabelled cDNA probes were generated using Prime-a-Gene® labelling system (Promega). Unincorporated [α-32P]dCTP was removed by spinning through Probequant™ G50 columns (Amersham Biosciences). Membranes were prehybridized at 65 °C for 1 h in Church Gilbert solution (0.5 M NaHPO4, pH 7.2, 1 mM EDTA and 7%, w/v, SDS) supplemented with denatured salmon sperm DNA (60 μg/ml) before the addition of the denatured radiolabelled probe and hybridization overnight. The membranes were then washed twice for 15 min in 2×SSC (where 1×SSC is 0.15 M NaCl/0.015 M sodium citrate) and 0.1% SDS at room temperature (25 °C) and once for 15 min in 0.2×SSC and 0.1% SDS at 60 °C. The hybridized cDNA probes were visualized by autoradiography.
Islet isolation and culturing
Pancreatic islets were isolated from 200–250 g Wistar rats by collagenase digestion and Histopaque density-gradient centrifugation as described previously [20]. Islets were used immediately. Details of treatments are provided in the Figure legends. During the final 10 min, cycloheximide was added to the cells at a concentration of 100 μg/ml to prevent ribosomal run-off. After treatment, islets were collected by centrifugation at 16000 g for 1 min. RNA for Northern-blot analysis was isolated using TRI reagent.
RESULTS
Glucose stimulates the selective recruitment of proinsulin mRNA to the ER
To determine the mechanism by which glucose specifically stimulates proinsulin synthesis in pancreatic β-cells, we initially investigated the effect of glucose on the recruitment of proinsulin mRNA to the ER, the site of proinsulin synthesis.
MIN6 cells, a mouse pancreatic β-cell line that synthesizes and secretes insulin in response to physiologically relevant glucose concentrations [21,22], were preincubated in KRB containing 0.5 or 2 mM glucose for 1 h followed by incubation in KRB containing the same glucose concentrations or 20 mM glucose for a further period of 1 h. The cells were then lysed and the lysates fractionated by consecutive centrifugation to generate a fraction enriched with ER and associated mRNAs (P6 fraction), a P200 fraction containing polysomal mRNA/mRNPs and an S200 fraction containing ‘free’ mRNAs (i.e. mRNA that is neither protein nor ribosome bound). The location of specific mRNAs within each fraction was determined by Northern-blot analysis (Figures 1a and 1b).
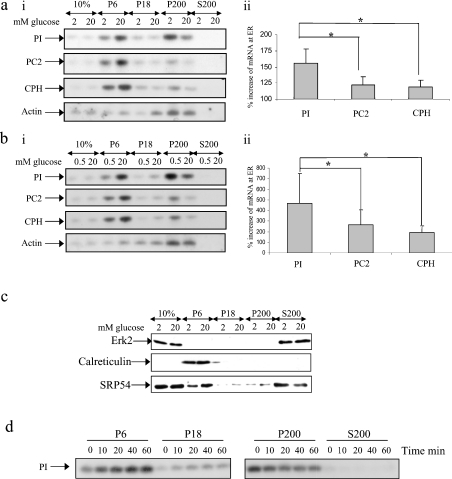
Figure 1. Glucose stimulates the recruitment of mRNAs encoding secretory membrane proteins to the ER: subcellular fractionation by consecutive centrifugation.
MIN6 cells were preincubated in KRB containing 0.5 or 2 mM glucose for 1 h followed by incubation for a further period of 1 h at the same glucose concentration or 20 mM glucose. Cells were fractionated by consecutive centrifugation resulting in a membrane fraction (P6), a residual membrane fraction (P18), a polysome fraction (P200) and a supernatant fraction containing free mRNAs (S200); 10% refers to 10% of total RNA/protein. (ai, bi) RNA isolated from each fraction was run on a 1% agarose formaldehyde gel, transferred on to a nylon membrane and probed for proinsulin (PI), PC2, CPH and actin mRNAs. (aii, bii) The percentage increase in mRNA levels at the ER in response to high glucose was determined by quantification of bands from Northern-blot analysis using ImageJ. The error bars indicate the S.E.M. (n=7). Significant differences are indicated by *P<0.05 (two sample t test). (c) Protein samples from each fraction (8% of the P6/P18/P200 fraction and 1% of the S200 fraction) were run on SDS/PAGE (12.5% polyacrylamide) and subjected to immunoblotting with anti-ERK2, calreticulin and SRP54 antibodies. Detection was by enhanced chemiluminescence. (d) Time course of the recruitment of proinsulin mRNA to the ER. MIN6 cells were incubated in KRB containing 0.5 mM glucose followed by incubation for the times indicated in KRB containing 20 mM glucose. Results presented are representative of three separate experiments.
At a low glucose concentration, the majority of proinsulin mRNA was found in the P200 fraction, whereas at high glucose concentration, the majority of proinsulin mRNA was found in the P6 fraction (Figures 1ai and 1bi). Therefore glucose stimulates the recruitment of proinsulin mRNA to the ER. At either low or high glucose concentration, no ‘free’ proinsulin mRNA was detected in the S200 fraction (Figures 1ai and 1bi). To ascertain whether glucose also caused a redistribution of other mRNAs encoding secretory proteins, the subcellular distributions of PC2 (prohormone convertase 2) and CPH (carboxypeptidase-H) mRNAs were also investigated (Figures 1a and 1b). In contrast with proinsulin mRNA, at either low or high glucose concentration, a large proportion of PC2 and CPH mRNAs were found to be associated with the ER (Figures 1a and 1b). As for proinsulin mRNA, an increase in glucose concentration stimulated the recruitment of both CPH and PC2 to the ER. However, the stimulatory effect of glucose on the recruitment of either PC2 or CPH mRNA to the ER was significantly less than the effect of glucose on the recruitment of proinsulin mRNA (Figures 1a and 1b). At either low or high glucose concentration, no PC2 or CPH mRNA was found free in the cytoplasm (S200 fraction; Figures 1ai and 1bi). mRNA encoding actin, which is translated in the cytoplasm, was found almost exclusively in the P200 fraction at either low or high glucose concentration (Figures 1ai and 1bi). However, glucose also stimulated the recruitment of a small proportion of the actin mRNA to the ER (Figures 1a and 1b).
