Abstract
The NCE103 gene of the yeast Saccharomyces cerevisiae encodes a CA (carbonic anhydrase) that catalyses the interconversion of CO2 and bicarbonate. It has previously been reported that nce103 null mutants require elevated CO2 concentrations for growth in batch cultures. To discriminate between ‘sparking’ effects of CO2 and a CO2 requirement for steady-state fermentative growth, we switched glucose-limited anaerobic chemostat cultures of an nce103 null mutant from sparging with pure CO2 to sparging with nitrogen gas. This switch resulted in wash-out of the biomass, demonstrating that elevated CO2 concentrations are required even under conditions where CO2 is produced at high rates by fermentative sugar metabolism. Nutritional analysis of the nce103 null mutant demonstrated that growth on glucose under a non-CO2-enriched nitrogen atmosphere was possible when the culture medium was provided with L-aspartate, fatty acids, uracil and L-argininine. Thus the main physiological role of CA during growth of S. cerevisiae on glucose-ammonium salts media is the provision of inorganic carbon for the bicarbonate-dependent carboxylation reactions catalysed by pyruvate carboxylase, acetyl-CoA carboxylase and CPSase (carbamoyl-phosphate synthetase). To our knowledge, the present study represents the first full determination of the nutritional requirements of a CA-negative organism to date.
Keywords: carbon dioxide, carbonic anhydrase, carboxylase, carboxylation, CPSase, Saccharomyces cerevisiae
Abbreviations: AIR, 5′-phosphoribosyl-5-aminoimidazole; CA, carbonic anhydrase; CP, carbamoyl-phosphate; CPSase, CP synthetase; TCA, tricarboxylic acid
INTRODUCTION
Carbonic anhydrase (CA; EC 4.2.1.1) is a ubiquitous enzyme present in organisms belonging to all branches of the evolutionary tree, from archaea to mammals. CA catalyses the reversible hydration of CO2 to HCO3−+H+. This reaction can also occur spontaneously when the CO2 concentration is sufficiently high. In nature, CO2 concentrations are strongly influenced by factors such as pH, temperature and presence of other solutes [1]. Based on protein structure, CAs are divided into four classes: α, β, γ and δ [2]. The differences in tertiary and quaternary structures among the four classes of CAs constitute a spectacular case of convergent evolution, in which the same catalytic activity has evolved via several independent, evolutionarily very ancient ways [2]. This suggests that CA-catalysed interconversion of CO2 and HCO3− is important for all forms of life.
Although universal, the α-class is best characterized in mammals, whereas isoenzymes of the β-class have been described in higher plants, algae, fungi, archaea and bacteria. Known members of the γ-class are only prokaryotes and the δ-class has only one member, the CA of the diatom Thalassiosira weissflogii [2,3]. Despite its widespread occurrence, CA has been deeply investigated in only a few organisms. Much research has been performed on mammalian cells because some CA isoenzymes are expressed at increased levels in tumours [4]. In mammals, CA is expressed in almost all tissues. Proposed functions include oxygen transport between lungs, red blood cells and tissues, pH regulation, ion exchange in the kidney and electrical activity in the retina and nervous system [5,6]. Another situation in which the role of CA has been extensively investigated is in autotrophic organisms, where CA is mostly related to the provision of bicarbonate for carbon fixation [7,8].
In contrast with the systems mentioned above, very little is known about the physiological role of CAs in heterotrophic microbes. Several studies on bacteria indicate that loss of CA activity impairs aerobic growth, unless the air is supplemented with extra CO2 [9–12]. Apparently, at elevated CO2 concentration, spontaneous hydration may obviate the need for CA-catalysed bicarbonate formation. This has led to the hypothesis that CA is involved in providing HCO3− for carboxylation reactions, such as the anaplerotic replenishment of C4 intermediates, citrulline biosynthesis or fatty acid elongation. Similar biosynthetic functions have been proposed for mammalian CA [13–15] but, to our knowledge, the nutritional requirements of CA-deficient mutants have hitherto not been fully investigated for any heterotrophic organism.
In the budding yeast Saccharomyces cerevisiae, CA is encoded by NCE103, a gene that was formerly reported to be involved in non-classical protein secretion [16]. Aerobic growth was also impaired in an S. cerevisiae nce103 null mutant, but could be restored by heterologous expression of CA-encoding genes from alfalfa (Msca1) or Homo sapiens (CAII) [17,18]. Furthermore, expression of the S. cerevisiae NCE103 gene in Escherichia coli yielded CA activity [18].
It was initially proposed that the aerobic growth defect of an S. cerevisiae nce103 null mutant reflected sensitivity to molecular oxygen [17]. However, a recent study [19] demonstrated that aerobic growth of an nce103 null mutant was possible when the air was enriched with CO2. We recently showed that NCE103 is transcriptionally down-regulated at high CO2 concentrations, irrespective of the oxygen concentration in the cultures [20]. This CO2-dependent transcriptional down-regulation of NCE103 and also of CA activity was recently confirmed [19]. High-throughput transcriptional studies revealed that NCE103 is induced by a variety of natural stresses, including high pH [21,22], as well as in respiratory-deficient mutants [23]. However, an nce103Δ mutant did not show sensitivity to growth at pH 7, the maximum pH value that enabled growth for both mutant and wild-type strains [17].
