Abstract
Neisseria gonorrhoeae is a strict human pathogen that invades and colonizes the urogenital tracts of males and females. Lipooligosaccharide (LOS) has been shown to play a role in gonococcal pathogenesis. The acyl transferase MsbB is involved in the biosynthesis of the lipid A portion of the LOS. In order to determine the role of an intact lipid A structure on the pathogenesis of N. gonorrhoeae, the msbB gene was cloned and sequenced, a deletion and insertion mutation was introduced into N. gonorrhoeae, and the mutant strain was designated 1291A11K3. Mass spectrometric analyses of 1291A11K3 LOS determined that this mutation resulted in a pentaacyl rather than a hexaacyl lipid A structure. These analyses also demonstrated an increase in the phosphorylation of lipid A and an increase in length of the oligosaccharide of a minor species of the msbB LOS. The interactions of this mutant with male urethral epithelial cells (uec) were examined. Transmission and scanning electron microscopy studies indicated that the msbB mutants formed close associations with and were internalized by the uec at levels similar to those of the parent strain. Gentamicin survival assays performed with 1291A11K3 and 1291 bacteria demonstrated that there was no difference in the abilities of the two strains to adhere to uec; however, significantly fewer 1291A11K3 bacteria than parent strain bacteria were recovered from gentamicin-treated uec. These studies suggest that the lipid A modification in the N. gonorrhoeae msbB mutant may render it more susceptible to innate intracellular killing mechanisms when internalized by uec.
Neisseria gonorrhoeae is a strict human pathogen, which invades and colonizes the epithelia of the urogenital tracts of both males and females (2, 12, 18). The lipooligosaccharide (LOS) of N. gonorrhoeae has been shown to play a role in the pathogenesis of human infections (19, 40). The LOS is composed of three major components: the oligosaccharide chain extensions, the core region, and lipid A. The oligosaccharide extensions of the LOS contain determinants which resemble human glycosphingolipid antigens that play a role in molecular mimicry. Studies have also shown that the oligosaccharide region of the LOS can be involved in receptor-mediated interactions (17, 19, 37). Lipid A of N. gonorrhoeae is similar in structure to lipid A from other gram-negative bacteria (31, 44, 46).
htrB (alternatively lpxL or waaM) is involved in the biosynthesis of lipid A of lipopolysaccharides (LPSs) and LOSs. HtrB is one of the 2-keto-3-deoxyoctulosonic acid (Kdo)-dependent acyl transferases responsible for the addition of a secondary acyl substitution on the lipid A portion of LPS or LOS. The htrB gene has been well characterized for Escherichia coli, Haemophilus influenzae, and Salmonella enterica serovar Typhimurium (6, 36, 44). Mutations in this gene have been shown to have a number of effects on the organism. One such effect is temperature sensitivity. E. coli, H. influenzae, and S. enterica serovar Typhimurium htrB mutants have all been shown to be initially sensitive to temperatures above 32.5°C (28, 31, 44). A number of different genes are able to subsequently suppress temperature sensitivity (27, 29) (D. M. B. Post and M. A. Apicella, unpublished data). msbB (alternatively lpxM or waaN) has been identified as one of these suppressors (27). Work done with E. coli has shown that MsbB is a late-functioning acyl transferase, which functions optimally after laurate incorporation by HtrB onto the E. coli KDO2-lipid IVA structure (9). Therefore, secondary acyl substitutions of the lipid A structure are thought to occur in a sequential manner.
Chemical analyses of LPS and LOS isolated from the serovar Typhimurium and H. influenzae htrB mutants demonstrated that the lipid A portion of LPS or LOS was modified (31, 44). Additional studies showed that LPS or LOS purified from these htrB mutants was reduced in its toxicity compared to LPS or LOS isolated from the parent strains (26, 36). Animal model studies using H. influenzae and serovar Typhimurium htrB mutants demonstrated that these organisms were less virulent than the parent strains (11, 26).
In this study, we investigated the role of an intact lipid A structure on the pathogenesis of N. gonorrhoeae. To perform these investigations, we created an msbB lipid A mutant of N. gonorrhoeae strain 1291. This mutation resulted in a pentaacyl rather than the hexaacyl lipid A structure found in the wild-type strain. The interactions of this mutant with male urethral epithelial cells (uec) were examined. These studies suggest that the lipid A modification in the N. gonorrhoeae msbB mutant may render it more susceptible to killing when internalized by the uec.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
All bacterial strains and plasmids used in this study are described in Table 1. E. coli strains were grown in Luria-Bertani (LB) medium at 37, 32, or 30°C and supplemented with appropriate antibiotics. For growth curves, an overnight culture was used to inoculate fresh medium. Cells were grown at 37°C with agitation, and readings were taken every 30 min. N. gonorrhoeae and Neisseria meningitidis organisms were grown on gonococcal agar (GCA) (Becton Dickinson, Sparks, Md.) supplemented with 1% IsoVitaleX or on brain heart infusion (BHI) agar (Becton Dickinson) supplemented with 2.5% heat-inactivated fetal calf serum (FCS) at 37°C in 5% CO2. Liquid cultures of N. gonorrhoeae were grown in BHI broth supplemented with 2.5% FCS or in gonococcal broth supplemented with 1% IsoVitaleX at 37°C. Kanamycin-resistant N. gonorrhoeae isolates were grown on supplemented BHI agar plates or in supplemented BHI broth with 50 μg of kanamycin per ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype, relevant phenotype, or selection marker | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F− φ80d lacZΔM15 Δ(lacZYA-argF)U169 deoR redA1 endA1 hsdR17(rk− mk+) phoAsupE44 λ thi-1 gyrA96 relA1 | Life Technologies |
| E. coli MLK2 | W3110 galE sup+ | Strain B178 (28) |
| E. coli MLK217 | MLK2 htrB1::mini Tn10 | 28 |
| E. coli MLK217A11 | MLK217, pNMBA11pUC19, ampicillin, tetracycline | This study |
| N. meningitidis strain NMB | Wild-type serogroup B | 42 |
| N. gonorrhoeae strain 1291 | Wild-type strain | 1 |
| N. gonorrhoeae strain 1291A11K3 | Kanamycin resistant, msbB mutant | This study |
| N. gonorrhoeae strain 24-1 | Serum-sensitive N. gonorrhoeae strain | Peter Rice |
| N. gonorrhoeae strain PID2 | Utilized as molecular weight standard | Our collection |
| Plasmids | ||
| pUC19 | Ampicillin, cloning vector | New England Biolabs |
| pCR2.1 | Ampicillin, cloning vector | Invitrogen |
| pNMBA11 | Ampicillin, msbB PCR product in pCR2.1 | This study |
| pNMBA11pUC19 | Ampicillin, XbaI, HindIII restricted msbB PCR product in XbaI, HindIII restricted pUC19 | This study |
| pNMBA11K3 | Ampicillin, kanamycin, insertion and deletion msbB mutant in pUC19 | This study |
| pUC18K3 | Ampicillin, kanamycin, aphA-3 gene used for insertion and deletion mutation | 34 |
Recombinant DNA and transformation methods.
Restriction and modifying enzymes were purchased from New England Biolabs (Beverly, Mass.) and Promega (Madison, Wis.). Standard DNA recombinant protocols were performed as previously described (38). Transformation of E. coli with plasmid DNA was done using the CaCl2 method (16). Transformation of N. gonorrhoeae was performed as previously described (43).
Complementation of an E.coli htrB mutant.
An overnight culture of E. coli MLK217 cells was inoculated into 500 ml of fresh LB medium. Cells were grown at 30°C with vigorous agitation to an optical density at 600 nm (OD600) of 0.5. The cells were chilled briefly on ice and centrifuged at 2,200 × g for 20 min at 2°C. The cell pellet was washed with ice-cold water and centrifuged at 2,200 × g for 20 min at 2°C. The cell pellet was washed a second time, centrifuged as described above, and resuspended in an equal volume of sterile 10% (vol/vol) glycerol-water (4). Thirty microliters of the cells was electroporated (4 kV, 330 μF, low-resistance ohms, fast charge rate) with 1 μl of the pNMBA11pUC19 DNA by using a Cell Porater (Invitrogen, Carlsbad, Calif.). Then, cells were incubated in 1 ml of SOC medium at 30°C for 90 min with agitation and plated on LB agar plates containing tetracycline (20 μg/ml) and ampicillin (100 μg/ml). Cells were grown in a 37°C incubator overnight. Colonies that were able to grow at 37°C were then tested using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analyses.
DNA isolation.
Plasmid DNA was prepared with the QIAprep Spin Miniprep kit or the QIAprep Midiprep kit according to the manufacturer's instructions (Qiagen Inc., Valencia, Calif.). Chromosomal DNA was isolated using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, Minn.).
Southern blot analyses.
Hybridization experiments were carried out according to the manufacturer's protocols (Boehringer Mannheim Corp., Indianapolis, Ind.). All probes were labeled by using either PCR labeling or random labeling with digoxigenin-labeled deoxynucleoside triphosphates (Boehringer Mannheim Corp.).
DNA sequencing and analyses.
DNA sequencing reactions were performed by using dye terminator cycle sequencing chemistry with AmpliTaq DNA polymerase and FS enzyme (PE Applied Biosystems, Foster City, Calif.). The reactions were run on and analyzed with an Applied Biosystems model 373A stretch fluorescence automated sequencer at the University of Iowa DNA Facility. All primers were either commercially available or purchased from either Genosys Corporation (Aldrich, Milwaukee, Wis.) or IDT Technologies (Coralville, Iowa). Sequence analysis was performed by using Assembly LIGN, version 1.0 (Oxford Molecular Group Inc., Oxford, United Kingdom), MacVector (Oxford Molecular Group Inc.), and Wisconsin Package, version 10.0 (Genetics Computer Group, Madison, Wis.).
