Abstract
The MUPs (major urinary proteins) of the house mouse, Mus domesticus, are lipocalins that bind and slowly release male-specific pheromones in deposited scent marks. However, females also express these proteins, consistent with a second role in encoding individual signatures in scent marks. We have purified and characterized an atypical MUP from the urine of male C57BL/6J inbred mice, which is responsible for the binding of most of the male pheromone, 2-sec-butyl-4,5-dihydrothiazole, and which is also responsible for the slow release of this pheromone from scent marks. This protein is absent from the urine of female mice of the same strain. The protein has been characterized by MS, leading to unequivocal identification as a previously uncharacterized gene product, providing compelling evidence for the expression of this gene in liver and manifestation in urine. These properties contrast strongly with those of the other MUPs in the same urine sample, and suggest that the requirement to manifest a male-specific pheromone has been met by evolution of a cognate protein specifically adapted to the binding and release of this ligand. This atypical MUP is also present in a random sample of wild-caught male mice, confirming that this protein is not specific to the inbred mouse strain but is present in natural populations also.
Keywords: ligand binding, major urinary protein (MUP), malespecific protein, pheromone, semiochemistry, urine
Abbreviations: DTT, dithiothreitol; ESI, electrospray ionization; IEF, isoelectric focusing; MALDI–TOF, matrix-assisted laser-desorption ionization–time of flight; MUP, major urinary protein; TCA, trichloroacetic acid; uMUP, urinary MUP
INTRODUCTION
The house mouse (Mus domesticus) exhibits an obligate proteinuria in the form of major urinary proteins (termed uMUPs to indicate that they are present in urine). These proteins, members of the lipocalin superfamily, are synthesized in the liver and pass through the bloodstream to the kidney where they evade renal uptake and are excreted in urine. This proteinuria is unusual on two counts. First, uMUP production represents a significant degree of protein synthesis; urine concentrations are typically 10–15 mg/ml but can attain levels in excess of 30 mg/ml. Secondly, the uMUP that is present in urine is not a single gene product, but the product of a highly polymorphic and polygenic MUP complex located on chromosome 4. Most wild mice produce approx. 10–15 discrete uMUP species, readily resolved by IEF (isoelectric focusing). Genetically homogeneous inbred laboratory mice, effectively having half of the MUP genes of a wild individual, produce approximately five to seven discrete uMUP species under the same conditions [1–3].
The uMUPs have at least two roles, both related to urine-mediated chemical communication. Lipocalins, of which MUPs are members, are a family of proteins that are extremely heterogeneous in structure and function, but which share a β-barrel structure that encloses a hydrophobic cavity, capable of predominantly binding apolar ligands. MUPs are eight-stranded β-barrels, and it has been demonstrated convincingly that they bind male signalling ligands, including 2-sec-butyl-4,5-dihydrothiazole and 3,4-dehydro-exo-brevicomin [4–6]. It is possible that protein binding serves to concentrate and protect the ligands from chemical degradation, but we have proved, through biochemical and behavioural studies, that the uMUPs are also able to delay the release of these volatile pheromones from urine scent marks, and that this delayed release elicits an appropriate behavioural response [7]. Thus one of the roles for uMUPs is to extend the time over which a deposited scent mark is effective in releasing pheromones.
However, this relatively simple role of ‘delayed release’ does not provide a satisfactory explanation for the polymorphism within the family, or the complexity of uMUP patterns within a single individual. In wild house mice, the complexity of individual MUP profiles is such that very few individuals are alike [8,9]. This dramatic polymorphism raised the possibility that the complex pattern of uMUPs could act as a unique ownership signature that was genetically stable. This signature could allow a receiver mouse to assess scent mark ownership, in the context of a range of social and physiological information, but in the absence of the scent owner. The receiver mouse would subsequently recognize the owner if encountered, or if further scent marks were discovered. By manipulation of uMUP profiles through breeding programmes in wild mice and by manipulation of uMUP profiles by addition of recombinant MUPs, we have demonstrated that uMUPs do indeed deliver a unique individuality signal ascribing ownership to a scent mark [10].
The demonstrable roles for uMUPs in slow release and individuality coding does not preclude additional outcomes of the molecular heterogeneity of the uMUP profile. In particular, there is good evidence for a sexual dimorphism in uMUP expression. Adult female mice express lower levels of uMUP compared with adult male mice [3,8] and there is evidence for sex-specific expression of some uMUP mRNA species [11]. However, these observations have not been translated into a detailed examination of specific protein expression in the two sexes. To pursue this further, we have isolated the uMUPs from an inbred strain and have examined their ability to bind and release natural pheromones. In particular, we have discovered a unique uMUP that is male-specific, responsible for a major component of the slow release of pheromones and which represents a novel uMUP, significantly different from those uMUPs previously characterized [16].
EXPERIMENTAL
Animals and urine collection
Urine was collected by bladder massage from male and female C57BL/6 mice (Charles River, Margate, Kent, U.K.), housed under standard laboratory conditions and with free access to food and water. Wild mice were caught live from seven sites within the U.K. and were brought to the laboratory where they were housed under similar conditions. Further samples were obtained from F1 mice derived from crosses in captivity of wild-caught mice. The wild mice usually urinate when picked up, facilitating collection of samples. Individual urine samples from inbred laboratory mice of each strain were pooled and stored at −20 °C until use.
Anion-exchange chromatography
Purification of individual MUP peaks from C57BL/6 urine was achieved by high-resolution anion-exchange chromatography on the Duo-Flow chromatography platform (Bio-Rad Laboratories, Hemel Hempstead, Herts., U.K.). Pooled urine was desalted on a 10 ml Sephadex G-25 column previously equilibrated with 50 mM Mes buffer (pH 5.0) and then passed through a 0.25 μm filter. The system was fitted with a BioRad UnoQ column (Vt=1 ml), equilibrated with several column volumes of 50 mM Mes buffer (pH 5.0). Typically, 1 mg of desalted protein was applied to the column. Bound protein was eluted from the column using a linear salt gradient of 0–200 mM NaCl. Fractions (1 ml) were collected, and those corresponding to individual MUP peaks were pooled and the protein content was determined by a dye-binding assay. Purity and mass were confirmed by MS.
