Abstract
In the presence of Zn2+, the Drosophila 26 S proteasome disassembles into RP (regulatory particle) and CP (catalytic particle), this process being accompanied by the dissociation of subunit Rpn10/p54, the ubiquitin receptor subunit of the proteasome. The dissociation of Rpn10/p54 induces extensive rearrangements within the lid subcomplex of the RP, while the structure of the ATPase ring of the base subcomplex seems to be maintained. As a consequence of the dissociation of the RP, the peptidase activity of the 26 S proteasome is lost. The Zn2+-induced structural and functional changes are fully reversible; removal of Zn2+ is followed by reassociation of subunit Rpn10/p54 to the RP, reassembly of the 26 S proteasome and resumption of the peptidase activity. After the Zn2+-induced dissociation, Rpn10/p54 interacts with a set of non-proteasomal proteins. Hsp82 (heat-shock protein 82) has been identified by MS as the main Rpn10/p54-interacting protein, suggesting its role in the reassembly of the 26 S proteasome after Zn2+ removal. The physiological relevance of another Rpn10/p54-interacting protein, the Smt3 SUMO (small ubiquitin-related modifier-1)-activating enzyme, detected by chemical cross-linking, has been confirmed by yeast two-hybrid analysis. Besides the Smt3 SUMO-activating enzyme, the Ubc9 SUMO-conjugating enzyme also exhibited in vivo interaction with the 5′-half of Rpn10/p54 in yeast cells. The mechanism of 26 S proteasome disassembly after ATP depletion is clearly different from that induced by Zn2+. Rpn10/p54 is permanently RP-bound during the ATP-dependent assembly–disassembly cycle, but during the Zn2+ cycle it reversibly shuttles between the RP-bound and free states.
Keywords: assembly–disassembly, catalytic particle, regulatory particle, 26 S proteasome, subunit Rpn10/p54, zinc
Abbreviations: CP, catalytic particle; DTT, dithiothreitol; Hsp, heat-shock protein; mAb, monoclonal antibody; MALDI–TOF, matrix-assisted laser-desorption ionization–time-of-flight; PIP, proteasome-interacting protein; PSD, post-source decay; RP, regulatory particle; SUMO, small ubiquitin-related modifier-1
INTRODUCTION
The ubiquitin–proteasome system is responsible for the controlled proteolysis of intracellular proteins. The first component of this system, the ubiquitinating enzyme cascade recognizes the different degradation signals present in short-lived proteins and, by attaching a multiubiquitin chain, marks these proteins for proteolysis (reviewed in [1]). Multiubiquitinated proteins are recognized and selectively degraded by the 26 S proteasome. In an ATP-dependent reaction, this large multiprotein complex is assembled from two subcomplexes: the CP (catalytic particle; reviewed in [2]) and the RP (regulatory particle). The orifices of the central channel of the CP, which are the sites of entry of substrate proteins [3], are situated at the bases of the barrel-shaped particle in Thermoplasma acidophilum [4]. In the crystal structure of the Saccharomyces cerevisiae CP, however, these orifices are missing, indicating that the channel is gated in eukaryotes [5,6]. The central channel has a narrow diameter, and is accessible only for completely unfolded proteins. The CP is a non-specific protease, which cannot discriminate between multiubiquitinated and non-ubiquitinated proteins.
The RPs, attached to the bases of the CP, ensure the selectivity of the 26 S proteasome for multiubiquitinated proteins (reviewed in [7]), unfold the substrate proteins by their chaperone-like activity [8,9], open the gated channel of the CP [10], reprocess the ubiquitin residues of the substrate proteins [11,12] and feed them into the CP. The activity of the RPs is strictly ATP-dependent. Besides the assembly of the 26 S proteasome from its subcomplexes, ATP is required for substrate unfolding, opening the gated channel of the CP, and most probably also for feeding the substrate proteins into the central channel of the CP. Six ATPase subunits of the RP, forming a heterohexameric ring, mediate all the ATP-dependent reactions [13]. The ATPase ring stacks to the base of the external α-rings of the CP, this configuration ensuring optimal access for the ATPase subunit Rpt2 to open the gated channel of the CP. During conventional chromatographic purification, the RP may be split into base and lid subcomplexes as a result of the artificially high ionic strength [14]. The ATPase ring, together with three non-ATPase subunits, forms the base subcomplex. The lid subcomplex is composed entirely of non-ATPase subunits. One of them, Rpn11, which contains a novel Zn2+-metalloprotease domain, is responsible for reprocessing the ubiquitin moieties of the multiubiquitinated substrate proteins. This deubiquitinating activity, which is strictly coupled with substrate degradation, is dependent on the unimpaired Zn2+-isopeptidase function of the subunit [11,12]. Removal of Zn2+, or mutation of the predicted active-site histidine residues, suspends the deubiquitinating activity and stabilizes the substrate protein. The roles of most of the lid subcomplex subunits are far less well known. Rpn1 and Rpn2, two non-ATPase subunits of the base, link the lid and base subcomplexes.
The selective recognition and binding of multiubiquitinated proteins are the primary and, from the aspect of cellular homoeostasis, the most critical functions of the RP. The earlier long debate on the identification of the ubiquitin receptor of the RP [15–18] seems to have been settled by two recent papers [19,20] that confirm the original idea that Rpn10/p54 (the nomenclature of the subunits of yeast, human and Drosophila RPs is presented in Table 1) fulfils all the criteria of an ubiquitin receptor. Although the co-operation of Rpn10/p54, Rad23 and other PIPs (proteasome-interacting proteins) in substrate recognition has been extensively analysed (reviewed in [21]), one feature of the mode of action of Rpn10/p54 in the substrate selection still awaits clarification. There are two alternative scenarios for the mode of substrate selection and binding. If it is assumed that Rpn10/p54 is located on the surface of the RP in an exposed configuration [22], substrate selection may proceed in the firmly bound state of this ubiquitin receptor subunit. However, since Rpn10/p54 is the only RP subunit that exists in RP-bound and free forms in most organisms [16,23,24], a shuttling cycle of this subunit may be presumed during substrate selection: after dissociation, the free subunit recruits multiubiquitinated substrates and, by reassociation with the RP, targets them for destruction. The reversible dissociation–association of Rpn10/p54, the first obvious requirement supporting this shuttling mechanism, is described in this paper.
Table 1. Human and yeast homologues of the Drosophila regulatory complex subunits.
| Drosophila | Human | Yeast | Subunit-specific antibody |
|---|---|---|---|
| p110 | S1 | Rpn2 | |
| p97 | S2 | Rpn1 | |
| p58 | S3 | Rpn3 | |
| p56 | S4 | Rpt2 | |
| p55 | S5b | Rpn5 | |
| p54 | S5a | Rpn10 | mAb 170, mAb 28 |
| p50 | S6′ | Rpt5 | mAb 112 |
| p48A | S6 | Rpt3 | |
| p48B | S7 | Rpt1 | |
| p42A | S10 | Rpn7 | mAb 123 |
| p42B | S9 | Rpn6 | |
| p42C | S8 | Rpt6 | |
| p42D | S10b | Rpt4 | |
| p39A | S11 | Rpn9 | mAb 50 |
| p39B | S12 | Rpn8 | |
| p37A | – | – | |
| p37B | S13 | Rpn11 |
EXPERIMENTAL
Purification of the proteasome
26 S proteasomes were purified to homogeneity from Drosophila embryos as described previously [25]. Partially purified 26 S proteasome fraction was purified to the DEAE-fractogel step, dialysed against 20 mM Tris/HCl (pH 7.6), 100 mM NaCl, 5 mM MgCl2, 1 mM ATP, 1 mM DTT (dithiothreitol) and 5% (v/v) glycerol and stored in aliquots at −80 °C. Just before Zn2+ addition, DTT was removed on a HiTrap Desalting column (Amersham Biosciences) equilibrated with 20 mM Tris/HCl (pH 7.6), 100 mM NaCl, 5 mM MgCl2, 1 mM ATP and 5% glycerol.
The conditions of protein cross-linking with disuccinimidyl suberate (Pierce, Rockford, IL, U.S.A.) and the analysis of the cross-linked proteasomal subunits by immunoblotting technique, as well as the characterization of the subunit-specific mAbs (monoclonal antibodies) have been described in [25].
