Abstract
GSH synthesis occurs via two enzymatic steps catalysed by GCL [glutamate–cysteine ligase, made up of GCLC (GCL catalytic subunit), and GCLM (GCL modifier subunit)] and GSS (GSH synthetase). Co-ordinated up-regulation of GCL and GSS further enhances GSH synthetic capacity. The present study examined whether TNFα (tumour necrosis factor α) influences the expression of rat GSH synthetic enzymes. To facilitate transcriptional studies of the rat GCLM, we cloned its 1.8 kb 5′-flanking region. TNFα induces the expression and recombinant promoter activities of GCLC, GCLM and GSS in H4IIE cells. TNFα induces NF-κB (nuclear factor κB) and AP-1 (activator protein 1) nuclear-binding activities. Blocking AP-1 with dominant negative c-Jun or NF-κB with IκBSR (IκB super-repressor, where IκB stands for inhibitory κB) lowered basal expression and inhibited the TNFα-mediated increase in mRNA levels of all three genes. While all three genes have multiple AP-1-binding sites, only GCLC has a NF-κB-binding site. Overexpression with p50 or p65 increased c-Jun mRNA levels, c-Jun-dependent promoter activity and the promoter activity of GCLM and GSS. Blocking NF-κB also lowered basal c-Jun expression and blunted the TNFα-mediated increase in c-Jun mRNA levels. TNFα treatment resulted in increased c-Jun and Nrf2 (nuclear factor erythroid 2-related factor 2) nuclear binding to the antioxidant response element of the rat GCLM and if this was prevented, TNFα no longer induced the GCLM promoter activity. In conclusion, both c-Jun and NF-κB are required for basal and TNFα-mediated induction of GSH synthetic enzymes in H4IIE cells. While NF-κB may exert a direct effect on the GCLC promoter, it induces the GCLM and GSS promoters indirectly via c-Jun.
Keywords: activator protein 1 (AP-1), glutamate–cysteine ligase (GCL), GSH synthetase (GSS), nuclear factor κB (NF-κB), tumour necrosis factor α (TNFα)
Abbreviations: AP-1, activator protein 1; ARE, antioxidant response element; ATF, activating transcription factor; ChIP, chromatin immunoprecipitation; EMSA, electrophoretic mobility-shift assay; GCL, glutamate–cysteine ligase; GCLC, GCL catalytic subunit; GCLM, GCL modifier subunit; GSS, GSH synthetase; IκB, inhibitory κB; IκBSR, IκB super-repressor; NF-κB, nuclear factor κB; Nrf2, nuclear factor erythroid 2-related factor 2; TNFα, tumour necrosis factor α; Z-VAD-FMK, benzyloxycarbonyl-valylalanyl-DL-aspartyl fluoromethane
INTRODUCTION
GSH is the main non-protein thiol in mammalian cells that participates in many critical cellular functions including antioxidant defence and cell growth [1–3]. GSH is synthesized in the cytosol of all mammalian cells via two ATP-requiring enzymatic steps: the formation of γ-glutamylcysteine from glutamate and cysteine, and formation of GSH from γ-glutamylcysteine and glycine. The first step of GSH biosynthesis is generally regarded as ratelimiting and catalysed by GCL (glutamate–cysteine ligase, also known as γ-glutamylcysteine synthetase), which is regulated physiologically by feedback competitive inhibition by GSH and the availability of cysteine [1,4]. The GCL enzyme is composed of a GCLC (GCL catalytic subunit; Mr∼73000) and a GCLM (GCL modifier subunit; Mr∼30000) that are encoded by different genes and dissociate under reducing conditions [5–7]. The catalytic subunit exhibits all of the catalytic activity of the isolated enzyme as well as feedback inhibition by GSH [7]. The modifier subunit is enzymatically inactive but plays an important regulatory function by lowering the Km of GCL for glutamate and raising the Ki for GSH [6,8]. Since GCL is a major determinant of the overall capacity of GSH synthesis, regulation of GCL subunits has been a topic of extensive research [1]. Changes in GCL activity can result from regulation at multiple levels affecting only the catalytic or modifier subunits or both. The 5′-flanking regions of the human GCL subunits have been cloned [9–11]. ARE (antioxidant response element, also known as electrophile response element, EpRE) and AP-1 (activator protein 1) are two cis-acting elements present in the promoter of both human GCL subunits that have been implicated in their transcriptional regulation by oxidants and β-naphthoflavone [1,11–15].
Our laboratory has described differential regulation of rat hepatic GCL subunit expression using both in vitro and in vivo treatments. GCLC, but not GCLM, expression increased during periods of rapid hepatocyte growth, after treatment of hepatocytes with hormones such as insulin or glucocorticoids, and after treatment of rats with ethanol [16–20]. On the other hand, treatment of hepatocytes with buthionine sulphoximine, t-butylhydroquinone or diethyl maleate, and treatment of rats with thioacetamide, induced the expression of both GCLC and GCLM [21,22]. Treatments that induced both GCLC and GCLM also induced the expression of GSS (GSH synthetase), which further enhanced the cell's ability to synthesize GSH [23]. To better understand transcription regulation of the rat GSH synthetic enzymes, we have cloned and characterized the 5′-flanking regions of rat GCLC and GSS [24,25]. Interestingly, ARE is not present in the cloned promoter region of either rat GCLC or GSS. Acetaldehyde induced rat GCLC promoter activity and nuclear binding activity of NF-κB (nuclear factor κB) and AP-1 to GCLC [24]. We also showed that AP-1 is required for basal expression and t-butylhydroquinone-mediated induction of rat GCLC, GCLM and GSS [25]. One area of controversy has been the role of NF-κB in regulating the expression of GCLC [1]. While some investigators observed that blocking NF-κB prevented up-regulation of GCLC induced by cytokines in mouse endothelial cells [26], oxidants in rat hepatocytes [21] or ionizing radiation in a human glioblastoma cell line [27], others did not find the NF-κB consensus element present in the human GCLC to be involved in either TNFα (tumour necrosis α) or okadaic acid-mediated induction of GCLC [28,29]. Whether NF-κB plays a role in the transcriptional regulation of GCLM and GSS is unknown. In the present study, we used TNFα, which is known to induce both NF-κB and AP-1, to address transcriptional regulation of the rat GSH synthetic enzymes and assess the involvement of both NF-κB and AP-1. To facilitate these studies, we have also cloned and characterized the 5′-flanking region of the rat GCLM.
MATERIALS AND METHODS
Materials
Cell culture media and fetal bovine serum were obtained from Gibco BRL Life Technologies (Grand Island, NY, U.S.A.). The Luciferase Assay System and recombinant human TNFα were obtained from Promega (Madison, WI, U.S.A.). All restriction endonucleases were obtained from either Promega or Gibco BRL Life Technologies. [32P]dCTP (3000 Ci/mmol) was purchased from DuPont (New England Nuclear, Boston, MA, U.S.A.). All other reagents were of analytical grade and were obtained from commercial sources.
Recombinant plasmids and adenoviral vectors
The rat GCLC and GSS promoter constructs were previously described [24,25] and subcloned in the sense orientation, upstream of the luciferase coding sequence of the pGL-3 enhancer vector (Promega). Recombinant, replication-defective adenovirus expressing dominant-negative c-Jun, TAM67 or IκBSR (IκBSR super-repressor, where IκB stands for inhibitory κB), which expresses a mutant IκB that cannot be phosphorylated and therefore irreversibly binds NF-κB and prevents its activation, were described previously [30]. Jun2-luciferase (Jun2-luc) construct, which contains one of the key AP-1 sites (TTACCTCA) of the human c-Jun promoter that is autoregulated by c-Jun [31], was kindly provided by Dr Z. Ronai (The Ruttenberg Cancer Center, Mount Sinai School of Medicine, NY, U.S.A.) [32]. pCMV-p50 and pCMV-p65 expression plasmids were kindly provided by Dr R. Rippe (University of North Carolina at Chapel Hill).