Analysis of bound ligands
Anion-exchange chromatography fractions were added in the ratio 1:1 (v/v) to AnalaR grade hexane (Fisher Scientific, Loughborough, Leics., U.K.) in 1 ml sealed glass vials. Vials were vortexed for 20 s and incubated for 20 min at room temperature (18–24 °C). The hexane layer was then removed to a sealed autosampler vial for analysis by GC-MS. Solvent extracts (1 μl of the hexane extract) were analysed using a DB-5MS 20 m×0.18 mm inner diameter ×0.18 μm film capillary column (J&W Scientific, Folsom, CA, U.S.A.) fitted to a Thermo Electron Trace GC fitted with a splitless injector. The detector was a Thermo Electron Polaris ion-trap mass spectrometer. Data acquisition, tabulation and analysis were controlled using Xcalibur software. All mass spectra were obtained by electron ionization at an ionization potential of 70 eV. Analysis of samples was in full scan mode (50–600 m/z). The extracted ion at 60 m/z was used to monitor 2-sec-butyl-4,5-dihydrothiazole. To measure the rates of release of thiazoles from individual uMUP proteins purified by ion-exchange chromatography, 10 μl uMUP solutions were spotted in replicate on to 20 mm glass-fibre discs [12]. At times thereafter, one of the replicate glass-fibre discs was removed to hexane (200 μl) and extracted to measure residual thiazoles as described above.
SDS/PAGE
SDS/PAGE was performed essentially as described by Laemmli [13]. Individual peaks from anion-exchange chromatography were mixed 1:1 (v/v) with 30% (w/v) TCA (trichloroacetic acid), incubated at 4 °C for 30 min and centrifuged at 11000 g for 10 min. The TCA was removed and the protein pellet washed twice in diethyl ether before being resuspended in sample buffer with or without 100 mM DTT (dithiothreitol), and then heated for 3 min in a boiling water bath. All samples were run on SDS/PAGE (15% polyacrylamide) gels. Protein bands were visualized with Coomassie Brilliant Blue. Typically, 1–10 μg of protein was applied to the gel.
IEF
IEF was performed on an Amersham Biosciences Multiphor flatbed system (Amersham Biosciences, Little Chalfont, Bucks., U.K.) using an Immobiline dry-plate gel, pH range 4.2–4.9, and cooled to 10 °C. Whole urine samples were diluted 1:10 with deionized water and 5 μl was applied to sample strips placed on the gel. Samples were loaded into the gel at 200 V, 5 mA and 15 W for 200 V·h. The sample strips were removed and the gel was electrophoresed at 3500 V, 5 mA and 15 W for 14.8 kV·h. After fixation in 20% TCA, the gel was stained with Coomassie Brilliant Blue.
Trypsin and endoproteinase LysC digestion
Protein-containing fractions from anion-exchange chromatography were concentrated and desalted to deionized water using Vivaspin centrifugal concentrators (3 kDa molecular mass cutoff; Vivascience, Hanover, Germany). The concentrated samples were then mixed with an equal volume of 30% TCA and incubated at 4 °C for 30 min. The protein precipitate was sedimented by centrifugation at 11000 g for 10 min at room temperature and washed twice with diethyl ether. Pellets were air-dried, 50 μl of 10 mM DTT stock solution was added and the mixture was incubated at 37 °C for 30 min. Iodoacetamide (50 μl, 55 mM) was added and the mixture was incubated for 1 h in the dark. The sample was then precipitated with 30% TCA and washed with diethyl ether as described above. Protein pellets were resuspended in 100 μl of digestion buffer (endopeptidase LysC: 25 mM Tris/HCl and 1 mM EDTA, pH 8.5; trypsin: 50 mM ammonium bicarbonate). Sequencing grade endoproteinase LysC or trypsin (1 μl and 0.1 μg/μl; Roche, Lewes, East Sussex, U.K.) was added and the digestion proceeded overnight at 37 °C.
For in-gel digests, plugs were removed from protein bands on the SDS/PAGE gel using a thin glass pipette and placed into microcentrifuge tubes. Each gel plug was destained using 100 μl of 50 mM ammonium bicarbonate, 50% (v/v) acetonitrile (trypsin) or 100 μl of 25 mM Tris/HCl, pH 8.5, 50% acetonitrile (endopeptidase LysC), and was incubated at 37 °C for 30 min. This step was repeated until no stain was visible. The plugs were then washed twice in 100 μl of 50 mM ammonium bicarbonate (trypsin) or 100 μl of 25 mM Tris/HCl, pH 8.5 (endopeptidase LysC), which was then discarded. The plugs were then incubated with 50 μl of 10 mM DTT. After 30 min at 37 °C, the DTT was discarded and 50 μl of 55 mM iodoacetamide was added to each tube and incubated for 1 h at room temperature in the dark. The iodoacetamide solution was then discarded and the plugs were washed twice as above before being dehydrated in 100% acetonitrile and then rehydrated in 9 μl of 50 mM ammonium bicarbonate (trypsin) or 25 mM Tris/HCl and 1 mM EDTA, pH 8.5 (endopeptidase LysC). Sequencing grade endoproteinase LysC or trypsin (1 μl, 0.1 μg/μl; Roche) was added and the digest was incubated overnight at 37 °C.
