Abstract
Envenomation by the brown recluse spider (Loxosceles reclusa) may cause local dermonecrosis and, rarely, coagulopathies, kidney failure and death. A venom phospholipase, SMaseD (sphingomyelinase D), is responsible for the pathological manifestations of envenomation. Recently, the recombinant SMaseD from Loxosceles laeta was demonstrated to hydrolyse LPC (lysophosphatidylcholine) to produce LPA (lysophosphatidic acid) and choline. Therefore activation of LPA signalling pathways may be involved in some manifestations of Loxosceles envenomation. To begin investigating this idea, we cloned a full-length cDNA encoding L. reclusa SMaseD. The 305 amino acid sequence of the L. reclusa enzyme is 87, 85 and 60% identical with those of L. arizonica, L. intermedia and L. laeta respectively. The recombinant enzyme expressed in bacteria had broad substrate specificity. The lysophospholipids LPC, LPI (18:1-1-oleyol lysophosphatidylinositol), LPS, LPG (18:1-1-oleoyl-lysophosphatidylglycerol), LBPA (18:1-1-oleoyl-lysobisphosphatidic acid) (all with various acyl chains), lyso-platelet-activating factor (C16:0), cyclic phosphatidic acid and sphingomyelin were hydrolysed, whereas sphingosylphosphorylcholine, PC (phosphatidylcholine; C22:6, C20:4 and C6:0), oxidized PCs and PAF (platelet-activating factor; C16:0) were not hydrolysed. The PAF analogue, edelfosine, inhibited enzyme activity. Recombinant enzyme plus LPC (C18:1) induced the migration of A2058 melanoma cells, and this activity was blocked by the LPA receptor antagonist, VPC32183. The recombinant spider enzyme was haemolytic, but this activity was absent from catalytically inactive H37N (His37→Asn) and H73N mutants. Our results demonstrate that Loxosceles phospholipase D hydrolyses a wider range of lysophospholipids than previously supposed, and thus the term ‘SMaseD’ is too limited in describing this enzyme.
Keywords: brown recluse spider, Loxosceles envenomation, lysophosphatidic acid (LPA), lysophosphatidylcholine (LPC), phospholipase D (PLD), sphingomyelinase D (SMaseD)
Abbreviations: ATX, autotaxin; C1P, ceramide-1-phosphate; DMEM, Dulbecco's modified Eagle's medium; faf-BSA, essentially fatty acid-free BSA; G3P, glycerol-3-phosphate; HRP, horseradish peroxidase; LBPA, 18:1-1-oleoyl-lysobisphosphatidic acid; LPA, lysophosphatidic acid; LPC, lysophosphati-dylcholine; LPE, 18:1-1-oleoyl-lysophosphatidylethanolamine; LPG, 18:1-1-oleoyl-lysophosphatidylglycerol; LPI, 18:1-1-oleoyl-lysophosphatidylinositol; LPS, 18:1-1-oleoyl-lysophosphatidylserine; PC, phosphatidylcholine; PLD, phospholipase D; lysoPLD, LPC-preferring PLD; ORF, open reading frame; PAF, platelet-activating factor; RACE, rapid amplification of cDNA ends; SMaseD, sphingomyelinase D; SPC, sphingosylphosphorylcholine, also known as lyso-sphingomyelin; TOOS, N-ethyl-N-(2-hydroxy-3-sulphopropyl)-m-toluidine; VBS2+, veronal-buffered saline
INTRODUCTION
Envenomation by the brown recluse spider (Loxosceles reclusa) can induce local dermonecrosis as well as systemic erythrocyte haemolysis, platelet aggregation and renal failure that, in rare cases, may lead to death [1]. Although the venom contains several proteins, the PLD (phospholipase D) ‘SMaseD’ (sphingomyelinase D) is responsible for dermonecrosis and complement-dependent haemolysis [2], blood vessel damage and fibrinogenolysis [3] as well as dysregulated endothelial cell-dependent neutrophil activation [4].
One of the products of Loxosceles SMaseD is C1P (ceramide-1-phosphate), which is known to be one component of the lipid bilayer but is not obviously a signalling molecule [5]. However, C1P is a bioactive lipid molecule capable of stimulating cell proliferation by increasing DNA synthesis, directly inhibiting acid sphingomyelinase and blocking ceramide synthesis and apoptosis, and, by directly activating phospholipase A2, it induces prostanoid synthesis in cells (reviewed in [6]). Recently, Moolenaar's group showed that the recombinant SMaseD from L. laeta cleaved LPC (lysophosphatidylcholine) to LPA (lysophosphatidic acid) and choline [7]. LPA is known to induce various biological and pathological responses such as platelet aggregation, endothelial hyperpermeability and pro-inflammatory responses by signalling through three G-protein-coupled receptors [8,9]. Thus some of the pathology associated with the envenomation by the brown recluse spider might be due to aberrant LPA production and signalling.
To initiate a study of the role of LPA production and LPA signalling pathways in the manifestations of envenomation by Loxosceles species, we cloned SMaseD from L. reclusa and characterized the recombinant SMaseD regarding substrate specificity and enzyme function. The results of this initial characterization are reported herein. The L. reclusa PLD cDNA sequence has been deposited with GenBank® (accession no. AY862486). The sequence was determined by automated DNA sequencing using the cDNA isolated by the authors of this paper, and this sequence is reported for the first time in this paper.
EXPERIMENTAL
Materials and reagents
Lipids were purchased from Avanti Polar Lipids (Alabaster, AL, U.S.A.). Essentially fatty acid-free BSA, referred to as faf-BSA, peroxidase (EC 1.11.1.7; hydrogen-peroxide oxidoreductase), choline oxidase (EC 1.1.3.17), glycerol-3-phosphate oxidase (EC 1.1.3.21), glycerol-3-phosphate dehydrogenase (EC 1.1.1.8) and 3-α-hydroxysteroid dehydrogenase (EC 1.1.1.50) were purchased from Sigma–Aldrich (St. Louis, MO, U.S.A.). Lysophospholipase (EC 3.1.1.5) was purchased from either Sigma–Aldrich or MP Biomedical (Irvine, CA, U.S.A.). A2058 (human melanoma) cells were a gift from Dr T. Clair (National Institutes of Health, Bethesda, MD, U.S.A.). S-protein–HRP conjugate (where HRP stands for horseradish peroxidase) was purchased from Novagen (San Diego, CA, U.S.A.).