Gel electrophoresis
Denaturing polyacrylamide gels (SDS/PAGE) were prepared by standard techniques. Native PAGE was performed on the single gel layer system described by Glickman et al. [26]. ATP was present or absent in the gel and in the running buffer as indicated. The 26 S proteasomes separated on native polyacrylamide gels were assayed for catalytic activity by the fluorigenic gel overlay technique using succinyl-Leu-Leu-Val-Tyr-amido-4-methylcoumarin (Bachem, Torrance, CA, U.S.A.) as described by Glickman et al. [26], or processed for immunoblotting after in-gel dissociation of the proteasomal subunits by soaking the gel for 5 min at room temperature (22 °C) in Western-blotting buffer (20 mM Tris base, 150 mM glycine and 20%, v/v, methanol; the pH of the solution is adjusted to 8.0 before the addition of methanol) supplemented with 1% SDS.
Zn2+ affinity chromatography was performed on a Fractogel® EMD Chelate (S) column (Merck, Gibbstown, NJ, U.S.A.) according to the manufacturer's instructions.
Phosphocreatine–creatine kinase (Sigma) was used as an ATP-regenerating system.
Purification and analysis of Rpn10/p54-interacting proteins
Immunoprecipitation was done with a mixture of two different mAbs (mAb 170 and mAb 28), which recognize two different epitopes of subunit Rpn10/p54. The mAbs were bound to Protein G–agarose (Sigma) and the beads were washed several times with PBS to remove unbound IgG and then incubated with the cross-linked protein fractions by mixing in the cold room overnight. After washing the beads five times with PBS and three times with PBS supplemented with 0.05% Tween 20 to remove unbound proteins, the immunoprecipitated proteins were recovered by SDS sample buffer and fractionated by SDS/PAGE.
For MS-based protein identification, colloidal Coomassie Brilliant Blue-stained protein bands were cut, diced and washed with 25 mM NH4HCO3 in 50% (v/v) acetonitrile/water. Disulphides were reduced with DTT (30 min, 56 °C) and alkylated with iodoacetamide (30 min, room temperature, in the dark). Proteins were in-gel-digested with side-chain-protected, porcine trypsin (Promega, Madison, WI, U.S.A.) for approx. 5 h. Tryptic digests were extracted and purified on C18 ZipTip (Millipore, Bedford, MA, U.S.A.). MS analysis was performed in positive-ion, reflectron mode, on a Reflex III MALDI–TOF (matrix-assisted laser-desorption ionization time-of-flight) mass spectrometer (Bruker, Karlsruhe, Germany), using 2,5-dihydroxybenzoic acid as the matrix. Two-point external calibration was applied, which guarantees mass accuracy within 200 p.p.m. Masses detected were submitted to a database search on the NCBI database using online software packages, such as MS-Fit in Protein Prospector (http://prospector.ucsf.edu). Some peptides were selected for PSD (post-source decay) analysis in order to obtain sequence information and, thus, to confirm or overwrite the results of the mass-fingerprint-based protein identification.
Cloning the Drosophila Hsp82 (heat-shock protein 82) cDNA
Hsp82 was cloned into the pFLAG-MAC expression vector (Sigma) after RT (reverse transcriptase)–PCR amplification using total RNA prepared from heat-shock treated Drosophila embryos (37 °C, 20 min). RevertAid first-strand cDNA kit (Fermentas, St. Leon-Rot, Germany) was used for reverse transcription and, for PCR amplification, the 5′-ACGAAGCTTATGCCAGAAGAAGCAGAGAC forward and the 5′-GTCGAATTCTTAATCGACCTCCTCCATGT reverse primers were used. The PCR product was digested with HindIII and EcoRI and cloned into the HindIII and EcoRI sites of pFLAG-MAC. The cloned Hsp82 cDNA was checked by DNA sequencing. Hsp82 protein was expressed and purified on anti-FLAG M2 affinity gel according to the manufacturer's instructions.
Yeast two-hybrid analysis
The full-length Drosophila Rpn10/p54 cDNA, its 5′-half (1–616 bp to the internal EcoRI site, [23]), or its 3′-half (616–1190 bp) were cloned into the pBTM 116 DNA-binding domain vector in frame with its Lex A DNA-binding sequence. pBTM 116 carries the trp1 nutritional marker gene. The reading frame was verified by DNA sequence analysis. Plasmids were transformed into S. cerevisiae L40 strain (trp1, leu2, his3, LYS2::lexAoplacZ), transformants were selected on minimal medium complemented with leucine and histidine and tested for self-activation by Lac Z assay. Since the full-length Rpn10/p54 and its 3′-half derivative exhibited self-activation, pBTM 116 carrying the 5′-half of the Rpn10/p54 cDNA was used for two-hybrid screening. It was co-transformed with a Drosophila melanogaster embryo Matchmaker cDNA activation domain library (ClonTech Laboratories, Palo Alto, CA, U.S.A.) according to the manufacturer's instructions. The cDNA library was cloned in the pACT2 vector, which carries the leu2 nutritional marker gene. Transformants carrying plasmids encoding interacting proteins were selected on minimal medium lacking histidine, and the interaction of the 5′-half of the Rpn10/p54 protein (fused to the Lex A DNA-binding domain) and the protein fused to the Gal4 activation domain was further verified by Lac Z assay. DNA sequencing and database analysis identified the selected cDNA present in the Gal4 activation domain fusion.
Pull-down experiments
The cDNAs of the Smt3 SUMO (small ubiquitin-related modifier-1)-activating enzyme and the DmUbc9 SUMO-conjugating enzyme were cloned into pFLAG-MAC vector, and the full-length Rpn10/p54 cDNA was cloned into pET28. The reading frame in the constructs was checked by DNA sequencing and the proteins were expressed. Anti-FLAG M2 affinity columns were charged with FLAG–Smt3 SUMO-activating enzyme, FLAG–DmUbc9 SUMO-conjugating enzyme or FLAG–Hsp82, and unbound proteins were removed by washing. The columns were loaded with 1 ml of a total Escherichia coli extract in which the full-length Rpn10/p54 was expressed, and the columns were washed extensively and eluted with an excess of FLAG peptide. The binding of Rpn10/p54 was analysed in immunoblots with mAb 170. In control experiments, uncharged anti-FLAG M2 affinity column was loaded with the same E. coli extract, and the non-specific binding of Rpn10/p54 was checked by the same technique.
RESULTS
Zn2+ induces disassembly of the 26 S proteasome and dissociation of the ubiquitin receptor subunit
The discovery that removal of the Zn2+ moiety from the Zn2+-isopeptidase domain of subunit Rpn11 not only destroys the deubiquitinating activity of Rpn11 but also suspends the whole proteasomal degradation cycle suggested that certain function(s) of the 26 S proteasome might be orchestrated through Zn2+-coordinated interactions of RP subunits. The removal of Zn2+ ions from its protein co-ordination site(s) by specific chelating agents seemed to be a plausible technique with which to approach these interactions. We attempted to trace these presumed Zn2+-dependent interactions of RP subunits by chemical cross-linking with bifunctional protein cross-linkers, which was demonstrated earlier to be a highly sensitive method that can be used to monitor the changes in the subunit interactions of the RP during the ATP-dependent assembly disassembly of the 26 S proteasome [25].
Highly purified or partially purified DEAE-fractogel fractions of Drosophila 26 S proteasomes were preincubated with increasing concentrations of 1,10-phenanthroline, followed by cross-linking with disuccinimidyl suberate. The proteins were fractionated on SDS/PAGE and the cross-linking pattern of the different RP subunits was visualized on immunoblots by using subunit-specific mAbs. Within the concentration range 0–2 mM, 1,10-phenanthroline did not induce any detectable change in the cross-linking pattern of the RP subunits (results not shown), indicating that either critical Zn2+ is firmly bound by RP subunits or its removal does not influence subunit interactions.