Despite the recent studies on the regulation of NCE103 and the phenotype of null mutants, the physiological role of CA in S. cerevisiae remains unclear. Therefore the aim of the present study was to determine the biochemical basis for the observed CO2 requirement of CA-deficient yeast strains. Moreover, we investigated whether this CO2 requirement merely occurs during the start-up phase of batch cultures, where metabolically generated CO2 is not sufficient to compensate for the high CO2 requirement of the mutant [9,24], or whether it also occurs during steady-state growth under conditions where fermentative sugar metabolism leads to sustained endogenous CO2 production.
EXPERIMENTAL
Strains and maintenance
The S. cerevisiae strain CEN.PK113-7D (MATa MAL2-8c SUC2) was kindly provided by Dr P. Kötter (der Johann Wolfgang Goethe-Universität Frankfurt, Frankfurt, Germany). It was maintained as 2 ml aliquots of frozen stock cultures in YPD medium (1%, w/v, yeast extract, 2%, w/v, peptone and 2%, w/v, glucose) supplemented with 30% (v/v) glycerol. One aliquot was used to inoculate precultures (100 ml) for chemostats and shake-flask cultivations.
Construction of an nce103Δ deletion strain
Construction of an NCE103 disruptant in the auxotrophic strain CEN.PK113-7D was carried out by PCR-targeting with the kanMX4 module, flanked by short homology regions of the gene [25]. The disruption cassette was obtained by amplifying the KanMX4 module from the plasmid pUG6 [25] with the oligonucleotides NCE103.D-F (5′-ATCAACTACAGCTAAGACTACAAATTTCAATTATTACACATCAGCAGCGAAGCTTCGTACGC-3′) and NCE103.D-R (5′-TTCTATTTCAATGAATATTATATAAGTATATCGGTGAGGCTAAAAGCATAGGCCACTAGTGGATCTG-3′). PCR was performed with Expand High Fidelity PCR system (Roche, Indianapolis, IN, U.S.A.) following the programme: 94 °C for 5 min; 30 cycles of 94 °C for 30 s, 58 °C for 1 min and 72 °C for 3 min and a final step of 72 °C for 7 min. Amplicons were purified with the GenElute kit (Sigma). Exponentially growing YPD cultures of the strain CEN.PK113-7D were transformed with 5 μg of the purified DNA cassette [26]. The recovery incubation after the heat shock was performed in 25 ml screw-cap tubes flushed with CO2 for 5 min (HoekLoos, Schiedam, The Netherlands), and the selection was on YPD plus G418 plates inside an airtight jar that contained an atmosphere with less than 0.1% O2 and 18% CO2 (Anaerocult A system; Merck, Darmstadt, Germany). Verification of correct NCE103 replacement was carried out by diagnostic PCR. The oligonucleotides used were NCE103.TEST-F (5′-TTTCTAACCATCACCCCGC-3′) and KanA (5′-CGCACGTCAAGACTGTCAAG-3′), for testing the correct insertion of the KanMX4 module in the 5′ flank of NCE103, and KanB (5′-TCGTATGTGAATGCTGGTCG-3′) and NCE103.TEST-R (5′-GGATCATGTGCCTTTGCC-3′), for the 3′ flank.
Batch and chemostat cultivation
Batch cultivations were performed in 250 ml shake flasks at 30 °C and 200 rev./min in orbital incubators. Cells were grown in a synthetic medium with (NH4)2SO4 as nitrogen source [27], supplemented with 20 g·l−1 glucose. When L-aspartic acid was used as the nitrogen source, it was added at 30 mM (final concentration) and the sulphate concentration was adjusted by addition of K2SO4 (38 mM). All media were adjusted to pH 6.5 with 2 M KOH.
Chemostat cultivation was performed at 30 °C in laboratory fermenters (Applikon Biotechnology, Schiedam, The Netherlands) with a working volume of 1 litre as described previously [28]. Cultures were fed with the same media as described above, but the glucose concentration was lowered to 7.5 g·l−1 to ensure glucose-limited growth [29]. The dilution rate was set to 0.10 h−1. The pH was measured on line and kept constant at 5.0 by the automatic addition of 2 M KOH with the use of an Applikon ADI 1030 biocontroller. Stirrer speed was 800 rev./min and the gas flow was 0.5 litre·min−1. For anaerobic cultivation, ergosterol (10 mg·l−1) and Tween 80 (420 mg·l−1) were added, and the medium vessel was flushed with N2. Cultures were sparged either with air, N2, CO2 or with a mixture containing 79% N2 and 21% O2 (HoekLoos, Schiedam, The Netherlands). Cultures were monitored as described previously [20]. In CO2-enriched cultures, respiratory quotients could not be calculated due to the absence of nitrogen (used as reference gas). Therefore oxygen consumption was estimated by assuming a 100% carbon balance.
