Abstract
In this study we examined the ability of Salmonella enterica serovar Typhimurium porins to activate activating protein 1 (AP-1) and nuclear factor κB (NF-κB) through the mitogen-activated protein kinase (MAPK) cascade, and we identified the AP-1-induced protein subunits. Our results demonstrate that these enzymes may participate in cell signaling pathways leading to AP-1 and NF-κB activation following porin stimulation of cells. Raf-1 was phosphorylated in response to the treatment of U937 cells with porins; moreover, the porin-mediated increase in Raf-1 phosphorylation is accompanied by the phosphorylation of MAPK kinase 1/2 (MEK1/2), p38, extracellular-signal-regulated kinase 1/2, and c-Jun N-terminal kinase. We used three different inhibitors of phosphorylation pathways: 2′-amino-3′-methoxyflavone (PD-098059), a selective inhibitor of MEK1 activator and the MAPK cascade; 4-(4-fluorophenyl)-2-(4-methylsulfinylphenyl)-5-(4-pyridyl)1H-imidazole (SB203580), a specific inhibitor of the p38 pathway; and 7β-acetoxy-1α,6β,9α-trihydroxy-8,13-epoxy-labd-14-en-11-one (forskolin), an inhibitor at the level of Raf-1 kinase. PD-098059 pretreatment of cells decreases AP-1 and NF-κB activation by lipopolysaccharide (LPS) but not by porins, and SB203580 pretreatment of cells decreases mainly AP-1 and NF-κB activation by porins; in contrast, forskolin pretreatment of cells does not affect AP-1 and NF-κB activation following either porin or LPS stimulation. Our data suggest that the p38 signaling pathway mainly regulates AP-1 and NF-κB activation in cells treated with S. enterica serovar Typhimurium porins. Antibody electrophoretic mobility shift assays showed that JunD and c-Fos binding is found in cells treated with porins, in cells treated with LPS, and in unstimulated cells. However, by 30 to 60 min of stimulation, a different complex including c-Jun appears in cells treated with porins or LPS, while the Fra-2 subunit is present only after porin stimulation. These data suggest different molecular mechanisms of activation induced by porins or by LPS.
Although lipopolysaccharide (LPS) has been clearly shown to play a major role in septic shock and in the induction of cytokine production, very little is known regarding other surface bacterial components of gram-negative bacteria. It has been reported that these components also play an important role in the pathway associated with infections by gram-negative bacteria (13).
LPS induces transcription of several genes encoding proinflammatory mediators (21, 52). In the past few years we have studied the various immunobiological effects induced by the outer membrane pore-forming proteins compared to those induced by LPS (16–20, 30). Porins are integral components of the outer membranes of all gram-negative bacteria and are intimately associated with the LPS; they induce many cellular responses, including cellular activation (23) and cytokine release (17, 19, 20, 28, 30). LPS and porins are released by several bacteria during both in vitro (10) and in vivo (59) growth, and this release is significantly enhanced when the bacteria are lysed following exposure to antibiotics or human serum (10, 12, 35). Active concentrations of both LPS and porins are often reached at infection sites from either gram-negative bacteria outer membrane blebbing or bacterial lysis as a consequence of host defense (59). Intracellular signaling pathways induced by LPS stimulation have been studied in detail (54, 56); in contrast, very little is known about the signaling pathways of other components derived from gram-negative bacteria.
Mitogen-activated protein kinase (MAPK) cascades are among the best known signal transduction systems and play a key role in the regulation of gene expression as well as cytoplasmic activities. MAPKs have also been shown to be involved in the regulation of cytokine responses (57). In mammalian systems, five different MAPK modules have been identified so far; single MAPK modules generally can signal independently of one another, and this specificity is manifest in distinct physiologic responses (49). MAPKs, with the exception of extracellular-signal-regulated kinase 3 (ERK3), are activated upon phosphorylation of both tyrosine and threonine residues by MAPK kinase (MEK) (49). Many different MEKs have been described, and in vitro assays indicate that each has one or at most two specific targets in the MAPK pathways: MEK1 and MEK2 act on ERK1 and ERK2, respectively. As shown in various cell types, LPS induces activation of ERK1 and ERK2 (4), c-Jun N-terminal kinases (JNKs) (25), and p38 (26).
The MAPK cascade activates transcription factors such as activating protein 1 (AP-1) and nuclear factor κB (NF-κB). The contribution of AP-1 family members to transcriptional regulation is controlled by a number of well-characterized mechanisms that have been reviewed recently (3, 32, 33). The genes encoding AP-1 proteins (fos and jun) are often among the first genes to be transcribed after stimulation of a cell and are usually only transiently expressed. The AP-1 protein function is regulated primarily by phosphoregulation (55). The AP-1 family members differ in their abilities to transactivate or repress transcription (14, 53).