Peptide mass fingerprinting
Peptide mixtures from the proteolytic digestion reactions were analysed on a MALDI–TOF (matrix-assisted laser-desorption ionization–time of flight)-MS (Waters Micromass, Manchester, U.K.), operated in reflectron mode with positive ion detection. Samples were mixed 1:1 (v/v) with a saturated solution of αcyano-4-hydroxycinnamic acid in ACN (acetonitrile)/water/ethanol/TFA (0.4%, by vol.) in equal volumes, before being spotted on to the MALDI target and air-dried. Spectra were gathered between m/z 1000 and 3500, the laser frequency was 5 Hz and data acquisition was for 2 s. Spectra typically consisted of up to 50 summed acquisitions (equivalent to 500 laser shots) at a laser energy of 30–50% of the maximum energy. External mass calibration was determined using a mixture of des-Arg bradykinin, neurotensin, ACTH (corticotropin) and oxidized insulin B chain (2.4, 2.4, 2.6 and 30 pmol/μl respectively) each in 50% acetonitrile and 0.1% (v/v) trifluroacetic acid. Peptide mass fingerprints were searched against the MSDB database (http://csc-fserve.hh.med.ic.ac.uk/msdb.html, version 30062004) using the MASCOT search engine [14].
Tandem MS (MS/MS)
All analyses were performed on a QToFmicro mass spectrometer (Waters Micromass) fitted with a nanoflow ESI (electrospray ionization) source. Samples were introduced as a static nanospray from a metal-coated capillary held at a potential of 1 kV relative to the sample cone. Peptides from tryptic in-solution digest were concentrated and desalted into a 50:50 solution of acetonitrile and 0.1% (v/v) formic acid using C18 ZipTips (Millipore, Watford, Herts., U.K.). Candidate multiply charged ions were identified by a survey scan from m/z 300–1500. Fragmentation energy was optimized to effect complete fragmentation of the precursor ion, and the MS/MS spectrum was acquired for 50–80 scans with a 2.4 s/0.1 s duty cycle. Spectra were processed using MaxENT 3 software and the peptide sequence was determined by manual interpretation using PepSeq software (Waters Micromass).
ESI–MS of intact proteins
Individual peaks from anion-exchange chromatography were concentrated and desalted to deionized water using Vivaspin centrifugal concentrators (3 kDa molecular mass cut-off). The samples were diluted 1:10 in 50% acetonitrile/0.1% formic acid. All analyses were performed on a QToFmicro mass spectrometer (Waters, Manchester, U.K.), fitted with an ESI source. Samples were introduced by either a continuous 5 μl/min infusion or static nanospray. Data gathered between 700 and 1400 Th was processed and transformed to a true mass scale using MaxENT 1 maximum entropy software (Waters Micromass). All data sets were processed at 1 Da/channel over a mass range of 18300–19000 Da, and a peak width of 0.75 Da was used to construct the damage model. The instrument was calibrated using a 500 fmol/μl solution of Glu fibrinopeptide in 50% acetonitrile and 0.1% formic acid.
RESULTS AND DISCUSSION
Identification of a novel MUP
Inbred strains of mice differ in their uMUP phenotype. However, most uMUPs migrate on SDS/PAGE with an apparent molecular mass of approx. 18 kDa, consistent with the masses predicted from cDNA sequences and measured by MS. In contrast, animals of the C57 lineage (including the C57BL/6 strain) have a pattern of protein expression distinct from many other strains [15,16]. Even on one-dimensional SDS/PAGE, urinary proteins from this strain exhibit a band migrating at a position corresponding to a mass of approx. 16 kDa (Figure 1). If the mobility of this protein on the gel was commensurate with its molecular mass, it would have been substantially smaller than a ‘typical’ MUP. To investigate this 16 kDa protein further, it was purified from the urine of male C57BL/6J mice. The C57BL/6J mouse is the strain for which a complete genome sequence is available [17] and which therefore offered the highest probability of urinary protein identification. At the time of writing of this paper, contiguous genome sequence data for this region of chromosome IV was not available (J. Mudge, personal communication), and it is not yet possible to definitively relate all proteins to known genes.
Figure 1. Chromatographic resolution of uMUPs from C57BL/6J mice.
Urinary proteins from adult male C57BL/6J mice were separated by strong anion-exchange chromatography, monitored by UV absorbance (shaded profile) and fractionated. Each fraction was assessed for the content of 2-sec-4,5-butyl-dihydrothiazole by GC-MS (a, ○). The four reproducible peaks (labelled I–IV) were each analysed by one-dimensional SDS/PAGE (b) and narrow-range IEF (pI range 4.1–4.9, c) and compared with the starting material (SM). The material in peak IV was also analysed by reducing (R) and non-reducing (NR) SDS/PAGE (b).
The protein content of mouse urine is almost exclusively uMUPs, and we have shown previously that it is feasible to purify individual uMUPs on a single dimension of high-resolution ionexchange chromatography [9]. Urine was desalted and fractionated by strong anion-exchange chromatography. Four major discrete peaks were resolved, labelled as I–IV (Figure 1a). When analysed on reducing SDS/PAGE, the first three peaks (I–III) yielded single bands on the SDS/PAGE of a mobility commensurate with that expected for uMUPs (18–19 kDa). However, peak IV contained a protein of unusually high mobility, as well as trace quantities of a protein of the similar mobility to peaks I–III uMUP (Figure 1b). Under non-reducing conditions, the mobility of the peak IV material was significantly higher than under reducing SDS/PAGE, as indeed was that of uMUPs from peaks I–III. We attribute this change to the compact structure maintained by disulphide bonds that can penetrate the polyacrylamide gel more readily. For most uMUPs, there is a single disulphide bond between residues 64 and 157 of the mature protein sequence [18]. The shift in mobility of the peak IV material was consistent with the presence of disulphide bond(s) in this protein also.
The uMUPs are readily resolved by narrow-range IEF (pI from 4.2 to 4.9 [8]). When unresolved C57BL/6J urine was analysed by IEF, the pattern of bands revealed four major MUP isoforms (Figure 1c). When the ion-exchange fractions were analysed by IEF, peaks II and III showed some overlap of charged variants, whereas peaks I and IV predominantly consisted of a single species. As expected, the most anionic protein on IEF (peak IV) was the last to elute from the anion-exchange column. The peak IV material was essentially homogeneous and was characterized further.