To determine the efficiency of the subcellular fractionation procedure, protein samples were analysed by Western blotting using antibodies against calreticulin, an ER luminal protein, and ERK2, a protein primarily located in the cytoplasm (Figure 1c). Calreticulin was only found in the P6 fraction, whereas ERK2 was only detected in the S200 fraction (Figure 1c). As the recruitment of mRNAs encoding secretory proteins to the ER is mediated by the SRP [16], we also investigated the subcellular distribution of SRP using an antibody directed against SRP54 (Figure 1c). At both low and high glucose concentrations, SRP54 was predominantly found free in the S200 fraction (Figure 1c). No consistent recruitment of SRP54 from the cytosol (S200) to the ER membrane (P6) in response to an increase in glucose concentration was detected (Figure 1c).
It has been reported that there is a 20 min lag phase between the addition of glucose and an increase in proinsulin synthesis [3,20]. To discover whether this lag period correlated with the time taken for the proinsulin mRNA to be recruited to the ER, a time course of glucose-stimulated recruitment of proinsulin mRNA to the ER was determined (Figure 1d). Glucose was shown to stimulate rapidly the recruitment of proinsulin mRNA to the ER (within 10–20 min). Therefore it is possible that the time taken for the recruitment of proinsulin mRNA to the ER is responsible for the previously observed delay in glucose-stimulated proinsulin synthesis.
Given the importance of these observations in our understanding of how glucose specifically stimulates proinsulin synthesis, we investigated the subcellular localization of proinsulin mRNA using an alternative technique in which the cells were permeabilized with digitonin and the cytosol washed out, leaving mRNAs bound to the ER associated to the permeabilized cells (Figure 2). This technique has been previously used to successfully fractionate cytosolic mRNAs from ER-bound mRNAs [23].
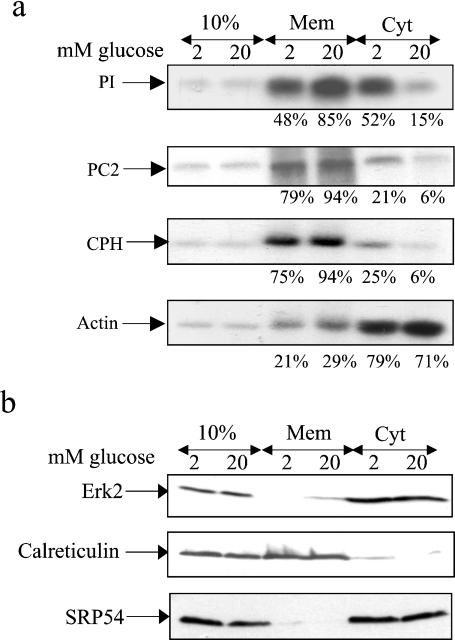
Figure 2. Glucose stimulates the recruitment of mRNAs encoding secretory membrane proteins to the ER: subcellular fractionation using the digitonin method.
MIN6 cells were preincubated in KRB containing 2 mM glucose for 1 h followed by incubation in KRB containing 2 or 20 mM glucose for a further period of 1 h. (a) Cells were permeabilized with digitonin and pelleted resulting in a membrane fraction (Mem) and a cytosolic fraction (Cyt). The 10% refers to 10% of total RNA/protein. RNA was isolated from each fraction and run on a 1% agarose formaldehyde gel, transferred on to a nylon membrane and probed for proinsulin (PI), PC2, CPH and actin mRNAs. The intensities of the bands were quantified using ImageJ and the mRNA levels were expressed as a percentage of total mRNA. (b) Protein samples from each fraction were run on SDS/PAGE (20% of membrane fraction and 20% of cytosolic fraction) and subjected to immunoblotting with anti-ERK2, calreticulin and SRP54 antibodies. Detection was by enhanced chemiluminescence. The results presented are representative of three separate experiments.
MIN6 cells were preincubated in KRB containing 2 mM glucose for 1 h followed by incubation in KRB containing either 2 or 20 mM glucose for a further 1 h. The cells were then fractionated using the digitonin fractionation technique. At low glucose concentration, proinsulin mRNA was located primarily in the cytosol, whereas at high glucose concentration, the majority of proinsulin mRNA was found to be associated with the ER (membrane fraction) (Figure 2a). In contrast, CPH and PC2 mRNAs, at either low or high glucose concentration, were primarily located at the ER (membrane fraction). However, glucose still stimulated the recruitment of PC2 and CPH mRNAs to the ER (Figure 2a). As expected, the actin mRNA was mainly found in the cytosolic fraction (Figure 2a). However, glucose stimulated the recruitment of a small fraction of the actin mRNA to the ER (Figure 2a).
To determine the efficiency of the subcellular fractionation procedure, the localization of calreticulin and ERK2 was determined by Western-blot analysis (Figure 2b). Calreticulin was only found in the membrane fraction, whereas ERK2 was only detected in the cytosolic fraction (Figure 2b), confirming the efficiency of the fractionation process. We also investigated the subcellular distribution of SRP using an antibody directed against SRP54 (Figure 2b). At both low and high glucose concentrations, SRP54 was predominantly found free in the cytoplasmic fraction (Figure 2b).
Therefore, using two complementary techniques, we provide evidence that: (i) glucose may stimulate the recruitment of all mRNAs encoding secretory proteins to the ER, (ii) the recruitment of proinsulin mRNA to the ER at low glucose concentration is selectively inhibited and (iii) glucose preferentially stimulates the recruitment of proinsulin mRNA to the ER at high glucose concentration.
Glucose-stimulated recruitment of proinsulin mRNA to the ER is important in glucose-stimulated proinsulin synthesis
To investigate whether the recruitment of proinsulin mRNA to the ER was an important mediator of glucose-stimulated proinsulin synthesis, membranes and associated mRNAs were isolated from MIN6 cells incubated at either low or high glucose concentration, the mRNA was translated in situ on the ER in a RRL cell-free translation system and the rate of proinsulin synthesis assessed (Figure 3a). The rate of proinsulin synthesis from the in situ translation reaction was 3.5-fold higher for membranes isolated from cells incubated at high glucose concentration compared with those incubated at low glucose concentration (Figure 3a). The depletion of ribosomes from the RRL had no effect on the rates of translation of proinsulin, indicating that the synthesis of proinsulin in situ is directed by endogenous ribosomes present within the ER fraction (results not shown). These results provide evidence that glucose-stimulated recruitment of proinsulin mRNA to the ER is sufficient to stimulate proinsulin synthesis significantly.