Cloning and mutagenesis of msbB gene.
The E. coli htrB gene sequence was used to search the N. gonorrhoeae strain FA1090 sequence at the University of Oklahoma website. The sequence that showed the highest homology to the E. coli gene (N. gonorrhoeae sequence bp 160985 to 162427) was used for the design of PCR primers. PCR amplification of this region was performed with N. meningitidis strain NMB genomic DNA and the primers gchtrB3 5"-CAACAGGCGGCGGTGGAACAG-3" and gchtrB4 5"-TTCGGCATCCACTCCCCTTTG-3". The 1,443-bp PCR product was cloned using the TA cloning vector pCR2.1 (Invitrogen), and this construct was designated pNMBA11. The XbaI and HindIII sites flanking the PCR product were used to clone the PCR fragment into XbaI- and HindIII-restricted pUC19 (New England Biolabs). This construct was ligated using T4 DNA ligase and subsequently transformed into E. coli DH5α cells (Invitrogen). This construct was designated pNMBA11pUC19 (Fig. 1). Restriction enzymes BclI and BssHII were used to make the deletion in the htrB gene, at positions 780 and 918 bp, respectively. These digests were done to completion, and 138 bp was removed from the msbB gene. The modifying enzyme T4 polymerase was used to blunt the BclI and BssHII ends. A kanamycin resistance cassette, encoded by the aphA-3 gene, was restricted with SmaI and gel purified using the QIAquick gel extraction kit (Qiagen). The modified pNMBA11pUC19 construct was ligated with the gel-purified aphA-3 gene and subsequently transformed into E. coli DH5α cells. This new construct was designated pNMBA11K3. The proper construct was confirmed with PCR and restriction enzyme digests. Plasmid DNA from pNMBA11K3 was used for the transformation with N. gonorrhoeae strain 1291.
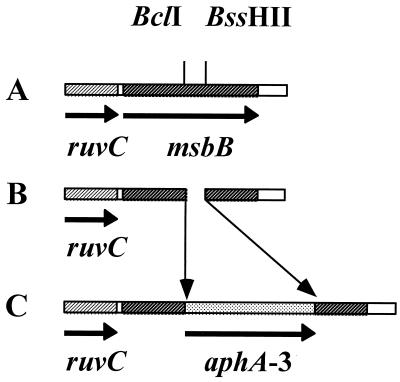
FIG. 1.
A deletion insertion mutant was made in the N. meningitidis msbB gene. (A) The N. meningitidis PCR product from the pNMBA11 plasmid was cloned into XbaI-HindIII-restricted pUC19. (B) A deletion was made in the msbB gene by restriction with BclI and BssHII. (C) The pNMBA11K3 plasmid was generated by ligating the kanamycin resistance gene, aphA-3, into the sites of deletion of the msbB gene.
SDS-PAGE analyses of LPS and LOS and immunoblot analyses of LOS.
LPS was isolated from E. coli bacteria by a modified proteinase K method. Cells were resuspended in phosphate-buffered saline (PBS) to an OD650 of 0.9. Cells were washed twice with PBS and resuspended in 200 μl of Buffer Part A (0.06 M Tris, 10 mM EDTA, 2.0% SDS [pH 6.8]). Samples were incubated in a 100°C water bath for 5 min. Then, 150 μl was transferred to a separate tube and 30 μl of a 2.5-mg/ml proteinase K solution was added. Samples were incubated at 37°C overnight. The sample was ethanol precipitated by adding 1/10 volume of 3 M sodium acetate and 2 volumes of ethanol. Samples were incubated at −20°C overnight. Samples were centrifuged at 16,000 × g for 5 min. The pellet was washed two times in 70% ethanol and dried overnight. LOS was isolated from 6 liters of BHI broth supplemented with 2.5% FCS for strain 1291 and 6 liters of BHI broth supplemented with 2.5% FCS and 50 μg of kanamycin per ml for strain 1291A11K3 by a modified hot-phenol-water preparation. Cells were centrifuged (∼2,500 × g), washed once with PBS, and centrifuged again (∼5,000 × g). The pellets were resuspended in 20 mM Tris MgCl2 and treated with lysozyme (final concentration, 0.2%) overnight on a tumbling rack. The samples were sonicated and treated with RNase (final concentration, 100 μg/ml) and DNase (40 U of micrococcal nuclease/50 ml) for 6 h on a tumbling rack at room temperature. SDS was added to each sample to a final concentration of 1%, and each sample was treated with proteinase K (10 μg/ml) overnight at 37°C. The sample was treated with pronase (10 μg/ml) for 6 h at 37°C. The samples were precipitated overnight at −20°C by adding 1/10 volume of 3 M sodium acetate and 3 volumes of ethanol. Next, each sample was centrifuged (3,200 × g), the pellet was resuspended in water, and the samples were spun again at high speed (17,000 × g). Then, the supernatants were reprecipitated with ethanol overnight at −20°C. The samples were centrifuged at low speed (3,200 × g), and the pellets were resuspended in water. Then, a traditional hot-phenol LOS extraction was performed on this material (24). The aqueous phases from this extraction were collected and precipitated overnight as previously described. The pellets were centrifuged at 3,200 × g, washed with 70% ethanol, and recentrifuged. These pellets were then resuspended in water and centrifuged at high speed (∼100,000 × g) for 2 h. The supernatants were removed, and the pellets were resuspended in water and lyophilized. SDS-PAGE was performed as previously described by Lesse et al. (32). Silver staining was done according to the protocol described by Tsai and Frasch (48). Western blot analysis was performed as previously described by Towbin et al. (47). The monoclonal antibody (MAb) 6B4 was diluted 1:2,000 in 1% bovine serum albumin (BSA) in Tris-buffered saline with Tween 20 (TBST), and the secondary antibody, goat anti-mouse immunoglobulin M (IgM) horseradish peroxidase was diluted 1:20,000 in 1% BSA TBST. The blot was developed with the Super Signal West Pico chemiluminescent substrate according to the manufacturer's instructions (Pierce, Rockford, Ill.).
O-Deacylation of LOS.
The 1291 wild-type and the 1291A11K3 mutant LOS samples (∼200 μg each) were treated with 50 μl of anhydrous hydrazine and incubated for 30 min at 37°C. After cooling to room temperature, the O-deacylated LOSs were precipitated with 250 μl of cold acetone and allowed to sit at −20°C for about 1 h. The samples were then centrifuged at 12,000 × g for 30 min at 4°C, and the supernatants were removed. Finally, the O-deacylated LOS pellets were redissolved in 100 μl of water, frozen, and lyophilized.
Preparation of lipid A and oligosaccharide fractions.
Approximately 500 μg of 1291 and 1291A11K3 LOS was treated with 250 μl of 1% acetic acid and heated at 100°C for 2 h. Samples were then centrifuged at 12,000 × g for 30 min at 4°C to pellet lipid A. After removal of the supernatants (oligosaccharide fractions), the lipid A pellets were washed with 250 μl of H2O and centrifuged again at 12,000 × g for 30 min at 4°C. The water washes were then added to the oligosaccharide fractions, frozen, and lyophilized. The lipid A pellets were dried in vacuo.
Lipid A fatty acid analysis.
For investigation of constituent fatty acids, lipid A from 1291 and 1291A11K3 was isolated. Two milligrams of LOS was resuspended in 0.02% triethylamine. Acetic acid was added to a final concentration of 1.5% (vol/vol). This mixture was heated at 100°C for 2 h. Tubes were cooled and spun at 16,000 × g in a microcentrifuge for 10 min at 4°C. The precipitate was washed with cold double-distilled water, the samples were spun at 16,000 × g for 5 min at 4°C, and the supernatant was removed. These washes were repeated two more times. The samples were dried overnight in a 37°C incubator. Further purification was performed using a standard acid-base hydrolysis procedure, with some minor modifications (13). To prepare derivatives for gas chromatography-mass spectrometry (GC-MS) analysis, the free fatty acids were first methylated with diazomethane in methanol (equal volumes), allowed to sit at room temperature for 10 min, and then dried under a stream of N2. This procedure was repeated, and then the methylated material was treated with 50 μl of ethyl nitride and 50 μl of bis(trimethylsiyl)trifluoroacetamide (BSTFA) and allowed to react overnight at room temperature. Samples were dried under a stream of N2 and resuspended in isooctane. Lipid A samples were measured by GC-MS using a gas chromatograph with DB5HT (30 m by 0.25 mm by 0.10 μm) and DBXLB (30 m by 0.25 mm by 0.25 μm) columns (J&W Scientific, Folsom, Calif.). Samples were run on a Hewlett-Packard series II model 5890 for GC samples and a model 5989A for MS samples. Lipid A samples were eluted with a temperature gradient of 70 to 280°C developed at a rate of 10°C/min. Samples were run at the Molecular Analysis Facility at the University of Iowa.
MALDI-TOF mass spectrometry.