One of the roles of uMUPs is to act as a binding/slow release vehicle for the male-specific pheromones 2-sec-butyl-4,5dihydrothiazole (‘thiazole’) and 3,4-dehydro-exo-brevicomin (‘brevicomin’) [7,19]. Mice of the C57BL/6J strain express very little brevicomin in urine (S. D. Armstrong, D. H. L. Robertson and R. J. Beynon, unpublished work), but there is a high level of output of thiazole. Accordingly, the material eluted from the ion-exchange chromatography medium was assessed for co-elution of the thiazole. Peaks I–III (collectively responsible for ∼87% of the total protein, determined by dye-binding assay) were able to bind approx. 40% of the total urinary thiazole, but peak IV (comprising just 13% of the total protein) contained over 40% of the proteinbound pheromone (Figure 1a). Based on this criterion, we presumed that the peak IV material was likely to be a MUP or a MUP-related protein, and was capable of delivering a similar functionality to the other MUP fractions, namely pheromone binding. Moreover, this protein seemed particularly adapted to bind thiazole.
The higher mobility of the peak IV material on SDS/PAGE was enigmatic. If the protein was similar to a typical uMUP, then the altered mobility could be due to atypical behaviour on electrophoresis, an N-terminal or C-terminal truncation, or, finally, an excision of a small central peptide segment with the two flanking peptides being linked by one or more disulphide bonds. The last explanation is unlikely, as under non-reducing conditions the mobility increased rather than decreased (Figure 1c). To assess any putative exoproteolysis, we measured the masses of the peak IV protein by MS together with the masses of the proteins present in fractions corresponding to peaks I–III (results not shown). For each peak, the material was concentrated and desalted before mass measurement using ESI–MS. The multiply charged envelopes corresponding to each peak were readily deconvoluted using maximum entropy software. Peak I contained a single protein of mass 18645±2 Da, peak II contained a protein of mass 18708±2 Da and peak III contained two proteins at 18695±2 and 18713±2 Da. Peak IV contained a single protein of mass 18894±2 Da. Exhaustive analysis of the ESI–MS spectra failed to reveal any evidence for an additional species of molecular mass of approx. 16 kDa. Thus the mass inferred from mobility on SDS/PAGE was not confirmed by measurement of the intact protein by MS.
Each of the four protein-containing peaks were reduced and subjected to in-solution endopeptidase LysC digestion. The endopeptidase LysC peptides were then analysed by MALDI–TOF-MS. We have previously shown that this proteinase is an effective tool for phenotype mapping of uMUPs, as virtually all of the protein sequence yields peptides analysable by MALDI–TOF-MS [9,16,20]. High-quality MALDI–TOF spectra were obtained for each of the proteins (see the Supplementary Figures 1 and 2, at http://www.BiochemJ.org/bj/391/bj3910343add.htm). For peaks I–III, the spectra were very similar, and allowed unequivocal identification of an MUP that was present in each fraction, by reference to known uMUPs (results not shown). The MALDI–TOF peptide mass fingerprint from peak I matched a MUP cDNA sequence (SwissProt accession no. P11588) but is a variant of this sequence (F56V) that we have previously identified [16] and which does not currently have a database entry. The predicted mass of the F56V variant of P11588 is 18645 Da, same as the measured mass of 18645±2 Da obtained by ESI–MS. The MALDI–TOF LysC PMF (peptide mass fingerprint) spectrum for peak II matched a MUP cDNA sequence [EMBL (European Molecular Biology Laboratory) accession no. CAC34259] of predicted mass 18708 Da, identical with the mass of the protein in this peak measured by ESI–MS (18708±2 Da). The equivalent spectrum for peak III matched a MUP [NCBI (Entrez Protein) accession no. AAB47130] with a predicted mass of 18695 Da, which also matched well with the measured mass of 18695±2 Da. However, even on cursory inspection, the endopeptidase LysC spectrum for peak IV was notably different from those obtained from the other three proteins (Supplementary Figure 1 at http://www.BiochemJ.org/bj/391/bj3910343add.htm). When the peptide mass fingerprint was searched against the comprehensive MSDB proteomics database (version 30062004), no high confidence matches were obtained, even though the spectrum was of very high quality.
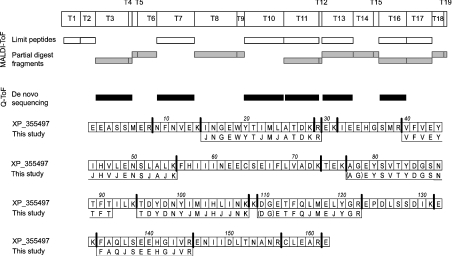
The uMUP family are highly conserved, and the failure to obtain a match by peptide mass fingerprinting raised the possibility that the peak IV protein is a novel thiazole-binding protein. To extend the characterization of this protein, tryptic digests of the peak IV protein were subjected to tandem MS to obtain de novo sequence information. Two peptides, both doubly charged [M+2H]2+ ions, at 779.9 and 937.5 Th could be sequenced over an extended region of the polypeptide chain. The sequence for the peptide of 779.9 Th was DGETFQJMEJYGR and that for the peptide of 937.5 Th was VFVEYJHVJENSJAJK (Supplementary Figure 2 at http://www.BiochemJ.org/bj/391/bj3910343add.htm) (‘J’ is used to indicate the ambiguity associated with the inability to discriminate between the isobaric pair leucine and isoleucine). The sequences were used to search the NCBI (National Center for Biotechnology Information) aggregated protein database using the BLAST alignment tool [21], and both sequences gave convincing hits to the same protein database entry, XP_355497 (‘similar to uMUP V’). This sequence was not present in MSDB30062004 and was therefore only identified as a result of de novo mass spectrometric sequencing. Having obtained a putative identity for this protein, several items of evidence confirm this being the correct match. First, the predicted average mass of the mature form of this protein is 18895 Da. This agrees favourably with the value of 18894±2 Da measured by ESI–MS, which should be 2 Da lower than the predicted mass to compensate for the putative oxidation of a pair of cysteine residues to form a disulphide bond. Secondly, the MALDI–TOF tryptic peptide mass fingerprint of this protein matches exceptionally well with the tryptic digest predicted from the cDNA inferred sequence, as does the fingerprint obtained with endopeptidase LysC (Figure 2). Indeed, when the partially digested fragments (one missed cleavage) are included, the MALDI–TOF spectra gave complete coverage of the protein sequence. Thus the unusual and unique protein in peak IV is this particular gene product. A further four peptides were sequenced by tandem MS and these also matched the putative sequence exactly (Figure 2). We therefore have provided direct molecular phenotypic evidence for expression of this protein, as inferred from cDNA and genomic data.
