Abstract
Ammodytoxin (Atx), an sPLA2 (secretory phospholipase A2), binds to γ and ε isoforms of porcine 14-3-3 proteins in vitro. 14-3-3 proteins are evolutionarily conserved eukaryotic regulatory proteins involved in a variety of biological processes, including cell-cycle regulation. We have now shown that Atx binds to yeast 14-3-3 proteins with an affinity similar to that for the mammalian isoforms. Thus yeast Saccharomyces cerevisiae can be used as a model eukaryotic cell, which lacks endogenous phospholipases A2, to assess the in vivo relevance of this interaction. Atx was expressed in yeast cells and shown to be biologically active inside the cells. It inhibited G2 cell-cycle arrest in yeast, which is regulated by 14-3-3 proteins. Interference with the cell cycle indicates a possible mechanism by which sPLA2s are able to cause the opposing effects, proliferation and apoptosis, in mammalian cells.
Keywords: secretory phospholipase A2, G2 cell-cycle arrest, cell proliferation and death, Saccharomyces cerevisiae, model organism, 14-3-3 protein
Abbreviations: Atx, ammodytoxin; GIIA, group IIA; sPLA2, secretory phospholipase A2; MALDI–TOF-MS, matrix-assisted laser-desorption ionization–time-of-flight MS
INTRODUCTION
Secretory phospholipases A2 (sPLA2s) belong to the superfamily of enzymes that catalyse the hydrolysis of the ester bond at the sn-2 position of phosphoglycerides. They are involved in several (patho) physiological processes, ranging from inflammation, cell injury and tumour resistance to neurotoxicity [1,2], and are thus important under physiologically normal conditions as well as in disease.
Atx (ammodytoxin) is a GIIA (group IIA) [3] neurotoxic sPLA2 from the venom of the long-nosed viper (Vipera ammodytes ammodytes). Both enzymatic activity and interactions with specific protein targets are required for its biological activity [4]. Only one cell-type-specific, Atx-interacting molecule has been identified to date, the neuronal M-type receptor [5]. Other molecular targets include calmodulin [6] and 14-3-3 proteins [7], both evolutionarily highly conserved regulatory cytoplasmic proteins present in all eukaryotes [8,9]. This suggests that the molecular mechanisms of the effects of Atx are very similar in all eukaryotic cells, once it enters the cell, and that it is the specificity of Atx binding to surface receptors on motor neurons that determines its specificity for neurotoxic action in mammals.
14-3-3 proteins are a family of conserved regulatory proteins expressed in all eukaryotic cells. They play an important role in a number of essential regulatory processes, including cell-cycle control [10,11]. GIIA sPLA2s are known to have effects that are connected to cell-cycle control. They can generate both proliferative and proapoptotic activities: the former were described for rat GIIA sPLA2 in kidney fibroblasts [12], whereas, for both snake and human GIIA sPLA2s, proapoptotic activities were observed in neurons [13,14]. The exact molecular mechanisms of these as well as some other effects of sPLA2s on cells are not known [15,16].
The wide range of physiological and pathophysiological effects exhibited by sPLA2s calls for simple experimental models [15]. The fact that Atx interacts with evolutionarily highly conserved proteins to produce its cellular effects enabled us to employ yeast for this purpose. Yeast has a number of advantages as a model system – most importantly, it can easily be genetically manipulated and it allows application of a wide range of whole-genome experimental approaches. Moreover, in Saccharomyces cerevisiae, there is no known homologue of sPLA2s that might interfere with Atx effects in yeast cells. Although at least three genes for phospholipase B, namely PLB1, PLB2 and PLB3, are present [17], they code for extracellular proteins that are not likely to interfere with the intracellularly acting Atx ([18] and the present study).
In the present study, we have shown that Atx binds in vitro to yeast homologues of 14-3-3 proteins. We expressed Atx in yeast cells and demonstrated its intracellular biological activity. To assess the in vivo interaction between Atx and 14-3-3 proteins, the effect of Atx on G2 cell-cycle arrest was investigated. For a global survey of changes in the Atx-expressing yeast cells, proteomic analysis was performed.
EXPERIMENTAL
Strains and media
The following strains were used: yUP10 (S288c-derived; MATα ura3 Met−), pex11Δ (S288c-derived, EUROSCARF accession no. Y17129; MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YOL147c::kanMX4) and the corresponding wild-type strain BY4742 (S288c-derived, EUROSCARF accession no. Y10000; MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0). BY4742 and pex11Δ strains were obtained from EUROSCARF (European Saccharomyces cerevisiae Archive for Functional Analysis, Frankfurt, Germany).