Figure 3. The recruitment of proinsulin mRNA on to the membranes at high glucose plays a significant role in glucose-stimulated proinsulin synthesis.
MIN6 cells were preincubated in KRB containing 0.5 mM glucose for 1 h followed by incubation in KRB containing 0.5 or 20 mM glucose for a further period of 1 h. Membranes were isolated by consecutive centrifugation as described in Figure 1. (a) Resuspended membranes were translated in situ in the presence of [35S]methionine. After translation, the membranes were pelleted at 13000 g for 10 min. Proinsulin (PI) was resolved by SDS/PAGE. (b) Poly(A)+ selected mRNA was isolated from the membranes and translated in vitro in the presence of [35S]methionine. Proinsulin (PI) was immunoprecipitated and resolved by SDS/PAGE. (c) MIN6 cells were preincubated in KRB containing 0.5 mM glucose for 1 h followed by incubation in KRB containing 0.5 or 20 mM glucose in the presence of [35S]methionine for a further period of 1 h. The cells were lysed and proinsulin (PI) resolved by SDS/PAGE. In all cases, proteins were visualized by autoradiography. In (a, c) the results are representative of three separate experiments and in (b) the results are representative of two separate experiments.
To determine whether this increase in the synthesis of proinsulin at the ER is solely the consequence of increased proinsulin mRNA levels, mRNA was purified from membrane fractions isolated from cells incubated at either low or high glucose concentration and translated in vitro, and the rates of proinsulin synthesis were determined (Figure 3b). The synthesis of proinsulin was found to be approx. 2–3-fold higher from membrane-associated mRNAs isolated from cells incubated at high glucose concentration compared with those incubated at low glucose concentration (Figure 3b). The differences in the rate of proinsulin synthesis at low versus high glucose concentration are similar to that seen in the in situ translation reactions (see Figure 3a).
In order to compare the rates of proinsulin synthesis from the in situ and in vitro translations with the rates of proinsulin synthesis in vivo, MIN6 cells were incubated at low or high glucose concentration in KRB in the presence of [35S]methionine and the rates of proinsulin synthesis were assessed (Figure 3c). In vivo, glucose stimulated approx. 5–6-fold increase in proinsulin synthesis. Glucose therefore stimulated an approx. 2-fold greater increase in proinsulin synthesis in vivo compared with that observed for in vitro or in situ translation reactions of membrane-associated proinsulin mRNA.
Therefore, taken together, these results provide evidence that glucose-stimulated recruitment of proinsulin mRNA to the ER plays a significant role in glucose-stimulated proinsulin synthesis. However, these results also indicate that additional mechanisms must also contribute to glucose-stimulated proinsulin synthesis.
Glucose stimulates the recruitment of ribosomes on to ribosome-associated proinsulin mRNA
To investigate which additional mechanisms may be important in glucose-stimulated proinsulin synthesis, we performed polysome profile analysis (Figure 4).
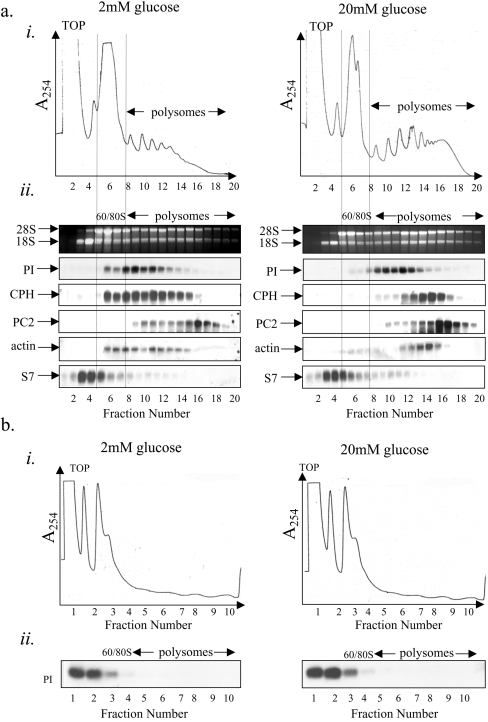
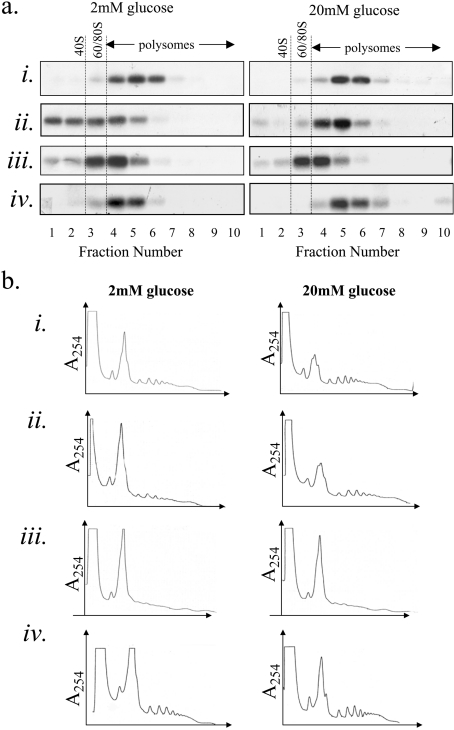
Figure 4. Glucose stimulates an increase in ribosome recruitment on to ribosome-associated proinsulin mRNA.
(a) MIN6 cells were preincubated in KRB containing 2 mM glucose for 1 h followed by incubation in KRB containing 2 or 20 mM glucose for a further period of 1 h. Cells were lysed and polysome analysis was carried out using 7–47% sucrose gradients. The gradients were fractionated from left (fraction 1) to right (fraction 20). (ai) Absorbance of the gradients was measured continually at 254 nm to give polysome profiles. (aii) RNA was isolated from each fraction and run on a 1% agarose formaldehyde gel. RNA was transferred on to a nylon membrane and probed for proinsulin (PI), CPH, PC2, actin and S7 mRNAs. The results presented are representative of three separate experiments. (b) Cells were treated and fractionated as described in (a) except that 15 mM EDTA was included in the lysis buffer and the sucrose gradients to dissociate the ribosomes.