Cell-culture conditions
The growth medium used for A2058 cells was DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) CD-FBS (charcoal dextran-stripped fetal bovine serum; Gemini Bioscience, Woodland, CA, U.S.A.) and antibiotics (100 units/ml penicillin, 100 μg/ml streptomycin and 250 ng/ml amphotericin B) (Invitrogen, Carlsbad, CA, U.S.A.).
Cloning of the SMaseD from venom glands of L. recluse
Total RNA was extracted from two pairs of L. reclusa venom glands purchased from SpiderPharm (Yarnell, AZ, U.S.A.) using the RNeasy kit (Qiagen, Valencia, CA, U.S.A.). A fragment [580–963 bp of the full translational ORF (open reading frame)] was amplified by using a degenerate primer set (sense, 5′-gggcatcccgagttaatgga-3′; anti-sense, 3′-ttaattcttgaatgtttccc-3′) designed from the nucleotide sequence of the related species, L. arizonica. The full mRNA sequence was identified by 3′- and 5′-RACE (3′- and 5′-rapid amplification of cDNA ends) using the Generacer kit (Invitrogen). The full ORF was subcloned DNA into pET30 (Novagen) for bacterial cell expression. The correctness of plasmid constructs was confirmed by automated DNA sequencing (Biomedical Research Core Laboratory, University of Virginia).
Introduction of point mutations into SMaseD
To construct mutant SMaseDs, the following primer sets were used for amplification of the plasmid construct. H37N (His37→Asn) (sense, 5′-cctatatggatcatggggaacatggtcaacgctatttat-3′; antisense, 5′-ataaatagcgttgaccatgttccccatgatccatatagg-3′), H73N (sense, 5′-aatcctgaatacacgtataacggcgttccatgtgattgc-3′; antisense, 5′-gcaatcacatggaacgccgttatacgtgtattcaggatt-3′), H108N (sense, 5′-ccaggcgattccaagtataatgcaaaattagtgttagtt-3′; antisense, 5′-aactaacactaattttgcattatacttggaatcgcctgg-3′), H164N (sense, 5′-tccataccagaccttaacaattataaattaattactgga-3′; antisense, 5′-tccagtaattaatttataattgttaaggtctggtatgga-3′), H180N (sense, 5′-acgctcaaaagcgaggggaatcccgagttaatggacaaa-3′; antisense, 5′-tttgtccattaactcgggattcccctcgcttttgagcgt-3′), H108A (sense, 5′-ccaggcgattccaagtatgctgcaaaattagtgttagtt-3′; antisense, 5′-aactaacactaattttgcagcatacttggaatcgcctgg-3′), T258V (sense, 5′-acagtggacaagcgcgcaacggttagagaggcactcgatgctgga-3′; antisense, 5′-tccagcatcgagtgcctctctaaccgttgcgcgcttgtccactgt-3′). Following amplification, the template DNA was digested with the restriction endonuclease DpnI and the plasmid was used to transform Escherichia coli TOP10F′ competent cells (Invitrogen). The correct mutations at the target sites were verified by automated DNA sequencing.
Purification of the recombinant SMaseD from bacteria
The SMaseD-pET30 plasmid was transformed into E. coli [strain: BL21(DE3)pLysS; Novagen.] and 1 mM isopropyl β-D-thiogalactoside was used for the induction of protein expression. The commercial bacterial cell lysis reagent (BugBuster; Novagen) including 25 unit/ml DNase I (Sigma, St. Louis, MO, U.S.A.) was used for the preparation of total cell lysates. Nickel-linked Sepharose was used for the purification of recombinant SMaseD from the bacterial cell lysates (Amersham Biosciences, Piscataway, NJ, U.S.A.) following the manufacturer's instructions. The SMaseD was found to be stable and resistant to bacterial protease; thus specific protease inhibitors were not used during the purification procedure. To increase the degree of purity, the S-protein–agarose (Novagen) was used as recommended by the manufacturer. The buffer used in the final elution was exchanged with PBS using a dialysis membrane (10 kDa cut-off) or a Centricon membrane (30 kDa cut-off). The concentration of the purified protein was determined by the FRETWorks S-Tag assay kit (Novagen) following the method recommended by the manufacturer.
Western blotting
Protein samples were subjected to SDS/PAGE (12–14% polyacrylamide) and transferred on to nitrocellulose paper (Bio-Rad Laboratories, Hercules, CA, U.S.A.) using typical electrotransfer procedures. For detection of recombinant proteins encoded by SMaseD-pET30, the S-protein–HRP conjugate was used (Novagen). Bound S-protein was detected by using the Lightning Chemiluminescence reagent Plus kit (PerkinElmer, Boston, MA, U.S.A.).
Choline release assay
In some cases, the hydrolytic activity of the recombinant SMaseD protein was determined by measuring the amount of choline released from the phospholipid substrate [7]. Briefly, 1 mM substrate in Hepes-buffered saline (10 mM Hepes, 140 mM NaCl, 0.5 mM KCl, 0.1 mM MgCl2 and 0.1% faf-BSA, pH 7.4) was incubated with the recombinant enzyme for the indicated times (standard, 1 h) at 37 °C in a total volume of 0.1 ml. The same volume of enzyme cocktail {50 mM Tris/HCl, 2.7 mM TOOS [N-ethyl-N-(2-hydroxy-3-sulphopropyl)-m-toluidine], 4.5 mM 4-aminoantipyrine, 47.7 units/ml peroxidase and 18 units/ml choline oxidase, pH 8.0} was added and incubated for a further 15 min at 37 °C. The amount of choline released from the reaction was calculated by measuring light absorbance A at 550 nm. In the case of SM (sphingomyelin), a 100 mM stock solution in 95% ethanol was diluted to the required concentration in Hepes-buffered saline. Ethanol at concentrations below 1.0% did not show significant inhibition of SMaseD enzyme activity or a decrease in Km value.