The gene coding for mature AtxA [19] was cloned in a pRS316-derived plasmid (ARS1 CEN4 URA3) under the control of the GAL1 promoter and transformed into the respective strains using the lithium acetate method. Strains transformed with the empty vector were used as the control in all experiments. Cells were grown in the synthetic minimum uracil dropout medium [YNB CSM−URA (where YNB stands for yeast nitrogen base and CSM−URA for complete supplement medium without uracil); 1.7 g of YNB+0.67 g of CSM−URA+2% (w/v) glucose or 2% (w/v) galactose in 1 litre of water], either with glucose (2%) as the carbon source, which represses Atx expression, or with galactose (2%) as the carbon source, which induces Atx expression. yUP10 strain was grown in rich [YPD (1%, w/v, yeast extract, 2%, w/v, peptone and 2% glucose)] medium for isolation of 14-3-3 proteins.
Isolation of Bmh1p and Bmh2p
Bmh1 and Bmh2 proteins (i.e. yeast 14-3-3 proteins) were isolated by the method of Toker et al. [20]. Frozen yeast (80 g), strain yUP10, was thawed and homogenized in 100 ml of buffer A (0.25 M sucrose, 20 mM Tris/HCl, pH 7.5, 2 mM EDTA, 10 mM EGTA and 1 mM dithiothreitol) to which proteinase inhibitors (31 μg/ml benzamidine, 25 μg/ml bacitracin, 2 μg/ml soya-bean trypsin inhibitor, 1.4 μg/ml pepstatin and 1 μg/ml leupeptin) had been added immediately before use. The final preparation was concentrated and dialysed against 50 mM Tris (pH 8.4) and 0.1 mM CaCl2. Western-blot analysis, using rabbit anti-14-3-3β polyclonal antibodies (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.), was performed to verify the identity of the isolated proteins as 14-3-3 homologues.
Surface plasmon resonance
Surface plasmon resonance experiments were performed at 25 °C using a Biacore® X system. Atx in 10 mM Mes (2-morpholinoethanesulphonic acid) buffer (pH 6.5) and 5 mM CaCl2 was immobilized on the CM5 sensor chip as described in [21]. The interaction between Bmh1/2 proteins and the chip-immobilized Atx was studied by injecting solutions of Bmh1/2 proteins in 50 mM Tris (pH 8.4) and 0.1 mM CaCl2 over the chip and following the changes in refractive index at the sensor surface. The sensor chip was regenerated with 5 mM NaOH between consecutive injections.
Determination of the mutation rate and of the ratio of G1/G2 phase cells
H2O2 treatment
Atx-expressing and control cells of the strain yUP10 were grown in the medium that induces Atx expression from early exponential phase until they reached late exponential phase. Cells were aliquoted and H2O2 was added to 5 mM of final concentration; the same volume of distilled water was added to mock-treated samples. The cells were rotated at 250 rev./min and 30 °C for an additional 3 h to determine the G1/G2 cell ratio, and for 16–48 h to determine the mutation rate. The viability of the cells was determined by plating appropriate dilutions on YPD plates and it was above 50% for wild-type cells after 3 h treatment under our experimental conditions.
Determination of the G1/G2 ratio
Ethanol (1 ml) was added to 0.5 ml of yeast culture and the cells were rotated overnight. Cells were pelleted and washed with PBS. RNase was added to 1 mg/ml concentration and the samples were incubated for 1 h at 37 °C, or alternatively overnight at 4 °C. Then, 1 μl of 20 mg/ml proteinase K in water was added and the samples were vortex-mixed and incubated for 1 h at 50–55 °C. The cells were pelleted, washed with PBS, resuspended with 200 μl of 100 μg/ml propidium iodide in PBS and incubated overnight in the dark at 4 °C. They were then analysed using an FACSCalibur (Becton Dickinson, San Jose, CA, U.S.A.) flow cytometer, and at least 100000 viable cells were counted for each measurement. Three independent experiments were performed for each strain under each condition and then average values and S.D. were calculated.
Determination of mutation rate
Cells were plated on to uracil and arginine dropout plates (−URA −ARG), incubated for 3 h at 30 °C and then overlaid with top agar containing canavanine (20 μg/ml final concentration). Colonies were counted after 72 h. Appropriate dilutions of the same cells were also plated on to YPD plates to determine the cell density, and the mutation rate was then calculated.
Proteomic analysis
Atx-expressing and control cells of the strain yUP10 were grown from early to late exponential phase in the medium that induces Atx expression and harvested by centrifugation. Proteins were isolated and 50 μg of protein from each sample was labelled with the Cy2 and Cy5 dyes, as described in [22]. An additional 250 μg of the proteins was added for enrichment, and two-dimensional gel electrophoresis was used for their separation, as described in [23].
Two-dimensional gels were scanned directly between glass plates; Sypro Ruby staining (Bio-Rad Laboratories, Hercules, CA, U.S.A.) was then performed. Differentially expressed proteins were determined and all those proteins that showed >2-fold increased expression were picked and trypsinized in-gel. Tryptic peptides were analysed by MALDI–TOF-MS (matrix-assisted laser-desorption ionization–time-of-flight MS), identified by peptide ‘fingerprinting’ using the Mascot program (http://www.matrixscience.com/) and confirmed by matching the mass and pI with the theoretical values of the identified polypeptides.