The O-deacylated LOS and lipid A samples were analyzed on a Voyager matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) instrument (Applied Biosystems) equipped with a nitrogen laser (337 nM). All spectra were recorded in the negative-ion mode using delayed extraction conditions as described in detail elsewhere (14). O-Deacylated LOSs were dissolved in 40 μl of H2O, and 4-μl aliquots were desalted by drop dialysis on VSWP 0.025-μm-pore-size nitrocellulose membranes (Millipore Corp., Bedford, Mass.) over deionized water for 45 min. Desalted O-deacylated LOS samples were mixed 1:1 with the matrix solution (a saturated solution of 2,5-dihydroxybenzoic acid in acetone) and allowed to dry at room temperature on a MALDI plate. Lipid A samples were dissolved in 100 μl of CH2Cl2/CH3OH (3:1) and mixed 1:1 with super DHB matrix solution (2,5-dihydroxybenzoic acid/5-methoxysalicylic acid [9:1, wt/wt] in a saturated solution in CH2Cl2/CH3OH [3:1, vol/vol]) (15). Approximately 100 laser shots were recorded for each sample. The spectra were then smoothed with a 19-point Savitsky-Golay function and mass calibrated with an external mass calibrant consisting of renin substrate tetradecapeptide, insulin chain B (oxidized), and bovine insulin (all from Sigma, St. Louis, Mo.). For external calibrations under these conditions, a mass accuracy of ≤0.1% was obtained. For comparison purposes, a two-point correction was made to the spectra of the O-deacylated LOS using the expected fragment ions for diphosphoryl lipid A (m/z 952.0) and the N. gonorrhoeae nonasaccharide (m/z, 1,839.6). All masses measured under these MALDI-TOF conditions were average mass values.
ESI-MS/MS.
Electrospray ionization tandem mass spectrometry (ESI-MS/MS) was run on an API 300 triple quadrupole mass spectrometer (PE-Sciex, Concord, Ontario, Canada) equipped with a Protana nanospray ion source (MDS Proteomics A/S, Odense, Denmark). Lipid A samples were dissolved in CH3OH/CH2Cl2, 2:1, (∼2 to 5 μg of lipid A/μl), and 3 μl of each was placed in a nanospray needle. Samples were run in the negative-ion mode with a typical nanospray needle voltage of −900 V. Singly charged precursor ions were selected in the first quadrupole, fragmented with nitrogen as the collision gas in the second quadrupole, and analyzed in the third quadrupole. The instrument was externally calibrated with human [glu1]-fibrinopeptide B (Bachem, Torrance, Calif.) in the positive-ion MS/MS mode and checked with maltoheptaose (Sigma) in the negative-ion MS/MS mode. All masses measured under these electrospray conditions were average mass values.
Bactericidal assays.
Ten milliliters of gonococcal broth with supplements was inoculated with bacteria to an OD600 of 0.03. These cultures were grown for 2 h at 37°C with agitation. Then, samples were diluted to a final concentration of 108 bacteria/ml in a phosphate-buffered salt solution consisting of 10 mM K2HPO4, 10 mM KH2PO4, 136 mM NaCl, 5 mM KCl, 1 mM CaCl2, 0.3 mM MgCl2 · 6H2O, 1 mM MgSO4 · 7H2O, and 0.01% BSA, pH 7.0. The assay was performed as previously described by Zaleski et al., with some modifications (50). Pooled normal human serum (PNHS) was diluted to 10%. The reaction mixtures were plated and allowed to grow for 1 to 2 days at 37°C in 5% CO2. Colonies were counted at that time, and the bactericidal effect was determined.
Growth of immortalized uec.
Human papillomavirus (HPV) was used to transduce male uec (H. A. Harvey, D. M. B. Post, and M. A. Apicella, unpublished data). HPV-transduced uec were grown in prostate epithelial growth medium (PrEGM) (Clonetics, San Diego, Calif.) on 100-mm-diameter tissue culture-treated petri dishes (Corning Inc., Corning, N.Y). When cultures were 10 to 14 days old, uec were passaged to 24-well tissue culture dishes (Corning Inc.) for invasion assays or to BioCoat membranes (Becton Dickinson) for scanning electron microscopy (SEM) and transmission electron microscopy (TEM). Cells were lifted from 100-mm-diameter dishes by treatment with a trypsin solution (0.25% trypsin, 0.1% EDTA) for 4 min at room temperature, removal of the trypsin solution, and incubation of the cells at 37°C for 4 min. Then, uec were suspended in 5% FCS/PrEGM, centrifuged for 2 min at 1,380 × g, and resuspended in the desired volume of PrEGM prior to seeding. When uec were >90% confluent, gonococcal challenges were performed as described below.
Assay for the invasion of male uec.
Invasion assays were performed as previously described with some modifications (25). Once uec were >90% confluent, cells were grown for at least 48 h prior to infection in antibiotic-free PrEGM. All challenges were performed using piliated colonies, based on colony morphology. N. gonorrhoeae organisms were grown overnight on GCA plates and resuspended in gonococcal broth to an OD600 of 0.16 (approximately 108 gonococci/ml). These cultures were then used to inoculate antibiotic-free PrEGM media to a final concentration of approximately 100 gonococci/epithelial cell. Aliquots of these suspensions were plated to determine the exact numbers of bacteria present in these suspensions. One milliliter of the appropriate suspension was added to each well and incubated for 4 h at 37°C. After incubation, cells were washed two times with antibiotic-free PrEGM media. Wells measuring invasion were incubated in 50 μg of gentamicin (Mediatech Inc., Herndon, Va.) per ml diluted 1:1,000 in antibiotic-free PrEGM for 45 min. A duplicate well was prepared for each sample to measure both adherence and invasion. These wells were incubated in antibiotic-free PrEGM for 45 min. Cells were washed two times with antibiotic-free PrEGM media, lifted with trypsin as previously described, and lysed with a 2% saponin solution. Then, dilutions of the lysates were plated and counted as previously described (45). Lysates were plated on GCA plates and grown at 37°C in 5% CO2 for 2 days. The percent invasion and percent adherence and invasion were calculated as percentages of the initial inoculum. Statistical analysis of the data was performed using the paired t test from the Statview program version 10.0 (Abacus Concepts Inc., Berkeley, Calif.).
Analysis of infected uec by SEM and TEM.
Cells were grown as described above. Bacterial suspensions were prepared as described above. Uec were infected with bacteria for 4 h at 37°C. Cells were washed twice with antibiotic-free PrEGM and fixed with 2% paraformaldehyde for 30 min. Membranes were kept in paraformaldehyde at 4°C until used for microscopy. SEM samples were mounted and processed according to previously described techniques (30) and viewed on a Hitachi S-4000 scanning electron microscope (Hitachi, Mountain View, Calif.). TEM samples were processed using standard techniques and embedded in Epon acrylic resin. Samples were sectioned as previously described and viewed with a Hitachi H-7000 transmission electron microscope (2). All samples were viewed with microscopes located in the Central Microscopy Research Facility at the University of Iowa.
Nucleotide sequence accession numbers.
The nucleotide sequences of the msbB genes from N. meningitidis strain NMB and N. gonorrhoeae strain 1291 are available in the GenBank database under accession numbers AF428103 and AY057903, respectively.
RESULTS
Homology analysis.
The National Center for Biotechnology Information website was used to obtain the amino acid sequences of HtrB from H. influenzae and MsbB from E. coli. The MsbB protein sequences of E. coli, N. gonorrhoeae, and N. meningitidis and the HtrB protein sequence from H. influenzae were compared with alignment programs from Genetics Computer Group. This alignment showed that these proteins had two regions that were highly conserved among all of the organisms. These segments were designated conserved regions one and two. A BLAST search was performed using these conserved regions, and the only proteins that were found to show high homology with them were lipid A acyl transferases from a number of bacteria (Fig. 2A and B). The two conserved regions showed high homology among a number of bacteria. These results suggest that these regions are possible active sites for these enzymes.
FIG. 2.
Homology analyses of predicted amino acid sequences of HtrB and MsbB from various bacteria. Residues with homology are shaded in black, and residues with similarity are shaded in grey. The consensus sequence is shown at the bottom of each panel. (A) Alignment of conserved region one (N. meningitidis strain NMB MsbB amino acids 186 to 203). (B) Alignment of conserved region two (N. meningitidis strain NMB MsbB amino acids 272 to 290). N.gono1291 and N.gono2, N. gonorrhoeae MsbB (GenBank accession no. AY057903); N.mening and N.mening2, N. meningitidis MsbB (GenBank accession no. AF428103); S. typhimurium, S. enterica serovar Typhimurium MsbB (GenBank accession no. AAD03801); E.coliHtrB-MsbB and E.coliHtrB2, E. coli strain K-12 HtrB and MsbB (SwissProt accession no. P24187 and P24205); S.flexerniMsbB, Shigella flexerni MsbB (GenBank accession no. AAB58154); H.influRdHtrB, H. influenzae strain Rd HtrB (GenBank accession no. AAC23173); H.influ2019 and H.influ2, H. influenzae strain 2019 HtrB (GenBank accession no. AAC43515); H.ducreyi and H.ducreyiHtrB2, Haemophilus ducreyi HtrB (GenBank accession no. AAF34642); H.influRdMsbB, H. influenzae strain Rd MsbB (GenBank accession no. AAC21868); V.cholerae and V.cholerae2, Vibrio cholerae strain N16961 lauroyl transferase (HtrB) (GenBank accession no. AAF93389); H.ducreyiMsbB, H. ducreyi MsbB (GenBank accession no. AAF33777); E.coliLpxP, E. coli strain K-12 LpxP (GenBank accession no. AAB66658); X.fastidiosa, Xylella fastidiosa lauroyl transferase (GenBank accession no. AAF82917); H.pylori2, Helicobacter pylori strain 26695 IbpB (GenBank accession no. AAD07343).