Analysis of nutritional requirements on agar plates
For growth on plates, precultures grown at elevated CO2 concentrations were washed with water and adjusted to A600=0.2. Then, 5 μl was spotted on to agar plates (2%) of the indicated media and extended across a 9 cm2 surface with an inoculation loop. Spotted dilution experiments [30] were not possible due to a hydrophobic layer formed on top of fatty-acid-supplemented plates. Plates were incubated at 30 °C for 3–5 days under a normal air atmosphere or, instead, in a jar with an atmosphere consisting of 5–7% O2 and 7–9% CO2 (Anaerocult C system; Merck). Where indicated, the synthetic medium was supplemented with the following nutritional complements: adenine (20 mg·l−1), uracil (20 mg·l−1), L-arginine (20 mg·l−1) and a fatty acid mixture containing myristic, stearic and palmitic acids (30 mg·l−1 each) and Tween 80 (10 g·l−1) as surfactant. Adenine, uracil and L-arginine were added from filter-sterilized concentrated stock solutions [31]. The fatty acid mixture was added from a 100-fold-concentrated stock solution prepared under sterile conditions.
RESULTS
Metabolic CO2 cannot fulfil the inorganic carbon demand of an nce103 null mutant
S. cerevisiae mutants lacking CA activity (encoded by NCE103) require a CO2-enriched atmosphere for growth [17]. This CO2 requirement is independent of the availability of oxygen [19]. Similar CO2 requirements have been reported for several CA-deficient bacteria [10–12] and even for wild-type E. coli [24].
Attempts to start up chemostat cultures of the nce103Δ strain by sparging with air (aerobic cultures) or nitrogen gas (anaerobic cultures) were unsuccessful. However, glucose-limited chemostat cultures of nce103Δ grown in CO2-enriched chemostat cultures (sparged either with a mixture containing 79% CO2 and 21% O2 or with pure CO2) exhibited similar biomass yields as corresponding cultures of the parental strain CEN.PK113-7D (NCE103) (Table 1). Apparently, as previously demonstrated in shake-flask cultures [19], elevated CO2 concentrations are sufficient to sustain glucose-limited growth of nce103Δ S. cerevisiae in glucose-limited chemostats.
Table 1. Biomass yields and biomass specific rates of alcoholic fermentation (qethanol) of the S. cerevisiae strain CEN.PK113-7D (wild-type) and the isogenic mutant nce103Δ in aerobic and anaerobic chemostat cultures sparged (0.5 litre·min−1) with high concentrations of CO2.
Data represent the means±S.D. for two independent chemostat cultures.
| Genotype | Sparging gas | Biomass yield (gbiomass/gglucose) | qethanol (mmol·h−1·g−1) |
|---|---|---|---|
| Wild-type | 79% CO2+21% O2 | 0.38±0.05 | 0.0±0.0 |
| nce103Δ | 79% CO2+21% O2 | 0.33±0.018 | 0.0±0.0 |
| Wild-type | CO2 | 0.09±0.004 | 10.4±1.1 |
| nce103Δ | CO2 | 0.09±0.00 | 10.0±0.1 |
In some studies, the CO2 requirement of CA-deficient micro-organisms has been interpreted in terms of growth initiation: at low inoculum densities and in lag-phase cultures, endogenous CO2 production may be insufficient to build up the critical level required for growth [32,33]. Consequently, in the absence of CA, the threshold HCO3− concentration required to ‘spark’ growth is not reached. This model is supported by the observation that high-density inocula relieved the CO2 requirement of a CA-deficient mutant of Ralstonia eutropha [9] and shortened the lag phase of E. coli in minimal medium [32].
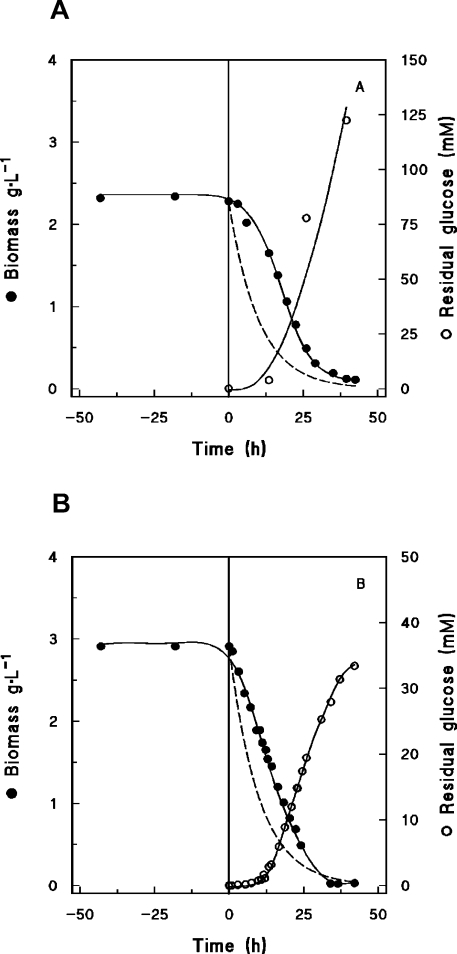
To investigate whether the phenotype of S. cerevisiae nce103 null mutant is due to a ‘sparking’ effect of CO2, chemostat cultures sparged with a high concentration of CO2 were switched to nitrogen gas (anaerobic cultures) or air (aerobic cultures). In anaerobic as well as aerobic cultures, this switch resulted in termination of growth, as evident from the wash-out of the biomass and the increase in the residual glucose concentration (Figure 1). This indicates that, even in cultures that are vigorously respiring or fermenting, the inorganic carbon requirement of nce103Δ S. cerevisiae cannot be met by endogenous CO2. We also observed a delay between the biomass decay and the theoretical wash-out curve (Figure 1), an effect that probably responded to the time needed to replace all the dissolved CO2 in the culture broth by nitrogen or air.