RESULTS
Binding of Atx to yeast 14-3-3 homologues
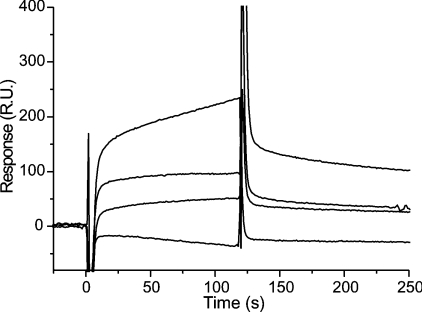
The two yeast homologues of 14-3-3 proteins, Bmh1p and Bmh2p, were isolated from yeast and their interaction with Atx was assayed using surface plasmon resonance (Figure 1). The apparent Kd for this interaction was in the μM range, similar to that for the interaction between Atx and mammalian 14-3-3 proteins [7].
Figure 1. Interaction between Bmh1/2 proteins and Atx.
The interaction was observed by surface plasmon resonance. Atx was immobilized on a CM5 chip and various concentrations (0, 32, 80 and 160 nM) of Bmh1/2 protein were passed across the surface of the chip at 20 μl/min in 50 mM Tris (pH 8.4) and 0.1 mM CaCl2 at 25 °C. RU, response unit.
Expression of active Atx in yeast cells
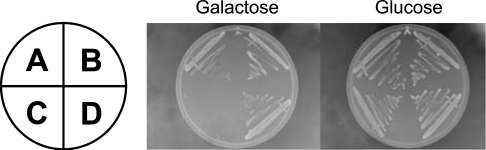
Atx expressed in yeast S. cerevisiae under the control of the galactose-inducible GAL1 promoter was biologically active. This was shown by expressing it in the pex11Δ strain that is defective in peroxisome biogenesis and inheritance [24,25]. The pex11Δ strain harbouring the GAL1-Atx plasmid grew on galactose medium significantly slower than the wild-type BY4742 strain or the respective strains harbouring empty plasmids (Figure 2). Growth rates of the wild-type strain harbouring the empty vector, the wild-type strain harbouring the GAL1-Atx plasmid and the pex11Δ strain harbouring the empty vector did not differ significantly under any of the conditions tested, and expression of an inactive Atx mutant had no effect on the pex11Δ strain (results not shown), confirming that the observed inhibition of growth was a consequence of active Atx expression. Subsequent replacement of galactose by glucose in the medium resulted in the pex11Δ cells harbouring the GAL1-Atx plasmid starting to grow again at the wild-type growth rate, indicating that the effect of Atx on the pex11Δ strain was cytostatic.
Figure 2. Atx expression inhibits growth of the pex11Δ strain.
Growth of wild-type and pex11Δ, GAL1-Atx harbouring and control strains (A, pex11Δ with empty vector; B, BY4742 with empty vector; C, pex11Δ GAL1-Atx; and D, BY4742 GAL1-Atx) on galactose and glucose media was compared. The plates were incubated for 48 h at 30 °C.
Effect of intracellular Atx on H2O2-mediated cell-cycle arrest
Bmh1p and Bmh2p are required for a proper response to DNA damage, in particular by involvement in DNA damage checkpoints [26]. We therefore considered the possible effects of Atx on the cell cycle of the wild-type strain of S. cerevisiae. Different growth perturbations were used to compare the responses of Atx-expressing and control yUP10 strains. After treatment of cells in late exponential phase with 5 mM H2O2, we observed an approx. 2-fold lower survival of Atx-expressing yeast cells compared with control cells. Since, in S. cerevisiae, H2O2 is known to cause cell-cycle arrest specifically in the G2 phase [27], inhibition of the arrest was considered as a possible cause of decreased survival of the Atx-expressing strain. The ratios of the number of cells in the G1 and G2 phases of Atx-expressing versus control cells were therefore determined after treating the cells with H2O2, using propidium iodide staining and flow cytometry. Under non-perturbed growth conditions, the ratio was the same for the two strains, i.e. 1:1.04±0.13 for the Atx-expressing strain and 1:1.03±0.07 for the control strain. However, when the cells were treated with 5 mM H2O2 for 3 h, the Atx-expressing strain showed no significant change in the G1/G2 ratio (1:1.08±0.04), whereas the control strain showed a decrease (1:1.23±0.06), indicative of G2 cell-cycle arrest, as reported in [27]. These observations suggest that Atx inhibited the G2 cell-cycle arrest in yeast S. cerevisiae.
Inhibition of the G2 cell-cycle arrest leads to increased mutation rate [28]. In keeping with this, when the yUP10 cells were exposed to 5 mM H2O2 for periods between 16 and 48 h, the mutation rate, measured by the occurrence of canavanine resistance, was 5–50-fold higher in Atx-expressing cells than in control cells, confirming the effect of Atx on the regulation of the cell cycle, and explaining the observed decreased survival rate of the Atx-expressing strain after H2O2 treatment. No difference in mutation rate was observed between non-treated Atx-expressing and control strains.