LPA assay
To measure the hydrolysis of lysophospholipids with head groups other than choline, the amount of LPA released was measured by a colorimetric LPA assay [10]. Briefly, a 1 mM substrate was incubated with recombinant SMaseD for the indicated time (standard, 1 h) in a total volume of 0.1 ml of Hepes-buffered saline. The same volume of enzyme cocktail was then added to the reaction. The enzyme cocktail consisted of 20 units/ml lysophospholipase, 1.3 units/ml peroxidase, 100 units/ml glycerol-3-phosphate oxidase, 10 units/ml glycerol-3-phosphate dehydrogenase (added just before the assay), 10 units/ml 3-α-hydroxysteroid dehydrogenase, 0.01 mM NADH, 1.0 mM cholic acid, 0.5 mM TOOS, 1 mM 4-AAP (4-aminoantipyrine), 0.01% Triton X-100 and 100 mM Hepes (pH 7.6). After incubation for 15 min to allow colour development, the light absorbance at 570 nm was measured and the amount of LPA was calculated from the standard curve generated using LPA18:1. The colour development was linear for more than 15 min using LPA18:1 concentrations up to 1 mM. To measure the level of contaminating G3P (glycerol 3-phosphate) in lipid substrates, the same enzyme cocktail without lysophospholipase was used and the amount of G3P was calculated from the standard curve generated using G3P. The amount of contaminating G3P (generally <5%) was subtracted when constructing the LPA standard curve.
Migration assay
The lower surface of transwell membranes (8 μm pore size; 24 mm diameter; Corning Costar, Corning, NY, U.S.A.) was coated with 0.01 mg/ml matrigel (growth factor-reduced; BD Biosciences, Bedford, MA, U.S.A.) following the manufacturer's instructions. To prepare the cell suspension, 90% confluent A2058 cell monolayers were detached by a brief treatment with 10× trypsin/EDTA solution (Invitrogen). Detached cells were washed once with normal growth media and one more time with the assay medium (DMEM without Phenol Red, 0.1% faf-BSA, no serum), and finally resuspended in assay media at 1.5×106 cells/ml. Cell suspension (1 ml) was added to the upper chamber and 2 ml of the assay media was placed in the lower chamber. After waiting 1 min for volume equilibration between the chambers, 0.02 ml of compounds (100× stock in 3% faf-BSA) was added to the lower chamber. After 4 h of incubation at 37 °C to allow cell migration, the assay media in both chambers were discarded and the cells from the lower surface of the membrane were detached by adding 0.8 ml of 10× trypsin/EDTA solution to the lower chamber. To confirm complete detachment of the migrated cells from the lower surface of the membrane, unmigrated cells were discarded by swabbing the upper membrane surface, and the lower surface of the membrane was examined by microscopy after staining with Diffquick staining kit (IMEB Inc., San Marcos, CA, U.S.A.). A 0.1 ml aliquot of the migrated cell suspension from the lower chamber was mixed with the same volume of 2× CyQuant assay solution (Molecular Probes, Eugene, OR, U.S.A.), and the cell mass was calculated by measuring the fluorescence at 480/520 nm using a FlexStation fluorimeter (Molecular Devices, Sunnyvale, CA, U.S.A.).
Haemolysis assay
Human erythrocytes (freshly donated) were washed three times with VBS2+ (veronal-buffered saline, pH 7.4; 10 mM sodium barbitone, 0.15 mM CaCl2, 0.5 mM MgCl2 and 145 mM NaCl) and resuspended in VBS2+ at 2%. The cells were incubated with the recombinant enzyme for 30 min at 37 °C. Control samples were incubated with VBS2+ buffer alone. After washing five times with VBS2+, the cells were resuspended to the original volume of VBS2+ and analysed for haemolysis [11]. Briefly, 0.1 ml of 2% erythrocytes pretreated with purified toxin was mixed with 0.1 ml of autologous plasma. Background or total cell lysis was evaluated by incubation of erythrocytes with VBS2+ or water respectively. After incubation for 1 h at 37 °C, non-lysed cells were discarded by centrifugation; the light absorbance of the released haemoglobin was measured at 405 nm.
RESULTS
Cloning of the SMaseD from L. reclusa
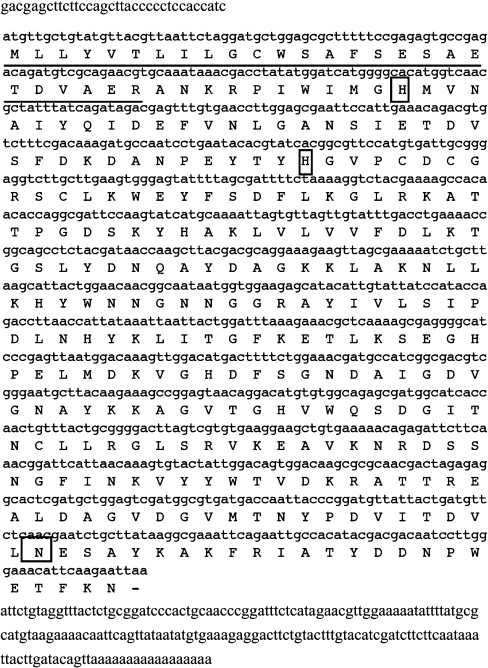
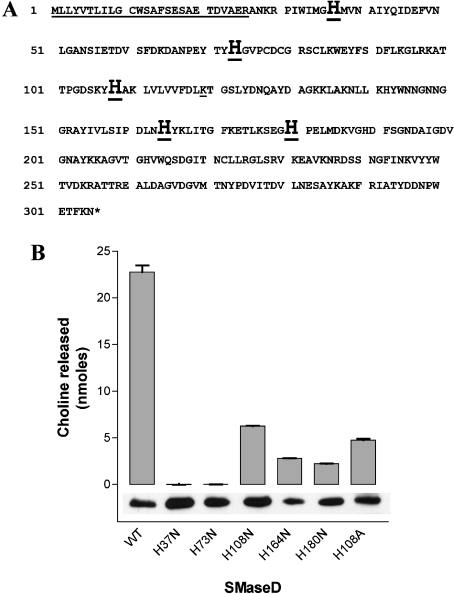
The full sequence of mRNA encoding SMaseD was identified from cDNA made from the total RNA extracted from the venom glands of L. reclusa (Figure 1). The mRNA sequence includes a 918 bp full translational ORF, 34 bp of 5′-UTR (5′-untranslated region) and 181 bp of 3′-UTR. The conceptionalized peptide consisted of 305 amino acids (molecular mass, 34 kDa). Peptide sequence analysis showed one possible N-glycosylation site (Asn282).