Figure 2. Identification strategy for the peak IV uMUP.
The sequence data of uMUP peak IV peptides obtained by tandem MS (‘Q-ToF’) were used to interrogate the NCBI protein database using protein BLAST tool. A highly significant match was obtained for the GenBank® sequence XP_355497. From this sequence, the theoretical tryptic or endopeptidase LysC digestion maps were constructed, and used to identify those peptides (both partial and complete digestion fragments) observed during MALDI–TOF peptide mass fingerprinting. Tryptic peptides sequenced by tandem MS are aligned to the sequence – the symbol ‘J’ is used to express ambiguity between isobaric pair leucine and isoleucine.
Because the predicted mass and the mass measured by ESI–MS are exactly in agreement at 18895 Da, one enigmatic feature of this protein remains unexplained, namely, the anomalous migration of this protein on SDS/PAGE. Most uMUPs have three cysteine residues (mature sequence numbering C64, C138 and C157). Two residues, C64 and C157, are oxidized to a disulphide bond. The peak IV uMUP lacks C138 and a serine residue is substituted at this position. However, there is no reason, a priori, why C64 and C157 in peak IV uMUP should not also form a disulphide bond. Indeed, the difference in mobility of the protein on reducing and non-reducing SDS/PAGE (Figure 1) is entirely consistent with the formation of this post-translational modification. However, this does not explain why the protein possesses enhanced mobility on the gel system. There are no dramatic differences between this peak IV protein and archetypical uMUPs, such as uMUP-I (SwissProt accession no. P11588) in terms of overall hydrophobicity or amino acid content. The primary distinguishing feature of the peak IV protein is the low isoelectric point. It is possible that the SDS fails to completely suppress the underlying charge such that the protein has an overall higher charge density. Compared with uMUP-I, the peak IV MUP has an additional two acidic residues and one less basic residue – a relatively modest difference to explain the shift in mobility. The calculated pI values of peak IV MUP and uMUP-I are 4.52 and 4.68 respectively, which is not a major difference. Finally, post-translational modifications other than disulphide bond formation are unlikely, given the precise agreement between the predicted mass and that observed by ESI–MS. At present, we are unable to explain the aberrant mobility, which at the least serves to emphasize the imperfect relationship between mobility and mass on SDS/PAGE.
Ligand binding characteristics
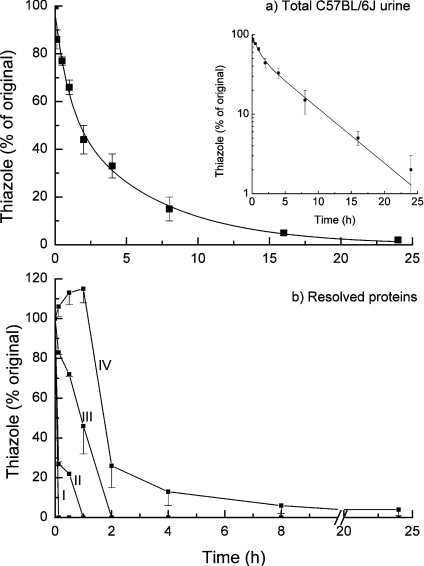
The marked tendency of thiazole to remain bound to peak IV (Figure 1) suggested that the association between this variant protein and its cognate ligand was tighter than that for the other MUP species in the urine sample. MUPs were therefore purified by anion-exchange chromatography and the four proteins were individually deposited in replicate samples on to glass-fibre discs and allowed to air-dry to simulate a deposited scent mark. At different times, the discs were removed and the residual bound thiazole was assessed by hexane extraction and GC-MS (Figure 3). When the release of thiazole from total C57BL/6J urine was analysed, the loss could not be explained by a single exponential (Figure 3a and semi-logarithmic plot inset) and a better fit was achieved with a biexponential curve, such that approx. 40% of the thiazole was lost with a first-order rate constant of 1.0 h−1, and approx. 60% of the thiazole was lost from the drying urine sample with a lower rate constant of 0.16 h−1. To assess whether this was attributable to the different proteins, the different fractions were assessed for their ability to release thiazole in the same experiment (Figure 3b). For proteins recovered in peaks I and II, the thiazole was lost very rapidly (within minutes) from the dried protein, such that it was not possible to obtain reliable estimates of the first-order rate constants for the loss of the pheromone. It is difficult to reconcile such rapid loss in a dried urine sample with a putative role of delayed release of this pheromone, and it may be necessary to search for additional roles for these proteins. Peak III material released the pheromone at a rate that is intermediate between peak II and peak IV. The peak IV material, however, exhibited a rather different behaviour. For the first 1–2 h, the loss was less and even manifested as an increase in thiazole recovery, after which the ligand was progressively lost more slowly than from the other proteins. We do not have an explanation for the delay phase at present, but it is possible that this reflects a change in state of the protein as the sample becomes progressively drier. The glass-fibre filters containing a deposited urine sample are apparently dry to touch within a few minutes, but a slower loss of tightly associated water could elicit a change in the structure of the peak IV protein and weaken the association with the ligand. Certainly, this is consistent with the ability of this protein to retain thiazole during exhaustive purification. These results point to a role for the peak IV protein in a slow phase of ligand release from total urine, consistent with the needs of territorial scent marking.