Complementation of an E.coli htrB mutant with pNMBA11pUC19.
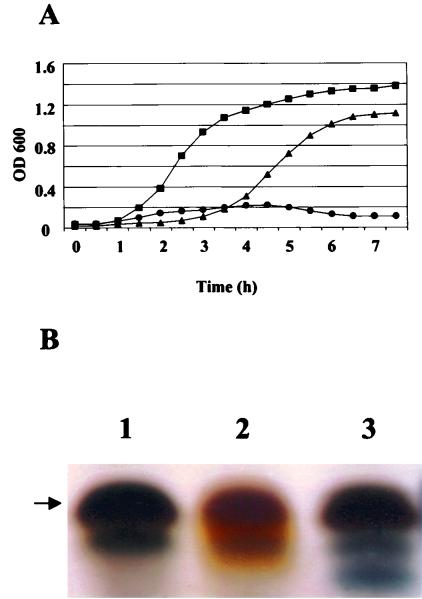
Complementation of an E. coli htrB mutant, MLK217, was performed. The complemented strain was designated MLK217A11. Growth curves (Fig. 3A) demonstrated that MLK217 was unable to grow at 37°C, consistent with previous reports of htrB mutants (28, 31). Additionally, these studies showed that the MLK217A11 bacteria were able to grow almost as well as the parent strain, MLK2. The longer lag phase in the growth of the MLK217A11 may be due to lower recognition of the neisserial promoter in E. coli. Analysis of a silver-stained SDS-PAGE gel (Fig. 3B) demonstrated that the LPS from both MLK2 and MLK217A11 stained black, whereas the LPS isolated from MLK217 stained brown. The brown staining pattern was consistent with previous reports of htrB mutants (31, 39). These results demonstrated that the pNMBA11pUC19 was able to complement the E. coli htrB mutant.
FIG.3.
Complementation of an E. coli htrB mutant with pNMBA11pUC19. (A) Growth curves, at 37°C, of MLK2 (▪), MLK217 (•), and MLK217A11 (▴). Data are representative of three separate experiments. (B) Silver-stained SDS-PAGE of LPS from MLK217A11 (lane 1), MLK217 (lane 2), and MLK2 (lane 3). Arrow, 4-kDa band.
Mutagenesis of the msbB gene.
A deletion of the msbB gene was made in the pNMBA11pUC19 construct using the restriction enzymes BclI and BssHII. These restriction enzyme reactions removed 138 nucleotides from the msbB gene. A kanamycin resistance gene, aphA-3, was ligated into the site of deletion. This construct was transformed into E. coli DH5α cells. Restriction enzyme analysis and PCRs were used to confirm that the proper construct had been made. This construct was designated pNMBA11K3.
Since the N. meningitidis msbB gene was used to construct the msbB mutation in N. gonorrhoeae, the putative msbB gene sequence from N. gonorrhoeae strain 1291 was compared with the putative msbB gene sequence from the N. meningitidis strain NMB using the Fasta program from Genetics Computer Group. This comparison showed a divergence of 54 nucleotides (94% homology). Analysis of the predicted protein sequences showed a difference in 15 amino acids (94.8% homology). Transformation of pNMBA11K3 into N. gonorrhoeae strain 1291 was performed as previously described. Selection for transformants was done on gonococcal medium base plates containing kanamycin. Transformants were able to grow at 37°C. PCR and Southern blot analyses demonstrated that the proper mutation had been incorporated into the N. gonorrhoeae strain 1291 genomic DNA (data not shown). This transformant was subsequently designated 1291A11K3.
Characterization of the LOS by SDS-PAGE and Western blot analyses.
Silver staining showed that the 1291A11K3 LOS migrated through the gel slightly faster than the 1291 LOS and that the 1291A11K3 LOS stained brown instead of black (Fig. 4A). This staining pattern was consistent with previous reports of LPS and LOS isolated from htrB mutants of E. coli and H. influenzae (31, 39). It has been previously demonstrated that MAb 6B4 binds to N. gonorrhoeae LOS through the terminal N-acetyl-lactosamine (3). Western blot analysis was performed using LOS isolated from N. gonorrhoeae strains 1291 and 1291A11K3 and the MAb 6B4. This blot showed that this antibody reacted with both the 1291 LOS and the 1291 A11K3 LOS (Fig. 4B). These results indicated that the oligosaccharide portion of the 1291A11K3 LOS maintained the terminal N-acetyl-lactosamine. This blot also showed a higher-molecular-weight band in addition to the expected LOS band.
FIG. 4.
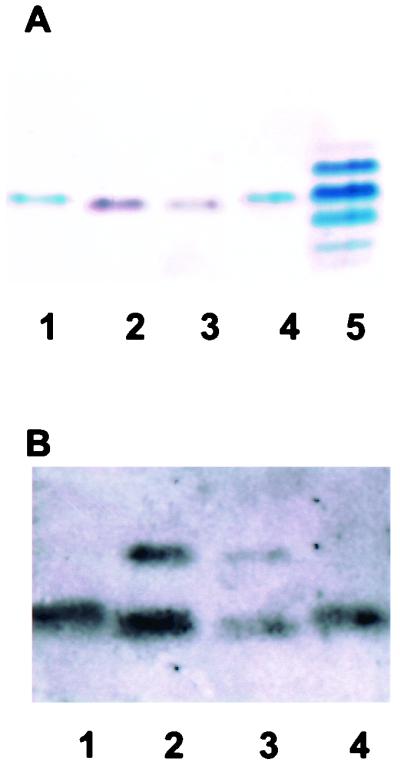
Characterization of 1291A11K3 LOS by SDS-PAGE analyses. (A) Silver-staining analysis of an SDS-PAGE gel. Lanes 1 and 4, 1291 LOS; lanes 2 and 3, 1291A11K3 LOS; lane 5, PID2 LOS. PID2 LOS is included as a molecular weight standard. (B) Western blot analysis using MAb 6B4. Lanes 1 and 4, 1291 LOS; lanes 2 and 3, 1291A11K3 LOS.
Mass spectrometric analyses of N.gonorrhoeae 1291A11K3 LOS.
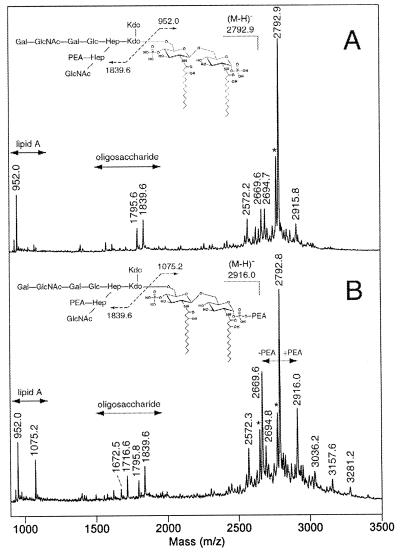
To determine the effect of the msbB mutation on the N. gonorrhoeae 1291 LOS structure, LOSs from strains 1291 and 1291A11K3 were O-deacylated to remove O-linked fatty acids from the lipid A moiety. When analyzed by MALDI mass spectrometry in the negative-ion mode, the 1291 O-deacylated LOS (Fig. 5A) gave a major deprotonated molecular ion, (M − H)−, at m/z 2,792.9, consistent with the previously published structure of the wild-type N. gonorrhoeae strain 1291 LOS (22). In addition to molecular ions (Table 2), the spectrum contains prompt fragments for the oligosaccharide (m/z 1,839.6 and 1,795.6) and lipid A (m/z 952.0) moieties. The lipid A fragment corresponds to a diphosphoryl, diglucosamine structure bearing two N-linked 3-hydroxymyristic acid residues. The oligosaccharide fragment at m/z 1,839.6 arises from the wild-type nonasaccharide structure bearing a single phosphoethanolamine (PEA) group (Fig. 5A, inset). Further loss of CO2 from this species (−44 Da) gives the fragment at m/z 1,795.6.
FIG. 5.
Negative-ion MALDI spectra of the O-deacylated LOS from N. gonorrhoeae strain 1291 (A) and N. gonorrhoeae strain 1291A11K3 (B). The major component of the 1291 O-deacylated LOS mixture. (M − H)− at m/z 2,792.9 contains a diphosphoryl lipid A and a nonasaccharide moiety bearing a single PEA group (see inset, top), consistent with the published structure of N. gonorrhoeae 1291 wild-type LOS (22). In the O-deacylated LOS mixture from the 1291A11K3 mutant, this species is also predominant but there is additional PEA heterogeneity in the sample. When present, the extra PEA group exists on the lipid A moiety (see inset, bottom), primarily on the reducing terminal phosphate (see text). Peaks marked with asterisks are (M − H-H2O)− species.
TABLE 2.
Molecular weights and proposed compositions of the O-deacylated LOS
| Observed Mr for strain: | Calculated Mr | Proposed compositions
|
||||||
|---|---|---|---|---|---|---|---|---|
| 1291 wild type | 1291A11K3 | Hex | HexNAc | Hep | Kdo | PEA | Lipid A forma | |
| 3,282.2 | 3,281.9b | 4 | 3 | 2 | 2 | 1 | DPLA + PEA | |
| 3,158.6 | 3,158.9b | 4 | 3 | 2 | 2 | 1 | DPLA | |
| 3,037.2 | 3,035.8b | 4 | 3 | 2 | 2 | DPLA | ||
| 2,916.8 | 2,917.0 | 2,916.6 | 3 | 2 | 2 | 2 | 1 | DPLA + PEA |
| 2,793.9 | 2,793.8 | 2,793.6 | 3 | 2 | 2 | 2 | 1 | DPLA |
| 2,695.7 | 2,695.8 | 2,695.6 | 3 | 2 | 2 | 2 | 1 | MPLA-H2O |
| 2,670.6 | 2,670.6 | 2,670.5 | 3 | 2 | 2 | 2 | DPLA | |
| 2,573.2 | 2,573.3 | 2,572.5 | 3 | 2 | 2 | 2 | MPLA-H2O | |
MPLA, monophosphorylated O-deacylated lipid A; DPLA, diphosphorylated O-deacetylated lipid A.