Proteomic analysis
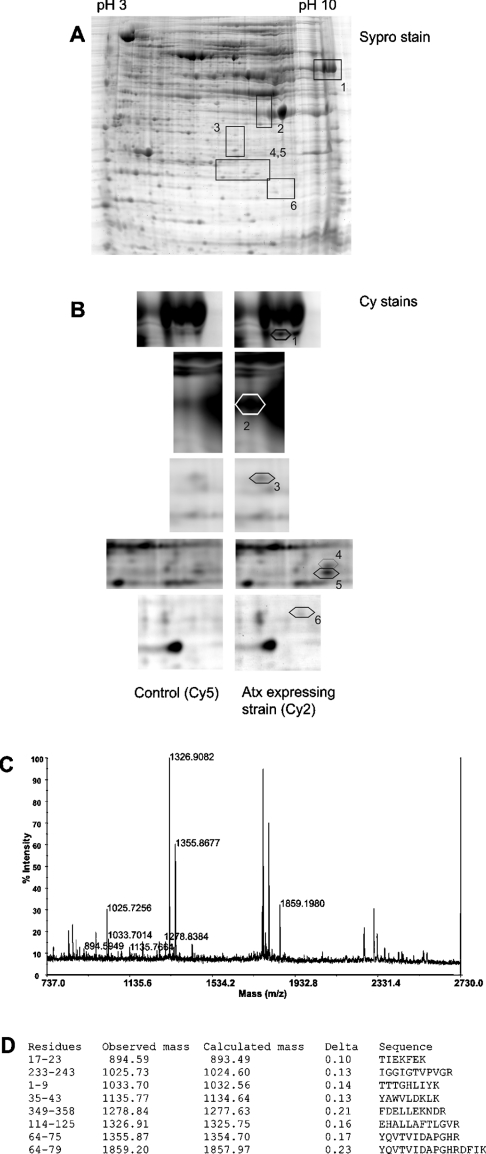
Changes in protein expression in the Atx-expressing wild-type yUP10 strain were determined by two-dimensional electrophoresis. Six polypeptides with >2-fold higher expression in Atx-expressing cells than in the control strain were detected using fluorescent labelling of proteins with Cy2 and Cy5, and identified by MALDI–TOF-MS (Table 1; Figure 3). No underexpressed proteins were found.
Table 1. Overexpressed proteins in the Atx-expressing yeast strain.
Proteins were analysed by two-dimensional electrophoresis and identified after trypsin digestion and peptide map ‘fingerprinting’. MM, molecular mass.
| Spot no. | Protein | Apparent MM/pI | Protein function | Fold overexpression |
|---|---|---|---|---|
| 1 | Tef1/2p* | 50 kDa/9.5 | Translation elongation factor EF 1α | 2.1 |
| 2 | Tdh3p | 36 kDa/7.0 | Glyceraldehyde-3-phosphate dehydrogenase | 2.4 |
| 3 | Ydl086w | 25 kDa/6.0 | Unknown | 2.2 |
| 4 | Adh1p (N-terminal peptide CAA24600) | 14 kDa/6.5 | Alcohol dehydrogenase | 2.3 |
| 5 | Tdh2/3p* (degradation product) | 13 kDa/6.4 | Glyceraldehyde-3-phosphate dehydrogenase | 2.2 |
| 6 | Rad2p (N-terminal peptide CAA69080) | 6 kDa/7.0 | Involved in nucleotide excision repair | 2.1 |
* From the data, it could not be determined which of the two isoforms was overexpressed.
Figure 3. Two-dimensional image of yeast proteins from Atx-expressing and control yUP10 strains and protein identification by MS.
(A) A representative Sypro-stained gel with boxed segments that are shown enlarged in (B) and the corresponding spot numbers. (B) Enlarged views after fluorescence scanning to identify the differentially expressed proteins from the two strains: proteins from the Atx-expressing strain were stained with Cy2 and those from the control strain were stained with Cy5. The differentially expressed proteins are marked spots 1–6 (see Table 1 for details). (C) MALDI–TOF-MS spectrum obtained from tryptic digest of spot 1. (D) Masses of ionized peaks were matched with those calculated for tryptic peptides of translation elongation factor Tef1/2p. Peptide sequences matching the peaks in the spectrum are shown.
Spot 1 corresponded to Tef1p and/or Tef2p. Since they have identical amino acid sequences, it was not possible by this method to determine which of the two proteins was overexpressed. Tdh3p was clearly identified in spot 2, despite its 96% identity with Tdh2p isoform, as was the Ydl086w protein of unknown function. In the case of the degradation product in spot 5, it was impossible to distinguish between the two isoforms. Adh1p and Rad2p were identified as degradation products, corresponding to partial sequences in the databases with GenBank® entries CAA24600 and CAA69080 respectively.