NF-κB is present in the cytoplasm of resting cells bound to its inhibitor, IκBα. Only as a consequence of cellular activation and proteolytic degradation of IκBα, NF-κB is released and translocated to the nucleus, where it transactivates several genes (50).
In contrast to the case for LPS, no data currently exist regarding the signaling mechanism and transcriptional activation by porins. In this study, we used a well-characterized monocytic cell line, U937, to investigate the mechanisms by which porins induce activated transcription factors AP-1 and NF-κB by activation of the MAPK cascade.
MATERIALS AND METHODS
Cells lines.
U937 monocytes (ATCC CRL-1593.2) were grown at 37°C in 5% CO2 in RPMI 1640 (Labtek, Eurobio) with HEPES supplemented with 10% heat-inactivated fetal calf serum, glutamine (2 mM), penicillin (100 U/ml), and streptomycin (100 U/ml) (Labtek, Eurobio) in 150-cm2 tissue culture flasks (Corning, New York, N.Y.). Before treatment of the cells, the serum concentration was reduced to 5% for 24 h at 37°C and then further reduced to zero for at least 10 to 12 h. This should prevent any interference from serum factors on the phosphorylation state of the proteins in the signaling cascade.
Preparation of macrophages from C3H/HeJ mice.
Murine macrophages from female C3H/HeJ mice (Harlan UK Limited Shaws Farm, Blackthon, Bicester, United Kingdom) were prepared from the peritoneal cavity according to conventional procedures. Briefly, resident macrophages were washed from the peritoneal cavity with RPMI 1640. After centrifugation at 170 × g for 10 min at 4°C, the cell pellet was resuspended in RPMI 1640 supplemented with 10% heat-inactivated fetal calf serum, glutamine (2 mM), penicillin (100 U/ml), and streptomycin (100 U/ml) (complete medium) at a concentration of 5 × 106 cells/ml. Adherent macrophage monolayers were obtained by plating the cells in six-well plastic trays at 5 × 106 cells/well for 2 h at 37°C in 5% CO2. Nonadherent cells were removed by suction, and freshly prepared complete medium was added with the indicated experimental reagents.
Bacterial strain.
The bacterial strain used was S. enterica serovar Typhimurium SH5014 grown in nutrient broth (Difco, Detroit, Mich.) for 18 to 24 h at 37°C under agitation. The cells were harvested at the end of the exponential growth phase, and outer membranes were prepared from cell envelopes according to the protocols described by Nurminen et al. (43).
Preparation of porins and LPS.
The porins were extracted as described by Nurminen (44). Briefly, 1 g (wet weight) of cell envelopes was suspended in 2% Triton X-100 in 0.01 M Tris-HCl (pH 7.5), containing 10 mM EDTA; after the addition of trypsin (10 mg/g of envelopes), the pellet was dissolved in a sodium dodecyl sulfate (SDS) buffer (4% [wt/vol] in 0.1 M sodium phosphate [pH 7.2]) and applied to an Ultragel ACA34 column (Pharmacia, Uppsala, Sweden) equilibrated with 0.25% SDS buffer. The fraction containing proteins, identified by measuring the absorption at 280 nm, was extensively dialyzed and checked by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) as described by Laemmli (34). The protein content of the porin preparation was determined by the method of Lowry et al. (38). LPS contamination was revealed on SDS-polyacrylamide gels stained with silver nitrate as described by Tsai and Frasch (51) and quantified by the Limulus amoebocyte lysate assay (Limulus test) as described by Yin et al. (58).
LPS was isolated from S. enterica serovar Typhimurium SH5014 using the phenol-chloroform-ether method described by Galanos et al. (15). Briefly, bacteria were washed sequentially with water, 95% ethanol, acetone, and diethyl ether at 4°C. The dry bacterial powder was treated with a mixture of liquid phenol-chloroform-petroleum ether in a volume ratio of 2:5:8. After centrifugation, the aqueous layer was collected and dialyzed to remove phenol. Subsequently, the dialyzed material was concentrated in a rotary evaporator at 35 to 40°C and centrifuged to remove any insoluble material. The supernatant was treated with RNase for 2 h at 55°C. After centrifugation, the pellet was collected, resuspended in water, and lyophilized.
Cell stimulation with porins and preparation of cell lysates.