Figure 1. Complete cDNA and translated peptide sequence of L. reclusa SMaseD.
Total RNA was extracted from the venom glands of L. reclusa (SpiderPharm). The degenerate primers designed from the sequence of SMaseD from related species, L. arizonica, were used for amplification of the initial fragment of SMaseD (580–963 bp of full ORF). The full cDNA sequence was identified by 3′- and 5′-RACE. Underlined, putative signal peptide; boxed, catalytic histidine residues, potential N-glycosylation site.
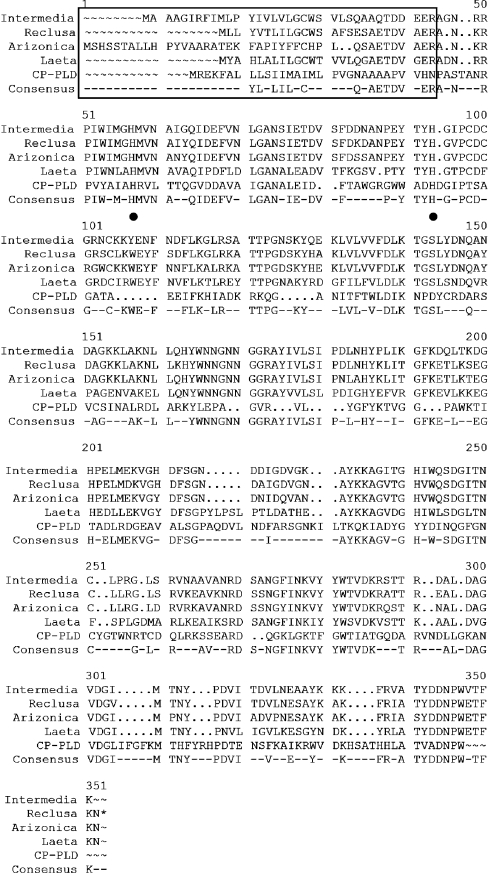
Recently, amino acid sequences of SMaseD from several Loxo-sceles species were reported [12–14]. Sequence alignment showed that the L. reclusa SMaseD shares 87% (L. arizonica), 60% (L. laeta), 85% (L. intermedia) and 84 and 46% (L. boneti SMaseD1 and SMaseD2) identical amino acids with SMaseD from these other Loxosceles species. The SMaseDs from L. arizonica and L. intermedia have longer signal peptide sequences than either L. laeta or L. reclusa. The location of the signal peptide in L. reclusa is inferred from the reported amino acid sequence of L. reclusa SMaseD protein purified from the venom [14]. The only other protein with significant amino acid sequence similarity to these spider enzymes is a bacterial (Corynebacterium pseudotuberculosis) PLD [7]. The alignment of these proteins is displayed in Figure 2.
Figure 2. Peptide sequence alignment of SMaseDs.
Intermedia, L. intermedia SMaseD; Reclusa, L. reclusa SMaseD; Arizonica, L. arizonica SMaseD; Laeta, L. laeta SMaseD; CP–PLD, C. pseudotuberculosis PLD. The GCG program, PRETTY, was used for the analysis. Solid line box, putative signal sequences; ●, histidine residues essential for enzyme activity. The 305 amino acid peptide sequence of L. reclusa enzyme is 87, 85 and 60% identical with those of L. arizonica, L. intermedia and L. laeta respectively.
Expression of the recombinant SMaseD in bacterial cells
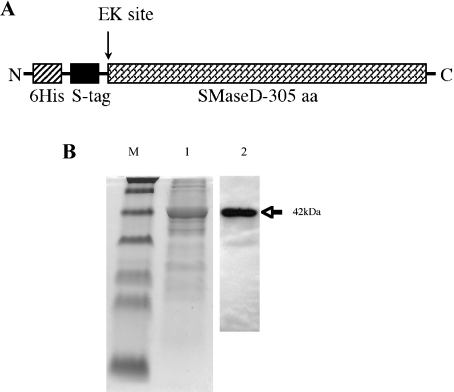
Forced expression of epitope-tagged recombinant SMaseD in bacteria resulted in a soluble protein that migrated at a relative molecular mass of 42 kDa (Figure 3). We purified the recombinant SMaseD from bacterial cell lysates using sequential affinity columns of Ni-linked Sepharose resin and S-protein–agarose. This purified protein was used to examine substrate specificity and for further characterization.
Figure 3. The expression of recombinant SMaseD in a bacterial expression system.
(A) Schematic diagram of recombinant SMaseD construct expressed in bacteria. EK, enterokinase cleavage site. (B) Coomassie Blue staining (lane 1, 2.4 μg of SMaseD loaded) and Western blotting with S-protein–HRP conjugate (lane 2, 0.1 μg of SMaseD loaded) of the SMaseD purified from the bacterial cell lysates by sequential affinity purification using Ni-Sepharose (Amersham Biosciences). Lane M, molecular mass standards.