MIN6 cells were incubated for 1 h in KRB containing either 2 or 20 mM glucose before cell lysis and sucrose sedimentation gradient centrifugation.
Increasing the glucose concentration from 2 to 20 mM led to a large decrease in the 80 S monosomal peak along with an increase in the number of polysomes (Figure 4ai). This indicates that glucose stimulates the recruitment of ribosomes on to mRNAs and hence stimulates an increase in the rate of initiation. Indeed, we have recently shown that glucose stimulates an increase in the initiation step of protein synthesis in MIN6 cells [19]. At low glucose concentration, proinsulin mRNA was found to co-sediment with either monosomes or small polysomes (Figures 4ai and 4ii). Upon an increase in glucose concentration from 2 to 20 mM, there was a small but significant shift of the proinsulin mRNA on to heavier polysomes, indicative of a recruitment of ribosomes on to the proinsulin mRNA (Figures 4ai and 4ii). At either low or high glucose concentration, no proinsulin mRNA was detected at the top of the gradient, the predicted position of free proinsulin mRNA (non-ribosome associated, see Figure 4b) (Figures 4ai and 4ii) and is in agreement with the results presented in Figures 1 and 2.
Glucose also stimulated the recruitment of ribosomes on to other secretory protein-encoding mRNAs, PC2 and CPH mRNAs, and on to a cytoplasmic mRNA, actin (Figure 4aii). However, glucose had little effect on the recruitment of ribosomes on to S7 mRNA, a 5′-tract of oligopyrimidine mRNA.
To establish that the sedimentation properties of proinsulin mRNA were determined by its association with ribosomes, sucrose sedimentation gradient centrifugations were also performed in the presence of EDTA, which causes the dissociation of ribosomes from mRNA but does not, in general, disrupt non-ribosomal RNA–protein complexes (mRNPs) [24]. In the presence of EDTA, the proinsulin mRNA sedimented at the top of the gradient, the position expected for free mRNA (Figures 4bi and 4bii). These results provide evidence that, at low glucose, all of the detectable proinsulin mRNA is ribosome-associated and that, upon an increase in glucose concentration, additional ribosomes are recruited on to this mRNA.
Analysis of ER-associated proinsulin mRNA
Polysome analysis of MIN6 cell extracts indicated that all proinsulin mRNAs are associated with ribosomes at either low or high glucose concentration and that glucose stimulates a small increase in ribosome recruitment on to ribosome-associated proinsulin mRNA. However, from results presented in Figure 1, it is clear that a large percentage of proinsulin mRNA is cytosolic and therefore is unlikely to be actively translated. Therefore an increase in the recruitment of ribosomes on to actively translating proinsulin mRNA at the ER may be masked by the population of non-translating, ribosome-associated, cytosolic proinsulin mRNA. Consequently, the sedimentation profiles of proinsulin mRNA, associated only with ER membranes at both low and high glucose concentrations, were determined by polysome profile analysis (Figure 5a).
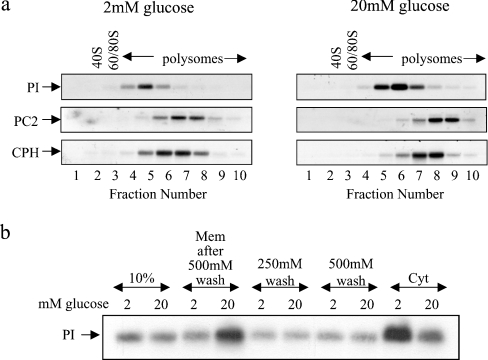
Figure 5. Glucose stimulates the recruitment of ribosomes on to membrane-bound proinsulin mRNA.
MIN6 cells were preincubated in KRB containing 2 mM glucose for 1 h followed by incubation in KRB containing 2 or 20 mM glucose for a further period of 1 h. (a) Cells were then lysed and fractionated by consecutive centrifugation. The membrane fractions were layered on to 20–50% sucrose gradients, centrifuged and fractionated from left (fraction 1) to right (fraction 10). RNA was isolated from each fraction and run on a 1% agarose formaldehyde gel. RNA was then transferred on to a nylon membrane and probed for proinsulin (PI), PC2 and CPH mRNAs. (b) Cells were then lysed and fractionated by the digitonin cell permeabilization method (using modified polysome buffer containing 130 mM KCl), resulting in a membrane fraction (Mem) and a cytosolic fraction (Cyt). The membrane fraction was consecutively washed with polysome buffer containing 250 mM KCl (250 mM wash) and 500 mM KCl (500 mM wash). RNA isolated from each fraction and washed was run on a 1% agarose formaldehyde gel, transferred on to a nylon membrane and probed for proinsulin (PI) mRNA; 10% refers to 10% of total RNA.
MIN6 cells were incubated for 1 h at either 2 or 20 mM glucose. The cells were then fractionated into cytosolic or membrane fractions and the membrane fraction was centrifuged through a sucrose sedimentation gradient. Analysis of the sedimentation profile of ER-associated proinsulin mRNA confirmed that, at low glucose concentration, all proinsulin mRNA sedimented with either monosones or polysomes. However, at high glucose concentration, there was a large increase in ER-associated proinsulin mRNA and a shift of proinsulin mRNA on to heavier polysomes (Figure 5a). These changes confirm that glucose stimulates the recruitment of proinsulin mRNA to the ER and also demonstrates that glucose stimulates the recruitment of ribosomes on to ribosome-associated proinsulin mRNA (Figure 5a). Glucose also stimulated the recruitment of ribosomes on to PC2 and CPH mRNAs; results were similar to those presented in Figure 4 (Figure 5a). Therefore glucose probably stimulates the recruitment of ribosomes on to all ER-associated mRNAs.