U937 cells (3 × 106 cells/ml) were stimulated with different porins or LPS concentrations for different time periods (3, 10, 20, and 60 min) in 96-well polypropylene plates. In some experiments, cells were preincubated for 60 min at 37°C with different inhibitors before stimulation with porins or LPS. In another assay, polymyxin B (Sigma-Aldrich S.r.l., Milan, Italy) was mixed with porins to neutralize the biological activity of possible traces of LPS that could be present in the preparation. Porins were incubated with polymyxin B at room temperature for 1 h at a ratio of 1:100 (41). In all of the tests performed, the porins plus polymyxin B, in a concentration range which is nontoxic for the cells, gave the same results as the porins alone (data not shown).
After incubation, the cells were washed twice with ice-cold phosphate-buffered saline (PBS) without Ca2+ and Mg2+ and resuspended in 250 μl of the appropriate lysis buffer (buffer A [150 mM NaCl, 50 mM Tris-HCl {pH 7.5}, 5 mM EDTA, 1% Nonidet P-40, 1 mM Na2MoO4, 40 μg of phenylmethylsulfonyl fluoride {PMSF} per ml, 0.2 mM Na3VO4, 1 mM dithiothreitol, 10 μg of aprotinin per ml, 2 μg of leupeptin per ml, 0.7 μg of pepstatin per ml, and 10 μg of soybean trypsin inhibitor per ml] or buffer B [137 mM NaCl, 20 mM Tris-HCl {pH 8.0}, 5 mM EDTA, 1 mM Na6P2O7, 10% glycerol, 1% Triton X-100, 1 mM EGTA, 1 mM PMSF, 1 mM Na3VO4, 10 mM NaF, 10 mM β-glycerophosphate, 20 μg of aprotinin per ml, and 20 μg of leupeptin per ml]), and the sample was spun down at 16,000 × g for 2 min at 4°C. The clarified cell lysates were used for enhanced chemiluminescence Western blot analysis. For kinase assays, buffers C (25 mM HEPES [pH 7.6], 20 mM MgCl2, 20 mM β-glycerophosphate, 1 mM Na3VO4, and 2 mM dithiothreitol) and D (20 mM HEPES [pH 7.2], 5 mM MgCl2, 1 mM EGTA, 5 mM β-mercaptoethanol, 2 mM Na3VO4, 10 μg of aprotinin per ml, and 1 mM PMSF) were used.
Kinase assays.
Cell lysates were precleared with protein A/G PLUS-Agarose (Santa Cruz Biotechnology, Inc.) (20 μl) for 45 min. Immunoprecipitation was done with the appropriate antibodies and beads at 4°C overnight with gentle rotation. After incubation the beads were pelleted (6,000 × g), washed three time with 500 μl of specific lysis buffer, and boiled in 20 μl of Laemmli sample buffer with 5% β-mercaptoethanol (34) for 5 min. The samples were pelleted, and the supernatant, containing cell phosphorylated proteins, was resolved by SDS-PAGE as described by Laemmli (34). Equal amounts of cell lysates, typically 50 μg, were separated on SDS-15% polyacrylamide gels using the buffer system described by Laemmli (34). Following electrophoresis, the separating gel was soaked in transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol) for 5 min, and then the proteins were transferred to polyvinylidene difluoride membranes (pore size, 0.45 μm) overnight at 30 V and 4°C. Blots were blocked for 1 h at room temperature in Tris-buffered saline (TBS) (150 mM NaCl, 20 mM Tris-HCl [pH 7.5]) containing 1% bovine serum albumin (BSA) plus 1% blotting-grade blocker nonfat milk (Bio-Rad Laboratories), and subsequently membranes were washed twice with TBS containing 0.05% Tween 20 (TTBS) before incubation with antiphosphorylated and nonphosphorylated kinase antibodies diluted 1:2,000 in TBS containing 1% BSA for 1 h at room temperature. After being washed six times with TTBS for 3 min, the polyvinylidene difluoride membranes were incubated at room temperature for 2 h with anti-mouse or anti-rabbit immunoglobulin G (IgG)-horseradish peroxidase secondary antibodies diluted 1:3,000. They were then washed six times with TTBS and twice with PBS for 5 min. Thereafter, proteins were visualized with enhanced chemiluminescence on Kodak X-OMAT LS film. For Western blotting analysis, the following phosphorylated antibodies were used: phospho-p44/42 MAPK (Thr202/Tyr204) E10 monoclonal antibody (MAb) (isotype, mouse IgG1) (anti-p44/42) (New England Biolabs, Beverly, Mass.), which detects doubly phosphorylated threonine 202 and tyrosine 204 of p44 and p42 MAPKs (ERK1 and ERK2) and is produced by immunizing mice with a synthetic phospho-Thr202 and phospho-Tyr204 peptide corresponding to residues around Thr202/Tyr204 