Recombinant SMaseD has broad substrate specificity
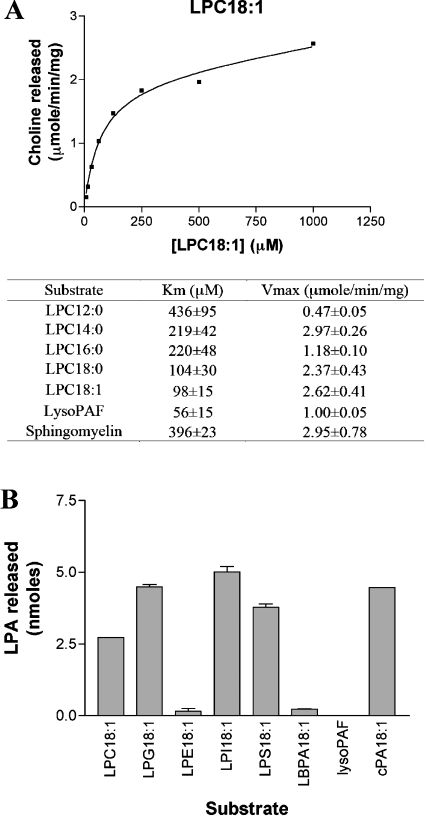
As shown in Figure 4(A), the purified recombinant protein hydrolysed sphingomyelin as detected by the appearance of choline. Furthermore, sphingomyelin with artificially short fatty acids (C2:0 and C6:0) was also hydrolysed (results not shown). Thus, as expected, the cDNA that we cloned from venom gland RNA encodes a functional SMaseD. Also, as expected from a previous report using L. laeta SMaseD, LPC with various acyl chains (C18:1, C18:0, C16:0, C14:0 and C12:0) was a substrate [7]. Compared with the previous kinetic values of mammalian lysoPLD (LPC-preferring PLD), ATX (autotaxin; Km=250 μM and Vmax=9.0 μmol·min−1·mg−1 for LPC18:1) [15], the recombinant SMaseD showed approx. 3-fold lower Km and Vmax values. In contrast, PC (phosphatidylcholine) with various acyl chains (C22:6, C20:4 and C6:0) did not show any detectable hydrolysis (results not shown). We conclude that the SMaseD from L. reclusa, like that of L. laeta, has intrinsic lysoPLD activity. Interestingly, the L. reclusa enzyme also hydrolysed lyso-PAF (where PAF stands for platelet activating factor) but not PAF, which has an acetyl group at the sn2 position. We examined the hydrolysis of edelfosine (a PAF analogue) and miltefosine (an antimetastatic agent) [16,17] using the enzymatic choline detection method, but the purified enzyme did not release choline from either of these molecules (results not shown). To examine the hydrolysis of other lysophospholipids without a choline head group, we measured the amount of LPA released as described in the Experimental section [10]. As documented in Figure 4(B), LPI (18:1-1-oleoyl-lysophosphatidylinositol), LPG (18:1-1-oleoyl-lysophosphatidylglycerol), LPE (18:1-1-oleoyllysophosphatidylethanolamine), LBPA (18:1-1-oleoyl-lysobisphosphatidic acid) and LPS (18:1-1-oleoyl-lysophosphatidylserine) were all hydrolysed by the recombinant enzyme. At a concentration of 1 mM, the substrate preference was LPI > LPG > LPS>LPC≫LPE, LBPA. Furthermore, the recombinant enzyme also hydrolysed cyclic phosphatidic acid to LPA. In contrast with the mammalian lysoPLD (ATX) [18], we did not detect hydrolysis of SPC (sphingosylphosphorylcholine, also known as lyso-sphingomyelin) (results not shown). The SMaseD did not exhibit detectable PLA1/2 activity, which was examined by using bis-BODIPY FL C11-PC (Molecular Probes) as a potential substrate (results not shown).
Figure 4. Substrate specificity of the purified recombinant SMaseD.
(A) LysoPLD activity was assayed by measuring hydrolysis of various lysophospholipids with choline head groups. Briefly, the substrate in Hepes-buffered saline was incubated with SMaseD (125 ng) for 1 h at 37 °C in a total volume of 0.1 ml. The amount of choline released from the reactions was measured as described in the Experimental section. Km and Vmax values were calculated using the Michaelis–Menten equation. Results shown are from a representative experiment of at least three independent assays. (B) Hydrolysis of non-choline lysophospholipids and cyclic phosphatidic acid by SMaseD was measured. The LPA produced from the reaction of 1 mM substrate with SMaseD (1 ng) was measured by direct LPA assay described in the Experimental section. LPA18:1 was used as a positive control, whereas lysoPAF was used as negative control for the assay. (Although SMaseD hydrolyses lysoPAF, the product, ether LPA, cannot be detected because lysophospholipase in LPA assay enzyme cocktail does not cleave ether-linkage.) The results shown are the means±S.E.M. for triplicate experiments.
His37 and His73 are essential for enzyme activity of the recombinant SMaseD
The peptide sequence analysis of the spider SMaseD suggested one putative GDPD (glycerol phosphodiester phosphodiesterase) domain (amino acids 246–276) and a potential HKD PLD catalytic domain (HXKXXXD, amino acids 108–117) [19]. However, the point mutation of amino acids in these consensus sequences (T258V, H108N and H108A) did not abolish enzyme activity (Figure 5). Because previous studies of the catalytic mechanism of PLD suggested the involvement of histidine residue(s) [20], we systematically mutated histidine residues conserved among Loxosceles SMaseDs (Figure 5A). The H37N and H73N mutations abolished detectable hydrolysing activity using LPC18:1 as a substrate (Figure 5B). Likewise, the H37N and H73N mutant proteins did not hydrolyse SM (results not shown). We could not find any peptide domain previously reported that has significant similarity to the peptide sequence of amino acids 37–73 of the spider enzyme.
Figure 5. His37 and His73 are required for enzyme activity of the recombinant L. reclusa SMaseD.
(A) The mutated amino acids are indicated by large bold letters. Potential signal peptide is underlined. (B) LysoPLD activity of the wild-type and mutant SMaseD proteins (2 ng/reaction) expressed in bacteria. Briefly, 1 mM LPC18:1 in Hepes-buffered saline was incubated with SMaseD for 1.5 h at 37 °C in a total volume of 0.1 ml. The amount of choline released from the reactions was measured as described in the Experimental section. The presence of the wild-type and mutant proteins (100 ng/lane) was confirmed by Western blotting using S-protein–HRP conjugate. The results shown are the means±S.E.M. for triplicate experiments.