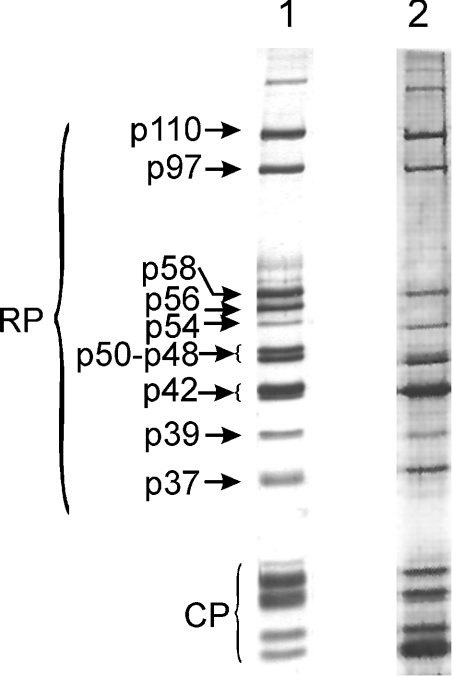
However, there is an alternative strategy through which the presumed Zn2+-dependent subunit interactions may be influenced. It is known that excess Zn2+ can severely inhibit the catalytic activity of several Zn2+-proteases, by binding to secondary Zn2+-binding sites and creating a bridging interaction with the catalytic Zn2+ ion [27–31]. The densely packed multiprotein structure of the RP may support such bridging interactions among their subunits. To exploit this approach, we first had to prove that there are RP subunits other than Rpn11, which possess Zn2+-binding sites. For this purpose, subunits of highly purified 26 S proteasome were dissociated in 6 M guanidinium chloride and loaded on to a Zn2+-charged metal chelating fractogel column equilibrated with 6 M guanidinium chloride in PBS. Unbound proteins were removed by washing the column with 6 M guanidinium chloride. The column was developed with a downward guanidinium chloride gradient (from 6 to 0 M in PBS) to allow the on-column refolding of Zn2+-bound proteasome subunits, which were finally eluted with an imidazole gradient. Several column-bound RP and CP subunits were recovered; all of them eluted at approx. 0.2 M imidazole, indicating the presence of strong Zn2+-binding sites on several proteasomal subunits (Figure 1). Among the RP subunits bound by the zinc column, p110, p97, p58 and p54 were identified by their molecular mass; a single polypeptide is present in these bands [25]. Immunoblots with subunit-specific antibodies [25] revealed that, of the 48, 42 and 39 kDa bands, which carry several different subunits, the p48A, p48B, p42A, p42C and p39A subunits were bound to the zinc column (results not shown). Proteasome subunits did not bind to the Zn2+-free metal chelating fractogel column (results not shown).
Figure 1. Zn2+-binding sites on proteasomal subunits.
Lane 1, silver-stained one-dimensional SDS/PAGE pattern of highly purified Drosophila 26 S proteasome; lane 2, proteasomal subunits recovered with 0.2 M imidazole from Zn2+-charged fractogel metal chelate column.
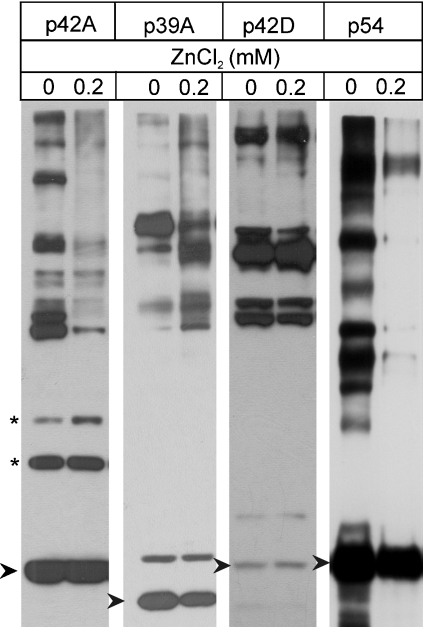
To test the effect of exogenous Zn2+ on the subunit interactions of the RP, DTT was removed from a highly purified 26 S proteasome preparation, which was then preincubated for 20 min at 25 °C with increasing concentrations (0–200 μM) of ZnCl2. To maintain the structural integrity of the 26 S proteasome, all the reactions were carried out in the presence of 1 mM ATP and an ATP-regenerating system. After this preincubation period, cross-linking of the RP subunits was initiated by the addition of disuccinimidyl suberate. The cross-linking reaction performed at 25 °C was halted 20 min later by the addition of an excess amount of glycine, and the cross-linking patterns of the RP subunits were analysed on immunoblots developed with different RP subunit-specific mAbs. As shown in Figure 2, excess Zn2+ induced characteristic rearrangements in the subunit interactions of the lid subcomplex. While certain subunit contacts were maintained (Figure 2, asterisks), others disappeared or new contacts were formed. The subunit interactions within the ATPase ring were not influenced, whereas all the contacts of the ubiquitin receptor subunit (Rpn10/p54) characteristic of the native proteasome were suspended. The rearrangements were Zn2+ concentration-dependent: the changes were already detectable at 50 μM Zn2+ concentration and reached a plateau at 200 μM (results not shown).
Figure 2. Zn2+-induced changes in proteasomal subunit interactions, detected by chemical cross-linking and immunoblot analysis.
Highly purified Drosophila 26 S proteasome was preincubated in the presence of ATP, with or without 200 μM ZnCl2, and cross-linked with disuccinimidyl suberate. The cross-linking patterns, representing subunit interactions, were analysed in immunoblots with subunit-specific mAbs. Changes in the subunit interactions of Rpn7/p42A and Rpn9/p39A lid subcomplex subunits were analysed with mAb 123 and mAb 50 respectively. The Rpt5/p50 ATPase subunit and the ubiquitin receptor subunit Rpn10/p54 were analysed with mAb 112 and mAb 170 respectively. The asterisk denotes subunit interactions not influenced by Zn2+ treatment. Arrow-heads mark the non-cross-linked subunits.
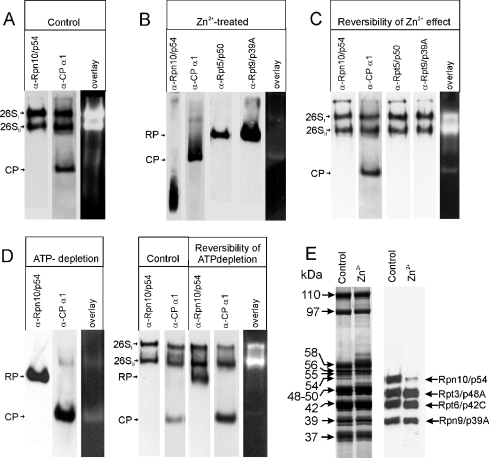
The data presented in Figure 2 suggest that subunit Rpn10/p54 dissociated from the RP in the presence of exogenous Zn2+, subsequently suspending all its interactions with other RP subunits. This assumption was tested on a partially purified 26 S proteasome (see below). DTT-free DEAE-fractogel fraction of the 26 S proteasome was preincubated for 20 min at 25 °C in the presence of 1 mM ATP and an ATP-regenerating system, with or without 200 μM ZnCl2, and then fractionated on native polyacrylamide gel containing 1 mM ATP (no DTT) and prepared with or without 200 μM ZnCl2. Immunoblot analysis of the control sample with mAbs specific for Rpn10/p54 (mAb 170, Figure 3A, lane 1) or the α1 CP subunit (mAb IIG7, Figure 3A, lane 2) revealed the conventional native polyacrylamide gel pattern: the 26SI and 26SII bands, corresponding to doubly capped and singly capped 26 S proteasomes respectively, reacted with both the RP- and the CP-specific antibodies, while the free CP present in the preparation had much faster electrophoretic mobility and reacted only with mAb IIG7. Both 26 S bands exhibited strong peptidase activity in overlay assays, while the peptidase activity of the free CP, in agreement with published data [26], was negligible (Figure 3A, lane 3). A completely different pattern was obtained in the presence of 200 μM Zn2+ (Figure 3B). The 26SI and 26SII forms of the 26 S proteasome were not observed, while mAb 50 and mAb 112, specific for the lid subunit Rpn9/p39A and the ATPase subunit Rpt5/p50 respectively, light up a single band which should correspond to the free RP, because it did not react with the CP-specific mAb IIG7. The total amount of CP was present as a free particle with an electrophoretic mobility higher than that of the free RP. As expected from the cross-linking pattern, Rpn10/p54 dissociated in toto from the RP and appeared in the front of the gel. Overlay assay revealed the loss of peptidase activity in the presence of Zn2+ (Figure 3B, lane 5). The loss of peptidase activity is not due to the inactivation of the catalytic activity of the CP, but it is a consequence of the dissociation of the RP. This follows from the observation that the peptidase activity of the zinc-treated proteasomes, measured by fluorimetry using succinyl-Leu-Leu-Val-Tyr-amido-4-methylcoumarin as substrate [26], can be enhanced 8-fold in the presence of 0.02% SDS. This is comparable with the increase in the catalytic activity of a highly purified CP in the presence of 0.02% SDS [26].