of human p44 MAPK; rabbit polyclonal phospho-MEK1/2 antibody (anti-p-MEK1/2) (New England Biolabs), which detects MEK1/2 only when activated by phosphorylation at Ser217 and Ser221 and does not cross-react with other related family members; anti-phospho-Raf-1 MAb (anti-p-Raf-1) (Upstate Biotechnology, Lake Placid, N.Y.), which recognizes Raf-1 phosphorylated at Ser338 and Tyr341 and does not bind to inactive Raf-1 or Raf-1 peptides phosphorylated only at Ser338 or Tyr341; anti-phospho-p38 (New England Biolabs), which is a rabbit polyclonal antibody raised against a peptide mapping at the amino terminus of p38 of mouse origin identical to the corresponding human sequence and directed against Thr180- and Tyr182-phosphorylated p38; and phospho-JNK antibody (anti-p-JNK) (Santa Cruz Biotechnology, Inc.), which is a mouse IgG1 MAb raised against a peptide corresponding to a short amino acid sequence phosphorylated on Thr183 and Tyr185 of JNK of human origin. For determination of the shift in mobility due to phosphorylation, Western blotting analysis was performed with the following antibodies: anti-ERK1/2 (Upstate Biotechnology), which is a rabbit polyclonal antibody raised against a peptide corresponding to residues 333 to 367 of rat ERK1 recognizing ERK1 and ERK2 at 44 and 42 kDa; anti-MEK1 (Santa Cruz Biotechnology, Inc.), which is a mouse IgG2b MAb raised against a recombinant protein corresponding to amino acids 1 to 393 representing full-length MEK1 of human origin; anti-MEK2 (Santa Cruz Biotechnology, Inc.), which is a rabbit polyclonal antibody raised against a peptide mapping at the amino terminus of MEK2 of human origin identical to the corresponding rat sequence; anti-Raf-1 (Santa Cruz Biotechnology, Inc.), which is provided as a mouse IgG1 MAb epitope mapping at the carboxy terminus of Raf-1 p74 of human origin; anti-p38 (Santa Cruz Biotechnology, Inc.), which is a rabbit polyclonal IgG1 antibody epitope corresponding to amino acids 213 to 360 mapping at the carboxy terminus of p38 of human origin; and anti-JNK (Santa Cruz Biotechnology, Inc.), which is a rabbit polyclonal IgG antibody produced by immunization with full-length human JNK (amino acids 1 to 384) produced in Escherichia coli. Immunoreactivity was localized with horseradish peroxidase-conjugated secondary antibodies and visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom).
Preparation of nuclear extract.
Crude nuclear extracts were prepared from cells by the method of Andrews and Faller (2). U937 cells, unstimulated or stimulated for different lengths of time (30, 60, and 120 min) were centrifuged and washed once in cold PBS. Cells were pelleted, resuspended in 400 μl of cold hypotonic lysis buffer E (10 mM HEPES-KOH [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 1 mM dithiothreitol, 2 mM PMSF, and 10 μg of aprotinin per ml), allowed to swell on ice for 10 min, and then vortexed for 10 s. Nuclei were sedimented at 3,000 × g for 10 s at 4°C, and the pellet was resuspended in 20 μl of cold buffer F (20 mM HEPES-KOH [pH 7.9], 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM dithiothreitol, 2 mM PMSF, and 10 μg of aprotinin per ml) and incubated on ice for 20 min for high-salt extraction. Insoluble debris were sedimented at 13,000 × g for 2 min, and the supernatant was collected and assayed for protein content using a colorimetric assay (Bio-Rad Laboratories) against a BSA standard. Aliquots were stored at −80°C.
Electrophoretic mobility shift assay (EMSA) and supershift assay.
Nuclear extracts (5 μg of protein) were incubated with 70,000 cpm of a 21-bp oligonucleotide containing the AP-1 (c-jun) consensus sequence (5′CGCTTGATGAGTCAGCCGGAA3′) (Roche Diagnostic GmbH, Roche Molecular Biochemicals, Mannheim, Germany) (36) and a 22-bp oligonucleotide containing the NF-κB oligonucleotide sequence (5′AGTTGAGGGGACTTTCCCAGG3′) (Roche Diagnostic GmbH, Roche Molecular Biochemicals) (39) that had been previously labeled with [γ-32P]ATP by use of T4 polynucleotide kinase (Roche Diagnostic GmbH, Roche Molecular Biochemicals). Incubations were performed for 20 min at 25°C in the presence of 10 mM Tris-HCl, 1 mM MgCl2, 0.5 mM EDTA, 50 mM NaCl, 4% glycerol, and 0.5 mM dithiothreitol.