Cell culture and TNFα treatment
H4IIE cells (a rat hepatoma cell line) were obtained from the Cell Culture Core of the USC Liver Disease Research Center and grown according to instructions provided by the A.T.C.C. (Manassas, VA, U.S.A.). Before treatment with TNFα, the medium was changed to withhold serum overnight. Cells were then treated with TNFα (1–60 ng/ml) for 4–12 h for various assays as described below.
Cloning of the 5′-flanking region of the rat GCLM gene and construction of 5′-deletion constructs
Overlapping clones containing the GCLM gene were isolated from a rat genomic library EMBL 3 (ClonTech, Palo Alto, CA, U.S.A.). The library was screened using a random 32P-labelled probe of the rat GCLM cDNA [6]. Five positive plaques were selected, DNA was isolated and digested with EcoRI. The insert fragment was subcloned into Bluescript KSII (+/−) vector (Stratagene, La Jolla, CA, U.S.A.) and sequenced in both directions using the automated ABI prism dRhodamine terminator cycle sequencer performed by the Sequencing and Genetic Analysis Core Facility (Norris Cancer Institute, Keck School of Medicine USC). The initial primers were universal primers for the Bluescript KSII (+/−) vector; all subsequent primers were nested primers designed using the available sequence information and the MacVector software program. The nucleotide sequence was verified by multiple bi-directional sequencing reactions. Sequences were aligned and a consensus sequence was generated using the ASSEMBLIGN software program. A 1.8 kb 5′-flanking region of the rat GCLM was cloned into the SmaI site of promoterless pGL-3 basic vector (Promega), creating the recombinant plasmid −1803/+3 GCLM-LUC. The desired 5′-deletion constructs (−902/+3, −323/+3 and −157/+3) were generated using restriction nucleases StuI, HpaI and ApaI. The sequence was deposited in GenBank® under the accession no. AF311745.
Transcription start sites of the rat GCLM
Transcription start sites were determined using the GeneRacer™ kit (Invitrogen, CA, U.S.A.). The 5′-ends of cDNA using the reverse primer 5′-TGTCGGTGCCCATGGCAGCTGCC-3′, which is complementary to −10 to +13 of the rat GCLM [6], were obtained. Rapid amplification of cDNA ends PCR products were cloned using Topo TA cloning for sequencing and sequencing analysis was performed by the Sequencing Facility, (Norris Cancer Center, Keck School of Medicine USC).
Analysis of promoter constructs in cell culture
To study the relative transcriptional activities of the GCLM promoter fragments, H4IIE cells (1×106 cells in 2 ml medium) were transiently transfected with 2.5 μg of GCLM promoter luciferase gene construct and promoterless pGL3-basic vector (as negative control) using the Superfect transfection reagent (Qiagen, Valencia, CA, U.S.A.) as we described previously [30]. To control the transfection efficiency, cells were co-transfected with the Renilla phRL-TK vector (Promega), which is a plasmid containing Renilla luciferase gene driven by HSV-TK promoter. After 10 h, cells were harvested and lysed in 200 μl of reporter lysis buffer (Luciferase Assay System, Promega). Aliquots of the cell lysates were sequentially assessed for firefly and Renilla luciferase activities using a TD-20/20 Luminometer (Promega). The luciferase activity driven by the GCLM promoter constructs was normalized to Renilla luciferase activity. Each experiment was performed with triplicate samples.
Effect of TNFα on the expression of GSH synthetic enzymes and recombinant promoter activity
H4IIE cells were treated with 0–15 ng/ml TNFα for 8 h or 7.5 ng/ml for 0–12 h. In some experiments, cells had been infected with adenovirus encoding IκBSR, TAM67 or adenovirus vector alone for 24 h before TNFα treatment as we described previously [30]. At the end of the TNFα treatment, total RNA was extracted and Northern-hybridization analysis was performed using specific GCLC, GCLM, GSS and c-Jun cDNA probes as described in [25]. To ensure equal loading of RNA samples and transfer in each of the lanes, before hybridization, membranes were rinsed with ethidium bromide and photographed and the same membranes were also rehybridized with a 32P-labelled β-actin cDNA probe as described in [25]. Autoradiography and densitometry (Gel Documentation System; Scientific Technologies, Carlsbad, CA, U.S.A. and NIH Image 1.60 software program) were used to quantitate relative RNA. Results of Northern-blot analysis were normalized to β-actin.
To assess the effect of TNFα on promoter activity, H4IIE cells were transfected with recombinant rat GCLC, GCLM or GSS promoter constructs and treated with TNFα (15 ng/ml) during the last 4 h of the transfection. During this short duration, there was no detectable apoptosis or toxicity. Luciferase activity driven by these promoter luciferase gene constructs was measured as described above.
To assess the effect of mutation of the ARE on TNFα-induced increase in GCLM promoter activity, H4IIE cells were transfected with rat GCLM promoter construct −329/+3-LUC, wild-type or mutated in the putative ARE (–295 to −285) (from 5′-TGCTTAGTCAT-3′ to 5′-TACATTGTCAT-3′) and treated with TNFα (15 ng/ml) during the last 4 h of the transfection. Luciferase activity driven by these promoter luciferase gene constructs was measured as described above.
Effect of p50 and p65 expression vectors on GCLM, GSS and Jun2-driven luciferase activities
To verify whether overexpression of p50 or p65 can influence the promoter activity of GCLM, GSS and reporter activity driven by Jun2, H4IIE cells were first transfected with either p50 or p65 expression vector (1.5 μg/well for 12 h) and then transfected with the recombinant promoter luciferase constructs. Luciferase activity was measured as described above.
EMSA (electrophoretic mobility-shift assay) and supershift assay
EMSAs for ARE were performed as described in [25]. Nuclear proteins (10 μg) from H4IIE cells treated with TNFα (15 ng/ml for 4 h) or vehicle control were preincubated with 2 μg of poly(dI-dC)·(dI-dC) in a buffer containing 10 mM Hepes (pH 7.6), 50 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol, 5 mM MgCl2 and 10% (v/v) glycerol for 10 min on ice. 32P-end labelled double-stranded DNA fragments containing the ARE (5′-GTTTCTGCTTAGTCATTGTCTTC-3′, with the ARE-binding site underlined) were then added with or without a 100-fold excess of unlabelled specific probe. Mixtures were incubated for 20 min on ice, loaded on to a 4% (w/v) non-denaturing polyacrylamide gel and subjected to electrophoresis in 50 mM Tris, 45 mM borate and 0.5 mM EDTA (pH 8.0). Gels were dried and subjected to autoradiography. Further confirmation of the identity of the binding proteins was performed by antibody supershift assays for c-Jun and Nrf2 (NF erythroid 2-related factor 2; Upstate Biotechnology, Lake Placid, NY, U.S.A. or Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.) as we described previously [25].
In an independent experiment, EMSA was performed to evaluate the effect of mutation of the ARE on nuclear-binding activity. Nuclear proteins from control H4IIE cells were used for these experiments. ARE site was mutated from 5′-TGCTTAGTCAT-3′ to 5′-TACTTAGTCAT-3′ (1 bp mutation), 5′-TACATAGTCAT-3′ (2 bp mutation) and 5′-TACATTGTCAT-3′ (3 bp mutation).