Figure 1. Anaerobic (100% CO2) (A) and aerobic (79% CO2/21% O2 mixture) (B) chemostat cultivations of S. cerevisiae nce103Δ null mutant.
At zero time (vertical line), the gas supply was switched to 100% N2 (A) or air (B). ●, biomass (dry weight); ○, residual glucose concentration; -----, theoretical wash-out curve. Independent replicate experiments gave identical results.
Nutritional complementation of nce103 null mutants at atmospheric CO2 pressures
The CO2 requirement of CA-deficient microorganisms has led to the suggestion that the enzyme may be needed to provide bicarbonate for heterotrophic carboxylation reactions [10,18]. Indeed, non-identified CO2-requiring mutants of several microbial species could be nutritionally complemented by addition of TCA (tricarboxylic acid) cycle intermediates, amino acids, purines and other metabolites whose biosynthesis require HCO3−-dependent carboxylations [33]. However, addition of these compounds did not restore growth at atmospheric CO2 partial pressures of several CA-deficient bacterial strains [9,12,24].
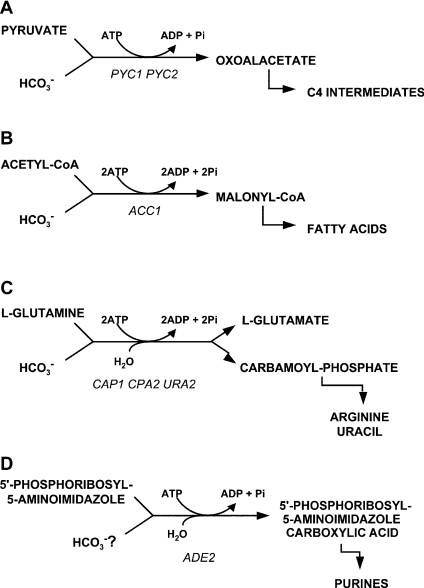
Before experimentally addressing the nutritional complementation of S. cerevisiae nce103Δ strains, we made an inventory of possible bicarbonate-dependent carboxylations during growth on glucose (Scheme 1). The two pyruvate carboxylases (Scheme 1A), encoded by the PYC1 and PYC2 genes, are exclusively responsible for replenishing the TCA cycle intermediates used for biosynthesis during growth of S. cerevisiae on sugars [34,35]. Addition of L-aspartate can nutritionally complement pyc1 pyc2 null mutants by providing an alternative source of C4-compounds [35]. Acetyl-CoA carboxylase (Scheme 1B), encoded by the ACC1 gene [36], provides malonyl-CoA required for fatty acid biosynthesis. Acetyl-CoA carboxylase-deficient mutants can be nutritionally complemented by addition of long-chain (C14–C16) unsaturated fatty acids [36,37]. CPSase (carbamoyl-phosphate synthetase; Scheme 1C) catalyses the HCO3−-dependent formation of CP from glutamine. CP is a precursor for L-citrulline, L-arginine and pyrimidines. Consequently, S. cerevisiae mutants that are unable to synthesize CP require either L-arginine or L-arginine and uracil for growth [38]. A final reaction that was considered is the carboxylation of AIR (5′-phosphoribosyl-5-aminoimidazole), a step in the pathway of purine biosynthesis (Scheme 1D). It is unclear if the substrate for AIR carboxylation is HCO3− or CO2. However, in adenine-deficient mutants unable to catalyse this reaction, adenine can be substituted by gassing the cultures with 30% CO2 [39]. We therefore decided to also take into account a possible purine requirement of nce103Δ S. cerevisiae.
Scheme 1. Metabolic reactions involving inorganic carboxylations in S. cerevisiae (see text).
(A) Pyruvate carboxylase (EC 6.4.1.1), (B) acetyl-CoA carboxylase (EC 6.3.4.14), (C) CPSase (EC 6.3.5.5) and (D) AIR carboxylase (EC 4.1.1.21). The genes encoding for the different enzymes are depicted in italics.