These proposed compositions represent reasonable fits to the observed molecular weights for these minor components, but they could not be verified by independent analyses of the free oligosaccharide fractions.
In the spectrum of the O-deacylated LOS from strain 1291A11K3 (Fig. 5B), the same major component is present at (M − H)− 2,792.8. However, there is evidence for considerably more heterogeneity in the number of PEA groups attached to both the oligosaccharide and lipid A portions of the molecule. In the molecular ion region, the species at (M − H)− 2,916.0 arises from the addition of a PEA (+123 Da) moiety to the major component, whereas the species at (M − H)− 2,669.6 arises from the loss of a PEA moiety from the major component. Inspection of the lipid A and oligosaccharide prompt fragments for 1291A11K3 reveals that when present, the additional PEA group is attached to lipid A (Fig. 5B, inset), as indicated by the new fragment at m/z 1,075.2. When the major component has lost a PEA group, it is missing from the oligosaccharide region, as indicated by the new oligosaccharide fragments at m/z 1,716.6 and 1,672.5. In addition to this effect on overall phosphorylation, there is also evidence for extensions of the LOS oligosaccharide structure in strain 1291A11K3. The high-molecular-weight species at (M − H)− 3,036.2, 3,157.6, and 3,281.2, which differ from one another by the number of PEA moieties present, appear to arise from the further addition of a hexose (Hex) and an N-acetylhexosamine (HexNAc) moiety to the major glycoform (Table 2).
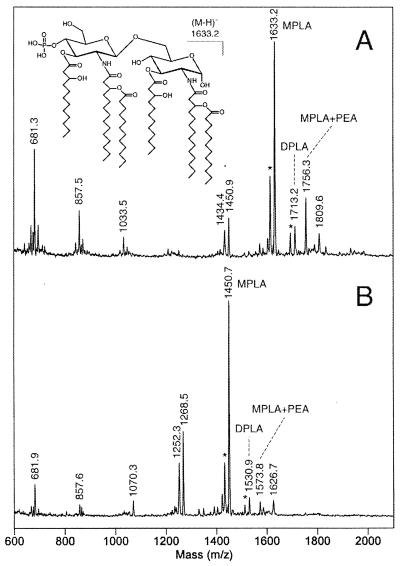
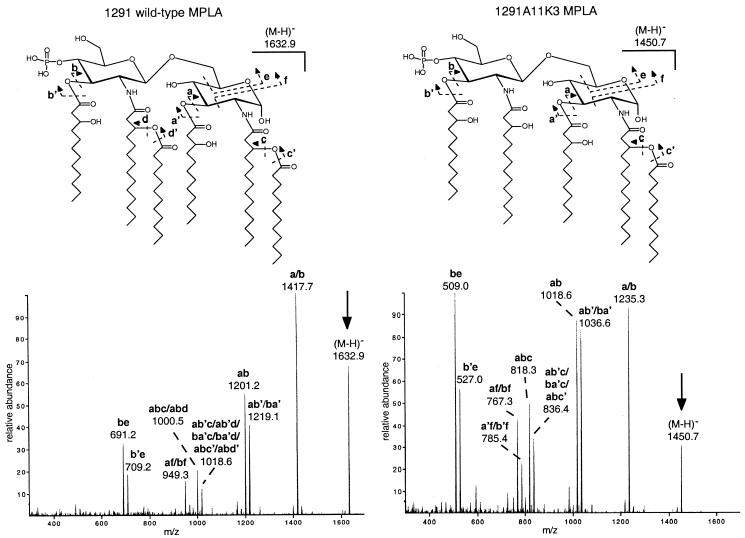
To compare the intact lipid A structures from the N. gonorrhoeae 1291 and 1291A11K3 strains, the LOS samples were subjected to mild acid hydrolysis to cleave the lipid A from the oligosaccharide moiety. Lipid A fractions were analyzed by MALDI mass spectrometry in the negative-ion mode. As seen in Fig. 6A, the 1291 lipid A preparation gave a major deprotonated molecular ion at (M − H)− 1,633.2, corresponding to a hexaacyl, monophosphoryl structure (46). In addition to this major component, a diphosphoryl species at (M − H)− 1,713.2 and a monophosphorylated species bearing an additional PEA group at (M − H)− 1,756.3 are also present. Prompt fragments seen in the spectrum at m/z 1,450.9 and 1,434.4 arise from losses of lauric acid (−182 Da) and 3-hydroxylauric acid (−198 Da), respectively.
FIG. 6.
Negative-ion MALDI spectra of the lipid A fractions from N. gonorrhoeae strain 1291 (A) and N. gonorrhoeae strain 1291A11K3 (B). The major peaks in each spectrum correspond to monophosphoryl lipid A (MPLA) species. Under the acetic acid hydrolysis conditions used, the more labile reducing terminal phosphate of lipid A is partially removed (7). Minor amounts of diphosphoryl lipid A (DPLA) species and MPLA species bearing a PEA group are also present. The mass difference between the corresponding peaks in the two spectra is 182 Da, indicating that one of the two lauric acid residues present in the 1291 structure (inset) is missing in the 1291A11K3 lipid A. The peaks at m/z 1809.6 and 1626.7 in the spectra of the 1291 and 1291A11K3 lipid A molecules, respectively, are 53 Da above the corresponding MPLA+PEA peaks and are likely iron adducts, (M−3H+FeII)−. Peaks marked with asterisks are (M − H-H2O)− species.
In the MALDI spectrum of the 1291A11K3 lipid A, the major component gave a molecular ion at (M − H)− 1,450.7, which is shifted to lower mass by 182.5 Da. This mass difference is consistent with the loss a single lauric acid residue (calculated residue mass, 182.3 Da) from the 1291 monophosphorylated lipid A structure. In addition to this species, the spectrum of the 1291A11K3 lipid A also contains peaks for a diphosphoryl species at (M − H)− 1,530.9 and a monophosphoryl species bearing an additional PEA at (M − H)− 1,573.8. Prompt fragments for the losses of lauric acid (m/z 1,268.5) and 3-hydroxylauric acid (m/z 1,252.3) are also present. The fact that mild acid hydrolysis of the 1291A11K3 LOS also gave mainly monophosphorylated lipid A suggests that the additional PEA moiety seen in the O-deacylated LOS (Fig. 5B) is linked primarily to the reducing terminal phosphate.
Strong acid hydrolysis of the lipid A samples was conducted to release all of the N-linked and O-linked fatty acids from the diglucosamine backbone. Following derivatization, the fatty acids were analyzed by GC-MS (Table 3). From the 1291 lipid A sample, lauric acid, 3-hydroxylauric acid, and 3-hydroxymyristic acid were detected in a 1.2:1.2:1.0 ratio. The 1291A11K3 lipid A sample contained lauric acid, 3-hydroxylauric acid, and 3-hydroxymyristic acid in a 0.5:1.3:1.0 ratio. Consistent with the MALDI data described above, these results suggest that the lipid A of the 1291A11K3 mutant has one less lauric acid residue than the lipid A from the parental 1291 strain.
TABLE 3.
Fatty acid analysis of the lipid A samples
| Compound | Relative % in:
|
|
|---|---|---|
| 1291 wild type | 1291A11K3 | |
| Lauric acid (C12:0) | 29.8 | 15.6 |
| Palmitic acid (C16:0)a | 1.6 | 2.0 |
| Stearic acid (C18:0)a | 2.5 | 3.6 |
| 3-OH-lauric acid (3-OH-C12:0) | 31.8 | 39.3 |
| 3-OH-myristic acid (3-OH-C14:0) | 25.7 | 29.3 |
| Other componentsb | 8.5 | 10.3 |
These fatty acids were detected in the lipid A preparations but were not observed in our analyses of intact lipid A species by mass spectrometric methods.
This category includes minor amounts of C12:1 and C14:1 fatty acids and a few unidentified components.
To establish which of the two possible lauric acids was missing in the 1291A11K3 mutant strain, the monophosphorylated lipid A samples were analyzed by ESI-MS/MS in the negative-ion mode. The major molecular ions for the 1291 (m/z 1,632.9) and 1291A11K3 (m/z 1,450.7) lipid A species were selected for collision-induced dissociation. As seen in Fig. 7, the lipid A samples gave similar fragmentation patterns, with the fragment series for 1291A11K3 shifted to lower masses by 182 Da. Most of the fragments arise from single and multiple losses of O-linked fatty acids from the parent ions, as indicated on the spectra and the structures shown (Fig. 7, insets). However, the low-mass fragments in the spectra of the 1291 (m/z 691.2 and 709.2) and 1291A11K3 (m/z 509.0 and 527.0) samples would appear to arise from two-bond cleavages: the loss of the 3-hydroxylauric acid residue on the distal glucosamine, as a free acid or a ketene, and a cross-ring cleavage of the reducing terminal glucosamine (indicated as an e-type cleavage in Fig. 7). In the nomenclature of Costello and Vath (10), this e-type cross-ring cleavage is designated a 0,4A2-type fragment. As this cross-ring cleavage releases all of the substituent-bearing positions from the reducing terminal glucosamine, the low-mass fragment ions thus contain only the lipids on the distal glucosamine. In this case, the fact that the fragments for the 1291A11K3 lipid A are 182 Da lower in mass than the fragments for the 1291 lipid A suggests that in strain 1291A11K3, a lauric acid is missing from the distal glucosamine of the lipid A structure.