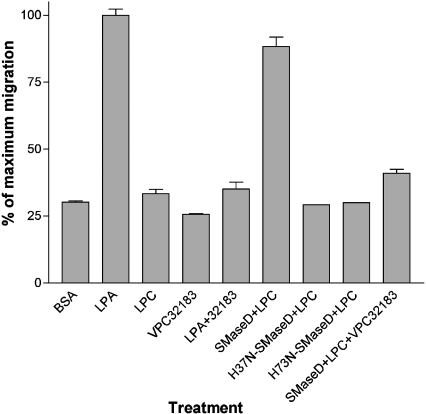
The recombinant SMaseD induced the migration of human melanoma A2058 cells through LPA receptor signalling
LPA is known to induce the chemotactic migration of the human A2058 melanoma cells via the LPA1 receptor [21–23]. ATX, which is a recently identified mammalian lysoPLD, evokes A2058 cell migration by releasing LPA from LPC [23]. Initially, we were unable to observe migration when recombinant spider SMaseD and LPC were added to the lower chamber of the migration well without pre-incubation. However, when the enzyme and substrate were pre-incubated, migration was observed and this migration was sensitive to the LPA1 receptor antagonist, VPC32183 (Figure 6). This result suggests that the migration assay buffer (DMEM+0.1% faf-BSA) might not be optimal for LPA generation by the L. reclusa SMaseD.
Figure 6. Recombinant L. reclusa SMaseD induced the migration of human melanoma A2058 cells.
Purified wild-type, H37N and H73N recombinant SMaseDs were used for the induction of A2058 cell migration as described in the Experimental section. Lipid concentrations were: LPA18:1, 50 nM; LPC18:1, 2.5 μM; and VPC32183, 10 μM. WT and mutant SMaseDs (5 ng) and LPC18:1 (1 mM) were pre-incubated for 2 h at 37 °C in a total volume of 20 μl after which 5 μl of the final reaction mixture was added to the 2 ml of assay buffer in the lower chamber of transwell. The results shown are the means±S.E.M. for duplicate experiments.
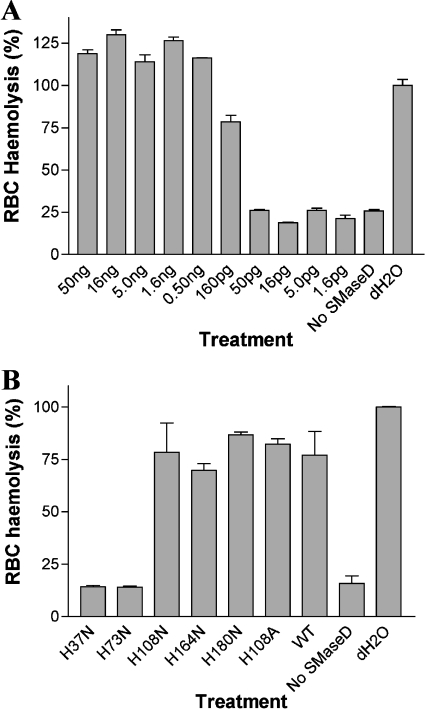
The purified recombinant SMaseD induced the erythrocyte haemolysis
Among the effects of Loxosceles PLD is erythrocyte haemolysis [11]. As shown in Figure 7(A), treatment of the human erythrocytes with purified recombinant of SMaseD from L. reclusa also induced haemolysis in a dose-dependent manner. This activity was not observed with the catalytically inactive (H37N and H73N) SMaseD protein (Figure 7B).
Figure 7. The purified recombinant L. reclusa SMaseD showed dose-dependent erythrocyte lysis.
Human erythrocytes (freshly donated) were washed three times with VBS2+ and resuspended in the same buffer at 2%. The cells were incubated with the purified recombinant SMaseDs [indicated amounts of wild-type SMaseD (A) and 2 ng of wild-type and mutant SMaseD (B)] for 30 min at 37 °C. Control samples were incubated with VBS2+ buffer alone. After washing with VBS2+, the cells were resuspended to the original volume of VBS2+ (0.1 ml) and mixed with 0.1 ml of autologous plasma. After incubation for 1 h at 37 °C, non-lysed cells were discarded by centrifugation (1000 g and 1 min); the light absorbance of the released haemoglobin was measured at 405 nm. Background or total cell lysis was evaluated by incubation of erythrocytes with VBS2+ (no SMaseD) or distilled water (dH2O) respectively. The results shown are the means±S.E.M. for duplicate experiments.
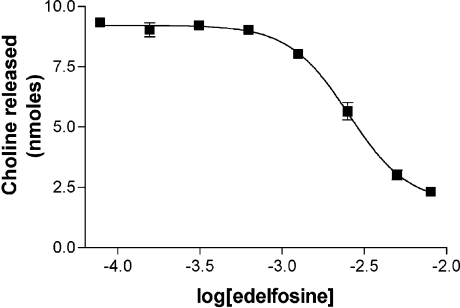
Edelfosine inhibited the lysoPLD enzyme activity of the recombinant SMaseD
Finally, we examined each lipid that was not a substrate for inhibitory activity. In doing so, we found that edelfosine inhibited the enzyme activity of recombinant L. reclusa SMaseD (Figure 8).
Figure 8. Edelfosine inhibits lysoPLD activity of the recombinant L. reclusa SMaseD.
Wild-type SMaseD (1 ng) was incubated with 1 mM LPC18:1 and various concentrations of edelfosine for 1 h at 37 °C in a total volume of 0.1 ml of Hepes-buffered saline. The amount of choline released from the reactions was measured by the enzymatic choline detection assay as described in the Experimental section. The results shown are the means±S.E.M. for triplicate experiments.
DISCUSSION
Using several Loxosceles sp. SMaseD DNA sequences as a guide, we cloned a similar cDNA from RNA of L. reclusa venom glands. As expected, the L. reclusa protein is closely related (60–87% identical amino acids) to the SMaseD enzymes of other Loxosceles species and we found that the recombinant L. reclusa protein releases choline from sphingomyelin, i.e. it has ‘SMaseD’ activity. We confirmed also for L. reclusa SMaseD the seminal finding of Moolenaar's laboratory that L. laeta SMaseD is a lysoPLD [7]. We extended that finding by demonstrating that lysophospholipids with a variety of headgroups are hydrolysed by the L. reclusa enzyme to release LPA. Thus this enzyme, as well as presumably related proteins from other Loxosceles sp., is both an SMaseD and a broad-spectrum lysoPLD. Like the L. laeta enzyme, the L. reclusa enzyme displays Km values for albumin-presented LPC that are well within the range of LPC levels in blood [24].