The glucose-stimulated recruitment of proinsulin mRNA to the membrane is probably mediated by promoting the association of the proinsulin mRNA RNC–SRP complex with the SR, located on the ER. However, more distal steps in the synthesis of proinsulin may also be regulated by glucose, such as the transfer of proinsulin mRNA RNC from the SR to the translocon. To investigate this, MIN6 cells, incubated at either low or high glucose concentration, were lysed and fractionated by the digitonin cell-permeabilization method (Figure 5b). The membrane fractions were then washed with increasing salt concentration in order to differentiate proinsulin mRNA tightly associated with the ER membrane (i.e. probably due to the insertion of the proinsulin nascent chain in the translocon) from proinsulin mRNA loosely associated with the ER (i.e. those whose nascent chain is yet to be transferred to the translocon) [25]. As shown in Figure 5(b), the two salt washes performed resulted in the loss of exactly the same amount of proinsulin mRNA from membranes isolated at either low or high glucose concentration. Therefore the same quantity of proinsulin mRNA at either low or high glucose concentration is loosely associated with the membranes (Figure 5b). Thus it is unlikely that the transfer of proinsulin RNC from the SR to the translocon is regulated by glucose.
Evidence that glucose may stimulate an increase in the rate of ribosome recycling/recruitment on to proinsulin mRNA
It has been reported previously that glucose stimulates the recruitment of proinsulin mRNA from a large pool of free mRNAs on to polysomes, i.e. an increase in proinsulin de novo initiation [3,7–9]. However, we were unable to detect any pool of free proinsulin mRNA or detect any increase in the rate of de novo initiation on the proinsulin mRNA. One explanation for the differences observed between this report and others is that we add cycloheximide to the cells before lysis, a step included to prevent ribosomal run-off. Omission of cycloheximide may result in increased ribosomal run-off and the formation of a pool of free proinsulin mRNA.
To ascertain whether the omission of cycloheximide could lead to the formation of a pool of free proinsulin mRNA, polysome analysis was performed on MIN6 cells incubated at either low or high glucose concentration, in which cycloheximide was either added or omitted from the method (Figures 6ai/ii and 6bi/ii). As previously demonstrated (see Figures 1 and 4), when cycloheximide is added 10 min before cell lysis, no free proinsulin mRNA is detected at either low or high glucose concentration (Figure 6ai). However, if no cycloheximide is added to the cells incubated at low glucose concentration before cell lysis, a large proportion of proinsulin mRNA sediments at the top of the gradient (i.e. as a free mRNA pool), possibly a consequence of ribosomal run-off (Figure 6aii). At high glucose concentration, in the absence of cycloheximide, only a small proportion of proinsulin mRNA sediments at the top of the gradient (Figure 6aii). The results presented here, in the absence of cycloheximide, appear to be similar to those reported previously [3,7–9]. However, in these previous studies, the results were interpreted as evidence for the presence of a large pool of free proinsulin mRNA that is recruited on to polysomes in response to glucose.
Figure 6. Evidence for increased ribosome recycling/recruitment on to proinsulin mRNA at high glucose.
MIN6 cells were preincubated in KRB containing 2 mM glucose for 1 h followed by incubation in KRB containing 2 or 20 mM glucose for a further period of 1 h. During the final 10 min of incubation, the following were added: (ai, bi) 0.1 mg/ml cycloheximide, (aii, bii) no treatment, (aiii, biii) 0.1 μg/ml pactamycin and (aiv, biv) 0.1 mg/ml cycloheximide and 0.1 μg/ml pactamycin. Cells were lysed and polysome analysis was carried out on 20–50% sucrose gradients. Gradients were fractionated from left (fraction 1) to right (fraction 10). (a) Northern-blot analysis: RNA was isolated from each fraction and run on a 1% agarose formaldehyde gel. RNA was then transferred on to a nylon membrane and probed for proinsulin (PI) mRNA. (b) Polysome profiles.
As it has been previously reported that glucose can stimulate the rate of elongation [7], we would have expected more ribosomal run-off at high versus low glucose concentration. However, we observe more ribosomal run-off at low glucose concentration than at high glucose concentration (Figure 6aii). One possibility for this apparent contradiction is that glucose stimulates an increase in ribosome recruitment on to the proinsulin mRNA (see Figures 4 and 5) (i.e. at high glucose concentration, ribosomal run-off from a proinsulin mRNA is counteracted by the loading of additional ribosomes on to that mRNA). To investigate this possibility, pactamycin, an inhibitor of a late step in initiation and dipeptide-bond formation [26–28], was added to MIN6 cells incubated at either low or high glucose concentration for 10 min before lysis to prevent ribosomal recruitment on to mRNA. Under these conditions, proinsulin mRNA sediments with either monosomes or disomes at either low or high glucose concentration, presumably due to the combination of ribosomal run-off and the inhibition of dipeptide-bond formation (Figure 6aiii). As at high glucose concentration, ribosomal run-off has definitely occurred in the presence of pactamycin, therefore ribosomal run-off must also occur in its absence (see Figures 6aii and 6aiii). Therefore this provides evidence that at high glucose concentration, in the absence of inhibitors (Figure 6aii), ribosomal run-off is coincident with the recruitment of ribosomes on to proinsulin mRNA. This could be mediated through the observed increase in ribosome recruitment on to ribosome-associated proinsulin mRNA (see Figures 4 and 5), or perhaps an increase in the rate of ribosome recycling. The addition of both pactamycin and cycloheximide to MIN6 cells incubated at either 2 or 20 mM glucose for 10 min before lysis to freeze the mRNA–ribosome complex generates a sedimentation profile of proinsulin mRNA similar to that generated in the presence of cycloheximide alone (Figures 6ai and 6aiv). Similar sedimentation profiles were observed for other mRNAs investigated, including PC2 and actin (results not shown). Taken together, these results provide evidence that glucose may stimulate a rapid increase in the rate of ribosome recycling/recruitment. However, other explanations for these results could also be possible.
Investigation of glucose-stimulated proinsulin synthesis in islets of Langerhans
To ascertain whether the principal results obtained in the clonal β-cell line MIN6 also occur in primary β-cells, changes in the subcellular distribution and sedimentation profiles of the proinsulin mRNA in response to glucose were investigated in islets of Langerhans.