FIG. 7.
Negative-ion electrospray MS/MS spectra of the monophosphorylated lipid A species from N. gonorrhoeae strain 1291 (left) and N. gonorrhoeae strain 1291A11K3 (right). The parent (M − H)− ions selected for collision-induced dissociation are indicated with arrows. Fragment ions are labeled on the spectra and indicated on the structures using a letter code. O-linked fatty acids are lost as free acids (a, b, c, and d cleavages) or ketenes (a', b', c', and d' cleavages). Fragments of type e and f are cross-ring cleavages.
Bactericidal activity of PNHS against N.gonorrhoeae strains 24-1, 1291, and 1291A11K3.
To determine if the msbB mutation affects serum susceptibility, three N. gonorrhoeae strains, 24-1, 1291, and 1291A11K3, were tested in triplicate in a serum bactericidal assay. Strain 24-1 was used as a serum-sensitive control. All three strains were able to grow in the presence of heat-inactivated normal human serum (data not shown). Strain 24-1 was highly serum sensitive in the presence of PNHS, while the serum sensitivity of strain 1291A11K3 was unchanged from that of the parent strain 1291 (data not shown). These studies indicated that modifications of the lipid A structure had no effect on susceptibility to killing by PNHS. DeMaria et al. reported similar findings with an htrB mutant of H. influenzae (11).
TEM and SEM analyses of uec infected with either 1291 or 1291A11K3.
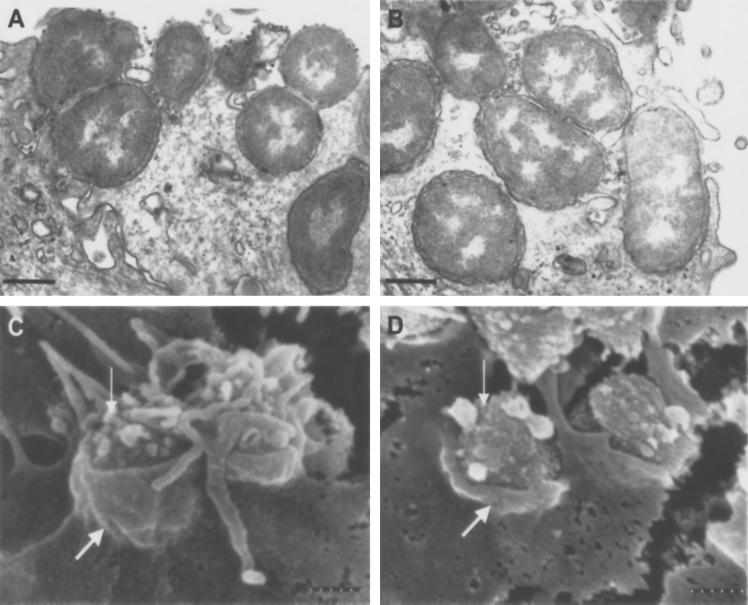
TEM studies showed cells infected with 1291A11K3 and 1291 and showed bacteria on the surfaces of the uec beginning to be internalized into the cells (Fig. 8A and B). These studies also showed both 1291 and 1291A11K3 bacteria entering uec and being internalized within vacuoles. In Fig. 8A and B, the tight association between the plasma membrane and the gonococci, indicative of a clathrin-dependent receptor-mediated endocytic process, is seen. SEM demonstrated a close association between the uec's plasma membrane and both 1291A11K3 and 1291 bacteria (Fig. 8C and D). The close association between the plasma membrane and the bacteria suggests internalization by clathrin-independent receptor-mediated endocytosis. Additionally, the frequency at which the 1291A11K3 mutant was found within uec, on analysis of multiple fields using TEM, suggested that it was internalized at a rate similar to that of the parent strain.
FIG. 8.
TEM of uec infected for 4 h with 1291A11K3 bacteria (A) or 1291 bacteria (B) (bar, 4 μm). SEM of uec infected for 4 h with 1291A11K3 bacteria (C) or 1291 bacteria (D) (bar, 300 nm). The thick arrows point to the uec plasma membrane, and the thin arrows point to the bacteria. The close association between the uec's plasma membrane and the bacteria is characteristic of clathrin-dependent receptor-mediated endocytosis.
Invasion assays performed with 1291 and 1291A11K3.
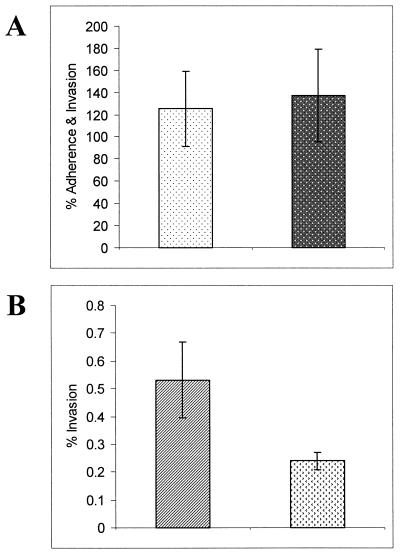
In order to study the effects of the msbB mutation on gonococcal invasion of and adherence to uec, standard gentamicin survival invasion assays were performed. The results from the experiment shown in Fig. 9 are representative of what was consistently seen in six separate experiments. These results showed that there was no significant difference in the abilities of strains 1291 and 1291A11K3 to adhere to uec (P = 0.6947). However, there was a significant difference between the numbers of 1291 and 1291A11K3 bacteria recovered from the gentamicin-treated uec (P = 0.0130). These data, combined with the electron microscopic analyses, suggest that strain 1291A11K3 enters cells in a fashion similar to that of strain 1291 but may be more susceptible to innate intracellular killing mechanisms found within urethral epithelial cells.
FIG. 9.
A representative example of the invasion assays performed on HPV-transduced uec using N. gonorrhoeae strains 1291 and 1291A11K3. (A) Mean values of percent adherence and invasion of 1291 ( ) and 1291A11K3 (
) and 1291A11K3 ( ) (P = 0.6947). (B) Mean values of percent invasion of 1291 (
) (P = 0.6947). (B) Mean values of percent invasion of 1291 ( ) and 1291A11K3 (
) and 1291A11K3 ( ) (P = 0.0130). Data are representative of six separate experiments. Adherence and invasion data were measured as the numbers of bacteria recovered from infected uec not treated with gentamicin. The invasion data were measured as the numbers of bacteria recovered from infected uec treated with gentamicin.
) (P = 0.0130). Data are representative of six separate experiments. Adherence and invasion data were measured as the numbers of bacteria recovered from infected uec not treated with gentamicin. The invasion data were measured as the numbers of bacteria recovered from infected uec treated with gentamicin.
DISCUSSION
HtrB and MsbB are Kdo-dependent acyl transferases involved in the biosynthesis of LPS and LOS (6, 9, 36). These transferases are thought to act in a sequential manner, with HtrB acting first and MsbB acting second (9). The mass spectrometric analysis of lipid A isolated from mutants supports this: lipid A from an H. influenzae htrB mutant was determined to be 90% tetraacyl and 10% pentaacyl (31), whereas lipid A from an E. coli msbB mutant was determined to be pentaacyl (41). This suggests that HtrB is able to be fully functional in the absence of a functional MsbB acyl transferase. Mutants in htrB isolated from E. coli, H. influenzae, and serovar Typhimurium have all been shown to be initially sensitive to temperature (28, 31, 44); however, the msbB mutant isolated from E. coli was not temperature sensitive (41). It has been proposed that if the lipid A is at least pentaacyl, it is able to grow at higher temperatures (9).
Discerning whether the gene identified in these studies is an htrB or msbB homologue is difficult due to the high similarity of these acyl transferases at both the amino acid and DNA levels. The gene identified in these studies was able to complement an E. coli htrB mutant. However, previous studies have shown that both htrB and msbB can complement an htrB mutant (27, 28, 31). We may, however, be able to draw some conclusions based on the phenotypes of the mutant. Previous studies of an msbB mutant demonstrated that these bacteria were not temperature sensitive (41). The mutant, 1291A11K3, developed in these studies grew normally at 37°C, both in liquid broth and on solid medium (data not shown). Additionally, mass spectrometric analyses revealed that the lipid A isolated from 1291A11K3 was pentaacyl. Previous studies have suggested that the addition of the secondary acyl substitutions occurs in a sequential manner, with HtrB acting first and MsbB acting second (9). Therefore, the lipid A of an htrB mutant would be expected to be primarily tetraacyl, and the lipid A from an msbB mutant would be expected to be pentaacyl. Therefore, it seems likely that the gene identified in these studies is an msbB homologue. In addition, a recent study by van der Ley et al. (49) demonstrated that mutations in two htrB/msbB homologues in N. meningitidis resulted in lipid A structures that were pentaacyl and tetraacyl. Their studies suggest that Neisseria bacteria may assemble their lipid A in a sequential manner, and therefore it seems likely that the gene identified in these studies acts in a fashion similar to that of the msbB gene from E. coli.