DISCUSSION
Atx was shown to bind in vitro to evolutionarily conserved mammalian proteins, γ and ε isoforms of 14-3-3 proteins and calmodulin [6,7]. There are two yeast homologues of 14-3-3 proteins, Bmh1p and Bmh2p, which are 92% identical in amino acid sequence and 72 and 73% identical with the human ε isoform respectively. We demonstrated in the present study that Atx binds in vitro to yeast homologues of 14-3-3 proteins, Bmh1p and Bmh2p, with apparent Kd values similar to those measured for the interaction between Atx and mammalian 14-3-3 proteins (Figure 1). On the other hand, Atx does not bind in vitro to yeast calmodulin, Cmd1p (U. Petrovič and I. Križaj, unpublished work). We took advantage of these facts to use yeast S. cerevisiae, which has no endogenous, intracellularly active PLA2s, as a model eukaryotic system to elucidate the effect of binding of Atx to 14-3-3 proteins in vivo.
The mature form of Atx was expressed in yeast cells. Comparison of the phospholipolytic activity of lysates of the Atx-expressing yUP10 strain under the GAL1 promoter with those of the control strain showed that Atx is enzymatically active and that the number of active Atx molecules per cell is approx. 105 [18], which is probably more than the amount of Atx that enters the mammalian cell on envenomation in vivo. Here, the biological activity of Atx inside the yeast cell was confirmed by expressing the phospholipase in peroxisome-deficient strains. The expression of Atx inhibited the growth of pex11Δ yeast cells (Figure 2), as well as of pex3Δ and pex5Δ strains (M. Mattiazzi, U. Petrovič and I. Križaj, unpublished work). One possible explanation for the requirement of functional peroxisomes for the viability of Atx-expressing strains is its intracellular phospholipolytic activity generating free fatty acids that are toxic to S. cerevisiae [29,30]. In wild-type yeast, free fatty acids are known to exert oxidative stress by uncoupling the respiratory chain [31], causing a transient arrest in cell multiplication before metabolic adaptation that includes fatty acid β-oxidation in peroxisomes [30]. Yeast strains with impaired peroxisome biogenesis and inheritance have decreased levels of fatty acid β-oxidation [24,32]. According to this hypothesis, in peroxisome-deficient strains, the metabolic adaptation to free fatty acids produced by enzymatically active Atx would be hampered; therefore in the Atx-expressing strain the growth arrest would be persistent. The fact that Atx exerts a cytostatic rather than a cytotoxic effect on the pex11Δ strain is in line with this hypothesis. Our experimental results support the notion that it is the intracellular activity of Atx that causes the observed effects. First, no phospholipolytic activity was detected in the extracellular medium of Atx-expressing yeast cells [18] and secondly, addition of Me-indoxam, a cell-impermeable inhibitor of sPLA2s [16,33] that also inhibits Atx activity, to the medium did not suppress the growth inhibition of Atx-expressing pex11Δ strain (results not shown).
In S. cerevisiae, Bmh1p and Bmh2p are involved in the induction of G2 cell-cycle arrest [26]. GIIA sPLA2s cause proliferative and proapoptotic effects in mammalian cells and it has been proposed that separate pathways lead to these opposing effects [12–14]. Alternatively, these effects could be caused by a common effect on cell-cycle regulation, and the final outcome of the sPLA2 activity would depend on the cell type and/or environmental factors. In keeping with this latter possibility, we checked whether Atx affects G2 cell-cycle arrest. Unlike in wild-type S. cerevisiae, where H2O2 causes specific G2 cell-cycle arrest [27], addition of H2O2 did not affect the G1/G2 phase ratio in the Atx-expressing wild-type strain. This could either mean that the Atx-expressing strain is resistant to H2O2-mediated DNA damage [34] or that G2 cell-cycle arrest is inhibited in the presence of this sPLA2. The increase in mutation rate and decrease in viability of the Atxexpressing strain demonstrated in the presence of H2O2 strongly favour the second explanation. Moreover, we observed no significant difference in the rate of H2O2 degradation between Atx-expressing and control strains (results not shown), further pointing to inhibition of G2 cell-cycle arrest in the Atx-expressing strain.