To investigate whether, at atmospheric CO2 pressures, Nce103p is required to provide bicarbonate for any or all of the processes mentioned above, the prototrophic reference strain S. cerevisiae CEN.PK113-7D (NCE103) and the isogenic nce103Δ mutant were tested for aerobic growth on glucose synthetic medium plates supplemented with all possible combinations of the end-products corresponding to the pathways mentioned above: L-aspartic acid (used as nitrogen source), long-chain fatty acids, L-arginine, uracil and adenine (see the Experimental section). Growth of the reference strain was not affected by the addition of these additional nutrients. However, under atmospheric pressure, good growth of the nce103Δ mutant was observed only when L-aspartate, fatty acids, L-arginine and uracil were all present (Figure 2, panel 31). The additional supply of adenine did not have a clear influence (Figure 2, panel 32), indicating that purine biosynthesis is not affected during growth of the nce103Δ strain at atmospheric CO2 pressure. Very slow residual growth was observed with L-aspartate, fatty acids and L-arginine (i.e. without inclusion of uracil; Figure 2, panel 15).
Figure 2. Aerobic growth of the prototrophic S. cerevisiae reference strain CEN.PK113-7D (wt; lower) and the isogenic mutant nce103Δ (upper) in MM-glucose plus different nutritional supplements.
Stationary phase precultures were diluted to A600=0.2, plated and incubated at 30 °C for 5 days. For each panel, the included components are indicated by closed squares. Asp, L-aspartic acid used as nitrogen source; FA, fatty acids; ADE, adenine; URA, uracil; Arg, L-arginine (see the Experimental section).
DISCUSSION
In previous studies, CO2 dependence of nce103Δ mutants was shown in aerobic, but not in anaerobic cultures [17,19]. However, in those studies, anaerobic growth was investigated in closed systems, from which CO2 produced during fermentation could not escape, thus leading to a CO2-enriched environment [17]. Our experiments with anaerobic chemostat cultures demonstrate that CA is essential for growth in anaerobic cultures sparged with nitrogen gas. Apparently, even in these vigorously fermenting yeast cells (fermentation rates of 10 mmol·g−1·h−1 [40]), loss of CO2 from the cells via diffusion is too fast to allow for a sufficient rate of spontaneous intracellular formation of bicarbonate. These observations show that, in S. cerevisiae, absence of CA yeast does not merely result in a requirement for ‘sparking’ amounts of CO2, as reported for many microorganisms [24,32,33,41], but instead causes a true CO2 requirement for steady-state growth at atmospheric CO2 pressures.
Although biosynthetic bicarbonate requirements have often been implicated in the phenotype of CA-deficient microorganisms [10,18,24], we are not aware of previous studies in which the biosynthetic requirements of a CA-negative microorganism have been fully determined. We have demonstrated that the requirement of CA-deficient S. cerevisiae for elevated CO2 concentrations originates from three bicarbonate-dependent carboxylation reactions: pyruvate decarboxylase, acetyl-CoA carboxylase and CPSase (Scheme 1 and Figure 2). CA-deficient mutants could only be grown at atmospheric CO2 partial pressures when products of the pathways in which these three enzymes participate were added to synthetic growth media (Figure 2). These observations demonstrate that, as long as cultures are not enriched with CO2, yeast CA is a key biosynthetic enzyme during growth on glucose.
S. cerevisiae nce103Δ strains showed residual growth in the absence of uracil, but not in the absence of L-arginine (Figure 2). In S. cerevisiae, as well as in many other organisms, there are two enzymes for CP synthesis: CPSase A, which is primarily regulated based on L-citrulline and L-arginine status of the cells, and CPSase P, which is mainly regulated by pyrimidine requirement [38]. However, CP from the two enzymes can be used in both pathways [38]. Therefore the residual growth of the nce103Δ strain in the absence of uracil may reflect a low rate of CP biosynthesis driven by spontaneous bicarbonate formation, combined with a higher affinity of pyrimidine biosynthesis for CP [42]. CA-deficient mutants did not show an adenine auxotrophy for aerobic growth (Figure 2), suggesting that the carboxylation of AIR may depend on molecular CO2 rather than on HCO3−.
Even after ‘full’ nutritional supplementation (i.e. addition of L-aspartic acid, fatty acids, uracil and L-arginine), growth of nce103 null mutants was slower than that of the reference strain. We observed a similar reduction in specific growth rate when acc1 mutants (lacking the main acetyl-CoA carboxylase in S. cerevisiae [43]) were supplemented with fatty acids (results not shown). This suggests that the reduced growth rate of nutritionally supplemented nce103 mutants was due to limitations in fatty acid uptake or processing. However, these observations should be interpreted with some caution as a mutant lacking the whole ACC1 gene could not be rescued by fatty acid supplementation, indicating that its role extends beyond fatty acid synthesis [44].