Figure 3. Zn2+-induced structural and functional changes in the 26 S proteasome, analysed by native PAGE.
(A) The DEAE-fractogel fraction of the Drosophila 26 S proteasome was fractionated on 1 mM ATP-containing native polyacrylamide gel and analysed by immunoblotting with mAb 170 (lane 1) and mAb IIG7 (lane 2), or its peptidase activity was tested in a fluorigenic overlay assay (lane 3). (B) The DEAE-fractogel fraction of the Drosophila 26 S proteasome was preincubated in the presence of ATP with 200 μM ZnCl2 and fractionated on 1 mM ATP and 200 μM ZnCl2-containing native polyacrylamide gel. Immunoblot analysis was performed with mAb 170 (lane 1), mAb IIG7 (lane 2), mAb 112 (lane 3) and mAb 50 (lane 4). The peptidase activity of the Zn2+-treated proteasomes was tested in a fluorigenic overlay assay (lane 5). (C) Reversibility of Zn2+-induced changes: Zn2+ was removed with 1,10-phenanthroline from the Zn2+-treated proteasome shown in (B), and analysed on 1 mM ATP-containing native gel. Immunoblot analysis was performed with mAb 170 (lane 1), mAb IIG7 (lane 2), mAb 112 (lane 3) and mAb 50 (lane 4). The peptidase activity was tested in a fluorigenic overlay assay (lane 5). (D) Reassembly of the 26 S proteasome after ATP depletion. Lanes 1–3: the ATP-depleted DEAE-fractogel fraction of the Drosophila 26 S proteasome was fractionated on native polyacrylamide gel prepared without ATP. Immunoblot analysis was performed with mAb 170 (lane 1) and mAb IIG7 (lane 2); the peptidase activity was tested in a fluorigenic overlay assay (lane 3). Lanes 4–5: the DEAE-fractogel fraction of the Drosophila 26 S proteasome was analysed on 1 mM ATP-containing native polyacrylamide gel. Immunoblots were tested with mAb 170 (lane 4) and mAb IIG7 (lane 5). Lanes 6–8: the ATP-depleted DEAE-fractogel fraction was incubated in the presence of ATP and fractionated on 1 mM ATP-containing native polyacrylamide gel. Immunoblot analysis was performed with mAb 170 (lane 6) and mAb IIG7 (lane 7); the peptidase activity was tested in a fluorigenic overlay assay (lane 8). (E) Integrity of the RP after zinc treatment. RP of control and zinc-treated DEAE-fractogel fraction was purified on a Superose 6 sizing column and their subunit pattern was analysed on silver-stained denaturing gel (lanes 1 and 2) and by immunoblotting (lanes 3 and 4) with a mixture of mAbs specific for the subunits indicated on the right side.
Thus excess Zn2+ induces disassembly of the 26 S proteasome into RP and CP, in parallel with full dissociation of the ubiquitin receptor subunit from the RP. The mobilities of the bands lit up by lid and base subunit-specific mAbs after zinc treatment cannot be distinguished, indicating that the integrity of the RP is not compromised; the RP does not fall apart into lid and base subcomplexes. These structural changes are accompanied by a complete loss of the peptidase activity. The cross-linking experiments depicted in Figure 2 revealed that the structure of the ATPase ring of the RP remains intact during this process, but major subunit rearrangements occur in the lid subcomplex.
To prove that Rpn10/p54 is the only subunit of the RP which dissociates in the presence of Zn2+, the RP of the zinc-treated DEAE-fractogel fraction was purified on a Superose 6 sizing column as described previously [32], and its subunit pattern was analysed on silver-stained SDS gel and by immunoblotting. As it is shown in Figure 3(E), besides the absence of the Rpn10/p54 subunit, the subunit patterns of the control and the zinc-treated RP are indistinguishable.
Reversibility of the Zn2+-induced structural and functional changes in the 26 S proteasome
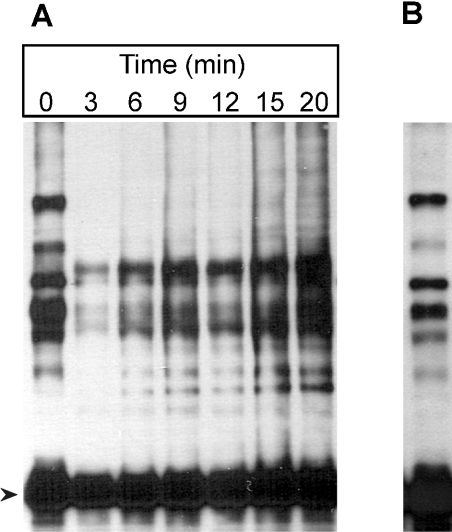
Reversal of the Zn2+-induced changes in the 26 S proteasome was first tested on a DTT-free highly purified 26 S proteasome. After a 20 min preincubation in the presence of 1 mM ATP, an ATP-regenerating system and 200 μM ZnCl2, 1,10-phenanthroline (200 μM final concentration) was added to the proteasomes and the incubation was continued for an additional 20 min. Reversal of the Zn2+-induced structural changes was tested on native polyacrylamide gels and by the cross-linking test. No detectable reversal occurred after the removal of the Zn2+ ions (results not shown). On the assumption that the reassociation of subunit Rpn10/p54 and the reassembly of the 26 S proteasome require non-proteasomal assembly factor(s), the reversibility of the Zn2+ effect was tested on partially purified 26 S proteasome fractions. DTT-free DEAE-fractogel fraction was preincubated in the presence of 1 mM ATP and an ATP-regenerating system with 200 μM ZnCl2 for increasing periods of time at 25 °C, and the association of Rpn10/p54 with potential non-proteasomal assembly factor(s) was tested by the cross-linking assay, using mAb 170 specific for subunit Rpn10/p54. As shown in Figure 4(A), the Zn2+-induced dissociation of Rpn10/p54 from the RP is very fast: a 3 min preincubation was sufficient to disrupt all subunit interactions characteristic of the native RP. Additionally, during the incubation, a completely new set of cross-linking products appeared, indicating the formation of a set of specific interactions of Rpn10/p54 with cellular protein(s) that do not occur in the highly purified 26 S proteasome preparation (Figure 2). The roles of these new, specific protein–protein interactions of Rpn10/p54 in the reversal of the Zn2+-induced structural changes were studied by native gel electrophoresis and cross-linking techniques. The DTT-free DEAE-fractogel fraction was preincubated in the presence of 1 mM ATP and an ATP-regenerating system with 200 μM ZnCl2 for 20 min at 25 °C. Removal of Zn2+ was achieved by the addition of 200 μM 1,10-phenanthroline, and cross-linking by disuccinimidyl suberate was started 10 min later. After a 20 min cross-linking period, the reversal of the Zn2+-induced changes was analysed by immunoblotting with mAb 170. Removal of Zn2+ resulted in the disappearance of all the Zn2+-specific cross-linking products and reformation of the major cross-linking products characteristic of the native 26 S proteasome (Figure 4B).
Figure 4. Detection of Rpn10/p54-interacting proteins.
(A) The DTT-free DEAE-fractogel fraction of the Drosophila 26 S proteasome was preincubated in the presence of ATP with 200 μM ZnCl2 for the time periods indicated. The preincubation was followed by cross-linking with disuccinimidyl suberate. Rpn10/p54-interacting proteins were detected by immunoblotting with mAb 170. (B) The DEAE-fractogel sample preincubated for 20 min with 200 μM ZnCl2 (A) was supplemented with 200 μM 1,10-phenanthroline, incubated for an additional 10 min, cross-linked and analysed as described in (A). Arrowhead marks the non-cross-linked Rpn10/p54 protein.
Native PAGE demonstrated that both electrophoretic variants of the 26 S proteasome were reformed upon Zn2+ removal (Figure 3C). Subunit Rpn10/p54 is incorporated into the reassembled 26 S proteasome, because the distribution of Rpn10/p54 between the 26SI and 26SII variants is indistinguishable from that of the Rpt5/p50 ATPase or Rpn9/p39A lid subunits (Figure 3C, lanes 1, 3 and 4) and no free monomer p54 was detected. Concomitantly with the structural reversal, there is a functional renaturation of the peptidase activity of the Zn2+-treated 26 S proteasomes after Zn2+ removal. As revealed by fluorigenic overlay assays, complete loss of peptidase activity occurred in the presence of 200 μM Zn2+ (Figure 3B, lane 5), but the peptidase activity was restored after Zn2+ removal (Figure 3C, lane 5).