The proposal that interaction of GIIA sPLA2 with 14-3-3 proteins affects cell-cycle regulation could also apply to mammals. 14-3-3 proteins are involved in the onset of the G2 cell-cycle arrest in mammalian cells [35,36], and molecules that interfere with their function are potent abrogators of G2 cell-cycle arrest [37,38]. Human GIIA sPLA2, which induces apoptosis in neurons [14], binds to 14-3-3 proteins in vitro (G. Faure and I. Križaj, unpublished work). In addition, besides the human GIIA sPLA2 and all the Atx isoforms, AtxA, AtxB and AtxC, other GIIA sPLA2s bind in vitro to yeast 14-3-3 proteins: β-bungarotoxin, taipoxin, and ammodytin L, a non-neurotoxic paralogue of Atx (M. Mattiazzi, U. Petrovič, G. Anderluh and I. Križaj, unpublished work). Two of these sPLA2s, β-bungarotoxin and ammodytin L, also have demonstrated cell-cycle-related effects on mammalian cells [13,39]. Although further experiments are required to establish this putative connection in mammalian cells, we can speculate, on the basis of our results from the yeast system, that binding to 14-3-3 proteins and consequent inhibition of the G2 cell-cycle arrest is the mechanism behind the proliferative and proapoptotic effects of GIIA sPLA2s on mammalian cells [12–14]. According to this hypothesis, in dividing cells without potentially lethal mutations, inhibition of cell-cycle arrest would lead to faster growth and pronounced proliferation, whereas, in other instances, such as cells with potentially lethal mutations, the inability to repair DNA damage would result in non-permissive genomic changes and would result in (apoptotic) cell death.
Proteomic analysis was used for a global-view comparison of Atx-expressing and control strains. The common denominator connecting three of the proteins shown to be overexpressed (Table 1), Tdh3p, Adh1p and Tef1/2p, is that they have been reported as the prime targets of S-thiolation in response to H2O2 [40]. In the case of glyceraldehyde-3-phosphate dehydrogenase, three isoforms are present in yeast, namely Tdh1p, Tdh2p and Tdh3p, of which only Tdh3p is the major target of S-thiolation [41]. Similarly, of the five isoforms of alcohol dehydrogenase in yeast, only Adh1p was identified as a target protein [40]. Translation elongation factors, including Tef1/2p, are the third functional group of the S-thiolation target proteins [40]. According to the ‘guilt-by-association’ paradigm, this suggests that most of the observed changes in the proteome are not the consequence of a specific effect of Atx on the metabolic processes in which the overexpressed proteins are involved, but rather are related to the putative oxidative stress caused by Atx expression. The oxidative stress could be a direct consequence of the free fatty acids generated by Atx, as described above. Another overexpressed protein was identified as Rad2p, a homologue of xeroderma pigmentosum group G protein, involved in DNA nucleotide excision repair [42,43]. In addition, Ydl086w, a protein with undetermined function and also identified as being overexpressed in the Atx-expressing strain (Table 1), is known to interact with two nuclear proteins, TFIIB of the RNA II polymerase holoenzyme Sua7p [44], and a DEAD-box helicase Dbp8p [45], indicating its involvement in nuclear processes, possibly including DNA repair. None of the identified overexpressed proteins is known to be involved in any other process affected by Atx in yeast. It is therefore reasonable to conclude that Atx expression in yeast activates the DNA damage repair machinery, which is in agreement with the effect of Atx on the inhibition of the G2 cell-cycle arrest, since this checkpoint is also important when the cells are growing in the absence of any mutagenic effect, and its impairment in mammalian cells results in increased DNA damage [46].
Acknowledgments
This work was supported by grant Z1-4468 from the Ministry of Education, Science and Sports of the Republic of Slovenia. We thank Dr M.H. Gelb (Department of Chemistry and Biochemistry, University of Washington, Seattle, WA 98195-1700, U.S.A.) for the gift of Me-indoxam and Dr R.H. Pain (Department of Biochemistry and Molecular Biology, Jožef Stefan Institute) for critically reading this paper.
References
- 1.Kini R. M. Chichester, U.K.: John Wiley & Sons; 1997. Venom Phospholipase A2 Enzymes. [Google Scholar]
- 2.Kudo I., Murakami M. Phospholipase A2 enzymes. Prostaglandins Other Lipid Mediat. 2002;68–69:3–58. doi: 10.1016/s0090-6980(02)00020-5. [DOI] [PubMed] [Google Scholar]
- 3.Dennis E. A. Diversity of group types, regulation, and function of phospholipase A2. J. Biol. Chem. 1994;269:13057–13060. [PubMed] [Google Scholar]