Through site-directed mutation of histidine codons, we identified two histidine residues (His37 and His73) that, when changed to asparagine residues, resulted in catalytically inactive proteins. While this manuscript was being revised, Murakami et al. published a high-resolution structure obtained from L. laeta SMaseD crystals [25]. We were gratified to find that the histidine residues identified as essential for catalysis by our mutation studies (i.e. His37 and His73) were found to be integral to the active site predicted by the crystal structure. A distantly related bacterial PLD from C. pseudotuberculosis has a similar substrate pattern [7]. Mutation of His44 of the bacterial enzyme, which corresponds to His37 in L. reclusa (Figure 2), results in loss of enzyme activity [26]. This concordance strengthens the argument that the spider and bacterial enzymes are related. The mutant proteins allow testing of the presumption, widely held but heretofore not verified, that catalytic activity is necessary for the pathological activities ascribed to Loxosceles PLD. Indeed, in one assay of biological activity, haemolysis, we found that catalytic activity is required.
We discovered that the PAF analogue, edelfosine, is a low-affinity inhibitor of the enzyme. Although a variety of glycerol-containing phospholipids are hydrolysed by the L. reclusa enzyme, our data suggest a strict preference for a lyso structure, i.e. an hydroxyl at the sn2 position of the glyceryl moiety. This sensitivity was demonstrated most directly with the ether lipid series where lysoPAF (sn2 hydroxyl) is a substrate while PAF (sn2 acetyl) is not and edelfosine (sn2 methoxy) is an inhibitor. A curious feature of the Loxosceles SMaseD enzymes is that, whereas only glycerol lysophospholipids are recognized, sphingoid lipids are not hydrolysed as the lyso form (e.g. SPC). Rather, sphingolipids are substrates only when the nitrogen at the second carbon is in an amide linkage, i.e. sphingomyelin (ceramide-1-phosphorylcholine). It would be interesting to know whether the SM analogues with different head groups (e.g. dihydroceramide-1-phosphorylinositol from yeast) are substrates of the spider enzyme.
Finally, we endorse Moolenaar's suggestion that the name ‘SMaseD’ is overly restrictive and thus not accurate. The ‘lysoPLD’ designation, although broader, is also not fully accurate. Although ‘SMaseD/lysoPLD’ is accurate, it seems rather too cumbersome. We suggest that the Loxosceles enzyme be referred to simply as ‘PLD’ to avoid the connotations inherent in other names.
Acknowledgments
This work was supported by a grant from the U.S. Public Health Service (NIH R01 GM052722) to K. R. L. We are grateful to Dr T. Clair (National Institutes of Health) for his gift of A2058 cells and to Dr B. H. Heasley and Dr T. L. Macdonald (Department of Chemistry, University of Virginia) for their gift of VPC32183, which is available now from Avanti Polar Lipids.
References
- 1.Sams H. H., Dunnick C. A., Smith M. L., King L. E., Jr Necrotic arachnidism. J. Am. Acad. Dermatol. 2001;44:561–573. doi: 10.1067/mjd.2001.112385. [DOI] [PubMed] [Google Scholar]
- 2.Tambourgi D. V., Magnoli F. C., van den Berg C. W., Morgan B. P., de Araujo P. S., Alves E. W., Da Silva W. D. Sphingomyelinases in the venom of the spider Loxosceles intermedia are responsible for both dermonecrosis and complement-dependent hemolysis. Biochem. Biophys. Res. Commun. 1998;251:366–373. doi: 10.1006/bbrc.1998.9474. [DOI] [PubMed] [Google Scholar]
- 3.Zanetti V. C., da Silveira R. B., Dreyfuss J. L., Haoach J., Mangili O. C., Veiga S. S., Gremski W. Morphological and biochemical evidence of blood vessel damage and fibrinogenolysis triggered by brown spider venom. Blood Coagul. Fibrinolysis. 2002;13:135–148. doi: 10.1097/00001721-200203000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Patel K. D., Modur V., Zimmerman G. A., Prescott S. M., McIntyre T. M. The necrotic venom of the brown recluse spider induces dysreguated endothelial cell-dependent neutrophil activation. J. Clin. Invest. 1994;94:631–642. doi: 10.1172/JCI117379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gulbins E., Kolesnick R. Raft ceramide in molecular medicine. Oncogene. 2003;22:7070–7077. doi: 10.1038/sj.onc.1207146. [DOI] [PubMed] [Google Scholar]
- 6.Gomez-Munoz A. Ceramide-1-phosphate: a novel regulator of cell activation. FEBS Lett. 2004;562:5–10. doi: 10.1016/s0014-5793(04)00211-x. [DOI] [PubMed] [Google Scholar]
- 7.van Meeteren L. A., Frederiks F., Giepmans B. N., Pedrosa M. F., Billington S. J., Jost B. H., Tambourgi D. V., Moolenaar W. H. Spider and bacterial sphingomyelinases D target cellular lysophosphatidic acid receptors by hydrolyzing lysophosphatidylcholine. J. Biol. Chem. 2004;279:10833–10836. doi: 10.1074/jbc.C300563200. [DOI] [PubMed] [Google Scholar]
- 8.Anliker B., Chun J. Lysophospholipid G protein-coupled receptors. J. Biol. Chem. 2004;279:20555–20558. doi: 10.1074/jbc.R400013200. [DOI] [PubMed] [Google Scholar]
- 9.Moolenaar W. H., van Meeteren L. A., Giepmans B. N. The ins and outs of lysophosphatidic acid signaling. BioEssays. 2004;26:870–881. doi: 10.1002/bies.20081. [DOI] [PubMed] [Google Scholar]