For the competition assay, a 100- to 200-fold molar excess of NF-κB and AP-1 consensus oligonucleotide was added to the binding reaction mixture prior to the addition of probe and incubated for 10 min at 25°C. In the supershift assay, the binding reaction was performed as described above, followed by a 2- to 3-h incubation on ice with the specific antibody.
Supershift assays were performed with the following antibodies purchased from Santa Cruz Biotechnology: anti-c-Jun/AP-1 (mouse MAb against residues 56 to 69 of human c-Jun), anti-JunB (goat polyclonal antibody against the amino-terminal domain of JunB p39 of murine JunB), anti-JunD (goat polyclonal antibody against the carboxy-terminal domain of JunD p39 of murine JunD), anti-c-Fos (goat polyclonal antibody against the amino-terminal domain of human c-Fos p62), anti-FosB (goat polyclonal antibody against an amino acid sequence mapping within a central domain of murine FosB), anti-Fra-1 (mouse MAb against an amino acid sequence mapping at the amino terminus of murine Fra-1), and anti-Fra-2 (rabbit polyclonal antibody against an amino acid sequence within the carboxy terminus of human Fra-2). In supershift studies, binding reactions were performed as described above, followed by a 2- to 3-h incubation on ice with the specific antibody.
After incubation, the samples were loaded onto a 4% polyacrylamide gel (with 0.5× Tris-borate-EDTA) and electrophoresed for 2 h at 200 V. The gel was dried and exposed to X-ray film (Kodak) at −80°C for 2 to 3 days.
LDH assay.
The lactate dehydrogenase (LDH) assay was carried out according to the manufacturer’s instructions with a cytotoxicity detection kit (Boehringer, Mannheim, Germany). LDH is a stable cytoplasmic enzyme present in all cells and is rapidly released into the cell culture supernatant upon damage of the plasma membrane. LDH activity was determined by a coupled enzymatic reaction whereby the tetrazolium salt was reduced to formazan. An increase in the number of dead or damaged cells resulted in an increase in LDH activity in the culture supernatant. The amounts of LDH showed that both treated and untreated cells were healthy.
Reproducibility.
The results obtained were confirmed by quantitation of the data with Sigma Gel software. The results shown are from a single experiment typical of at least three giving identical results.
RESULTS
Purity of porin preparation.
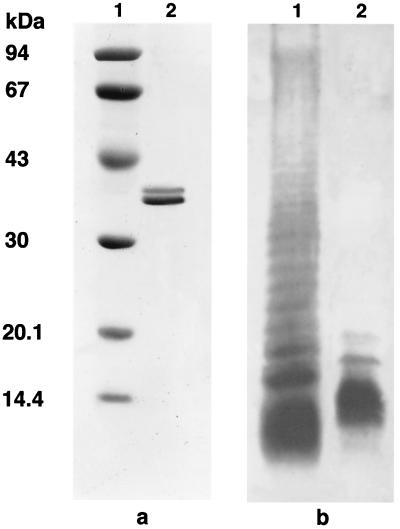
The purity of the porin preparation and the eventual LPS contamination have been extensively addressed in previous works (16, 19). Any possible trace of LPS, quantified by the Limulus test, was estimated to be <0.005% (wt/wt) compared with a standard S. enterica serovar Typhimurium LPS solution. These traces of LPS did not show any biological activity under our experimental conditions (data not shown). The purity of the porin preparation was checked by SDS-PAGE (Fig. 1a), which revealed the presence of two bands with molecular masses of 34 and 36 kDa. The pattern of LPS was revealed on SDS-polyacrylamide gels stained with silver nitrate (Fig. 1b).
FIG. 1.
Pattern of S. enterica serovar Typhimurium SH5014 porins and LPS on SDS-PAGE. (A) Lane 1, molecular mass standards (Amersham Pharmacia Biotech, Milan, Italy) (phosphorylase b, 94 kDa,; albumin, 67 kDa; ovalbumin, 43 kDa; carbonic anhydrase, 30 kDa; trypsin inhibitor, 20.1 kDa; α-lactalbumin, 14.4 kDa); lane 2, S. enterica serovar Typhimurium porins (10 μg). (B) Lane 1, S. enterica serovar Typhimurium LPS standard (Sigma-Aldrich S.r.l.); lane 2, S. enterica serovar Typhimurium SH5014 LPS (10 μg).
Porin treatment activates the Raf-1-MEK1-MEK2-MAPK pathway.