Histidine and cysteine side chains, which are responsible for zinc co-ordination, can also bind several other bivalent cations. This is true for the proteasomal subunits as well: the same set of subunits bound by a zinc column (Figure 1) also binds to a nickel column (results not shown). The specificity of zinc on the structural and functional rearrangement of the 26 S proteasome, however, is supported by the observation that Ca2+, Ni2+, Co2+ and Mn2+ in the concentration range 50–200 μM can neither induce the disassembly of the 26 S proteasome and dissociation of the ubiquitin-receptor subunit nor disrupt the peptidase activity of the 26 S proteasome (results not shown).
Two different routes in the assembly–disassembly of the 26 S proteasome
It has long been known that ATP is essential for maintenance of the structural integrity of the 26 S proteasome. In the absence of ATP, the 26 S proteasome dissociates into RP and CP [33]. In partially purified preparations, this process is reversible, re-addition of ATP resulting in reassembly of the 26 S proteasomes [32]. To test the fate of subunit Rpn10/p54 after ATP depletion, the partially purified DEAE-fractogel fraction was run through a HiTrap desalting column equilibrated with 20 mM Tris/HCl (pH 7.6), 100 mM NaCl, 5 mM MgCl2 and 5% glycerol. The ATP- and DTT-depleted sample was fractionated on a native polyacrylamide gel prepared without ATP and DTT. Samples were blotted and the assembly state of the proteasomes and the dissociation of subunit Rpn10/p54 were analysed with mAb 170 and mAb IIG7. Although the 26 S proteasome dissociated into free RP and CP in the absence of ATP, subunit Rpn10/p54 did not dissociate from the RP (Figure 3D, lanes 1 and 2). Disassembly of the 26 S proteasome is accompanied by the loss of peptidase activity (Figure 3D, lane 3). Disassembly of the 26 S proteasome is partially reversed when ATP- and DTT-depleted proteasome is incubated again with ATP and an ATP-regenerating system, and fractionated on ATP containing native gel (Figure 3D, lanes 4–8). Addition of DTT did not improve the efficiency of reassembly (results not shown). Thus the mechanism of ATP-dependent assembly–disassembly of the 26 S proteasome is completely different from that observed after Zn2+ addition/removal. During the ATP cycle, subunit Rpn10/p54 is permanently RP-bound, whereas during the Zn2+-cycle it exhibits a reversible shuttling.
Identification of Rpn10/p54-interacting proteins
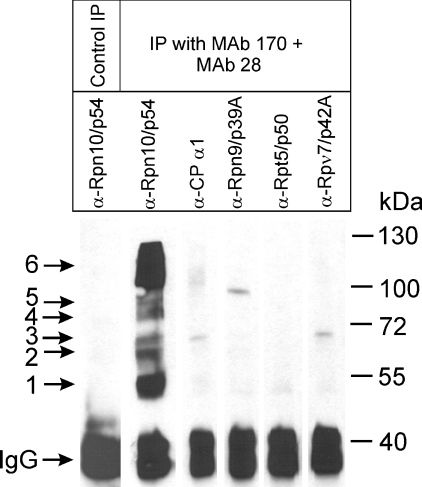
It appeared reasonable to suppose that proteins interacting with subunit Rpn10/p54 after its Zn2+-induced dissociation are involved in the structural and functional restoration of the intact 26 S proteasome accompanying Zn2+ removal. For the identification of these interacting proteins, DTT-free DEAE-fractogel fraction was incubated in the presence of 1 mM ATP and an ATP-regenerating system with 200 μM ZnCl2 for 20 min, and cross-linked with disuccinimidyl suberate. The cross-linking reaction was halted by the addition of an excess of glycine, and the sample was fractionated on a Superose 6 sizing column to separate the RPs, the CP, the Rpn10/p54 subunit cross-linked with its interacting partners, the free Rpn10/p54 subunit and other monomer proteins. The fractions from Superose 6 chromatography were analysed by immunoblot assay, using mAb 170 to localize the elution position of the cross-linked Rpn10/p54-interacting protein fractions. These fractions were pooled and immunoprecipitated with a mixture of mAb 170 and mAb 28. A non-specific mAb developed against potato leghaemoglobin was used as control in the immunoprecipitation. As shown in Figure 5, mAb 170 and mAb 28 precipitated the complex pattern of cross-linked Rpn10/p54 derivatives that appear after Zn2+ treatment (lane 2). These bands are completely missing from the control immunoprecipitate obtained with a non-specific antibody (Figure 5, lane 1). Immunoblot analysis of the precipitated proteins with several proteasome-specific mAbs further confirmed the specificity of the immunoprecipitation: apart from Rpn10/p54, no other RP or CP subunit was present in the precipitated fraction (Figure 5, lanes 3–6). The immunoprecipitated proteins were separated by SDS/PAGE and stained with Coomassie Blue; the bands were excised from the gel, and after in-gel trypsin digestion, the unfractionated digest mixtures were subjected to MALDI—TOF MS. Proteins were first tentatively identified from database searches with the monoisotopic MH+ values observed. These IDs were further confirmed by sequence information provided by PSD spectra of selected peptides. Besides Rpn10/p54, four different proteins were identified in the immunoprecipitated material as cross-linked partners: band 2: heat-shock cognate protein Hsc 70Cb (NCBI accession no. 4753683, confirmed by PSD of: N594KLQGGPFER603); band 3: Smt3-activating enzyme 2 (NCBI accession no. 6934296, confirmed by PSD of: Q62FLFHR67 and F278FNEDITYLLR288); band 4: Drosophila protein with a function of glutamyl amidopeptidase (NCBI accession no. 24646518, confirmed by PSD of: Y773INWAWDESNVR784); and bands 5 and 6: Hsp82 (NCBI accession no. 8127, confirmed by PSD of: A324LLFIPR330 and H313FSVEGQLEFR323). The presence of Hsp82 both in band 5 and in the broad band 6 may be due to its well-known oligomerization tendency, which is enhanced by bivalent cations (reviewed in [34]). IgM, derived from the ascites fluid of mAb 170 was identified in band 1.
Figure 5. Purification of Rpn10/p54-interacting proteins by immunoprecipitation.
The cross-linked DEAE-fractogel sample shown in Figure 4(A) (20 min) was immunoprecipitated with a mixture of mAb 28 and mAb 170. mAb developed against potato leghaemoglobin was used for control immunoprecipitation. Aliquots of the precipitated materials were analysed by immunoblotting. Lane 1: the immunoprecipitate obtained using potato leghaemoglobin antibody was analysed with mAb 170. The immunoprecipitate produced with a mixture of mAb 28 and mAb 170 was analysed with mAb 170 (lane 2), mAb IIG7 (lane 3), mAb 50 (lane 4), mAb 112 (lane 5) and mAb 123 (lane 6).