ChIP (chromatin immunoprecipitation) assay
To verify whether c-Jun and Nrf2 bind to the ARE of the rat GCLM promoter in an endogenous chromatin configuration, ChIP assay was performed according to the ChIP assay kit method provided by Upstate Biotechnology (Waltham, MA, U.S.A.). H4IIE cells were treated with TNFα (15 ng/ml for 4 h) or vehicle control and processed for ChIP assay according to manufacturer's instructions except for minor modifications. Briefly, proteins were cross-linked to DNA by treating cells with 1% formaldehyde at 37 °C for 10 min. After fixation, cells were washed with ice-cold PBS, resuspended and lysed in SDS lysis buffer (Upstate Biotechnology) and centrifuged for 5 min at 110 g. Cell lysates were sonicated at 25–30% power, 5×10 s, using a Sonic Dismembrator Model F60 (Fisher Scientific, Pittsburgh, PA, U.S.A.) to fragment the chromatin to approx. 1 kb or less. The sonicated cell lysates were spun in a microcentrifuge at 13000 g for 10 min at 4 °C. The supernatant containing the soluble chromatin sample was treated with 1 μl of 5 M NaCl and heated at 65 °C for 4 h to reverse the protein–DNA cross-links. Reversed soluble chromatin sample (20 μl) was removed and used as input control (total chromatin fraction) for final PCR reaction. The remaining chromatin solution was split into equal fractions and subjected to immunoprecipitation in the presence or absence of specific antibodies. Antibodies used for immunoprecipitation were anti-c-Jun and Nrf2 antibodies (Santa Cruz Biotechnology). PCR of the mouse GCLM promoter region across the ARE (TGCTTAGTCAT, −295 to −285 bp relative to the ATG start codon) was performed using the forward primer 5′-AGTTAACGGTTACGAAGCACTTTC-3′ (bp −333 to −310 relative to the ATG start codon) and reverse primer 5′-AGTTGAGCAGGTTCCCGGTCTGC-3′ (bp +60 to +82 relative to the ATG start codon). All PCR products were run on 8% (v/v) acrylamide gels and stained with ethidium bromide for 15–30 min.
Statistical analysis
Data are given as means±S.E.M. Statistical analysis was performed using ANOVA followed by Fisher's test for multiple comparisons. Significance was defined as P<0.05.
RESULTS
Cloning and sequencing of the 5′-flanking region of the rat GCLM
The sequence of the 1.80 kb product is shown in Figure 1. Analysis of the transcription factor-binding site was performed using Transcription factor search (http://www.cbrc.jp/research/db/TFSEARCH.html) and MatInspector (http://www.genomatix.de/cgi-bin/eldorado/main.pl). The 5′-flanking region of the rat GCLM contains several consensus binding sites for AP-1, heat-shock transcription factor, CAAT enhancer-binding protein and myeloid zinc-finger 1. In addition, consensus binding sites for Sp1, hepatocyte-enriched nuclear factor, c-Myb and ARE (5′-RTKAYNNNGCR-3′) are also present.
Figure 1. Nucleotide sequence of the 5′-flanking region of the rat GCLM gene.
Sequence is numbered relative to the translational start site. The putative regulatory elements are indicated in bold letters above the underlined sequences. The arrows denote transcriptional start sites. HSF, heat-shock transcription factor; C/EBP, CCAAT/enhancer-binding protein; STATx, signal transducer and activator of transcription x; MZF1, myeloid zinc finger 1; Sp1, stimulating protein 1; AML-1a, acute myeloid leukaemia-1a.
Transcriptional start site
Two transcriptional start sites are located at 275 and 152 nt upstream of the translational start site respectively. These are shown by arrows in Figure 1. There is a great deal of similarity between the rat and mouse GCLM 5′-flanking region (GenBank® accession no. AF149054), especially in the first 300 bp upstream of the translational start site (Figure 2). Both contain an ARE very close to the proximal transcriptional start site [15].
Figure 2. Alignment of the rat and mouse GCLM 5′-flanking regions [15].
The numbers above the rat sequence denote position relative to the translational start site (ATG). Note the similarity in the 5′-flanking sequence and transcriptional start sites (shown as forward arrows, top for rat and bottom for mouse).
Functional analysis of the 5′-flanking region of rat GCLM
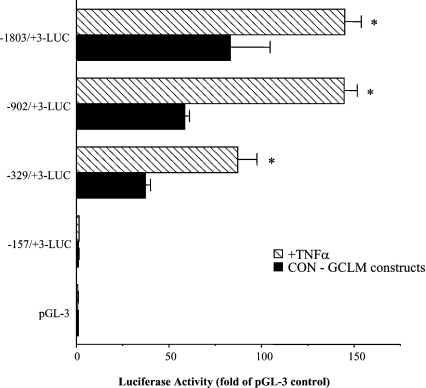
To delineate sequences that drive the expression of the rat GCLM, deletion mutants ranging from −1803/+3 to −157/+3 were cloned into the promoterless luciferase reporter gene vector pGL3-basic. The promoterless construct pGL3-basic served as the background control. Luciferase activity was measured after transient transfection of H4IIE cells with these constructs. Figure 3 shows that the rat GCLM promoter was able to drive efficiently luciferase expression in H4IIE cells. Construct −157/+3 has essentially no activity, whereas construct −902/+2 has near maximal promoter activity, with a 60-fold increase over vector control. Inclusion of additional upstream sequences increased the promoter activity only minimally.
Figure 3. Transient transfection analysis of the rat GCLM promoter–luciferase constructs in H4IIE cells.
Progressive 5′-deletions of the GCLM promoter extending from −1803 to +3 bp were generated and fused to the promoterless luciferase pGL-3 basic vector as described in the Materials and methods section. Numbering is defined relative to the translational start site. Results represent means±S.E.M. from four independent experiments performed in triplicates. TNFα treatment (15 ng/ml) was only during the last 4 h of the transfection. Data are expressed as relative luciferase activity compared with that of pGL-3 basic vector control, which is assigned a value of 1.0. *P<0.05 versus respective control GCLM (CON-GCLM) constructs (ANOVA followed by Fisher's test).
Effect of TNFα on GCLC, GCLM and GSS expression and promoter activity
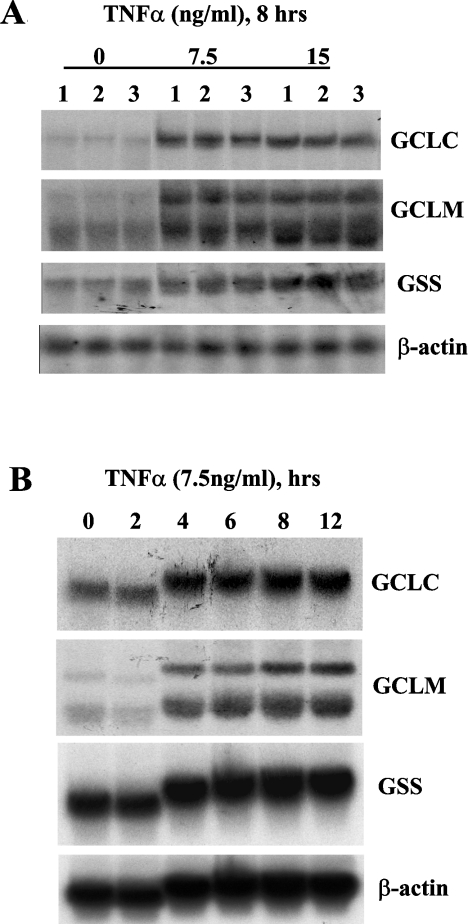
To study the effect of NF-κB and AP-1 on the expression and promoter activity of the rat GSH synthetic enzymes, we used TNFα, which is well known for its ability to induce both [30]. Figure 4 shows that TNFα increased GCLC, GCLM and GSS expression in H4IIE cells in a dose- and time-dependent manner. Maximal induction of GCLC and GCLM was seen with 7.5 ng/ml TNFα for 8 h (332±7% of control for GCLC, 233±9% for GCLM, results represent means±S.E.M. from three experiments, P<0.05 versus control); while maximal induction of GSS was seen with 15 ng/ml for 8 h (221±3%, results represent means±S.E.M. from three experiments, P<0.05 versus control).
Figure 4. Effect of TNFα on expression of GCLC, GCLM and GSS in H4IIE cells.
(A) H4IIE cells were treated with varying concentrations (0–15 ng/ml) of TNFα for 8 h. Total RNA (20 μg/lane) was subjected to Northern-blot analysis using cDNA probe for GCLC as described in the Materials and methods section. The same membrane was sequentially rehybridized with cDNA probes for GCLM, GSS and β-actin to ensure equal loading. (B) H4IIE cells were treated with TNFα (7.5 ng/ml) for varying duration (0–12 h) and processed for Northern-blot analysis as described above.