To establish the acylation pattern of the pentaacyl lipid A in 1291A11K3, we analyzed the monophosphoryl lipid A molecules from 1291 and 1291A11K3 using tandem mass spectrometry. In monophosphorylated lipid A molecules, the single phosphate group exists on the distal glucosamine and thus creates asymmetry in the structures. Furthermore, in negative-ion MS analysis, charge is expected to be retained on the lone phosphate group, facilitating interpretation of fragmentation pathways. The low-mass fragments observed in these studies were attributed to two-bond cleavages comprised of the loss of a single fatty acid moiety and a diagnostic cross-ring cleavage. The cross-ring cleavages established that the distal nonreducing terminal glucosamine of the pentaacyl 1291A11K3 lipid A sample is missing a lauric acid compared to the 1291 hexaacyl lipid A structure. In ESI-MS/MS investigations of the lipid A molecules from Enterobacter agglomerans, Salmonella minnesota, and Shigella flexneri, low-mass fragment ions corresponding to those seen in the 1291 lipid A spectrum (m/z 691 and 709) were also observed. However, in those species, the fragment ions were assigned as nonreducing terminal acylium ions (5, 8). As acylium ions of that composition could not arise from the N. gonorrhoeae 1291 lipid A structure, we assigned the fragments differently. Comparison of the lipid A structures, however, suggests that in fact our assignments could also fit for E. agglomerans, S. minnesota, and S. flexneri, since the lipid A molecules from those species have the same N-linked acyloxyacyl fatty acids on their distal glucosamines as N. gonorrhoeae 1291. Consequently, the shift of those fragment ions to lower mass in the 1291A11K3 mutant provides strong evidence that a lone 3-hydroxymyristyl moiety exists in amide linkage on the distal glucosamine of the lipid A.
Modification of the acylation pattern of lipid A was not the only change that was found in the LOS of 1291A11K3. Changes in the phosphorylation pattern as well as the addition of higher-molecular-weight species of LOS were also seen. Previous studies by Lee et al. (31) showed similar modifications in the LOS of an H. influenzae htrB mutant. The high-molecular-weight species detected in the 1291A11K3 LOS appear to correspond to species that have been previously observed in the LOS from N. gonorrhoeae MS11mkC, a variant of N. gonorrhoeae strain MS11 isolated from men with gonorrhea (23). It is unclear if the bacteria use these modifications of their LOS to compensate for the changes in lipid A, or if the changes in the phosphorylation pattern and oligosaccharide portion of the LOS are directly regulated by HtrB and MsbB. Since HtrB and MsbB have both been shown to be acyl transferases, it seems unlikely that they have a direct role in the modifications of the oligosaccharide chain length or the amount of PEA present on the LOS (6, 9, 36). However, further studies are needed to try and determine more clearly the roles that htrB and msbB play in these modifications of the LOS.
Recent studies have focused on the factors involved in the entry of N. gonorrhoeae into eukaryotic cells. A number of bacterial and host factors have been identified. Piliation of the gonococcus, the presence of the Opa protein, and an intact LOS appear to be some of the prerequisite factors necessary for gonococcal invasion (20, 35). Additionally, studies involving the neisserial IgA1 protease have suggested that it may play a role in intracellular survival of the gonococcus (33). Our laboratory has been studying the role of gonococcal LOS in genital epithelial cell invasion. Our laboratory has shown that with male uec this process involves the asialoglycoprotein receptor (ASGP-R) (17). The ASGP-R is expressed on the surface of male uec and recognizes ligands with terminal galactose or N-acetylgalactosamine residues (17, 18). Studies performed using human sperm cells, HepG2 cells, and uec have shown that the terminal galactose of the gonococcal LOS binds specifically to the ASGP-R expressed on the surfaces of these human cells (17, 19, 37). The internalization of this complex has been shown to be a clathrin-dependent receptor-mediated endocytic process (17). Studies of both male urethral exudates and primary uec infected with N. gonorrhoeae have suggested that receptor-mediated endocytosis may be responsible for the internalization of a high percentage of these bacteria into male uec (2, 17, 18).
The invasion assays performed in this study suggested that there was no difference in the abilities of the 1291A11K3 bacteria and of the parent strain bacteria to adhere to uec. Additionally, the microscopy data clearly showed that both strains of bacteria were internalized by the uec and both were able to form close associations with the uec's plasma membrane at similar rates. This association was indicative of entry by means of a receptor-mediated endocytosis mechanism. Therefore, the mutation in the msbB gene did not seem to have any apparent effects on the ability of this strain to be internalized by the uec.
The invasion assay data also showed that significantly fewer 1291A11K3 bacteria than parent strain bacteria were recovered from gentamicin-treated uec. At least two possibilities exist for explaining these results. First, the 1291A11K3 bacteria may invade the uec at a lower rate than the wild-type bacteria. Second, the 1291A11K3 organisms are more susceptible to intracellular killing mechanisms once they have been internalized by the uec. Since the bacteria seem to be internalized at similar rates the second explanation seems likely. After the male uec internalize the bacteria, the gonococci must be able to survive intracellularly. It is not known if the bacteria remain within a vacuole after internalization, and if so it is unclear how they are able to avoid endosomal degradation. Some studies have suggested that the neisserial IgA1 protease may be involved in intracellular survival of N. gonorrhoeae (33). IgA1 protease has been shown to cleave the host cell lysosome- associated membrane protein (LAMP-1) (33). LAMP-1 is thought to be involved in the maturation of late endosomes and lysosomes (33). The degradation of this glycoprotein is thought to contribute to the ability of the gonococcus to survive intracellularly (33). However, male volunteer studies demonstrated that a gonococcal IgA1 protease mutant was not compromised in its ability, compared to the parent strain, to cause urethritis (21).
Our studies presented here demonstrate that the mutation in the msbB gene in N. gonorrhoeae affects how these organisms interact with male uec. The modifications in the LOS may make the msbB organisms more susceptible to intracellular killing. Currently, the mechanisms by which epithelial cells are able to kill intracellular bacteria are not well understood. Further study of these killing mechanisms should enable future studies to determine more directly what role the LOS, and more specifically lipid A, plays in the survival of gonococci within male uec.
Acknowledgments
We thank all members of the Apicella lab for their useful discussions and technical assistance. Additionally, we thank Hillery Harvey for the use of the HPV-transformed urethral epithelial cells and UCSF Mass Spectrometry Resource for access to the triple quadrupole mass spectrometer (RR01614).
The University of Iowa DNA facility is supported in part by the Diabetes Endocrinology Research Center with National Institutes of Health grant DK25295 and by the College of Medicine. Research in M. A. Apicella's laboratory was supported by AI44642, AI45424, and AI45728. D. M. B. Post's work was in part supported by NIH training grant T32A107511. Research in B. W. Gibson's laboratory was supported by AI44642 and AI31254.
Editor: T. R. Kozel
REFERENCES
- 1.Apicella, M. A. 1974. Antigenically distinct population of Neisseria gonorrhoeae: isolation and characterization of the responsible determinants. J. Infect. Dis. 130:619-625. [DOI] [PubMed] [Google Scholar]
- 2.Apicella, M. A., M. R. Ketterer, F. K. N. Lee, D. Zhou, P. A. Rice, and M. S. Blake. 1996. The pathogenesis of gonococcal urethritis in men: confocal and immunoelectron microscopic analysis of urethral exudates from men infected with Neisseria gonorrhoeae. J. Infect. Dis. 173:636-646. [DOI] [PubMed] [Google Scholar]
- 3.Apicella, M. A., R. E. Mandrell, M. Shero, M. E. Wilson, J. McLeod Griffiss, G. F. Brooks, C. Lammel, J. F. Breen, and P. A. Rice. 1990. Modification of sialic acid of Neisseria gonorrhoeae lipooligosaccharide epitope expression in human urethral exudates: an immunoelectron microscopic analysis. J. Infect. Dis. 162:506-512. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology, vol. 1. John Wiley & Sons, New York, N.Y.