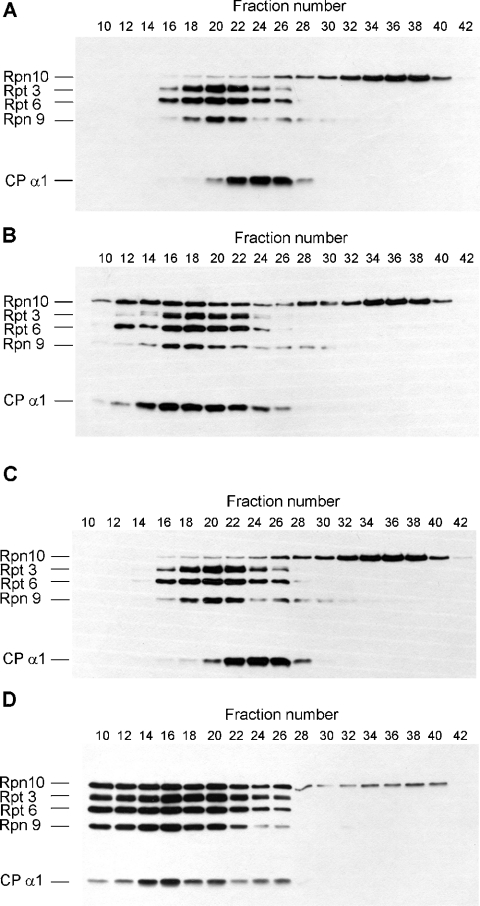
The identification of the Hsp82 chaperone as the main Rpn10/p54-interacting protein and its published role in the assembly of the yeast 26 S proteasome as well as in the maintenance of the structural integrity of the yeast 26 S proteasome [35] suggested that it might have a role in the reassembly of the Drosophila 26 S proteasome after zinc treatment. The failure of the reassembly of the highly purified 26 S proteasome after zinc removal, as opposed to the high efficiency of such reassembly in the partially purified DEAE-fractogel fraction, may be due to the lack of Hsp82 protein in the highly purified 26 S proteasome. To address the role of Hsp82 in the zinc-dependent reassembly process, highly purified 26 S proteasome was preincubated in the presence of ATP, an ATP-regenerating system and 200 μM Zn2+ for 20 min at 25 °C, zinc was then removed as described above and the proteasome was incubated for an additional 20 min in the absence or presence of 5-fold molar excess of purified Hsp82 protein. Reassembly of the 26 S proteasome and the incorporation of Rpn10/p54 subunit into the RP were analysed by immunoblot technique after fractionation of the proteins on a Superose 6 sizing column. Although the resolution of the Superose 6 column is far less than that of the native PAGE, the preparative Superose 6 column allowed easier detection of the dilute highly purified 26 S proteasome preparation. As it is shown in Figure 6(A), reassembly of the zinc-treated highly purified 26 S proteasome cannot be achieved by zinc removal alone. The elution peak of Rpn10/p54 (fraction 36) is in the position of the monomer proteins, the peak of the free CP is in fraction 24 and that of the free RP is in fraction 20. Purified Hsp82 protein supported the partial reassembly of the 26 S proteasome after zinc removal (Figure 6B). A substantial fraction of Rpn10/p54 is reincorporated into the RP, the shift between the elution position of the free CP and the free RP disappears, and their common elution position in fractions 16–18 corresponds to that of the singly capped 26 S proteasomes [32]. The Hsp82-supported reassembly of the highly purified 26 S proteasome, however, is far less complete than that observed in the partially purified DEAE-fractogel fraction (Figures 3C, 6C and 6D), suggesting that other assembly factor(s), like Hsc 70Cb, may also participate in this process.
Figure 6. Hsp82 supports the reassembly of zinc-treated highly purified 26 S proteasome.
(A) Highly purified Drosophila 26 S proteasome was preincubated in the presence of ATP with 200 μM ZnCl2. After zinc removal, incubation was continued for an additional 20 min and the proteasomes were fractionated on Superose 6 sizing column. Column fractions (200 μl) were concentrated by precipitation with 7 vol. of acetone, run on 10% (w/v) SDS gel and immunoblotted with a mixture of five mAbs specific for subunits indicated on the left. (B) Highly purified Drosophila 26 S proteasome was preincubated in the presence of ATP with 200 μM ZnCl2. After the removal of zinc, incubation was continued with 5-fold molar excess of purified Hsp82 protein for an additional 20 min and the proteasomes were fractionated on Superose 6 sizing column. Column fractions (200 μl) were concentrated by precipitation with 7 vol. of acetone, run on 10% SDS gel and immunoblotted with a mixture of five mAbs specific for subunits indicated on the left. (C) DEAE-fractogel fraction of Drosophila 26 S proteasome was preincubated in the presence of ATP with 200 μM ZnCl2 and fractionated on Superose 6 sizing column in the presence of 200 μM ZnCl2. Aliquots (20 μl) of the column fraction were run on 10% SDS gel and immunoblotted with a mixture of five mAbs specific for subunits indicated on the left. (D) DEAE-fractogel fraction of Drosophila 26 S proteasome was preincubated in the presence of ATP with 200 μM ZnCl2. After zinc removal, incubation was continued for an additional 20 min and the proteasomes were fractionated on Superose 6 sizing column. Aliquots (20 μl) of the column fraction were run on 10% SDS gel and immunoblotted with a mixture of five mAbs specific for subunits indicated on the left.
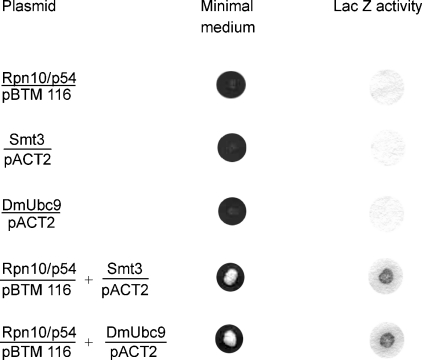
To explore further the physiological relevance of the protein–protein interactions of the zinc-dissociated Rpn10/p54 detected by chemical cross-linking, yeast two-hybrid screen was performed to select proteins that can in vivo interact with Rpn10/p54. As the C-terminal half of the Rpn10/p54 protein induced self-activation of the Gal4 system, only the N-terminal half of the subunit was used as bait, cloned into the pBTM 116 DNA-binding domain vector. This plasmid was co-transformed with a Drosophila embryonic cDNA library cloned in the pACT2 Gal4 activation-domain vector. Co-transformants carrying plasmids coding for interacting proteins were selected on minimal medium lacking histidine. The specificity of the interactions was further verified by the Lac Z test. Among the interacting proteins selected, two were highly relevant for the present study: in agreement with the cross-linking studies, we have selected the Drosophila Smt3 SUMO-activating enzyme. In addition, the DmUbc9 Drosophila SUMO-conjugating enzyme [36] has also been identified among the Rpn10/p54-interacting proteins (Figure 7).
Figure 7. In vivo interaction of the 5′-half of Rpn10/p54 with the Smt3 SUMO-activating and the DmUbc9 SUMO-conjugating enzymes.
Single- and double-transformant yeasts were grown on minimal medium and tested for Lac Z activity.
The interaction of the Smt3 SUMO-activating enzyme and the DmUbc9 Drosophila SUMO-conjugating enzyme with Rpn10/p54 has been confirmed by pull-down experiments. Anti-FLAG M2 affinity gel charged with FLAG-tagged Smt3 or FLAG-tagged DmUbc9 was loaded with recombinant Rpn10/p54. The binding of Rpn10/p54 was analysed in immunoblots after elution of the columns with FLAG peptide. As shown in Figure 8 (lane 1), Rpn10/p54 does not bind to uncharged affinity column, but binds to both Smt3-charged (lane 2) and DmUbc9-charged (lane 3) columns.
Figure 8. In vitro interaction of the Smt3 SUMO-activating enzyme, the DmUbc9 SUMO-conjugating enzyme and the Hsp82 protein with the full-length Rpn10/p54.
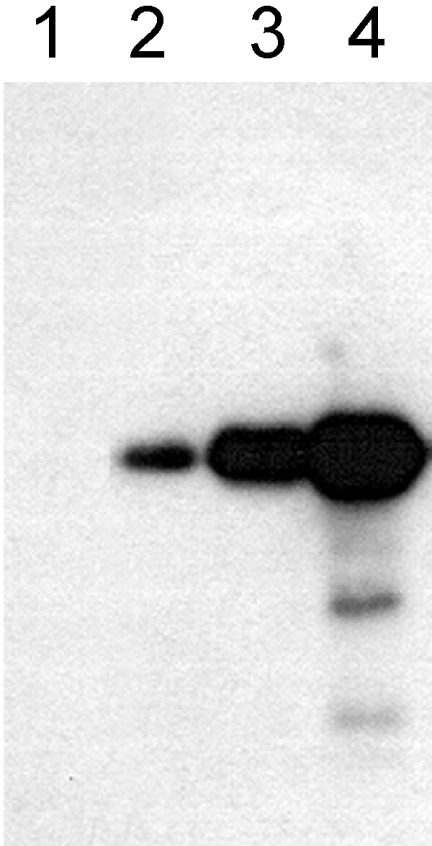
Uncharged anti-FLAG M2 affinity column (lane 1), or anti-FLAG M2 affinity columns charged with FLAG–Smt3 SUMO-activating enzyme (lane 2), FLAG–DmUbc9 SUMO-conjugating enzyme (lane 3) or FLAG–Hsp82, were loaded with Rpn10/p54. The columns were eluted with an excess of FLAG peptide and analysed by immunoblotting with mAb 170.