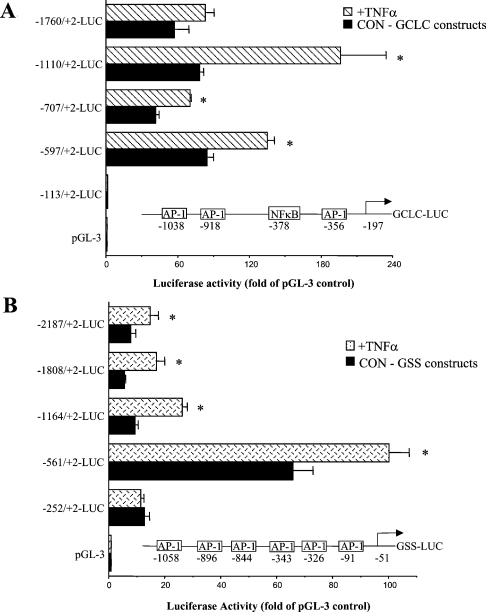
We next investigated the effect of TNFα treatment (15 ng/ml for 4 h) on the promoter activity of GCLC, GCLM and GSS. Figure 5 shows that TNFα treatment induced the reporter activity driven by the GCLC promoter (particularly the promoter construct −1110/+2), and the GSS promoter (particularly the promoter construct −1164/+2), where TNFα led to an approx. 2-fold increase in activity. Consensus AP-1 and NF-κB sites present in the rat GCLC and GSS 5′-flanking regions are shown in Figure 5. We have previously shown that acetaldehyde treatment of H4IIE cells increased AP-1 binding to the AP-1 site at −356 and NF-κB binding to the site at −378 of GCLC [24], and t-butylhydroquinone treatment increased AP-1 binding to the six AP-1 sites shown in the 5′-flanking region of GSS [25] (Figure 5). TNFα treatment also induced the rat GCLM promoter, particularly the −902/+2 promoter construct (Figure 3).
Figure 5. Effect of TNFα on luciferase activity driven by rat GCLC promoter constructs (A) or GSS promoter constructs (B).
H4IIE cells were transiently transfected with rat GCLC or GSS promoter constructs and treated with TNFα (15 ng/ml during the last 4 h) or vehicle control as described in the Materials and methods section. Positions of the consensus AP-1 and NF-κB sites present in the rat GCLC and GSS 5′-flanking regions are shown. Results represent means±S.E.M. from four independent experiments performed in triplicates. Data are expressed as relative luciferase activity compared with that of pGL-3 enhancer vector control, which is assigned a value of 1.0. *P<0.05 versus respective control GCLC (CON-GCLC) or GSS (CON-GSS) constructs (ANOVA followed by Fisher's test).
Role of NF-κB and AP-1 in basal and TNFα-mediated increase in GCLC, GCLM and GSS expression
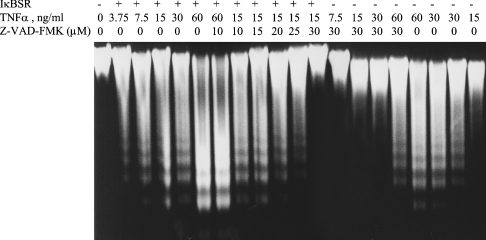
To evaluate the importance of NF-κB and AP-1 both in the basal expression and in the TNFα-mediated increase in GCLM, GCLC and GSS expression, H4IIE cells were infected with IκBSR, dominant negative c-Jun (TAM67) or empty vector, and the effect of blocking NF-κB or AP-1 activity on TNFα-mediated changes was examined. When H4IIE cells were treated with TNFα at high doses (>15 ng/ml) for long duration (≥8 h), apoptosis as measured by DNA fragmentation occurred as described previously [30] (Figure 6). This was made worse if NF-κB activation was blocked (Figure 6, compare TNFα alone versus TNFα+IκBSR). To make sure that the effect of TNFα on gene expression is not due to toxicity, cells were pretreated with Z-VAD-FMK (benzyloxycarbonyl-valylalanyl-DL-aspartylfluoromethane), a pancaspase inhibitor, for 1 h before treatment with TNFα. From the dose–response experiment, 30 μM of Z-VAD-FMK blocked apoptosis induced by treating H4IIE cells with 15 ng/ml TNFα for 8 h (Figure 6). This was the condition used to assess the effect of blocking NF-κB on gene expression induced by TNFα.
Figure 6. TNFα induces apoptosis in H4IIE cells.
H4IIE cells were treated with TNFα (0–60 ng/ml)±IκBSR±Z-VAD-FMK (0–30 μM) for 8 h, and apoptosis was determined by DNA fragmentation as described in the Materials and methods section. Note that there is no apoptosis at 15 ng/ml dose (first lane from the right). IκBSR greatly potentiated apoptosis (see DNA fragmentation at the 3.75 ng/ml dose, second lane from the left). This can be blocked in a dose-dependent manner by Z-VAD-FMK so that no fragmentation occurs at 15 ng/ml TNFα + IκBSR in the presence of 30 μM Z-VAD-FMK.
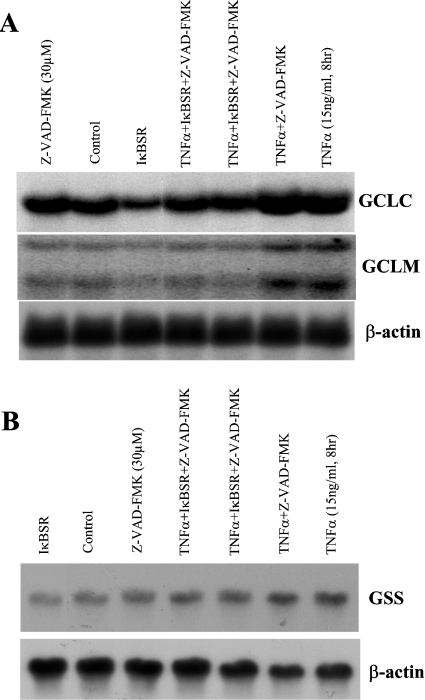
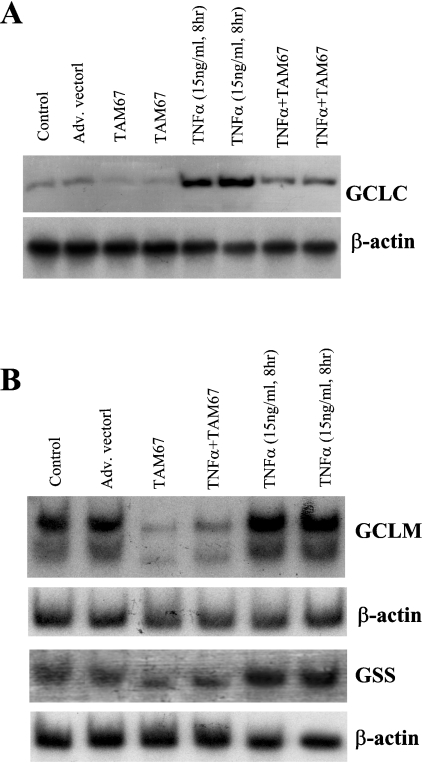
Figure 7 shows that the basal expression of GCLC, GCLM and GSS fell (40% for GCLC, 44% for GCLM and 35% for GSS) when NF-κB was blocked (compare IκBSR with control) and, although the TNFα-mediated induction in GCLC, GCLM and GSS was low, it was still much higher than IκBSR alone (by 46% for GCLC, 66% for GCLM and 77% for GSS). This suggests that TNFα induced GCLC, GCLM and GSS expressions by both NF-κB-dependent and -independent mechanisms. Figure 8 shows that the basal expression of GCLC, GCLM and GSS also requires AP-1 since the expression decreased (25% lower for GCLC, 73% lower for GCLM and 50% lower for GSS) when AP-1 was blocked. Dominant negative c-Jun also blunted the increase in GCLC, GCLM and GSS after TNFα treatment.