To verify whether porins stimulate the Raf-1-MEK1-MEK2-MAPK signal transduction pathway, U937 cells (3 × 106 cells/ml) were treated with 5 μg of porins per ml. Cell lysates were prepared at different time points after the beginning of the stimulation. The lysates obtained were immunoprecipitated with antibodies that specifically recognize the phosphorylated and nonphosphorylated forms of each enzyme. We had previously detected related proteins in untreated U937 cells using anti-Raf-1, anti-MEK1, anti-MEK2, anti-ERK1/2, anti-JNK, and anti-p38. A small difference was detected between cells incubated in the presence and absence of serum (data not shown).
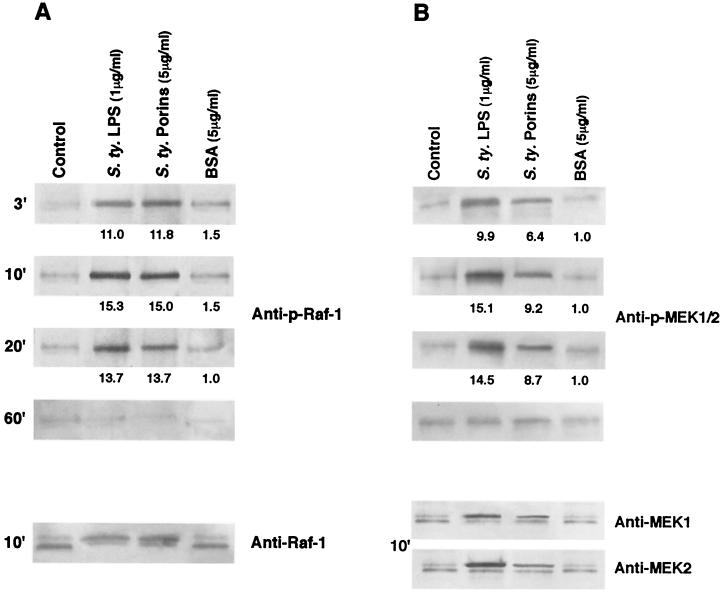
To investigate whether Raf-1 was involved in the transduction of activating signal, lysates from porin-treated U937 cells were analyzed by immunoblotting using anti-p-Raf-1 MAb and evaluating the shift in Raf-1 electrophoretic mobility by using anti-Raf-1. Increased phosphorylation was visible by 3 min after porin treatment; the peak of activity occurred in the first 10 min, and this persisted for at least 20 min thereafter, returning to baseline levels by 60 min. A gel shift assay was used to confirm phosphorylation at the optimal activation time (10 or 20 min). Activated kinases are phosphorylated and therefore migrate more slowly on SDS-PAGE. The shift obtained with anti-Raf-1 MAb confirms the result obtained with anti-p-Raf-1 MAb. The results reported in Fig. 2A demonstrate that porin stimulation increased Raf-1 phosphorylation; the increase was similar to that obtained using LPS (1 μg/ml). Raf-1 has been shown to function in the MAPK signal transduction pathway as a MEK kinase.
FIG. 2.
Effect of S. enterica serovar Typhimurium (S.ty) porins on Raf-1 (A) and MEK1/2 (B) activation after different stimulation times. Proteins from cell lysates (3 × 106 cells/ml) were analyzed by SDS-15% PAGE and immunoblotted with a Raf-1- or MEK1/2-phosphospecific antibody. Shifts in band mobility on SDS-PAGE due to phosphorylation were obtained with anti-Raf-1, anti-MEK1, and anti-MEK2. Fold increases in Raf-1 and MEK1/2 activation, compared to unstimulated cells (control), are shown below each lane for each blot. BSA represents a control of an unspecific stimulus.
To verify whether porin-induced Raf-1 phosphorylation was paralleled by phosphorylation of the entire MAPK pathway, we next examined the activation of MEK1-MEK2-MAPK in untreated and porin- or LPS-stimulated U937 cells. Using anti-p-MEK1/2 antibodies, we can observe immunoreactive bands of phosphorylated enzymes. MEK1/2 phosphorylation reaches its peak by 10 min and goes back to a baseline value by 60 min (Fig. 2B). The shift at 10 min in electrophoretic mobility of the bands obtained with anti-MEK1 and -MEK2 antibodies confirms the result obtained with anti-p-MEK1/2 antibodies.