- 5.Boue, S. M., and R. B. Cole. 2000. Confirmation of the structure of lipid A from Enterobacter agglomerans by electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 35:361-368. [DOI] [PubMed] [Google Scholar]
- 6.Brozek, K. A., and C. R. H. Raetz. 1990. Biosynthesis of lipid A in Escherichia coli. Acyl carrier protein-dependent incorporation of laurate and myristate. J. Biol. Chem. 265:15410-15417. [PubMed] [Google Scholar]
- 7.Caroff, M., A. Tacken, and L. Szabó. 1988. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr. Res. 175:273-282. [DOI] [PubMed] [Google Scholar]
- 8.Chan, S., and V. N. Reinhold. 1994. Detailed structural characterization of lipid A: electrospray ionization coupled with tandem mass spectrometry. Anal. Biochem. 218:63-73. [DOI] [PubMed] [Google Scholar]
- 9.Clementz, T., Z. Zhou, and C. R. H. Raetz. 1997. Function of the Escherichia coli msbB gene, a multicopy suppressor of htrB knockouts, in the acylation of lipid A. J. Biol. Chem. 272:10353-10360. [DOI] [PubMed] [Google Scholar]
- 10.Costello, C. E., and J. E. Vath. 1990. Tandem mass spectrometry of glycolipids. Methods Enzymol. 193:738-768. [DOI] [PubMed] [Google Scholar]
- 11.DeMaria, T. F., M. A. Apicella, W. A. Nichols, and E. R. Leake. 1997. Evaluation of the virulence of nontypeable Haemophilus influenzae lipooligosaccharide htrB and rfaD mutants in the chinchilla model of otitis media. Infect. Immun. 65:4431-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards, J., J. Q. Shao, K. A. Ault, and M. A. Apicella. 2000. Neisseria gonorrhoeae elicits membrane ruffling and cytoskeletal rearrangements upon infection of primary human endocervical and ectocervical cells. Infect. Immun. 68:5354-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giardina, P. C., J. A. Weiss, B. W. Gibson, and M. A. Apicella. Isolation and analysis of radiolabeled meningococcal endotoxin, p. 441-458. In A. J. Pollard and M. C. J. Maiden (ed.), Methods in molecular medicine, vol. 67. Meningococcal disease: methods and protocols, in press. Humana Press Inc., Totowa, N.J. [DOI] [PubMed]
- 14.Gibson, B. W., J. J. Engstrom, C. M. John, W. Hines, and A. M. Falick. 1997. Characterization of bacterial lipooligosaccharides by delayed extraction matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Am. Soc. Mass Spectrom. 8:645-658. [Google Scholar]
- 15.Guo, L., K. B. Lim, J. S. Gunn, B. Bainbridge, R. P. Darveau, M. Hackett, and S. I. Miller. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250-253. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 17.Harvey, H. A., M. P. Jennings, C. A. Campbell, R. Williams, and M. A. Apicella. 2001. Receptor-mediated endocytosis of Neisseria gonorrhoeae into primary human urethral epithelial cells: the role of the asialoglycoprotein receptor. Mol. Microbiol. 42:659-672. [DOI] [PubMed]
- 18.Harvey, H. A., M. R. Ketterer, A. Preston, D. Lubaroff, R. Williams, and M. A. Apicella. 1997. Ultrastructural analysis of primary human urethral epithelial cell cultures infected with Neisseria gonorrhoeae. Infect. Immun. 65:2420-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harvey, H. A., N. Porat, C. A. Campbell, M. Jennings, B. W. Gibson, N. J. Phillips, M. A. Apicella, and M. S. Blake. 2000. Gonococcal lipooligosaccharide is a ligand for the asialoglycoprotein receptor on human sperm. Mol. Microbiol. 36:1059-1070. [DOI] [PubMed] [Google Scholar]
- 20.Jerse, A. E., and R. F. Rest. 1997. Adhesion and invasion by the pathogenic neisseria. Trends Microbiol. 5:217-221. [DOI] [PubMed] [Google Scholar]
- 21.Johannsen, D. B., D. M. Johnston, H. O. Koymen, M. S. Cohen, and J. G. Cannon. 1999. A Neisseria gonorrhoeae immunoglobulin A1 protease mutant is infectious in the human challenge model of urethral infection. Infect. Immun. 67:3009-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.John, C. M., J. M. Griffiss, M. A. Apicella, R. E. Mandrell, and B. W. Gibson. 1991. The structural basis for pyocin resistance in Neisseria gonorrhoeae lipooligosaccharides. J. Biol. Chem. 266:19303-19311. [PubMed] [Google Scholar]
- 23.John, C. M., H. Schneider, and J. M. Griffiss. 1999. Neisseria gonorrhoeae that infect men have lipooligosaccharides with terminal N-acetyllactosamine repeats. J. Biol. Chem. 274:1017-1025. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, K. G., and M. B. Perry. 1975. Improved techniques for the preparation of bacterial lipopolysaccharides. Can. J. Microbiol. 22:29-34. [DOI] [PubMed] [Google Scholar]
- 25.Jones, B. D., C. A. Lee, and S. Falkow. 1992. Invasion by Salmonella typhimurium is affected by the direction of flagellar rotation. Infect. Immun. 60:2475-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones, B. D., W. A. Nichols, B. W. Gibson, M. G. Sunshine, and M. A. Apicella. 1997. Study of the role of the htrB gene in Salmonella typhimurium virulence. Infect. Immun. 65:4778-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karow, M., and C. Georgopoulos. 1992. Isolation and characterization of the Escherichia coli msbB gene, a multicopy suppressor of null mutations in the high-temperature requirement gene htrB. J. Bacteriol. 174:702-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karow, M., O. Fayet, A. Cegielska, T. Ziegelhoffer, and C. Georgopoulos. 1991. Isolation and characterization of the Escherichia coli htrB gene, whose product is essential for bacterial viability above 33°C in rich media. J. Bacteriol. 173:741-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karow, M., O. Fayet, and C. Georgopoulos. 1992. The lethal phenotype caused by null mutations in the Escherichia coli htrB gene is suppressed by mutations in the accBC operon, encoding two subunits of acetyl coenzyme A carboxylase. J. Bacteriol. 174:7407-7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ketterer, M. R., J. Q. Shao, D. B. Hornick, B. Buscher, V. K. Bandi, and M. A. Apicella. 1999. Infection of primary human bronchial epithelial cells by Haemophilus influenzae: macropinocytosis as a mechanism of airway epithelial cell entry. Infect. Immun. 67:4161-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, N., M. Sunshine, J. Engstrom, B. Gibson, and M. A. Apicella. 1995. Mutation of the htrB locus of Haemophilus influenzae nontypeable strain 2019 is associated with modifications of lipid A and phosphorylation of the lipooligosaccharide. J. Biol. Chem. 270:27151-27159. [PubMed] [Google Scholar]
- 32.Lesse, A. J., A. A. Campagnari, W. E. Bittner, and M. A. Apicella. 1990. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine sodium dodecyl sulfate polyacarylamide gel electrophoresis. J. Immunol. Methods 126:109-117. [DOI] [PubMed] [Google Scholar]
- 33.Lin, L., P. Ayala, J. Larson, M. Mulks, M. Fukuda, S. R. Carlsson, C. Enns, and M. So. 1997. The Neisseria type 2 IgA1 protease cleaves LAMP1 and promotes survival of bacteria within epithelial cells. Mol. Microbiol. 24:1083-1094. [DOI] [PubMed] [Google Scholar]
- 34.Menard, R., P. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naumann, M., T. Rudel, and T. F. Meyer. 1999. Host cell interactions and signaling with Neisseria gonorrhoeae. Curr. Opin. Microbiol. 2:62-70. [DOI] [PubMed] [Google Scholar]
- 36.Nichols, W. A., C. R. H. Raetz, T. Clementz, A. L. Smith, J. A. Hanson, M. R. Ketterer, M. Sunshine, and M. A. Apicella. 1997. htrB of Haemophilus influenzae: determination of biochemical activity and effects on virulence and lipooligosaccharide toxicity. J. Endotoxin Res. 4:163-172. [Google Scholar]
- 37.Porat, N., M. A. Apicella, and M. S. Blake. 1995. Neisseria gonorrhoeae utilizes and enhances the biosynthesis of the asialoglycoprotein receptor expressed on the surface of the hepatic HepG2 cell line. Infect. Immun. 63:1498-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Schnaitman, C., and J. D. Klena. 1993. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol. Rev. 57:655-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider, H., J. M. Griffiss, J. W. Boslego, P. J. Hitchcock, K. O. McJunkin, and M. A. Apicella. 1991. Expression of paragloboside-like lipooligosaccharides may be a necessary component of gonococcal pathogenesis in men. J. Exp. Med. 174:1601-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Somerville, J. E., Jr., L. Cassiano, B. Bainbridge, M. D. Cunningham, and R. P. Darveau. 1996. A novel Escherichia coli lipid A mutant that produces an anti-inflammatory lipopolysaccharide. J. Clin. Investig. 97:359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stephens, D. S., C. F. McAllister, D. Zhou, F. K. Lee, and M. A. Apicella. 1994. Tn 916-generated, lipooligosaccharide mutants of Neisseria meningitidis and Neisseria gonorrhoeae. Infect. Immun. 62:2947-2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stephens, D. S., J. S. Swartley, S. Kathariou, and S. A. Morse. 1991. Insertion of Tn 916 in Neisseria meningitidis resulting in loss of group B capsular polysaccharide. Infect. Immun. 59:4097-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sunshine, M., B. W. Gibson, J. J. Engstrom, W. A. Nichols, B. D. Jones, and M. A. Apicella. 1997. Mutation of the htrB gene in a virulent Salmonella typhimurium strain by intergeneric transduction: strain construction and phenotypic characterization. J. Bacteriol. 179:5521-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swords, W. E., B. A. Buscher, K. Ver Steeg II, A. Preston, W. A. Nichols, J. N. Weiser, B. W. Gibson, and M. A. Apicella. 2000. Nontypeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Mol. Microbiol. 37:13-27. [DOI] [PubMed] [Google Scholar]
- 46.Takayama, K., N. Qureshi, K. Hyver, J. Honovich, R. J. Cotter, P. Mascagni, and H. Schneider. 1986. Characterization of a structural series of lipid A obtained from the lipopolysaccharides of Neisseria gonorrhoeae. J. Biol. Chem. 261:10624-10631. [PubMed] [Google Scholar]
- 47.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 49.van der Ley, P., L. Steeghs, H. J. Hamstra, J. ten Hove, B. Zomer, and L. van Alphen. 2001. Modification of lipid A biosynthesis in Neisseria meningitidis lpxL mutants: influence on lipopolysaccharide structure, toxicity, and adjuvant activity. Infect. Immun. 69:5981-5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zaleski, A., N. K. Scheffler, P. Densen, F. K. N. Lee, A. A. Capagnari, B. W. Gibson, and M. A. Apicella. 2000. Lipooligosaccharide Pk (Galα1-4Galβ1-4Glc) epitope of Moraxella catarrhalis is a factor in resistance to bactericidal activity mediated by normal human serum. Infect. Immun. 68:5261-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]