Figure 7. Role of NF-κB in TNFα-mediated up-regulation of GSH synthetic enzymes.
H4IIE cells were infected with IκBSR or empty vector and treated with TNFα (15 ng/ml, 8 h) with or without Z-VAD-FMK. Northern-blot analysis was performed for GCLC, GCLM (A) and GSS (B) as described in the Materials and methods section. Z-VAD-FMK treatment alone had no influence on the mRNA levels of GCLC, GCLM and GSS. IκBSR treatment alone resulted in a significant lowering of baseline mRNA levels of all three genes by approx. 40–45%. It also blunted but did not eliminate the TNFα-mediated increase in mRNA levels of all three genes. Representative Northern blots from three independent experiments are shown.
Figure 8. Role of c-Jun in TNFα-mediated up-regulation of GSH synthetic enzymes.
H4IIE cells were infected with dominant negative c-Jun (TAM67) or empty vector and treated with TNFα (15 ng/ml, 8 h) or vehicle control. Northern-blot analysis was performed for GCLC (A), GCLM and GSS (B) as described in the Materials and methods section. TAM67 treatment alone resulted in a significant lowering of baseline mRNA levels of all three genes by approx. 30–75%. It also blocked significantly the TNFα-mediated increase in mRNA levels of all three genes. Representative Northern blots from three independent experiments are shown.
Effect of p50 and p65 expression vectors on GCLM and GSS promoters and c-Jun expression
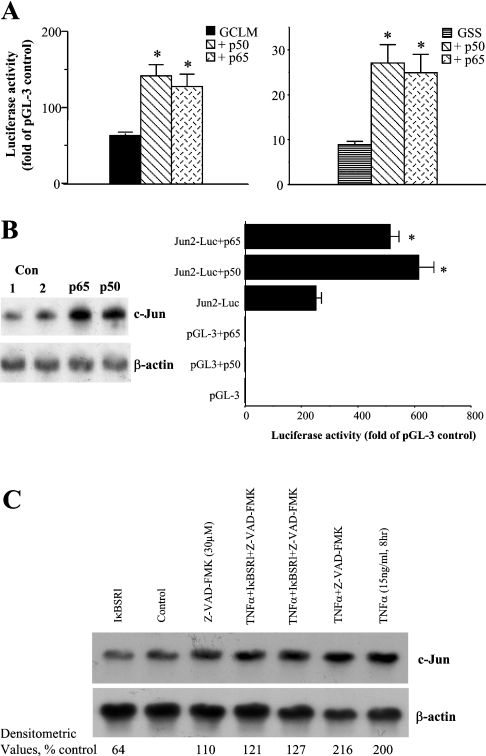
While GCLC 5′-flanking region contains a consensus NF-κB-binding site [24], there are no NF-κB-binding sites in the cloned GCLM or GSS promoter [25]. This prompted us to examine whether p50 and p65, two key members of the NF-κB family, can regulate the promoter activity of GCLM and GSS. Figure 9(A) shows that overexpression of either p50 or p65 in H4IIE cells induced the promoter activity of rat GCLM (−902/+3-LUC) and GSS (−1164/+2-LUC) by approx. 2-fold, despite the lack of NF-κB-binding sites. These promoter constructs were studied because they were maximally induced after TNFα treatment. Overexpression of either p50 or p65 increased c-Jun mRNA levels and induced the c-Jun promoter activity, which is known to be induced by c-Jun [31] (Figure 9B), suggesting that NF-κB activation can contribute to the induction of c-Jun-dependent target genes. Consistent with this notion, TNFα treatment of H4IIE cells doubled the c-Jun mRNA levels and this was significantly blocked if NF-κB activation was prevented by IκBSR (Figure 9C; TNFα treatment=208±8% of control, while IκBSR+TNFα=124±3% of control densitometric values). Furthermore, baseline c-Jun mRNA level was 36% lower if NF-κB was blocked by IκBSR (Figure 9C; IκBSR=64±2% of control densitometric values).
Figure 9. NF-kB positively regulates rat GCLM and GSS promoter activities, c-Jun expression and c-Jun-dependent promoter activity, and mediates c-Jun induction by TNFa.
Effect of p50 and p65 overexpression on GCLM and GSS promoter activity (A), c-Jun mRNA levels and c-Jun-dependent promoter activity (B). (C) The effect of blocking NF-κB with or without TNFα treatment on c-Jun mRNA level is shown. H4IIE cells were co-transfected with GCLM promoter construct −902/+3-LUC, GSS promoter construct −1164/+2-LUC or Jun2-Luc construct and p50 or p65 expression vector or empty vector control as described in the Materials and methods section. Overexpression of either p50 or p65 more than doubled the GCLM, GSS and c-Jun-dependent promoter activity (A, B), as well as increasing the steady state c-Jun mRNA levels (B). Baseline c-Jun level was 36% lower when NF-κB was blocked by IκBSR (C). TNFα treatment also increased the steady-state c-Jun mRNA level by 108%, which was significantly blocked by IκBSR (C). *P<0.05 versus respective controls (GCLM, GSS or Jun2-Luc). Con, control.
Effect of TNFα on c-Jun and Nrf2 binding to the GCLM ARE
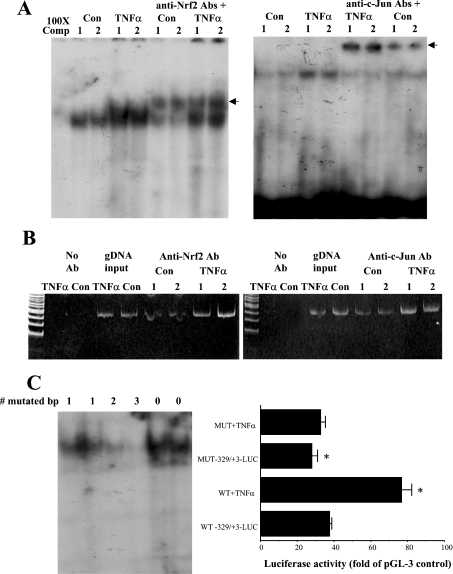
The GCLM construct −329/+3-LUC is efficiently induced by TNFα treatment, while the construct −157/+3-LUC has essentially no activity (Figure 3). This suggests that the effect of TNFα is exerted via key elements that lie between −329 and −157. Between −329 and −157 of the rat GCLM lies an ARE (Figure 1). We next investigated whether TNFα might exert its effect on GCLM via this element. The ARE in the human GCLC and GCLM is known to be transactivated by Nrf2, possibly in complex with c-Jun or Maf proteins [13,14]. We first examined whether TNFα treatment induces increased nuclear binding activity to this ARE element. Indeed, Figure 10(A) shows that TNFα treatment increased the nuclear-binding activity of both Nrf2 and c-Jun to the rat GCLM-ARE. This occurred not only with a recombinant probe for the ARE (EMSA with supershift), but also in the endogenous chromatin configuration as demonstrated using the ChIP assay (Figure 10B).
Figure 10. Effect of TNFα on c-Jun and Nrf2 binding to the GCLM ARE (A, B) and importance of the ARE in TNFα-mediated induction of GCLM promoter activity (C).