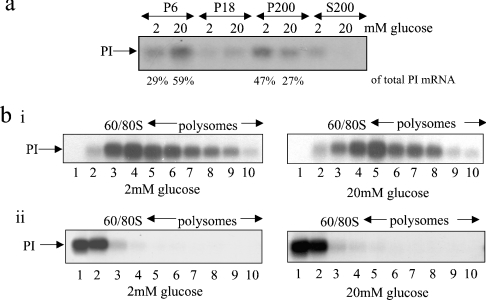
Islets of Langerhans were isolated from rats and incubated in KRB containing either 2 or 20 mM glucose for 1 h. The cells were then lysed and the lysates fractionated by consecutive centrifugation. The location of proinsulin mRNA within each fraction was determined by Northern-blot analysis (Figure 7a). At 2 mM glucose, a large proportion of the proinsulin mRNA was found in the P200 fraction (47%), whereas 29% was found to be associated with the ER (P6) (Figure 7a). In contrast, at 20 mM glucose, the majority of proinsulin mRNA (59%) was found to be associated with the ER (P6 fraction) while 27% was found in the P200 fraction (Figure 7a). Therefore, in response to an increase in glucose concentration, there is an approx. 200% increase in the amount of proinsulin mRNA at the ER (Figure 7a). At either low or high glucose concentration, little to no free proinsulin mRNA was detected (S200 fraction) (Figure 7a). These results are similar to those observed in MIN6 cells. Additionally, islets of Langerhans were incubated for 1 h in KRB containing 2 or 20 mM glucose before cell lysis and sucrose sedimentation gradient centrifugation. The sedimentation of proinsulin mRNA across the gradients was determined by Northern-blot analysis (Figure 7bi). In order to establish that the sedimentation properties of proinsulin mRNA are the consequence of its association with ribosomes, sucrose sedimentation gradient centrifugations were also performed in the presence of EDTA (Figure 7bii).
Figure 7. Glucose-stimulated proinsulin synthesis in islets of Langerhans.
Rat islets were preincubated in KRB containing 2 mM glucose for 2 h followed by incubation in KRB containing 2 or 20 mM glucose for a further period of 1 h. (a) Cells were fractionated by consecutive centrifugation as described in Figure 1. RNA was isolated from each fraction and run on a 1% agarose formaldehyde gel, then transferred on to a nylon membrane and probed for proinsulin (PI) mRNA; (bi) Cells were lysed and the lysates were layered on to 20–50% sucrose gradients, centrifuged and fractionated from left (fraction 1) to right (fraction 10). RNA was isolated from each fraction and run on a 1% agarose formaldehyde gel. RNA was then transferred on to a nylon membrane and probed for proinsulin (PI) mRNA. (bii) Cells were treated and fractionated as described in (bi) except that 15 mM EDTA was included in the lysis buffer and the sucrose gradients to dissociate the ribosomes but leave mRNPs intact.
In the absence of EDTA, all the proinsulin mRNA sedimented with either monosomes or polysomes (Figure 7bi). Upon an increase in glucose concentration, there was a small but significant shift of the proinsulin mRNA on to heavier polysomes, as observed in MIN6 cells (compare Figure 7bi with Figure 4). In the presence of EDTA, proinsulin mRNA sedimented at the top of the gradient, at either low or high glucose concentration, the expected position of free mRNA (Figure 7bii). These results confirm that, at low glucose concentration, all the detectable proinsulin mRNAs are ribosome-associated and that, upon an increase in glucose concentration, more ribosomes are recruited on to this mRNA. These results also show that there is neither a large pool of free proinsulin mRNA nor a perceivable increase in the de novo initiation rate on the proinsulin mRNA in response to glucose.
DISCUSSION
In response to an increase in extracellular glucose concentration, there is a rapid increase in proinsulin synthesis and a co-ordinate increase in the majority of other secretory proteins, including the proinsulin endopeptidases PC2 and PC3 [3,6,11,20,29]. These rapid increases in protein synthesis are mediated almost entirely at the translational level [3,5,6] and ensure that proteins released upon glucose-stimulated exocytosis are immediately replenished with newly synthesized proteins. Moreover, ongoing translation is necessary for glucose-stimulated proinsulin secretion as inhibition of translation leads to defects in insulin secretion [30]. As glucose stimulates a greater fold increase in proinsulin synthesis compared with that of the majority of other proteins, including other secretory granule proteins, it is believed that glucose stimulates proinsulin synthesis by a specific mechanism [5]. However, the mechanism by which glucose stimulates secretory granule protein synthesis, and specifically proinsulin synthesis, is poorly understood.
In the present study, we demonstrate, in both MIN6 cells and islets of Langerhans, that glucose stimulates the recruitment of ribosome-associated proinsulin mRNA, which is primarily located in the cytosol, to the ER. Additionally, we show, in MIN6 cells, that glucose stimulates the recruitment of other mRNAs encoding secretory proteins (i.e. CPH and PC2) from the cytosol to the ER. In MIN6 cells, the recruitment of proinsulin, CPH and PC2 mRNAs to the ER parallel increases in their synthesis (Figure 2; I. C. Greenman and T. P. Herbert, unpublished work; [22]). Indeed, it has been previously reported that the synthesis of the majority of secretory proteins are stimulated by glucose in islets [20]. Therefore it is likely that glucose stimulates the recruitment of all mRNAs encoding secretory membrane proteins to the ER and that this is an important mechanism by which glucose specifically stimulates the synthesis of secretory granule proteins.
Importantly, glucose has a greater stimulatory effect on the recruitment of proinsulin mRNA to the ER compared with its effect on the recruitment of either CPH or PC2 mRNAs to the ER (see Figures 1 and 2). This glucose-stimulated recruitment of proinsulin mRNA to the ER is sufficient to lead to a significant increase in proinsulin synthesis as assessed by either in vitro translation of proinsulin mRNA isolated from the membranes of MIN6 cells incubated at either low or high glucose concentration or from in situ translation of proinsulin mRNA on ER membranes isolated from MIN6 cells incubated at either low or high glucose concentration (see Figure 3). Therefore the recruitment of the proinsulin mRNA to the ER plays a significant role in glucose-stimulated proinsulin synthesis. Moreover, as proinsulin mRNA is preferentially recruited to the ER in response to glucose, this explains, at least in part, why glucose stimulates the synthesis of proinsulin above that of other secretory granule proteins, such as PC2 [6]. This selective recruitment of mRNAs encoding secretory proteins to the ER provides a novel mechanism to regulate secretory protein expression and may prove a paradigm for the regulated expression of secretory membrane proteins in other neuroendocrine cells.