Figure 3. Ligand release kinetics for different uMUP subforms.
Intact C57BL/6J male mouse urine (a) or individual uMUP subforms (peaks I–IV), purified from the same source by ion-exchange chromatography (b) were assessed for their ability to release bound thiazole. The urine or purified uMUPs were dried on glass-fibre discs, and the rate of loss of 2-sec-3,4-dihydrothiazole was measured by serial sampling, hexane extraction and GC/MS, as described in the Experimental section. For each experiment, the release curves were assessed four times and data are expressed as the average of the residual thiazole±S.E.M. (n=4). For the intact urine, the data were fitted to monoexponential (results not shown) or biexponential decay curves (solid line), and the data are plotted semi-logarithmically to emphasize the biphasic nature of the curve.
There are 34 amino acid differences between peak IV uMUP and uMUP-I (see Supplementary Figure 3 at http://www.BiochemJ.org/bj/391/bj3910343add.htm), distributed throughout the sequence. Of these, eight [22] or five [23,24] are defined as cavity-forming residues. Some substitutions, notably F38→M, A102→I and G118→E, have the greatest potential to change the binding of the thiazole. In uMUP-I, G118 is in proximity to the nitrogen atom of thiazole and it is feasible that substitution for an acidic residue would serve to enhance interaction with this atom, increasing affinity for the ligand. Precise details of the interactions leading to slow release of the thiazole must await solution of the three-dimensional structure.
Sexual dimorphism in uMUP expression
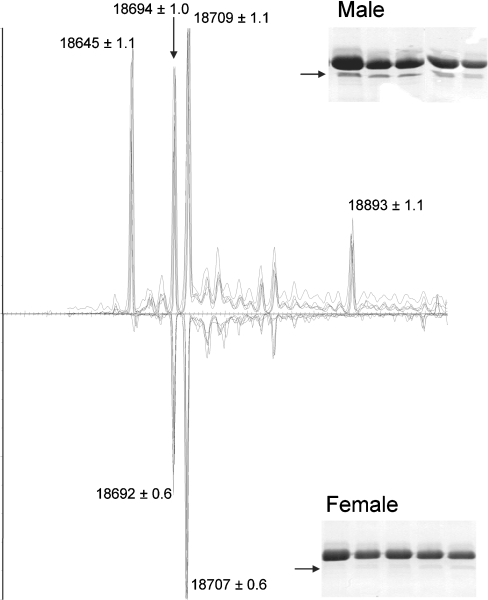
There is good evidence for sexual dimorphism in both the amount of protein and the specific uMUP gene products expressed by mice [8,25–28]. However, these analyses have not been defined in terms of the presence or absence of specific uMUPs. Accordingly, we compared urine samples from male and female C57BL/6J mice. Intact mass measurement of the urinary proteins from female mice yielded a very different pattern compared with males (Figure 4). Two major peaks, at 18645 and 18894 Da, were absent from the protein profile. As would be predicted, the high-mobility peak IV band, corresponding to the 18895 Da protein, was also absent from the one-dimensional SDS/PAGE analysis of the female samples. The male-specific protein at 18645 Da (uMUP X) also binds very little thiazole (Figure 1), consistent with the observation that it is incapable of retaining or effecting a slow release of this pheromone (Figure 3). This is consistent with the relatively high dissociation constant (Kd) for thiazole, measured for this protein by isothermal titration calorimetry [29] (the 18645 Da protein is referred to in this paper as MUP-VII). We have not explored any other role for this protein at present, but we have previously demonstrated that the protein isoforms are also capable of carrying information in their own right [10]. This 18645 Da protein, and that at 18895 Da, could form the involatile signature of sex of the owner in urine scent marks, the latter in combination with thiazole, a male-specific pheromone.
Figure 4. Differential expression of uMUPs from male and female C57BL/6J mice.
Total urinary protein from male or female C57BL/6J urine was desalted and analysed by ESI–MS. The envelope of multiply charged protein ions was deconvoluted using MAXENT 1 software to yield the true mass composition of the sample. For ease of comparison, the true mass spectrum for female urine is inverted. Spectra from five males and five females are overlaid to illustrate consistency between individuals. Additionally, the uMUPs from adult male and female individuals were analysed by one-dimensional SDS/PAGE (insets). The peak IV uMUP, migrating at an apparent molecular mass of 16 kDa, is indicated by an arrow.
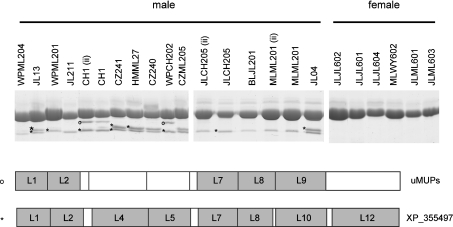
Inbred mice were originally derived from wild-caught individuals and it was necessary to test the possibility that the peak IV protein was a single-mutated gene product that was exclusive to this particular strain. Rather than test additional inbred strains, which also have a restricted genetic background, we conducted SDS/PAGE and peptide mass fingerprinting on urinary proteins from a range of adult wild-caught mice, trapped from different populations in the U.K. Of 84 wild-caught males or F1 male offspring derived from crosses between two wild-caught individuals, 78 expressed a clear band corresponding to peak IV. Of 15 females (a mixture of wild-caught and F1 animals), only one expressed a band that could be peak IV. Representative data are given in Figure 5. Of 14 male mice, at least one high-mobility band was in evidence, and in some individuals, a doublet was present. Peptide mass fingerprinting confirmed that both of these high-mobility bands corresponded to the peak IV 18895 Da protein. We do not know why this protein migrates as a doublet in some instances. In few animals, an additional band, migrating slightly faster than the major MUP band, yielded a peptide mass fingerprint consistent with it being a ‘typical’ uMUP – it is possible that this is a proteolytic fragment. In contrast, in all six of the female samples analysed by SDS/PAGE, the peak IV high-mobility band was absent. Thus the male-specific peak IV protein is widely distributed across the wild-mouse population, exhibiting a sexual dimorphism in expression.