H4IIE cells were treated with TNFα (15 ng/ml, 4 h) or vehicle control and subjected to EMSA with supershift (A) or ChIP analysis (B) as described in the Materials and methods section. (A) EMSA with supershift results: note that TNFα treatment increased the nuclear binding activity of both Nrf2 and c-Jun to the ARE of the rat GCLM. Arrows in (A) point to supershifted bands. 100X Comp, 100-fold unlabelled probe. (B) H4IIE cells were treated with TNFα or vehicle control, then processed for ChIP assay as described in the Materials and methods section. PCR products from amplification of the ARE site after immunoprecipitation with antisera against Nrf2 or c-Jun demonstrate that TNFα treatment led to increased Nrf2 and c-Jun binding to the ARE site. Input genomic DNA (gDNA input) was used as a positive control and a no antibody immunoprecipitation (no Ab) was used as a negative control. Representative results from two experiments are shown. (C) The functional significance of the ARE element. To confirm functionality of the ARE, EMSA was first performed to document the effect of site-directed mutagenesis on nuclear-binding activity (C, left panel, the number of mutated bases is shown on top; note that nuclear binding was completely abolished only when three bases were mutated). The right panel shows the effect of preventing ARE binding on baseline and TNFα-mediated increase in GCLM promoter activity. H4IIE cells were transfected with wild-type (WT) or mutated (MUT, mutated in three bases) GCLM −329/+3-LUC constructs and treated with TNFα (15 ng/ml, 4 h). Results represent means±S.E.M. from five to nine experiments (C, right panel). Data are expressed as relative luciferase activity compared with that of pGL-3 basic vector control, which is assigned a value of 1.0. Note that the mutant construct had lower baseline promoter activity and TNFα no longer was able to induce its reporter activity. *P<0.001 versus WT −329/+3-LUC (ANOVA followed by Fisher's test).
Importance of the ARE in basal and TNFα-mediated increase in GCLM promoter activity
To see if the ARE is functional and critical in mediating the effect of TNFα on the rat GCLM promoter activity, promoter activity was measured using wild-type or mutated construct −329/+3-LUC (containing 3 bp mutation). EMSA (Figure 10C, left panel) shows the effect of mutation on nuclear-binding activity to the ARE site. Note that binding was completely abolished only when three bases were mutated. The right panel of Figure 10(C) shows the effect of TNFα treatment on the GCLM promoter construct mutated at the ARE site that prevented nuclear binding. Note that the basal promoter activity fell by 26% but, more importantly, TNFα no longer induced the luciferase activity driven by this construct (Figure 10C, right panel).
DISCUSSION
GSH is the most abundant intracellular non-protein thiol, with multiple functions including antioxidant defence, modulation of cell proliferation and detoxification of xenobiotics [1]. One of the major determinants of the synthesis of GSH is the activity of GCL. Because of its importance, regulation of GCL has been a topic of extensive research. Regulation can occur transcriptionally or post-transcriptionally, affecting only the catalytic or modifier subunit or both [1]. Recently, we showed that the second enzyme, GSS, is also transcriptionally regulated in a co-ordinated manner as GCL subunits further enhance the GSH synthetic capacity [23,25]. Studies examining transcriptional regulation of the human GCL subunits revealed the importance of ARE and AP-1 in mediating the effect of oxidants on up-regulating these genes [9,13]. The rat GCL subunits are regulated similarly to the human GCL subunits [1]. t-Butylhydroquinone induces the expression of both GCL subunits in human and rat [12,14,25]. It also induces the expression of rat GSS [25]. Interestingly, the cloned 5′-flanking regions of the rat GCLC or GSS do not contain any consensus ARE element, but the reporter activity driven by recombinant rat GCLC and GSS promoter constructs is induced by t-butylhydroquinone treatment [25]. Increased AP-1 trans-activation was found to be the major underlying mechanism responsible for the up-regulation of the rat GCLC and GSS genes [25]. In the present study, we examined the role of NF-κB on regulation of the rat GCL subunits and GSS. This is an area of controversy as both positive and negative studies have been reported with regard to whether oxidants up-regulate GCLC via NF-κB [1]. No studies have examined whether NF-κB signalling regulates GCLM or GSS. We used TNFα, which up-regulates both AP-1 and NF-κB, to examine the role of these transcription factors in regulating the expression of rat GCL subunits and GSS and elucidate possible mechanisms.
To facilitate transcriptional studies of the rat GCLM subunit, we first cloned and characterized the 5′-flanking region. The 5′-flanking region shares a great deal of similarity to the mouse GCLM [15]. Both have two transcriptional start sites and an ARE that is very close to the proximal transcriptional start site. Transfection studies showed that the 5′-flanking sequence of the rat GCLM gene contains a functional promoter, which was able to drive luciferase expression in H4IIE cells efficiently. The region between −329 and −157 contains possible enhancer elements as inclusion of this region increased the reporter activity by approx. 30-fold. This region contains an ARE, which is the prime candidate responsible for this increase. Indeed, when the ARE is mutated to prevent nuclear binding, the basal promoter activity decreased by 26%. However, it also suggests that other enhancer elements exist in this region as the bulk of the activity still remained.
We next examined whether TNFα treatment alters endogenous gene expression or recombinant promoter activity. TNFα treatment led to a co-ordinated induction of the mRNA levels of endogenous GCL subunits and GSS in a time- and dose-dependent fashion. It also increased the promoter activity of all three genes. However, while the rat GCLC 5′-flanking region contains an NF-κB site, rat GCLM and GSS 5′-flanking regions do not. This prompted us to examine whether NF-κB activation is important in the TNFα-mediated up-regulation of GCLM and GSS. To address this, we used IκBSR to block NF-κB activation. To our surprise, the basal expression of GCLC, GCLM and GSS decreased when cells were treated with IκBSR. Blocking NF-κB activation also significantly inhibited the up-regulation of these genes in response to TNFα. However, TNFα was still able to increase the mRNA levels of GCLC, GCLM and GSS in the presence of IκBSR, suggesting that both NF-κB-dependent and NF-κB-independent pathways were involved. We had previously shown that AP-1 was required for basal expression of GCLC, GCLM and GSS [25]. In the present study, we confirmed our previous finding that blocking AP-1 with dominant negative c-Jun lowered the basal expression of GCLC, GCLM and GSS. Blocking AP-1 also inhibited the TNFα-mediated up-regulation, perhaps even more effectively than blocking NF-κB. Thus both NF-κB and AP-1 pathways are important for the TNFα-mediated induction of the GSH synthetic enzymes.
In our experiments, we used TAM67 (dominant-negative c-Jun) to inhibit AP-1 activity. However, AP-1 is a dimeric complex that comprises members of the Jun (c-Jun, JunB and JunD), Fos (c-Fos, FosB, Fra-1 and Fra-2), Maf (c-Maf, MafB, MafA, MafG/F/K and Nrl) and ATF (activating transcription factor) (ATF2/LRF1/ATF3, B-ATF, JDP1 and JDP2) families [33]. The main AP-1 proteins in mammalian cells are JUN and FOS [34], and c-Jun is the most potent transcriptional activator of all of the Jun family members [33]. While the Fos proteins can hetero-dimerize with members of the Jun family, the Jun proteins can both homo- and heterodimerize with Fos members [35]. Because of the key role that c-Jun plays, a very common practice has been to use dominant-negative c-Jun to block AP-1 activity [30,36–38]. Indeed, it has been well demonstrated by other investigators that TAM67 blocks AP-1-driven reporter activity [38]. Nevertheless, we cannot rule out the possibility that other family members listed above may also participate.
Why would NF-κB pathway be involved in the TNFα-mediated up-regulation of GCLM and GSS if these promoters lack NF-κB-binding sites? This prompted us to examine possible interactions between NF-κB and AP-1. NF-κB and AP-1 belong to families of transcription factors that exert pleiotropic regulation effects. Physical interaction of NF-κB subunit p65 with AP-1 subunit c-Jun was shown to synergize and potentiate each other's biological function [39]. However, it is unknown whether c-Jun expression can be influenced by NF-κB. First, we found that overexpression of either p50 or p65 subunit more than doubled the promoter activity of GCLM and GSS. Since AP-1 had been previously shown [25] to be critical for the expression of GCLM and GSS, we examined whether p50 and p65 overexpression can influence the expression of c-Jun and c-Jun-dependent promoter activity. Indeed, p50 and p65 overexpression nearly doubled the mRNA levels of c-Jun and the c-Jun-dependent promoter activity. Thus NF-κB can directly influence c-Jun gene expression and its trans-activating activity. Furthermore, lowering NF-κB by IκBSR reduced the basal c-Jun mRNA level and significantly inhibited the TNFα-mediated increase in c-Jun expression. Taken together, our results show that NF-κB can modulate GCLM and GSS despite the lack of NF-κB-binding sites because of its ability to modulate c-Jun, which is critical for the expression of both GCLM and GSS. This can also help to reconcile the contradictory findings with regard to the role of NF-κB in the regulation of GCLC expression. Thus NF-κB may exert its action indirectly via AP-1 to influence the ability of oxidants and cytokines to up-regulate the expression of the GSH synthetic enzymes. Since the rat GCLC has a NF-κB-binding site in its promoter, and we previously showed that acetaldehyde treatment increased NF-κB binding to this site [24], it is likely that both direct and indirect actions of NF-κB are involved in the TNFα-mediated induction of this gene.