- 4.Križaj I., Gubenšek F. Neuronal receptors for phospholipases A2 and β-neurotoxicity. Biochimie. 2000;82:807–814. doi: 10.1016/s0300-9084(00)01172-x. [DOI] [PubMed] [Google Scholar]
- 5.Vardjan N., Sherman N. E., Pungerčar J., Fox J. W., Gubenšek F., Križaj I. High-molecular-mass receptors for ammodytoxin in pig are tissue-specific isoforms of M-type phospholipase A2 receptor. Biochem. Biophys. Res. Commun. 2001;289:143–149. doi: 10.1006/bbrc.2001.5940. [DOI] [PubMed] [Google Scholar]
- 6.Šribar J., Čopič A., Pariš A., Sherman N. E., Gubenšek F., Fox J. W., Križaj I. A high affinity acceptor for phospholipase A2 with neurotoxic activity is a calmodulin. J. Biol. Chem. 2001;276:12493–12496. doi: 10.1074/jbc.C100048200. [DOI] [PubMed] [Google Scholar]
- 7.Šribar J., Sherman N. E., Prijatelj P., Faure G., Gubenšek F., Fox J. W., Aitken A., Pungerčar J., Križaj I. The neurotoxic phospholipase A2 associates, through a non-phosphorylated binding motif, with 14-3-3 protein γ and ε isoforms. Biochem. Biophys. Res. Commun. 2003;302:692–696. doi: 10.1016/s0006-291x(03)00228-6. [DOI] [PubMed] [Google Scholar]
- 8.Chin D., Means A. R. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 9.Fu H., Subramanian R. R., Masters S. C. 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 10.Mackintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem. J. 2004;381:329–342. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilker E., Yaffe M. B. 14-3-3 Proteins – a focus on cancer and human disease. J. Mol. Cell. Cardiol. 2004;37:633–642. doi: 10.1016/j.yjmcc.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y., Lemasters J., Herman B. Secretory group IIA phospholipase A2 generates anti-apoptotic survival signals in kidney fibroblasts. J. Biol. Chem. 2001;274:27726–27733. doi: 10.1074/jbc.274.39.27726. [DOI] [PubMed] [Google Scholar]
- 13.Herkert M., Shakhman O., Schweins E., Becker C. M. β-Bungarotoxin is a potent inducer of apoptosis in cultured rat neurons by receptor-mediated internalization. Eur. J. Neurosci. 2001;14:821–828. doi: 10.1046/j.0953-816x.2001.01699.x. [DOI] [PubMed] [Google Scholar]
- 14.Yagami T., Ueda K., Asakura K., Hata S., Kuroda T., Sakaeda T., Takasu N., Tanaka K., Gemba T., Hori Y. Human group IIA secretory phospholipase A2 induces neuronal cell death via apoptosis. Mol. Pharmacol. 2002;61:114–126. doi: 10.1124/mol.61.1.114. [DOI] [PubMed] [Google Scholar]
- 15.Chilton F. Would the real role(s) for secretory PLA2s please stand up. J. Clin. Invest. 1996;97:2161–2162. doi: 10.1172/JCI118654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mounier C. M., Ghomashchi F., Lindsay M. R., James S., Singer A. G., Parton R. G., Gelb M. H. Arachidonic acid release from mammalian cells transfected with human groups IIA and X secreted phospholipase A2 occurs predominantly during the secretory process and with the involvement of cytosolic phospholipase A2-α. J. Biol. Chem. 2004;279:25024–25038. doi: 10.1074/jbc.M313019200. [DOI] [PubMed] [Google Scholar]
- 17.Merkel O., Fido M., Mayr J. A., Pruger H., Raab F., Zandonella G., Kohlwein S. D., Paltauf F. Characterization and function in vivo of two novel phospholipases B/lysophospholipases from Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:28121–28127. doi: 10.1074/jbc.274.40.28121. [DOI] [PubMed] [Google Scholar]
- 18.Petrovič U., Šribar J., Pariš A., Rupnik M., Kržan M., Vardjan N., Gubenšek F., Zorec R., Križaj I. Ammodytoxin, a neurotoxic secreted phospholipase A2, can act in the cytosol of the nerve cell. Biochem. Biophys. Res. Commun. 2004;324:981–985. doi: 10.1016/j.bbrc.2004.09.144. [DOI] [PubMed] [Google Scholar]
- 19.Pungerčar J., Kordiš D., Štrukelj B., Liang N. S., Gubenšek F. Cloning and nucleotide sequence of a cDNA encoding ammodytoxin A, the most toxic phospholipase A2 from the venom of long-nosed viper (Vipera ammodytes) Toxicon. 1991;29:269–273. doi: 10.1016/0041-0101(91)90112-5. [DOI] [PubMed] [Google Scholar]
- 20.Toker A., Ellis C. A., Sellers L. A., Aitken A. Protein kinase C inhibitor proteins. Purification from sheep brain and sequence similarity to lipocortins and 14-3-3 protein. Eur. J. Biochem. 1990;191:421–429. doi: 10.1111/j.1432-1033.1990.tb19138.x. [DOI] [PubMed] [Google Scholar]
- 21.Faure G., Villela C., Perales J., Bon C. Interaction of the neurotoxic and nontoxic secretory phospholipases A2 with the crotoxin inhibitor from Crotalus serum. Eur. J. Biochem. 2000;227:4799–4808. doi: 10.1046/j.1432-1327.2000.01532.x. [DOI] [PubMed] [Google Scholar]