- 10.Kishimoto T., Matsuoka T., Imamura S., Mizuno K. A novel colorimetric assay for the determination of lysophosphatidic acid in plasma using an enzymatic cycling method. Clin. Chim. Acta. 2003;333:59–67. doi: 10.1016/s0009-8981(03)00165-7. [DOI] [PubMed] [Google Scholar]
- 11.Tambourgi D. V., De Sousa Da Silva M., Billington S. J., Goncalves De Andrade R. M., Magnoli F. C., Songer J. G., Van Den Berg C. W. Mechanism of induction of complement susceptibility of erythrocytes by spider and bacterial sphingomyelinases. Immunology. 2002;107:93–101. doi: 10.1046/j.1365-2567.2002.01483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandes Pedrosa M. F., Junqueira de Azevedo Ide L., Goncalves-de-Andrade R. M., van den Berg C. W., Ramos C. R., Ho P. L., Tambourgi D. V. Molecular cloning and expression of a functional dermonecrotic and haemolytic factor from Loxosceles laeta venom. Biochem. Biophys. Res. Commun. 2002;298:638–645. doi: 10.1016/s0006-291x(02)02521-4. [DOI] [PubMed] [Google Scholar]
- 13.Kalapothakis E., Araujo S. C., de Castro C. S., Mendes T. M., Gomez M. V., Mangili O. C., Gubert I. C., Chavez-Olortegui C. Molecular cloning, expression and immunological properties of LiD1, a protein from the dermonecrotic family of Loxosceles intermedia spider venom. Toxicon. 2002;40:1691–1699. doi: 10.1016/s0041-0101(02)00201-5. [DOI] [PubMed] [Google Scholar]
- 14.Ramos-Cerrillo B., Olvera A., Odell G. V., Zamudio F., Paniagua-Solis J., Alagon A., Stock R. P. Genetic and enzymatic characterization of sphingomyelinase D isoforms from the North American fiddleback spiders Loxosceles boneti and Loxosceles reclusa. Toxicon. 2004;44:507–514. doi: 10.1016/j.toxicon.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 15.Umezu-Goto M., Kishi Y., Taira A., Hama K., Dohmae N., Takio K., Yamori T., Mills G. B., Inoue K., Aoki J., et al. Autotaxin has lysophospholipase D activity leading to tumor cell growth and motility by lysophosphatidic acid production. J. Cell Biol. 2002;158:227–233. doi: 10.1083/jcb.200204026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardile V., Mascarucci P., De Simoni M. G., Jiang X., Bindoni M. The effect of ether lipid 1-O-octadecyl-2-O-methoxy-rac-glycero-3-phosphocholine and its analogues platelet activating factor and carbamyl-platelet activating factor on the biosynthesis of interleukin-6 in human thymic epithelial cells cultivated in vitro. Cytokine. 1996;8:698–701. doi: 10.1006/cyto.1996.0092. [DOI] [PubMed] [Google Scholar]
- 17.Wieder T., Orfanos C. E., Geilen C. C. Induction of ceramide-mediated apoptosis by the anticancer phospholipid analog, hexadecylphosphocholine. J. Biol. Chem. 1998;273:11025–11031. doi: 10.1074/jbc.273.18.11025. [DOI] [PubMed] [Google Scholar]
- 18.Clair T., Aoki J., Koh E., Bandle R. W., Nam S. W., Ptaszynska M. M., Mills G. B., Schiffmann E., Liotta L. A., Stracke M. L. Autotaxin hydrolyzes sphingosylphosphorylcholine to produce the regulator of migration, sphingosine-1-phosphate. Cancer Res. 2003;63:5446–5453. [PubMed] [Google Scholar]
- 19.Stuckey J. A., Dixon J. E. Crystal structure of a phospholipase D family member. Nat. Struct. Biol. 1999;6:278–284. doi: 10.1038/6716. [DOI] [PubMed] [Google Scholar]
- 20.Leiros I., McSweeney S., Hough E. The reaction mechanism of phospholipase D from Streptomyces sp. strain PMF. Snapshots along the reaction pathway reveal a pentacoordinate reaction intermediate and an unexpected final product. J. Mol. Biol. 2004;339:805–820. doi: 10.1016/j.jmb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Hama K., Aoki J., Fukaya M., Kishi Y., Sakai T., Suzuki R., Ohta H., Yamori T., Watanabe M., Chun J., et al. Lysophosphatidic acid and autotaxin stimulate cell motility of neoplastic and non-neoplastic cells through LPA1. J. Biol. Chem. 2004;279:17634–17639. doi: 10.1074/jbc.M313927200. [DOI] [PubMed] [Google Scholar]
- 22.Shida D., Kitayama J., Yamaguchi H., Okaji Y., Tsuno N. H., Watanabe T., Takuwa Y., Nagawa H. Lysophosphatidic acid (LPA) enhances the metastatic potential of human colon carcinoma DLD1 cells through LPA1. Cancer Res. 2003;63:1706–1711. [PubMed] [Google Scholar]
- 23.Koh E., Clair T., Woodhouse E. C., Schiffmann E., Liotta L., Stracke M. Site-directed mutations in the tumor-associated cytokine, autotaxin, eliminate nucleotide phosphodiesterase, lysophospholipase D, and motogenic activities. Cancer Res. 2003;63:2042–2045. [PubMed] [Google Scholar]
- 24.Aoki J., Taira A., Takanezawa Y., Kishi Y., Hama K., Kishimoto T., Mizuno K., Saku K., Taguchi R., Arai H. Serum lysophosphatidic acid is produced through diverse phospholipase pathways. J. Biol. Chem. 2002;277:48737–48744. doi: 10.1074/jbc.M206812200. [DOI] [PubMed] [Google Scholar]
- 25.Murakami M. T., Fernandes-Pedrosa M. F., Tambourgi D. V., Arni R. K. Structural basis for metal-ion coordination and the catalytic mechanism of sphingomyelinases D. J. Biol. Chem. 2005;280:13658–13664. doi: 10.1074/jbc.M412437200. [DOI] [PubMed] [Google Scholar]
- 26.Tachedjian M., Krywult J., Moore R. J., Hodgson A. L. Caseous lymphadenitis vaccine development: site-specific inactivation of the Corynebacterium pseudotuberculosis phospholipase D gene. Vaccine. 1995;13:1785–1792. doi: 10.1016/0264-410x(95)00144-p. [DOI] [PubMed] [Google Scholar]