To see if TNFα's stimulatory effect on GCLM resides directly at the level of the GCLM promoter, we investigated the possible involvement of the ARE located at 295 bp upstream of the translational start site. This ARE is a likely choice as TNFα treatment induced the GCLM promoter construct −329/+3 but not −157/+3. ARE has been shown to be important in the transcriptional regulation of human GCL subunits [13,14]. Nrf2, a member of the cap ‘n’ collar-basic leucine zipper proteins, is known to bind and trans-activate AREs present in the human GCLC and GCLM promoters in response to treatment with various oxidants, possibly in complexes with other Jun or Maf proteins [13,14,40]. Consistent with these previous findings, TNFα treatment induces nuclear binding of both Nrf2 and c-Jun to the ARE of the rat GCLM promoter. This occurred in the endogenous chromatin configuration as demonstrated by ChIP analysis. When binding to this element is prevented by site-directed mutagenesis, TNFα no longer was able to induce the promoter activity. This confirms the functional importance of the ARE in mediating the effect of TNFα. Given TNFα's ability to induce c-Jun nuclear-binding activity, it is plausible that this can lead to the increased Nrf2/c-Jun binding to the ARE. However, we cannot exclude the possibility that TNFα treatment can also increase the expression of Nrf2, which will require further study.
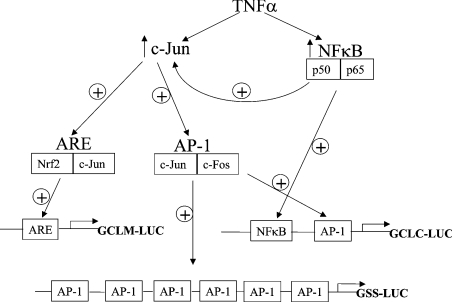
In summary, we have cloned and analysed the 5′-flanking region of the rat GCLM gene and defined the role of NF-κB and AP-1 in the TNFα-mediated co-ordinated up-regulation of the rat GSH synthetic enzymes. We found that both c-Jun and NF-κB are required for basal and TNFα-mediated induction of GCLC, GCLM and GSS expression in H4IIE cells. NF-κB modulates the promoter activity of GCLM and GSS despite the absence of NF-κB consensus elements. This occurs because NF-κB also modulates the expression of c-Jun. The ability of NF-κB to regulate c-Jun expression has not been previously reported and further illustrates the complexity of transcription factor interactions. Figure 11 shows a model that brings all of the information from the present study together.
Figure 11. Proposed model for the interactions between NF-κB, c-Jun, AP-1, ARE and their roles in TNFα-mediated induction of rat GCLC, GCLM and GSS.
⊕ indicates activation or induction (NF-κB induces the expression of c-Jun, whereas all other effects are transactivation of the respective binding sites).
Acknowledgments
This work was supported by National Institutes of Health grant DK-45334. H4IIE cells were provided by the Cell Culture Core of the USC Research Center for Liver Diseases (P30 DK48522).
References
- 1.Lu S. C. Regulation of hepatic glutathione synthesis: current concept and controversies. FASEB J. 1999;13:1169–1183. [PubMed] [Google Scholar]
- 2.Suthanthiran M., Anderson M. E., Sharma V. K., Meister A. Glutathione regulates activation-dependent DNA synthesis in highly purified normal human T lymphocytes stimulated via the CD2 and CD3 antigens. Proc. Natl. Acad. Sci. U.S.A. 1990;87:3343–3347. doi: 10.1073/pnas.87.9.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poot M., Teubert H., Rabinovitch P. S., Kavanagh T. J. De novo synthesis of glutathione is required for both entry into and progression through the cell cycle. J. Cell. Physiol. 1995;163:555–560. doi: 10.1002/jcp.1041630316. [DOI] [PubMed] [Google Scholar]
- 4.Richman P. G., Meister A. Regulation of γ-glutamylcysteine synthetase by nonallosteric feedback inhibition by glutathione. J. Biol. Chem. 1975;250:1422–1426. [PubMed] [Google Scholar]
- 5.Yan N., Meister A. Amino acid sequence of rat kidney γ-glutamylcysteine synthetase. J. Biol. Chem. 1990;265:1588–1593. [PubMed] [Google Scholar]
- 6.Huang C., Anderson M. E., Meister A. Amino acid sequence and function of the light subunit of rat kidney γ-glutamylcysteine synthetase. J. Biol. Chem. 1993;268:20578–20583. [PubMed] [Google Scholar]
- 7.Seelig G. F., Simondsen R. P., Meister A. Reversible dissociation of γ-glutamylcysteine synthetase into two subunits. J. Biol. Chem. 1984;259:9345–9347. [PubMed] [Google Scholar]
- 8.Huang C., Chang L., Anderson M. E., Meister A. Catalytic and regulatory properties of the heavy subunit of rat kidney γ-glutamylcysteine synthetase. J. Biol. Chem. 1993;268:19675–19680. [PubMed] [Google Scholar]
- 9.Mulcahy R. T., Wartman M. A., Bailey H. H., Gipp J. J. Constitutive and β-naphthoflavone-induced expression of the human γ-glutamylcysteine synthetase heavy subunit gene is regulated by a distal antioxidant response element/TRE sequence. J. Biol. Chem. 1997;272:7445–7454. doi: 10.1074/jbc.272.11.7445. [DOI] [PubMed] [Google Scholar]
- 10.Galloway D. C., Blake D. G., Shepherd A. G., McLellan L. I. Regulation of human γ-glutamylcysteine synthetase: co-ordinate induction of the catalytic and regulatory subunits in HepG2 cells. Biochem. J. 1997;328:99–104. doi: 10.1042/bj3280099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moinova H. R., Mulcahy R. T. An electrophile responsive element (EpRE) regulates β-naphthoflavone induction of the human γ-glutamylcysteine synthetase regulatory subunit gene. J. Biol. Chem. 1998;273:14683–14689. doi: 10.1074/jbc.273.24.14683. [DOI] [PubMed] [Google Scholar]
- 12.Galloway D. C., McLellan L. I. Inducible expression of the γ-glutamylcysteine synthetase light subunit by t-butylhydroquinone in HepG2 cells is not dependent on an antioxidant-responsive element. Biochem. J. 1998;336:535–539. doi: 10.1042/bj3360535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moinova H. R., Mulcahy R. T. Up-regulation of the human γ-glutamylcysteine synthetase regulatory subunit gene involves binding of Nrf-2 to an electrophile responsive element. Biochem. Biophys. Res. Commun. 1999;261:661–668. doi: 10.1006/bbrc.1999.1109. [DOI] [PubMed] [Google Scholar]
- 14.Wild A. C., Moinova H. R., Mulcahy R. T. Regulation of γ-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. J. Biol. Chem. 1999;274:33627–33636. doi: 10.1074/jbc.274.47.33627. [DOI] [PubMed] [Google Scholar]
- 15.Solis W. A., Dalton T. P., Dieter M. Z., Freshwater S., Harrer J. M., He L., Shertzer H. G., Nebert D. W. Glutamate-cysteine ligase modifier subunit: mouse Gclm gene structure and regulation by agents that cause oxidative stress. Biochem. Pharmacol. 2002;63:1739–1754. doi: 10.1016/s0006-2952(02)00897-3. [DOI] [PubMed] [Google Scholar]