Contrary to previous reports, we could not detect any significant pool of free proinsulin mRNA in both MIN6 cells and islets [3,7–9]. However, at either low or high glucose concentration, we observed that almost all detectable proinsulin mRNA is associated with ribosomes. At low glucose concentration, a large proportion of the ribosome-associated proinsulin mRNA is located in the cytosol and not associated with the ER, the site of proinsulin synthesis (Figures 1 and 2). Therefore the translation of proinsulin from this pool of mRNA is probably arrested possibly through an interaction between the signal peptide and SRP. In contrast, at either low or high glucose concentration, the majority of both CPH and PC2 mRNAs are associated with the ER (Figures 1 and 2). The mechanism by which proinsulin mRNA is prevented from being recruited to the ER at low glucose concentration is unclear, but this is a likely key in understanding the mechanism by which glucose stimulates proinsulin synthesis in pancreatic β-cells.
Previous reports examining the subcellular distribution of proinsulin mRNA in islets, incubated at either low or high glucose concentration, have observed a large pool of free proinsulin mRNA (47–62%) located in the cytosol and that, upon an increase in glucose concentration, there is a recruitment of approx. 15–38% of the total proinsulin mRNA on to monosomes/polysomes located either within the cytosol or associated with the ER, through an increase in the rate of de novo proinsulin mRNA initiation [3,7–9]. However, the interpretation of these previous reports should be re-evaluated as no cycloheximide, an inhibitor of elongation, was added to the cells before cell lysis or to the lysis buffer. Cycloheximide is routinely added to the media before cell lysis and to the lysis buffer to prevent ribosomal run-off. Indeed, we show that the failure to add cycloheximide leads to the appearance of a large pool of free proinsulin mRNA, presumably the result of ribosomal run-off (Figure 6).
The results presented in this paper clearly demonstrate that proinsulin mRNA recruitment to the ER is likely to play a significant role in glucose-stimulated proinsulin synthesis. Nonetheless, the recruitment of the proinsulin mRNA to the ER alone cannot fully explain the large increase in proinsulin synthesis observed in pancreatic β-cells in response to glucose. Indeed, we also demonstrate that glucose stimulates the recruitment of ribosomes on to ribosome-associated proinsulin mRNA (Figure 4). An increase in ribosome recruitment on to PC2, CPH and actin mRNAs was also observed, indicating that there is a probable global increase in the rate of initiation (Figure 4).
We also provide evidence that glucose may stimulate an increase in the rate of ribosome recycling (Figure 6). Indeed, the 5′- and 3′-UTRs of the proinsulin mRNA have been shown to act co-operatively to stimulate proinsulin synthesis in response to glucose [13], possibly via an increase in the rate of ribosome recycling from the 3′-UTR to the 5′-UTR.
Based on the evidence provided in this paper, we believe that the combinational effects of glucose on the recruitment of proinsulin mRNA to the ER and an increase in the rate of initiation may be sufficient to account for the glucose-stimulated increase in proinsulin synthesis in pancreatic β-cells.
Acknowledgments
I.C.G. was supported by a BBSRC case studentship. E.G. is supported by the Wellcome Trust. C.M. is supported by an MRC studentship.
References
- 1.Campbell I. L., Hellquist L. N., Taylor K. W. Insulin biosynthesis and its regulation. Clin. Sci. 1982;62:449–455. doi: 10.1042/cs0620449. [DOI] [PubMed] [Google Scholar]
- 2.Itoh N., Sei T., Nose K., Okamoto H. Glucose stimulation of the proinsulin synthesis in isolated pancreatic islets without increasing amount of proinsulin mRNA. FEBS Lett. 1978;93:343–347. doi: 10.1016/0014-5793(78)81136-3. [DOI] [PubMed] [Google Scholar]
- 3.Itoh N., Okamoto H. Translational control of proinsulin synthesis by glucose. Nature (London) 1980;283:100–102. doi: 10.1038/283100a0. [DOI] [PubMed] [Google Scholar]
- 4.Permutt M. A., Kipnis D. M. Insulin biosynthesis. I. On the mechanism of glucose stimulation. J. Biol. Chem. 1972;247:1194–1199. [PubMed] [Google Scholar]
- 5.Jahr H., Schroder D., Ziegler B., Ziegler M., Zuhlke H. Transcriptional and translational control of glucose-stimulated (pro)insulin biosynthesis. Eur. J. Biochem. 1980;110:499–505. doi: 10.1111/j.1432-1033.1980.tb04892.x. [DOI] [PubMed] [Google Scholar]
- 6.Alarcon C., Lincoln B., Rhodes C. J. The biosynthesis of the subtilisin-related proprotein convertase PC3, but not that of the PC2 convertase, is regulated by glucose in parallel to proinsulin biosynthesis in rat pancreatic islets. J. Biol. Chem. 1993;268:4276–4280. [PubMed] [Google Scholar]
- 7.Welsh M., Scherberg N., Gilmore R., Steiner D. F. Translational control of insulin biosynthesis. Evidence for regulation of elongation, initiation and signal-recognition-particle-mediated translational arrest by glucose. Biochem. J. 1986;235:459–467. doi: 10.1042/bj2350459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welsh N., Welsh M., Steiner D. F., Hellerstrom C. Mechanisms of leucine- and theophylline-stimulated insulin biosynthesis in isolated rat pancreatic islets. Biochem. J. 1987;246:245–248. doi: 10.1042/bj2460245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welsh N., Oberg C., Welsh M. GTP-binding proteins may stimulate insulin biosynthesis in rat pancreatic islets by enhancing the signal-recognition-particle-dependent translocation of the insulin mRNA poly-/mono-some complex to the endoplasmic reticulum. Biochem. J. 1991;275:23–28. doi: 10.1042/bj2750023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knight S. W., Docherty K. RNA-protein interactions in the 5′ untranslated region of preproinsulin mRNA. J. Mol. Endo. 1992;8:1–10. doi: 10.1677/jme.0.0080225. [DOI] [PubMed] [Google Scholar]