The contribution of CA to growth will depend on the growth substrates. For example, nce103 mutants did not require L-aspartic acid for growth on ethanol (results not shown). This result was anticipated, since the glyoxylate cycle rather than pyruvate carboxylase is responsible for the synthesis of C4-compounds during growth on ethanol and acetate. CA can be anticipated to play a key role when urea is the nitrogen source. Urea is metabolized to NH3 and CO2 in a carboxylation reaction catalysed by bicarbonate-dependent urea carboxylase [45]. Lack of NCE103 might therefore lead to insufficient HCO3− supply for urea utilization. This possibility was not followed up in detail as we anticipated interference of the presence of the nitrogen sources L-aspartate and L-arginine in the growth media.
The metabolic role of yeast CA can probably be extended to other organisms. CO2-requiring mutants (called ‘CO2 mutants’) of E. coli, Neurospora crassa and other microorganisms have been known for a long time [35]. In some of these mutants, addition of TCA-cycle intermediates, amino acids or other compounds could replace CO2 [46]. The yeast Cyniclomyces guttulatus (formerly Saccharomycopsis guttulatus), which occurs in the digestive tract of rabbit, mouse and other animals, also requires CO2 for growth [47]. Remarkably, this requirement can be circumvented by the use of an enriched growth medium [48]. On the basis of our results, we speculate that evolution in CO2-rich environments such as animal intestines may have yielded microorganisms that are naturally CA-deficient.
Studies with the green alga Chlamydomonas reinhardtii showed a correlation between CA gene expression and supply of NH4+, a nitrogen source that stimulates anaplerosis [49]. In addition, previous studies showed that exposure of mammalian cells to acetazolamide and other CA inhibitors led to a decrease in CP synthesis [13] and reduced carbon flux towards lipids and TCA cycle intermediates [14,15]. In addition to its proposed role in pH control, the biosynthetic role of CA may be relevant in cancer cells, in which CA is highly expressed [4]. In this respect, it is of interest to note that glucose dissimilation in many tumours occurs predominantly via homolactate fermentation [50] and thus does not lead to the production of large amounts of CO2.
Acknowledgments
The research group of J.T.P. is part of the Kluyver Centre for Genomics of Industrial Fermentation (Delft, The Netherlands), which is supported by The Netherlands Genomics Initiative. We thank our Kluyver Centre colleagues Dr J.-M. Daran for many helpful discussions, and M. Luttik and E. de Hulster for technical support. J.A. is supported by an EU Marie Curie postdoctoral fellowship.
References
- 1.Jones R. P., Greenfield P. F. Effect of carbon dioxide on yeast growth and fermentation. Enzyme Microb. Technol. 1982;4:210–223. [Google Scholar]
- 2.Liljas A., Laurberg M. A wheel invented three times. The molecular structures of the three carbonic anhydrases. EMBO Rep. 2000;1:16–17. doi: 10.1093/embo-reports/kvd016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tripp B. C., Smith K., Ferry J. G. Carbonic anhydrase: new insights for an ancient enzyme. J. Biol. Chem. 2001;276:48615–48618. doi: 10.1074/jbc.R100045200. [DOI] [PubMed] [Google Scholar]
- 4.Potter C. P., Harris A. L. Diagnostic, prognostic and therapeutic implications of carbonic anhydrases in cancer. Br. J. Cancer. 2003;89:2–7. doi: 10.1038/sj.bjc.6600936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dodgson S. J. New York: Plenum Press; 1991. The Carbonic Anhydrases; Cellular Physiology and Molecular Genetics. [Google Scholar]
- 6.Chegwidden W. R., Carter N. D., Edwards Y. H. Basel: Birkhauser; 2000. The Carbonic Anhydrases: New Horizons. [Google Scholar]
- 7.Sultemeyer D., Schmidt C., Fock H. P. Carbonic anhydrases in higher plants and aquatic microorganisms. Physiol. Plant. 1993;88:179–190. [Google Scholar]
- 8.Tsuzuki M., Miyachi S. The function of carbonic anhydrase in aquatic photosynthesis. Aquat. Bot. 1989;34:85–104. [Google Scholar]
- 9.Kusian B., Sultemeyer D., Bowien B. Carbonic anhydrase is essential for growth of Ralstonia eutropha at ambient CO2 concentrations. J. Bacteriol. 2002;184:5018–5026. doi: 10.1128/JB.184.18.5018-5026.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merlin C., Masters M., McAteer S., Coulson A. Why is carbonic anhydrase essential to Escherichia coli? J. Bacteriol. 2003;185:6415–6424. doi: 10.1128/JB.185.21.6415-6424.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashimoto M., Kato J. Indispensability of the Escherichia coli carbonic anhydrases YadF and CynT in cell proliferation at a low CO2 partial pressure. Biosci. Biotech. Biochem. 2003;67:919–922. doi: 10.1271/bbb.67.919. [DOI] [PubMed] [Google Scholar]