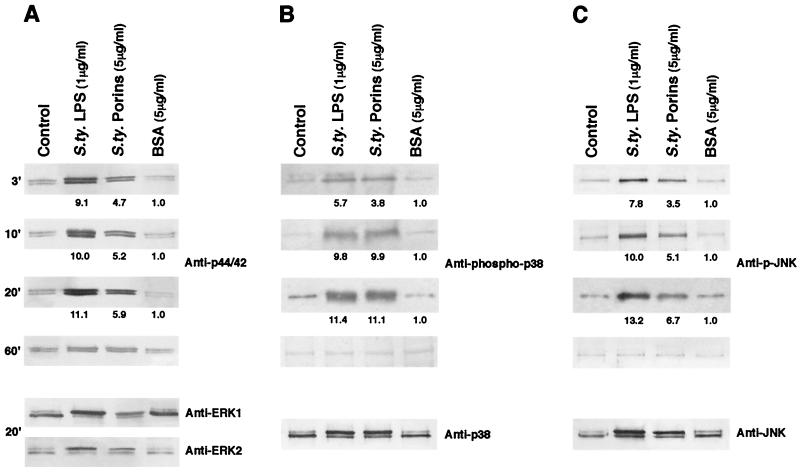
Using anti-p44/42 antibodies, we observed that porins increased the appearance of the phosphorylated form of this enzyme as well. A time course of ERK1/2 activation following porin treatment is shown in Fig. 3A. Anti-p44/42 reveals phosphorylated bands by 3 min; these reach the highest expression by 20 min and go back to baseline levels by 60 min. The shift at 20 min in electrophoretic mobility of the bands obtained with anti-ERK1/ERK2 antibodies confirmed the result obtained with anti-p44/42 antibodies.
FIG. 3.
Effect of S. enterica serovar Typhimurium (S.ty) porins on ERK1/2 (A), p38 (B), and JNK (C) activation after different stimulation times. Proteins from cell lysates (3 × 106 cells/ml) were analyzed by SDS-15% PAGE and immunoblotted with an ERK1/2-, p38-, or JNK-phosphospecific antibody. Shifts in band mobility on SDS-PAGE due to phosphorylation were obtained with anti-ERK1/2, anti-p38, and anti-JNK. ERK1 appears as the upper band (44 kDa); ERK2 appears as the lower band (42 kDa). Fold increases in ERK1/2, p38, and JNK activation are shown below each lane for each blot. BSA represents a control of an unspecific stimulus.
Using an anti-phospho-p38 antibody, we could observe similar immunoreactive bands in U937 cell lysates treated with porins or LPS. The shift at 20 min in electrophoretic mobility of the bands obtained with anti-p38 antibody confirmed the results obtained with anti-phospho-p38 antibody (Fig. 3B).
Using the anti-p-JNK MAb in Western blot analysis, we could observe an increased phosphorylation after stimulation with LPS compared to that in untreated and porin-stimulated cells. The shift in electrophoretic mobility of the bands obtained with anti-JNK antibody after stimulation for 20 min confirmed the result obtained with anti-p-JNK MAb (Fig. 3C).
Porin concentrations of from 500 ng/ml (about 0.02 μM) to 20 μg/ml (about 0.8 μM) are required to induce enzyme phosphorylation, while when stimulating cells with LPS the concentration required to observe phosphorylation induction ranges from 100 ng/ml (about 0.05 μM) to 10 μg/ml (about 5 μM) (data not shown).
The range of concentrations used is nontoxic for the cells. There was no significant amount of LDH (a measure of cell damage) in the cell supernatants. Also, the duration of treatment with porins or LPS at the indicated concentrations failed to induce any significant release of LDH from the cells (data not shown). The specificity of the effect was demonstrated using a different protein (BSA) as a stimulus; no phosphorylation of the enzymes investigated in our study was observed.
The possibility that traces of LPS contaminating the preparation of porins could be responsible for the biological effect observed can be excluded. The minimum concentration of LPS able to produce phosphorylation in our in vitro model is 100 ng/ml, which is much more than the quantity of LPS present in our preparation (about 50 pg/μg of porins).
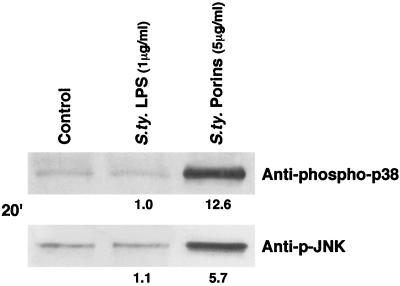
The results obtained when stimulating U937 cells with porins were not related to eventual traces of LPS present in the porin preparations. Stimulations performed with S. enterica serovar Typhimurium porins at a concentration of 5 μg/ml on LPS-hyporesponsive C3H/HeJ peritoneal macrophages induced JNK/p38 activation, whereas stimulation of C3H/HeJ cells with LPS (1 μg/ml) did not elicit JNK/p38 activation (Fig. 4).