- 16.Lu S. C., Ge J., Kuhlenkamp J., Kaplowitz N. Insulin and glucocorticoid dependence of hepatic γ-glutamate-cysteine ligase and GSH synthesis in the rat: studies in cultured hepatocytes and in vivo. J. Clin. Invest. 1992;90:524–532. doi: 10.1172/JCI115890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu S. C., Ge J. Loss of suppression of GSH synthesis under low cell density in primary cultures of rat hepatocytes. Am. J. Physiol. 1992;263:C1181–C1189. doi: 10.1152/ajpcell.1992.263.6.C1181. [DOI] [PubMed] [Google Scholar]
- 18.Huang Z. Z., Li H., Cai J., Kuhlenkamp J., Kaplowitz N., Lu S. C. Changes in glutathione homeostasis during liver regeneration in the rat. Hepatology. 1998;27:147–153. doi: 10.1002/hep.510270123. [DOI] [PubMed] [Google Scholar]
- 19.Cai J., Sun W., Lu S. C. Hormonal and cell density regulation of hepatic γ-glutamate-cysteine ligase gene expression. Mol. Pharmacol. 1995;48:212–218. [PubMed] [Google Scholar]
- 20.Lu S. C., Huang Z. Z., Yang J., Tsukamoto H. Effect of ethanol and high fat feeding on hepatic γ-glutamate-cysteine ligase subunit expression in the rat. Hepatology. 1999;30:209–214. doi: 10.1002/hep.510300134. [DOI] [PubMed] [Google Scholar]
- 21.Cai J., Huang Z. Z., Lu S. C. Differential regulation of γ-glutamate-cysteine ligase heavy and light subunit gene expression. Biochem. J. 1997;326:167–172. doi: 10.1042/bj3260167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu S. C., Huang Z. Z., Yang H., Tsukamoto H. Effect of thioacetamide on hepatic γ-glutamate-cysteine ligase subunit expression. Toxicol. Appl. Pharmacol. 1999;159:161–168. doi: 10.1006/taap.1999.8729. [DOI] [PubMed] [Google Scholar]
- 23.Huang Z. Z., Yang H. P., Chen C. J., Lu S. C. Inducers of γ-glutamylcysteine synthetase and their effects on glutathione synthetase expression. Biochim. Biophys. Acta. 2000;1493:48–55. doi: 10.1016/s0167-4781(00)00156-1. [DOI] [PubMed] [Google Scholar]
- 24.Yang H. P., Huang Z. Z., Wang J. H., Ou X. P., Lu S. C. Cloning and characterization of the 5′-flanking region of the rat glutamate-cysteine ligase catalytic subunit. Biochem. J. 2001;357:447–455. doi: 10.1042/0264-6021:3570447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H. P., Zeng Y., Lee T. D., Yang Y., Ou X. P., Chen L. X., Haque M., Rippe R., Lu S. C. Role of AP-1 in the co-ordinate induction of rat glutamate-cysteine ligase and glutathione synthetase by tert-butylhydroquinone. J. Biol. Chem. 2002;277:35232–35239. doi: 10.1074/jbc.M203812200. [DOI] [PubMed] [Google Scholar]
- 26.Urata Y., Yamamoto H., Goto S., Tsushima H., Akazawa S., Yamashita S., Nagataki S., Kondo T. Long exposure to high glucose concentration impairs the responsive expression of γ-glutamylcysteine synthetase by interleukin-1β and tumor necrosis factor-α in mouse endothelial cells. J. Biol. Chem. 1996;271:15146–15152. doi: 10.1074/jbc.271.25.15146. [DOI] [PubMed] [Google Scholar]
- 27.Iwanaga M., Mori K., Iida T., Urata Y., Matsuo T., Wasunaga A., Shibata S., Kondo T. Nuclear factor kappa B dependent induction of gamma-glutamylcysteine synthetase by ionizing radiation in T98G human gliobastoma cells. Free Radical Biol. Med. 1998;24:1256–1268. doi: 10.1016/s0891-5849(97)00443-7. [DOI] [PubMed] [Google Scholar]
- 28.Morales A., García-Ruiz C., Miranda M., Marí M., Colell A., Ardite E., Fernández-Checa J. C. Tumor necrosis factor increases hepatocellular glutathione by transcriptional regulation of the heavy subunit chain of γ-glutamylcysteine synthetase. J. Biol. Chem. 1997;272:30371–30379. doi: 10.1074/jbc.272.48.30371. [DOI] [PubMed] [Google Scholar]
- 29.Sekhar K. R., Meredith M. J., Kerr L. D., Soltaninassab S. R., Spitz D. R., Xu Z. Q., Freeman M. L. Expression of glutathione and γ-glutamylcysteine synthetase mRNA is Jun dependent. Biochem. Biophys. Res. Commun. 1997;234:588–593. doi: 10.1006/bbrc.1997.6697. [DOI] [PubMed] [Google Scholar]
- 30.Yang H. P., Sadda M. R., Yu V., Zeng Y., Lee T. D., Ou X. P., Chen L. X., Lu S. C. Induction of human methionine adenosyltransferase 2A expression by tumor necrosis factor alpha: role of NF-κB and AP-1. J. Biol. Chem. 2003;278:50887–50896. doi: 10.1074/jbc.M307600200. [DOI] [PubMed] [Google Scholar]
- 31.Van Dam H., Wilhelm D., Herr I., Steffen A., Herrlich P., Angel P. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 1995;14:1798–1811. doi: 10.1002/j.1460-2075.1995.tb07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhoumik A., Huang T. G., Ivanov V., Gangi L., Qiao R. F., Woo S. L. C., Chen S. H., Ronai Z. An ATF2-derived peptide sensitizes melanomas to apoptosis and inhibits their growth and metastasis. J. Clin. Invest. 2002;110:643–650. doi: 10.1172/JCI16081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaulian E., Karin M. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 34.Eferl R., Wagner E. F. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 35.Jochum W., Passegué E., Wagner E. F. AP-1 in mouse development and tumorigenesis. Oncogene. 2001;20:2401–2412. doi: 10.1038/sj.onc.1204389. [DOI] [PubMed] [Google Scholar]
- 36.Olive M., Krylov D., Echlin D. R., Gardner K., Taparowsky E., Vinson C. A dominant negative to activation protein-1 (AP-1) that abolishes DNA binding and inhibits oncogenesis. J. Biol. Chem. 1997;272:18486–18594. doi: 10.1074/jbc.272.30.18586. [DOI] [PubMed] [Google Scholar]
- 37.Li J. J., Cao Y., Young M. R., Colburn N. H. Induced expression of dominant-negative c-jun downregulates NFκB and AP-1target genes and suppresses tumor phenotype in human keratinocytes. Mol. Carcinog. 2000;29:159–169. doi: 10.1002/1098-2744(200011)29:3<159::aid-mc5>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 38.Thompson E. J., Gupta A., Stratton M. S., Bowden G. T. Mechanism of action of a dominant negative c-jun mutant in inhibiting activator protein-1 activation. Mol. Carcinog. 2002;35:157–162. doi: 10.1002/mc.10090. [DOI] [PubMed] [Google Scholar]
- 39.Stein B., Baldwin A. S., Ballard D. W., Greene W. C., Angel P., Herrlich P. Cross-coupling of the NF-kappa B p65 and Fos/Jun transcription factors produces potentiated biological function. EMBO J. 1993;12:3879–3891. doi: 10.1002/j.1460-2075.1993.tb06066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erickson A. M., Nevarea Z., Gipp J. J., Mulcahy R. T. Identification of a variant antioxidant response element in the promoter of the human glutamate-cysteine ligase modifier subunit gene. J. Biol. Chem. 2002;277:30730–30737. doi: 10.1074/jbc.M205225200. [DOI] [PubMed] [Google Scholar]