- 12.Mitsuhashi S., Ohnishi J., Hayashi M., Ikeda M. A gene homologous to beta-type carbonic anhydrase is essential for the growth of Corynebacterium glutamicum under atmospheric conditions. Appl. Microbiol. Biotechnol. 2004;63:592–601. doi: 10.1007/s00253-003-1402-8. [DOI] [PubMed] [Google Scholar]
- 13.Dodgson S. J., Forster R. E., Schwed D. A., Storey B. T. Contribution of matrix carbonic-anhydrase to citrulline synthesis in isolated guinea-pig liver-mitochondria. J. Biol. Chem. 1983;258:7696–7701. [PubMed] [Google Scholar]
- 14.Lynch C. J., Fox H., Hazen S. A., Stanley B. A., Dodgson S., Lanoue K. F. Role of hepatic carbonic anhydrase in de novo lipogenesis. Biochem. J. 1995;310:197–202. doi: 10.1042/bj3100197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hazen S. A., Waheed A., Sly W. S., Lanoue K. F., Lynch C. J. Differentiation-dependent expression of CA V and the role of carbonic anhydrase isozymes in pyruvate carboxylation in adipocytes. FASEB J. 1996;10:481–490. doi: 10.1096/fasebj.10.4.8647347. [DOI] [PubMed] [Google Scholar]
- 16.Cleves A. E., Cooper D. N., Barondes S. H., Kelly R. B. A new pathway for protein export in Saccharomyces cerevisiae. J. Cell Biol. 1996;133:1017–1026. doi: 10.1083/jcb.133.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Götz R., Gnann A., Zimmermann F. K. Deletion of the carbonic anhydrase-like gene NCE103 of the yeast Saccharomyces cerevisiae causes an oxygen-sensitive growth defect. Yeast. 1999;15:855–864. doi: 10.1002/(SICI)1097-0061(199907)15:10A<855::AID-YEA425>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 18.Clark D., Rowlett R. S., Coleman J. R., Klessig D. F. Complementation of the yeast deletion mutant DeltaNCE103 by members of the beta class of carbonic anhydrases is dependent on carbonic anhydrase activity rather than on antioxidant activity. Biochem. J. 2004;379:609–615. doi: 10.1042/BJ20031711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amoroso G., Morell-Avrahov L., Müller D., Klug K., Sültemeyer D. The gene NCE103 (YNL036w) from Saccharomyces cerevisiae encodes a functional carbonic anhydrase and its transcription is regulated by the concentration of inorganic carbon in the medium. Mol. Microbiol. 2005;56:549–558. doi: 10.1111/j.1365-2958.2005.04560.x. [DOI] [PubMed] [Google Scholar]
- 20.Aguilera J., Petit T., de Winde J. H., Pronk J. T. Physiological and genome-wide transcriptional responses of Saccharomyces cerevisiae to high carbon dioxide concentrations. FEMS Yeast Res. 2005;5:579–593. doi: 10.1016/j.femsyr.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., Brown P. O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Causton H. C., Ren B., Koh S. S., Harbison C. T., Kanin E., Jennings E. G., Lee T. I., True H. L., Lander E. S., Young R. A. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Epstein C. B., Waddle J. A., Hale W., Dave V., Thornton J., Macatee T. L., Garner H. R., Butow R. A. Genome-wide responses to mitochondrial dysfunction. Mol. Biol. Cell. 2001;12:297–308. doi: 10.1091/mbc.12.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozliak E. I., Fuchs J. A., Guilloton M. B., Anderson P. M. Role of bicarbonate CO2 in the inhibition of Escherichia coli growth by cyanate. J. Bacteriol. 1995;177:3213–3219. doi: 10.1128/jb.177.11.3213-3219.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wach A., Brachat A., Pohlmann R., Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 26.Guldener U., Heck S., Fiedler T., Beinhauer J., Hegemann J. H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verduyn C., Postma E., Scheffers W. A., van Dijken J. P. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 28.Van den Berg M. A., Jong-Gubbels P., Kortland C. J., van Dijken J. P., Pronk J. T., Steensma H. Y. The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties and transcriptional regulation. J. Biol. Chem. 1996;271:28953–28959. doi: 10.1074/jbc.271.46.28953. [DOI] [PubMed] [Google Scholar]
- 29.Boer V. M., de Winde J. H., Pronk J. T., Piper M. D. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J. Biol. Chem. 2003;278:3265–3274. doi: 10.1074/jbc.M209759200. [DOI] [PubMed] [Google Scholar]
- 30.Aguilera J., Rodriguez-Vargas S., Prieto J. A. The HOG MAP kinase pathway is required for the induction of methylglyoxal-responsive genes and determines methylglyoxal resistance in Saccharomyces cerevisiae. Mol. Microbiol. 2005;56:228–239. doi: 10.1111/j.1365-2958.2005.04533.x. [DOI] [PubMed] [Google Scholar]