- 11.Herbert T. P., Alarcon C., Skelly R. H., Bollheimer L. C., Schuppin G. T., Rhodes C. J. Regulation of prohormone conversion by co-ordinated control of processing endopeptidase biosynthesis with that of the prohormone substrate. In: Hook V. Y. H., editor. Proteolytic and Cellular Mechanisms in Prohormone and Proprotein Processing. Austin, TX: R. G. Landes Company; 1998. pp. 105–120. [Google Scholar]
- 12.Thomson A. M., Rogers J. T., Leedman P. J. Iron-regulatory proteins, iron-responsive elements and ferritin mRNA translation. Int. J. Biochem. Cell Biol. 1999;31:1139–1152. doi: 10.1016/s1357-2725(99)00080-1. [DOI] [PubMed] [Google Scholar]
- 13.Wicksteed B., Herbert T. P., Alarcon C., Lingohr M. K., Moss L. G., Rhodes C. J. Cooperativity between the preproinsulin mRNA untranslated regions is necessary for glucose-stimulated translation. J. Biol. Chem. 2001;276:22553–22558. doi: 10.1074/jbc.M011214200. [DOI] [PubMed] [Google Scholar]
- 14.Tillmar L., Carlsson C., Welsh N. Control of insulin mRNA stability in rat pancreatic islets. Regulatory role of a 3′-untranslated region pyrimidine-rich sequence. J. Biol. Chem. 2002;277:1099–1106. doi: 10.1074/jbc.M108340200. [DOI] [PubMed] [Google Scholar]
- 15.Knoch K. P., Bergert H., Borgonovo B., Saeger H. D., Altkruger A., Verkade P., Solimena M. Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nat. Cell Biol. 2004;6:207–214. doi: 10.1038/ncb1099. [DOI] [PubMed] [Google Scholar]
- 16.Keenan R. J., Freymann D. M., Stroud R. M., Walter P. The signal recognition particle. Annu. Rev. Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 17.Pool M. R. Getting to the membrane: how is co-translational protein targeting to the endoplasmic reticulum regulated? Biochem. Soc. Trans. 2003;31:1232–1237. doi: 10.1042/bst0311232. [DOI] [PubMed] [Google Scholar]
- 18.Frey S., Pool M., Seedorf M. Scp160p, an RNA-binding, polysome-associated protein, localizes to the endoplasmic reticulum of Saccharomyces cerevisiae in a microtubule-dependent manner. J. Biol. Chem. 2001;276:15905–15912. doi: 10.1074/jbc.M009430200. [DOI] [PubMed] [Google Scholar]
- 19.Gomez E., Powell M. L., Greenman I. C., Herbert T. P. Glucose-stimulated protein synthesis in pancreatic β-cells parallels an increase in the availability of the translational ternary complex (eIF2-GTP.Met-tRNAi) and the dephosphorylation of eIF2 alpha. J. Biol. Chem. 2004;279:53937–53946. doi: 10.1074/jbc.M408682200. [DOI] [PubMed] [Google Scholar]
- 20.Guest P. C., Rhodes C. J., Hutton J. C. Regulation of the biosynthesis of insulin-secretory-granule proteins. Co-ordinate translational control is exerted on some, but not all, granule matrix constituents. Biochem. J. 1989;257:431–437. doi: 10.1042/bj2570431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishihara H., Asano T., Tsukuda K., Katagiri H., Inukai K., Anai M., Kikuchi M., Yazaki Y., Miyazaki J. I., Oka Y. Pancreatic β cell line MIN6 exhibits characteristics of glucose metabolism and glucose-stimulated insulin secretion similar to those of normal islets. Diabetologia. 1993;36:1139–1145. doi: 10.1007/BF00401058. [DOI] [PubMed] [Google Scholar]
- 22.Skelly R. H., Schuppin G. T., Ishihara H., Oka Y., Rhodes C. J. Glucose-regulated translational control of proinsulin biosynthesis with that of the proinsulin endopeptidases PC2 and PC3 in the insulin-producing MIN6 cell line. Diabetes. 1996;45:37–43. doi: 10.2337/diab.45.1.37. [DOI] [PubMed] [Google Scholar]
- 23.Seiser R. M., Nicchitta C. V. The fate of membrane-bound ribosomes following the termination of protein synthesis. J. Biol. Chem. 2000;275:33820–33827. doi: 10.1074/jbc.M004462200. [DOI] [PubMed] [Google Scholar]
- 24.Calzone F. J., Angerer R. C., Gorovsky M. A. Regulation of protein synthesis in Tetrahymena: isolation and characterization of polysomes by gel filtration and precipitation at pH 5.3. Nucleic Acids Res. 1982;10:2145–2161. doi: 10.1093/nar/10.6.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolin S. L., Walter P. Discrete nascent chain lengths are required for the insertion of presecretory proteins into microsomal membranes. J. Cell Biol. 1993;121:1211–1219. doi: 10.1083/jcb.121.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kappen L. S., Goldberg I. H. Inhibition of globin chain initiation in reticulocyte lysates by pactamycin: accumulation of methionyl-valine. Biochem. Biophys. Res. Commun. 1973;54:1083–1091. doi: 10.1016/0006-291x(73)90804-8. [DOI] [PubMed] [Google Scholar]
- 27.Kappen L. S., Suzuki H., Goldberg I. H. Inhibition of reticulocyte peptide-chain initiation by pactamycin: accumulation of inactive ribosomal initiation complexes. Proc. Natl. Acad. Sci. U.S.A. 1973;70:22–26. doi: 10.1073/pnas.70.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kappen L. S., Goldberg I. H. Analysis of the two steps in polypeptide chain initiation inhibited by pactamycin. Biochemistry. 1976;15:811–818. doi: 10.1021/bi00649a013. [DOI] [PubMed] [Google Scholar]
- 29.Xu G., Marshall C. A., Lin T. A., Kwon G., Munivenkatappa R. B., Hill J. R., Lawrence J. C., Jr, McDaniel M. L. Insulin mediates glucose-stimulated phosphorylation of PHAS-I by pancreatic β cells. An insulin-receptor mechanism for autoregulation of protein synthesis by translation. J. Biol. Chem. 1998;273:4485–4491. doi: 10.1074/jbc.273.8.4485. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Barrado M. J., Ravier M. A., Rolland J. F., Gilon P., Nenquin M., Henquin J. C. Inhibition of protein synthesis sequentially impairs distinct steps of stimulus-secretion coupling in pancreatic β cells. Endocrinology. 2001;142:299–307. doi: 10.1210/endo.142.1.7910. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]