Figure 5. Expression of peak IV uMUP in wild-caught mice.
Urine from male and female wild mice, trapped from a number of different locations, was analysed by one-dimensional SDS/PAGE. Selected bands on the gels were subjected to in-gel digestion with endopeptidase LysC and MALDI–TOF-MS. ⋆, proteins in bands whose spectra matched with the uMUP, previously identified in peak IV; ○, bands whose mass spectra matched with uMUPs other than the peak IV protein (see text).
Each mouse expresses a complex pattern of uMUPs, which is particularly pronounced when wild-caught heterozygotic animals are analysed, rather than highly inbred genetically homogeneous laboratory mice. A typical mouse will, on an average, express 10–15 distinct uMUPs in urine, and no two unrelated animals from a population appear to have the same profile of uMUPs. The reasons for this complexity are not completely understood, but we have demonstrated that the pattern of proteins contributes to the involatile ownership or identity signal that is associated with a urine scent mark [10]. However, the present study also provides convincing evidence for a second class of uMUPs that contribute to the male-specific scent components. The peak IV uMUP appears to be the protein that binds most of the thiazole in urine, and which is also responsible for the slow release of the ligand. This slow release is critical for the longevity of territorial scent marks. In the absence of protein binding, the free ligand evaporates rapidly from a dried sample (with a half-life of less than 1 min). This rate of loss would be incompatible with a requirement to repeatedly mark a territory to signal sustained ownership and territorial dominance. The combination of a volatile ligand (ensuring transmission through the air) and tight protein binding can achieve the optimal balance of volatility and release characteristics. It seems as though the mouse has evolved a protein to control the rate of release of one of the male-specific dominance signalling chemicals [30]. It is also possible that this uMUP and the uMUP at 18645 Da are also semiochemicals in their own right, and that MUP-specific receptors in the vomeronasal organ are sensitive to these proteins, which communicate the sex of the scent mark owner. There is increasing evidence for receptors of urinary lipocalins in the rat [31], but, at present, this remains a tantalizing possibility. Naive females are only attracted to male pheromones in the context of involatile urinary components [32], consistent with the idea that MUP–ligand complexes may elicit the pheromonal signal in the context of additional information of the scent owner. It is highly likely that the individual MUPs present in a typical urine scent mark may each fulfil discrete roles, but also that the overall pattern of these proteins in urine provides an opportunity for communication of additional information, such as sex and individual identity, capitalizing on the combinatorial complexity that is a feature of this class of gene products. It is becoming increasingly possible to anticipate elucidation of the molecular mechanisms whereby incredible subtlety of information is communicated in a scent mark.
Online data
Acknowledgments
This work was supported by research grants to R. J. B. and J. L. H. from the Biotechnology and Biological Sciences Research Council. We are grateful to M. Thom for provision of some mouse samples and to R. Humphries and J. Waters for technical assistance.
References
- 1.Beynon R. J., Hurst J. L. Multiple roles of major urinary proteins in the house mouse, Mus domesticus. Biochem. Soc. Trans. 2003;31:142–146. doi: 10.1042/bst0310142. [DOI] [PubMed] [Google Scholar]
- 2.Hurst J. L., Beynon R. J. Scent wars: the chemobiology of competitive signalling in mice. BioEssays. 2004;26:1288–1298. doi: 10.1002/bies.20147. [DOI] [PubMed] [Google Scholar]
- 3.Beynon R. J., Hurst J. L. Urinary proteins and the modulation of chemical scents in mice and rats. Peptides. 2004;25:1553–1563. doi: 10.1016/j.peptides.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 4.Bacchini A., Gaetani E., Cavaggioni A. Pheromone binding proteins of the mouse, Mus musculus. Experientia. 1992;48:419–421. doi: 10.1007/BF01923448. [DOI] [PubMed] [Google Scholar]
- 5.Robertson D. H. L., Beynon R. J., Evershed R. P. Extraction, characterization and binding analysis of two pheromonally active ligands associated with major urinary protein of house mouse (Mus musculus) J. Chem. Ecol. 1993;19:1405–1416. doi: 10.1007/BF00984885. [DOI] [PubMed] [Google Scholar]
- 6.Novotny M. V., Ma W., Wiesler D., Zidek L. Positive identification of the puberty-accelerating pheromone of the house mouse: the volatile ligands associating with the major urinary protein. Proc. R. Soc. London Ser B. 1999;266:2017–2022. doi: 10.1098/rspb.1999.0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hurst J. L., Robertson D. H. L., Tolladay U., Beynon R. J. Proteins in urine scent marks of male house mice extend the longevity of olfactory signals. Anim. Behav. 1998;55:1289–1297. doi: 10.1006/anbe.1997.0650. [DOI] [PubMed] [Google Scholar]
- 8.Payne C. E., Malone N., Humphries R. E., Bradbrook C., Veggerby C., Beynon R. J., Hurst J. L. Heterogeneity of major urinary proteins in house mice: population and sex differences. In: Marchelewska-Koj A., Muller-Schwarze D., Lepri J., editors. Chemical Signals in Vertebrates, vol. 9. New York: Plenum Press; 2001. pp. 233–240. [Google Scholar]
- 9.Beynon R. J., Veggerby C., Payne C. E., Robertson D. H., Gaskell S. J., Humphries R. E., Hurst J. L. Polymorphism in major urinary proteins: molecular heterogeneity in a wild mouse population. J. Chem. Ecol. 2002;28:1429–1446. doi: 10.1023/a:1016252703836. [DOI] [PubMed] [Google Scholar]