On using the 5′-half of Rpn10/p54 as bait in the yeast two-hybrid screen, we could not detect the interaction of the N-terminal half of the subunit with the Hsp82 protein. In the pull-down experiments, however, on use of the full-length Rpn10/p54, a strong interaction was demonstrated (Figure 8, lane 4).
DISCUSSION
Zn2+ seems to be a more generalized effector of the proteasome as it was assumed previously [11,12]. Besides its essential role in the reprocessing of ubiquitin moieties from multiubiquitinated proteins and its involvement in the whole proteolytic degradation cycle, Zn2+ can induce an extensive structural rearrangement of the Drosophila 26 S proteasome. In the presence of excess Zn2+, the 26 S proteasome disassembles into RP and CP, this process being accompanied by the dissociation of subunit Rpn10/p54, the ubiquitin receptor of the proteasome. The dissociation of subunit Rpn10/p54 induces extensive rearrangements within the lid subcomplex, while the structure of the ATPase ring of the base subcomplex seems to be maintained, gross subunit rearrangements, detectable by chemical cross-linking, do not occur. Due to the dissociation of the RP, the peptidase activity of the proteasome is lost.
The intracellular free Zn2+ concentration in a eukaryotic cell is in the nanomolar range, but the total intracellular Zn2+ concentration is approx. 200 μM [37]. Thus the Zn2+ concentration required to induce the changes described above is much higher than the free intracellular Zn2+ concentration. The specificity of this Zn2+ effect, however, is supported by the observation that the Zn2+-induced structural and functional changes are reversible: removal of Zn2+ is followed by reassociation of the subunit Rpn10/p54 to the RP, reassembly of the intact 26 S proteasome and resumption of the peptidase activity. Inside the cell, most of the Zn2+ is bound by specific Zn2+ transporters, which deliver this essential metal element for a variety of biological processes [38,39]. The delivery of the Zn2+ ion by a specific Zn2+ transporter to a critical position of the acceptor molecule may substantially reduce the critical Zn2+ concentration required for the induction of a biological process. The effect of zinc seems to be highly specific; other bivalent cations tested cannot induce these structural changes.
We present experimental evidence that the assembly–disassembly of the 26 S proteasome may proceed via two distinct mechanisms. Subunit Rpn10/p54 is permanently RP-bound during the ATP-dependent assembly cycle, but during the Zn2+ cycle it reversibly shuttles between RP-bound and free states. The structural rearrangements accompanying these assembly–disassembly processes are clearly different. During the ATP-dependent cycle, the rearrangement of the subunit contacts of the ATPase ring predominates [25], while the Zn2+ cycle induces extensive reorganization of the subunits belonging to the lid subcomplex. Both assembly mechanisms demand non-proteasomal assembly factor(s): assembly does not occur in a highly purified 26 S proteasome preparation. The assembly factor(s) required during the ATP- or Zn2+-dependent cycles are probably not fully identical. In the partially purified DEAE-fractogel fraction of the Drosophila 26 S proteasome, the Zn2+-dependent assembly is almost complete, while the ATP-dependent process is only partial (Figure 3).
There are two different lines of evidence that support the assumption that the Rpn10/p54-interacting proteins detected by chemical cross-linking in the presence of zinc may have physiological relevance. In highly purified 26 S proteasome preparation, the structural and functional reversal of the zinc-induced changes do not occur after the removal of zinc. Drosophila Hsp82 protein, identified by MS as the main Rpn10/p54-interacting component, can support the partial reassembly of the zinc-treated highly purified Drosophila 26 S proteasome. The role of the yeast Hsp90 chaperone (the yeast orthologue of the Drosophila Hsp82) in the assembly and maintenance of the structural integrity of the 26 S proteasome is well-documented [35]: inactivation of a temperature-sensitive yeast mutant Hsp90 protein by incubation at a non-permissive temperature caused dissociation of the 26 S proteasome into RP and CP and the loss of peptidase activity. The dissociated subcomplexes reassembled in an Hsp90-dependent fashion both in vivo and in vitro. Moreover, genetic interactions have been found between Hsp90 and several RP non-ATPase subunits. Among these, the genetic interaction between Hsp90 and Rpn10/p54 caused the most severe phenotype. The ability of the Drosophila Hsp82 protein to support a partial reassembly of the 26 S proteasome indicates that the in vivo and in vitro assembly mechanisms are conserved between yeast and Drosophila. Our results, however, suggest that, besides Hsp82, other assembly factor(s), such as Hsc70Cb, are also involved in the refolding process required for a complete reassembly of the 26 S proteasome after zinc treatment.
The second line of evidence supporting the relevance of our chemical cross-linking method in the identification of Rpn10/p54-interacting proteins is the demonstration that, not only in vitro but also in vivo, Rpn10/p54 can interact with the Smt3 SUMO-activating enzyme. Our yeast two-hybrid screen gives further support for the physiological importance of this interaction by demonstrating that not only the Smt3 SUMO-activating enzyme but also the DmUbc9 SUMO-conjugating enzyme [36] can interact in vivo with the Rpn10/p54 subunit. Smt3 and DmUbc9 proteins catalyse the first two steps of protein sumoylation, and sumoylation of several regulatory complex subunits has recently been demonstrated by affinity purification–tandem MS techniques [40].
To explore the biological relevance of the Zn2+-induced changes observed in the 26 S proteasome in vitro, it will be obligatory to prove that Zn2+ induces similar changes in vivo, the Zn2+ transporter involved in the in vivo process should be identified, and the regulatory cascade that activates the process should be recognized. The recent observation that, in HeLa cells, the Zn2+ ionophore pyrrolidine dithiocarbamate in combination with Zn2+ inhibits the proteasome-dependent proteolysis of several key proteasomal substrates (p53, p21 etc.) encourages the supposition that Zn2+, similarly as in vitro, influences the 26 S proteasomes in vivo [41].
The shuttling theory of multiubiquitinated substrate recognition postulates that the ubiquitin receptor of the proteasome first dissociates from the RP, then recruits the multiubiquitinated protein and targets it to the proteasome for proteolysis. The ability of Rpn10/p54 to undergo triggered dissociation from the RP and its ability to recognize and bind multiubiquitinated proteins in vitro [15–17] lend strong support to this mechanism. Free Rpn10/p54, detected in freshly prepared crude extracts in several species [16,23,24], may represent the fraction of the multiubiquitin receptor dissociated in vivo.
While this paper was under review, Babbitt et al. [42] reported that FLAG-tagged 26 S proteasomes immunopurified on an anti-FLAG column eluted in association with a large set of PIPs. In the presence of ATP, PIPs are released from the 26 S proteasome, this process being accompanied by the disassembly of the 26 S proteasome into RP and CP, together with the dissociation of subunit Rpn10. 26 S proteasome particles that are not associated with PIPs (i.e. which were not engaged in vivo with proteolysis) do not disassemble in the presence of ATP. In the absence of ATP, however, these 26 S proteasome particles disassemble into RP and CP, but subunit Rpn10 remained stably associated with RP during this disassembly. These two distinct disassembly processes are very similar to that observed in our in vitro system. The demonstration of the role of zinc in this process may open up the way to further exploration of the exact mechanism of this complex process.
Acknowledgments
This work was supported by a National Scientific Research Grant (OTKA T 46177).