- 22.Tonge R., Shaw J., Middleton B., Rowlinson R., Rayner S., Young J., Pognan F., Hawkins E., Currie I., Davison M. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics. 2001;1:377–396. doi: 10.1002/1615-9861(200103)1:3<377::AID-PROT377>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 23.Gharbi S., Gaffney P., Yang A., Zvelebil M. J., Cramer R., Waterfield M. D., Timms J. F. Evaluation of two-dimensional differential gel electrophoresis for proteomic expression analysis of a model breast cancer cell system. Mol. Cell. Proteomics. 2002;1:91–98. doi: 10.1074/mcp.t100007-mcp200. [DOI] [PubMed] [Google Scholar]
- 24.Erdmann R., Blobel G. Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J. Cell Biol. 1995;128:509–523. doi: 10.1083/jcb.128.4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall P. A., Krimkevich Y. I., Lark R. H., Dyer J. M., Veenhuis M., Goodman J. M. Pmp27 promotes peroxisomal proliferation. J. Cell Biol. 1995;129:345–355. doi: 10.1083/jcb.129.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lottersberger F., Rubert F., Baldo V., Lucchini G., Longhese M. P. Functions of Saccharomyces cerevisiae 14-3-3 proteins in response to DNA damage and to DNA replication stress. Genetics. 2003;165:1717–1732. doi: 10.1093/genetics/165.4.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flattery-O'Brien J. A., Dawes I. W. Hydrogen peroxide causes RAD9dependent cell cycle arrest in G2 in Saccharomyces cerevisiae whereas menadione causes G1 arrest independent of RAD9 function. J. Biol. Chem. 1998;273:8564–8571. doi: 10.1074/jbc.273.15.8564. [DOI] [PubMed] [Google Scholar]
- 28.Nyberg K. A., Michelson R. J., Putnam C. W., Weinert T. A. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 29.Hunkova Z., Fencl A. Toxic effects of fatty acids on yeast cells: possible mechanisms of action. Biotechnol. Bioeng. 1978;20:1235–1247. doi: 10.1002/bit.260200809. [DOI] [PubMed] [Google Scholar]
- 30.Koerkamp M. G., Rep M., Bussemaker H. J., Hardy G. P., Mul A., Piekarska K., Szigyarto C. A., De Mattos J. M., Tabak H. F. Dissection of transient oxidative stress response in Saccharomyces cerevisiae by using DNA microarrays. Mol. Biol. Cell. 2002;13:2783–2794. doi: 10.1091/mbc.E02-02-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polčic P., Šabova L., Kolarov J. Fatty acids induced uncoupling of Saccharomyces cerevisiae mitochondria requires an intact ADP/ATP carrier. FEBS Lett. 1997;412:207–210. doi: 10.1016/s0014-5793(97)00778-3. [DOI] [PubMed] [Google Scholar]
- 32.Lazarow P. B., Kunau W. H. Peroxisomes. In: Pringle J. R., Broach J. R., Jones E. W., editors. The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae – Cell Cycle and Cell Biology. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 547–605. [Google Scholar]
- 33.Hagishita S., Yamada M., Shirahase K., Okuda T., Murakami Y., Ito Y., Matsuura T., Wada M., Kato T., Ueno M., et al. Potent inhibitors of secretory phospholipase A2: synthesis and inhibitory activities of indollizine and indene derivatives. J. Med. Chem. 1996;39:3636–3658. doi: 10.1021/jm960395q. [DOI] [PubMed] [Google Scholar]
- 34.Imlay J. A., Chin S. M., Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 35.Hermeking H., Lengauer C., Polyak K., He T. C., Zhang L., Thiagalingam S., Kinzler K. W., Vogelstein B. 14-3-3σ is a p53-regulated inhibitor of G2/M progression. Mol. Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 36.Peng C. Y., Graves P. R., Thoma R. S., Wu Z., Shaw A. S., Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 37.Hermeking H. The 14-3-3 cancer connection. Nat. Rev. Cancer. 2003;3:931–943. doi: 10.1038/nrc1230. [DOI] [PubMed] [Google Scholar]
- 38.Kawabe T. G2 checkpoint abrogators as anticancer drugs. Mol. Cancer Ther. 2004;3:513–519. [PubMed] [Google Scholar]
- 39.Rufini S., Cesaroni M. P., Balestro N., Luly P. Proliferative effect of ammodytin L from the venom of Vipera ammodytes on 208F rat fibroblasts in culture. Biochem. J. 1996;320:467–472. doi: 10.1042/bj3200467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shenton D., Grant C. M. Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem. J. 2003;374:513–519. doi: 10.1042/BJ20030414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grant C. M., Quinn K. A., Dawes I. W. Differential protein S-thiolation of glyceraldehyde-3-phosphate dehydrogenase isoenzymes influences sensitivity to oxidative stress. Mol. Cell. Biol. 1999;19:2650–2656. doi: 10.1128/mcb.19.4.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prakash S., Prakash L. Nucleotide excision repair in yeast. Mutat. Res. 2000;451:13–24. doi: 10.1016/s0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- 43.de Laat W. L., Jaspers N. G., Hoeijmakers J. H. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 44.Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature (London) 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 46.Abraham R. T. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]