- 10.Hurst J. L., Payne C. E., Nevison C. M., Marie A. D., Humphries R. E., Robertson D. H., Cavaggioni A., Beynon R. J. Individual recognition in mice mediated by major urinary proteins. Nature (London) 2001;414:631–634. doi: 10.1038/414631a. [DOI] [PubMed] [Google Scholar]
- 11.Held W. A., Gallagher J. F., Hohman C. M., Kuhn N. J., Sampsell B. M., Hughes R. G., Jr Identification and characterization of functional genes encoding the mouse major urinary proteins. Mol. Cell. Biol. 1987;7:3705–3712. doi: 10.1128/mcb.7.10.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robertson D. H. L., Marie A. D., Veggerby C., Hurst J. L., Beynon R. J. Characteristics of ligand binding and release by major urinary proteins. In: Marchlewska-Koj A., Muller-Schwarze D., Lepri J., editors. Chemical Signals in Vertebrates. New York: Plenum Press; 2001. pp. 169–176. [Google Scholar]
- 13.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 15.Evershed R. P., Robertson D. H., Beynon R. J., Green B. N. Application of electrospray ionization mass spectrometry with maximum-entropy analysis to allelic ‘fingerprinting’ of major urinary proteins. Rapid Commun. Mass Spectrom. 1993;7:882–886. doi: 10.1002/rcm.1290071005. [DOI] [PubMed] [Google Scholar]
- 16.Robertson D. H., Cox K. A., Gaskell S. J., Evershed R. P., Beynon R. J. Molecular heterogeneity in the major urinary proteins of the house mouse Mus musculus. Biochem. J. 1996;316:265–272. doi: 10.1042/bj3160265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waterston R. H., Lindblad-Toh K., Birney E., Rogers J., Abril J. F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature (London) 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 18.Bocskei Z., Groom C. R., Flower D. R., Wright C. E., Phillips S. E. V., Cavaggioni A., Findlay J. B. C., North A. C. T. Pheromone binding to two rodent urinary proteins revealed by X-ray crystallography. Nature (London) 1992;360:186–188. doi: 10.1038/360186a0. [DOI] [PubMed] [Google Scholar]
- 19.Beynon R. J., Robertson D. H. L., Hubbard S. J., Gaskell S. J., Hurst J. L. The role of protein binding in chemical communication: major urinary proteins in the house mouse. In: Johnston R. E., Muller-Schwarze D., Sorensen P., editors. Advances in Chemical Communication in Vertebrates. New York: Plenum Press; 1999. pp. 137–147. [Google Scholar]
- 20.Darwish Marie A., Veggerby C., Robertson D. H., Gaskell S. J., Hubbard S. J., Martinsen L., Hurst J. L., Beynon R. J. Effect of polymorphisms on ligand binding by mouse major urinary proteins. Protein Sci. 2001;10:411–417. doi: 10.1110/ps.31701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGinnis S., Madden T. L. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:20–25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lucke C., Franzoni L., Abbate F., Lohr F., Ferrari E., Sorbi R. T., Ruterjans H., Spisni A. Solution structure of a recombinant mouse major urinary protein. Eur. J. Biochem. 1999;266:1210–1218. doi: 10.1046/j.1432-1327.1999.00984.x. [DOI] [PubMed] [Google Scholar]
- 23.Timm D. E., Baker L. J., Mueller H., Zidek L., Novotny M. V. Structural basis of pheromone binding to mouse major urinary protein (MUP-I) Protein Sci. 2001;10:997–1004. doi: 10.1110/ps.52201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bingham R. J., Findlay J. B., Hsieh S. Y., Kalverda A. P., Kjellberg A., Perazzolo C., Phillips S. E., Seshadri K., Trinh C. H., Turnbull W. B., et al. Thermodynamics of binding of 2-methoxy-3-isopropylpyrazine and 2-methoxy-3-isobutylpyrazine to the major urinary protein. J. Am. Chem. Soc. 2004;126:1675–1681. doi: 10.1021/ja038461i. [DOI] [PubMed] [Google Scholar]
- 25.Hayakawa J., Nikaido H., Koizumi T. Components of major urinary proteins (MUP's) in the mouse. Sex, strain, and subspecies differences. J. Hered. 1983;74:453–456. doi: 10.1093/oxfordjournals.jhered.a109837. [DOI] [PubMed] [Google Scholar]
- 26.Shaw P. H., Held W. A., Hastie N. D. The gene family for major urinary proteins: expression in several secretory tissues of the mouse. Cell (Cambridge, Mass.) 1983;32:755–761. doi: 10.1016/0092-8674(83)90061-2. [DOI] [PubMed] [Google Scholar]
- 27.Sampsell B. M., Held W. A. Variation in the major urinary protein multigene family in wild-derived mice. Genetics. 1985;109:549–568. doi: 10.1093/genetics/109.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson D., al-Shawi R., Bishop J. O. Sexual dimorphism and growth hormone induction of murine pheromone-binding proteins. J. Mol. Endocrinol. 1995;14:21–34. doi: 10.1677/jme.0.0140021. [DOI] [PubMed] [Google Scholar]
- 29.Sharrow S. D., Vaughn J. L., Zidek L., Novotny M. V., Stone M. J. Pheromone binding by polymorphic mouse major urinary proteins. Protein Sci. 2002;11:2247–2256. doi: 10.1110/ps.0204202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harvey S., Jemiolo B., Novotny M. Pattern of volatile compounds in dominant and subordinate male-mouse urine. J. Chem. Ecol. 1989;15:2061–2072. doi: 10.1007/BF01207438. [DOI] [PubMed] [Google Scholar]
- 31.Krieger J., Schmitt A., Lobel D., Gudermann T., Schultz G., Breer H., Boekhoff I. Selective activation of G protein subtypes in the vomeronasal organ upon stimulation with urine-derived compounds. J. Biol. Chem. 1999;274:4655–4662. doi: 10.1074/jbc.274.8.4655. [DOI] [PubMed] [Google Scholar]
- 32.Moncho-Bogani J., Lanuza E., Hernandez A., Novejarque A., Martinez-Garcia F. Attractive properties of sexual pheromones in mice: innate or learned? Physiol. Behav. 2002;77:167–176. doi: 10.1016/s0031-9384(02)00842-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.