References
- 1.Pickart C. M. Back to the future with ubiquitin. Cell (Cambridge, Mass.) 2004;116:181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- 2.Groll M., Hubert R. Substrate access and processing by the 20S proteasome core particle. Int. J. Biochem. Cell Biol. 2003;35:606–616. doi: 10.1016/s1357-2725(02)00390-4. [DOI] [PubMed] [Google Scholar]
- 3.Wenzel T., Baumeister W. Conformational constrains in protein degradation by the 20S proteasome. Nat. Struct. Biol. 1995;2:199–204. doi: 10.1038/nsb0395-199. [DOI] [PubMed] [Google Scholar]
- 4.Löwe J., Stock D., Jap B., Zwickl P., Baumeister W., Huber R. Crystal structure of the 20S proteasome from the archeon T. acidophilum at 3.4 Å resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 5.Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H. D., Huber R. Structure of the 20SA proteasome from yeast at 2.4 Å resolution. Nature (London) 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 6.Groll M., Bajorek M., Köhler A., Moroder L., Rubin D. M., Huber R., Glickman M. H., Finley D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 7.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 8.Braun B. C., Glickman M., Kraft R., Dahlman B., Kloetzel P. M., Finley D., Schmidt M. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat. Cell Biol. 1999;1:221–226. doi: 10.1038/12043. [DOI] [PubMed] [Google Scholar]
- 9.Strickland E., Hakala K., Thomas P. J., DeMartino G. N. Recognition of misfolding proteins by PA700, the regulatory subcomplex of the 26S proteasome. J. Biol. Chem. 2000;275:5565–5572. doi: 10.1074/jbc.275.8.5565. [DOI] [PubMed] [Google Scholar]
- 10.Koehler A., Cascio P., Legget D. S., Woo K. M., Goldberg A. L., Finley D. The axial channel of the proteasome core particle is gated by Rpt2 ATPase and controls both substrate entry and product release. Mol. Cell. 2001;7:1143–1152. doi: 10.1016/s1097-2765(01)00274-x. [DOI] [PubMed] [Google Scholar]
- 11.Verma R., Aravind L., Oania R., McDonald W. H., Yates J. R., Koonin E. V., Deshaies R. J. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 12.Yao T., Cohen R. E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature (London) 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 13.Rubin D. M., Glickman M. H., Larsen C. N., Dhruvakumar S., Finley D. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 1998;17:4909–4919. doi: 10.1093/emboj/17.17.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glickman M. H., Rubin D. M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumeister W., Fried V. A., Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin conjugate degradation and related to the COP9 signalosome and eIF3. Cell (Cambridge, Mass.) 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 15.Deveraux Q., Ustrell V., Pickart C., Rechsteiner M. A 26S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 16.Van Nocker S., Sadis S., Rubin D. M., Glickman M., Fu H., Coux O., Wefes I., Finley D., Vierstra R. D. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol. 1996;11:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haracska L., Udvardy A. Mapping the ubiquitin-binding domains in the p54 regulatory complex subunit of the Drosophila 26S protease. FEBS Lett. 1997;412:331–336. doi: 10.1016/s0014-5793(97)00808-9. [DOI] [PubMed] [Google Scholar]
- 18.Lam Y. A., Lawson G. T., Velayutham M., Zweier J. L., Pickart C. M. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature (London) 2002;416:763–767. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- 19.Elsasser S., Chandler-Militello D., Müller B., Hanna J., Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- 20.Verma R., Oania R., Graumann J., Deshaies R. J. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell (Cambridge, Mass.) 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 21.Hartmann-Petersem R., Gordon C. Proteins interacting with the 26S proteasome. Cell. Mol. Life Sci. 2004;61:1589–1595. doi: 10.1007/s00018-004-4132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haracska L., Udvardy A. Dissection of the regulator complex of the Drosophila 26S protease by limited proteolysis. Biochem. Biophys. Res. Commun. 1996;220:166–170. doi: 10.1006/bbrc.1996.0375. [DOI] [PubMed] [Google Scholar]
- 23.Haracska L., Udvardy A. Cloning and sequencing a non-ATPase subunit of the regulatory complex of the Drosophila 26S protease. Eur. J. Biochem. 1995;231:720–725. doi: 10.1111/j.1432-1033.1995.tb20753.x. [DOI] [PubMed] [Google Scholar]
- 24.Wilkinson C. R. M., Ferrell K., Penney M., Wallace M., Dubiel W., Gordon C. Analysis of a gene encoding Rpn10 of the fission yeast proteasome reveals that the polyubiquitin-binding site of this subunit is essential when Rpn12/Mts3 activity is compromised. J. Biol. Chem. 2000;275:15182–15192. doi: 10.1074/jbc.275.20.15182. [DOI] [PubMed] [Google Scholar]
- 25.Kurucz É., Andó I., Sümegi M., Hölzl H., Kapelari B., Baumeister W., Udvardy A. Assembly of the Drosophila 26S proteasome is accompanied by extensive subunit rearrangements. Biochem. J. 2002;365:527–536. doi: 10.1042/BJ20011520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glickman M. H., Rubin D. M., Fried V., Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmquist B., Vallee B. L. Metal substitutions and inhibition of thermolysin: spectra of the cobalt enzyme. J. Biol. Chem. 1974;249:4601–4607. [PubMed] [Google Scholar]
- 28.Kerr M. A., Kenny A. J. The molecular weight and properties of a neutral metallo-endopeptidase from rabbit kidney brush border. Biochem. J. 1974;137:489–495. doi: 10.1042/bj1370489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bunning P., Holmquist B., Riordan J. F. Substrate specificity and kinetic characteristics of angiotensin converting enzyme. Biochemistry. 1983;22:103–110. doi: 10.1021/bi00270a015. [DOI] [PubMed] [Google Scholar]
- 30.Mallaya S. K., Van Wart H. E. Mechanism of inhibition of human neutrophil collagenase by Gold(I) chrysotherapeutic compounds. J. Biol. Chem. 1989;264:1594–1601. [PubMed] [Google Scholar]
- 31.Gomez-Ortiz M., Gomish-Rüth F. X., Huber R., Avilés F. X. Inhibition of carboxypeptidase a by excess Zn2+: analysis of the structural determinant by X-ray crystallography. FEBS Lett. 1997;400:336–340. doi: 10.1016/s0014-5793(96)01412-3. [DOI] [PubMed] [Google Scholar]
- 32.Udvardy A. Purification and characterization of a multiprotein component of the Drosophila 26S proteolytic complex. J. Biol. Chem. 1993;268:9055–9062. [PubMed] [Google Scholar]
- 33.Hoffman L., Pratt G., Rechsteiner M. Multiple forms of the 20S multicatalytic and the 26S ubiquitin/ATP-dependent proteases from rabbit reticulocyte lysate. J. Biol. Chem. 1992;267:22362–22368. [PubMed] [Google Scholar]
- 34.Csermely P., Schnaider T., Söti C., Prohászka Z., Nardai G. The 90-kDa molecular chaperone family: structure, function, and clinical application. A comprehensive review. Pharmacol. Ther. 1998;79:129–168. doi: 10.1016/s0163-7258(98)00013-8. [DOI] [PubMed] [Google Scholar]
- 35.Imai J., Maruya M., Yashiroda H., Yahara I., Tanaka K. The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J. 2003;22:3557–3567. doi: 10.1093/emboj/cdg349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joanisse D. R., Inaguma Y., Tanguay R. M. Cloning and developmental expression of a nuclear ubiquitin-conjugating enzyme (DmUbc9) that interacts with small heat shock proteins in Drosophila melanogaster. Biochim. Biophys. Res. Commun. 1998;244:102–109. doi: 10.1006/bbrc.1998.8214. [DOI] [PubMed] [Google Scholar]
- 37.Palmiter R. D., Findley S. D. Cloning and functional characterization of a mammalian Zn2+ transporter that confers resistance to Zn2+ EMBO J. 1995;14:639–649. doi: 10.1002/j.1460-2075.1995.tb07042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamashita S., Miyagi C., Fukada T., Kagara N., Che Y.-S., Hirano T. Zn2+ transporter LIVI controls epithelial-mesenchymal transition in zebrafish gastrula organizer. Nature (London) 2004;429:298–302. doi: 10.1038/nature02545. [DOI] [PubMed] [Google Scholar]
- 39.Kambe T., Yamaguchi-Iwai Y., Sasaki R., Nagao M. Overview of mammalian Zn2+ transporters. Cell. Mol. Life Sci. 2004;61:49–68. doi: 10.1007/s00018-003-3148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panse V. G., Hardeland U., Werner T., Kuster B., Hurt E. A proteome-wide approach identifies sumolyated substrate proteins in yeast. J. Biol. Chem. 2004;279:41346–41351. doi: 10.1074/jbc.M407950200. [DOI] [PubMed] [Google Scholar]
- 41.Kim I., Kim C. H., Kim J. H., Lee J., Choi J. J., Chen Z. A., Lee G. M., Chung K. C., Hsu C. Y., Ahn Y. S. Pyrrolidine dithiocarbamate and Zn2+ inhibit proteasome-dependent proteolysis. Exp. Cell Res. 2004;298:229–238. doi: 10.1016/j.yexcr.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 42.Babbitt S. E., Kiss A., Deffenbaugh A. E., Chang Y.-H., Bailly E., Erdjument-Bromage H., Tempst P., Buranda T., Sklar L. A., Baumler J., et al. ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle. Cell (Cambridge, Mass.) 2005;121:553–565. doi: 10.1016/j.cell.2005.03.028. [DOI] [PubMed] [Google Scholar]