- 31.Burke D., Dawson D., Stearns T. Plainview, NY: Cold Spring Harbor Laboratory Press; 2000. Methods in Yeast Genetics. [Google Scholar]
- 32.Neidhardt F. C., Bloch P. L., Smith D. F. Culture medium for enterobacteria. J. Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Repaske R., Repaske A. C., Mayer R. D. Carbon dioxide control of lag period and growth of Streptococcus sanguis. J. Bacteriol. 1974;117:652–659. doi: 10.1128/jb.117.2.652-659.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stucka R., Dequin S., Salmon J. M., Gancedo C. DNA sequences in chromosome II and chromosome VII code for pyruvate carboxylase isoenzymes in Saccharomyces cerevisiae – analysis of pyruvate carboxylase-deficient strains. Mol. Gen. Genet. 1991;229:307–315. doi: 10.1007/BF00272171. [DOI] [PubMed] [Google Scholar]
- 35.Brewster N. K., Val D. L., Walker M. E., Wallace J. C. Regulation of pyruvate-carboxylase isozyme (Pyc1, Pyc2) gene-expression in Saccharomyces cerevisiae during fermentative and nonfermentative growth. Arch. Biochem. Biophys. 1994;311:62–71. doi: 10.1006/abbi.1994.1209. [DOI] [PubMed] [Google Scholar]
- 36.Roggenkamp R., Numa S., Schweizer E. Fatty acid-requiring mutant of Saccharomyces cerevisiae defective in acetyl-CoA carboxylase. Proc. Natl. Acad. Sci. U.S.A. 1980;77:1814–1817. doi: 10.1073/pnas.77.4.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schweizer E., Bolling H. A Saccharomyces cerevisiae mutant defective in saturated fatty acid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 1970;67:660–666. doi: 10.1073/pnas.67.2.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lacroute F., Pierard A., Grenson M., Wiame J. M. Biosynthesis of carbamoyl phosphate in Saccharomyces cerevisiae. J. Gen. Microbiol. 1965;40:127–142. doi: 10.1099/00221287-40-1-127. [DOI] [PubMed] [Google Scholar]
- 39.Woods R. A. Response of ad-2 mutants of Saccharomyces cerevisiae to carbon dioxide. Mol. Gen. Genet. 1969;105:314–316. doi: 10.1007/BF00277586. [DOI] [PubMed] [Google Scholar]
- 40.Tai S. L., Boer V. M., Daran-Lapujade P., Walsh M. C., de Winde J. H., Daran J. M., Pronk J. T. Two-dimensional transcriptome analysis in chemostat cultures. Combinatorial effects of oxygen availability and macronutrient limitation in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:437–447. doi: 10.1074/jbc.M410573200. [DOI] [PubMed] [Google Scholar]
- 41.Repaske R., Clayton M. A. Control of Escherichia coli growth by CO2. J. Bacteriol. 1978;135:1162–1164. doi: 10.1128/jb.135.3.1162-1164.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Penverne B., Belkaid M., Herve G. In situ behavior of the pyrimidine pathway enzymes in Saccharomyces cerevisiae. 4. The channeling of carbamylphosphate to aspartate transcarbamylase and its partition in the pyrimidine and arginine pathways. Arch. Biochem. Biophys. 1994;309:85–93. doi: 10.1006/abbi.1994.1089. [DOI] [PubMed] [Google Scholar]
- 43.Mishina M., Roggenkamp R., Schweizer E. Yeast mutants defective in acetyl-coenzyme-A carboxylase and biotin-apocarboxylase ligase. Eur. J. Biochem. 1980;111:79–87. doi: 10.1111/j.1432-1033.1980.tb06077.x. [DOI] [PubMed] [Google Scholar]
- 44.Schneiter R., Hitomi M., Ivessa A. S., Fasch E. V., Kohlwein S. D., Tartakoff A. M. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol. Cell. Biol. 1996;16:7161–7172. doi: 10.1128/mcb.16.12.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitney P. A., Cooper T. G. Urea carboxylase and allophanate hydrolase. Two components of adenosine triphosphate:urea amido-lyase in Saccharomyces cerevisiae. J. Biol. Chem. 1972;247:1349–1353. [PubMed] [Google Scholar]
- 46.Charles H. P., Roberts G. A. Carbon dioxide as a growth factor for mutants of Escherichia coli. J. Gen. Microbiol. 1968;51:211–224. doi: 10.1099/00221287-51-2-211. [DOI] [PubMed] [Google Scholar]
- 47.Buecher E. J., Phaff H. J. Growth of Saccharomycopsis Schionning under continuous gassing. J. Bacteriol. 1970;104:133–137. doi: 10.1128/jb.104.1.133-137.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zierdt C. H., Detlefson C., Muller J., Waggie K. S. Cyniclomyces guttulatus (Saccharomycopsis guttulata). culture, ultrastructure and physiology. Antonie Van Leeuwenhoek. 1988;54:357–366. doi: 10.1007/BF00393526. [DOI] [PubMed] [Google Scholar]
- 49.Giordano M., Norici A., Forssen M., Eriksson M., Raven J. A. An anaplerotic role for mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol. 2003;132:2126–2134. doi: 10.1104/pp.103.023424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holm H., Staedt E., Schlickeiser G., Gunther H. J., Leweling H. Substrate balances across colonic carcinomas in humans. Cancer Res. 1995;55:1373–1378. [PubMed] [Google Scholar]