FIG. 4.
Western blotting analysis with pJNK and phospho-p38 antibodies of lysates from C3H/HeJ peritoneal macrophages (5 × 106 cells/ml) after stimulation with LPS (1 μg/ml) or porins (5 μg/ml) for 20 min. The experimental procedure was as described in Materials and Methods. S. ty., S. enterica serovar Typhimurium.
Porin stimulation results in the activation of AP-1 and NF-κB in U937 cells.
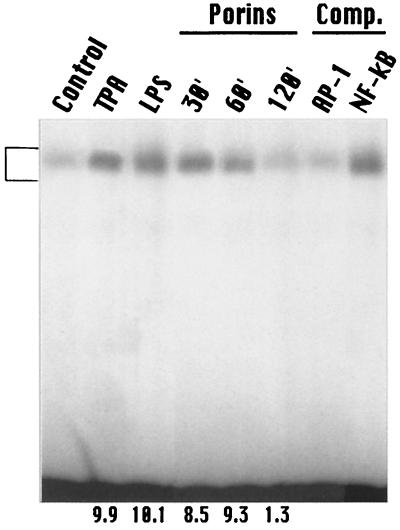
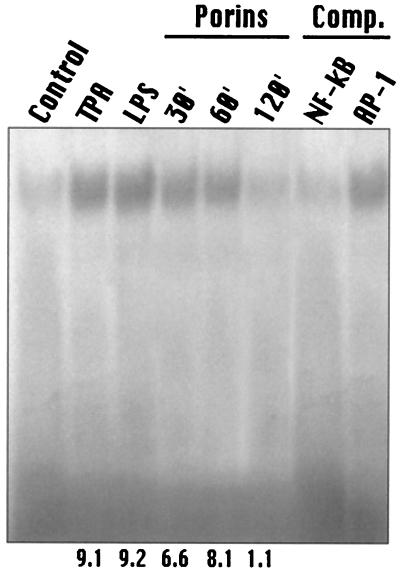
AP-1 is induced by porins of S. enterica serovar Typhimurium in the human monocytic cell line U937. For AP-1 and NF-κB consensus oligomers, a predominant DNA-protein complex was revealed by EMSA. A low level of predominant AP-1 and NF-κB consensus probe-binding complex was present in untreated nuclei. Following treatment with porins at 5 μg/ml, the level of this constitutive form of AP-1 and NF-κB significantly increased by 30 min, was maintained at the same level by 60 min, and returned to background levels by 120 min (Fig. 5 and 6). AP-1 and NF-κB induction by porins occurred at similar levels of LPS (1 μg/ml) 12-O-tetradecanoylphorbol-13-acetate (TPA) (100 ng/ml).
FIG. 5.
EMSA with AP-1 consensus oligonucleotide. U937 cells (3 × 106 cells/ml) were stimulated with porins (5 μg/ml), LPS (1 μg/ml), or TPA (100 ng/ml). Control, control nuclear extract (5 μg); TPA, TPA-induced nuclear extract (5 μg); LPS, LPS-induced nuclear extract (5 μg); Porins, porin-induced nuclear extract (5 μg) at 30, 60, and 120 min.; Comp., binding reactions with unlabeled excesses of AP-1 and NF-κB consensus oligonucleotides. A 100- to 200-fold molar excess of unlabeled probe was used for competition experiments. The bracket at the left indicates control and induced complexes. Fold increases in AP-1 activation are shown below each lane for each blot.
FIG. 6.
EMSA with NF-κB consensus oligonucleotide. U937 cells (3 × 106 cells/ml) were stimulated with porins (5 μg/ml), LPS (1 μg/ml), or TPA (100 ng/ml). Control, control nuclear extract (5 μg); TPA, TPA-induced nuclear extract (5 μg); LPS, LPS-induced nuclear extract (5 μg); Porins, porin-induced nuclear extract (5 μg) at 30, 60, and 120 min.; Comp., binding reactions with unlabeled excesses of NF-κB and AP-1 consensus oligonucleotides. The bracket at the left indicates control and induced complexes. Fold increases in NF-κB activation are shown below each lane for each blot.
To demonstrate that the DNA oligomer-protein complexes formed in the EMSA represent specific molecular interactions, unlabeled-probe competition experiments were performed. The corresponding unlabeled probe strongly competes for complex formation, whereas the unspecific probe does not. These data indicate that the proteins activated by porins and detected by EMSA were highly specific for the AP-1 and NF-κB probes and do not present unspecific binding. As a control we used LPS or TPA. In control experiments, using either LPS or TPA, activation of AP-1 and NF-κB by 30 min was observed.
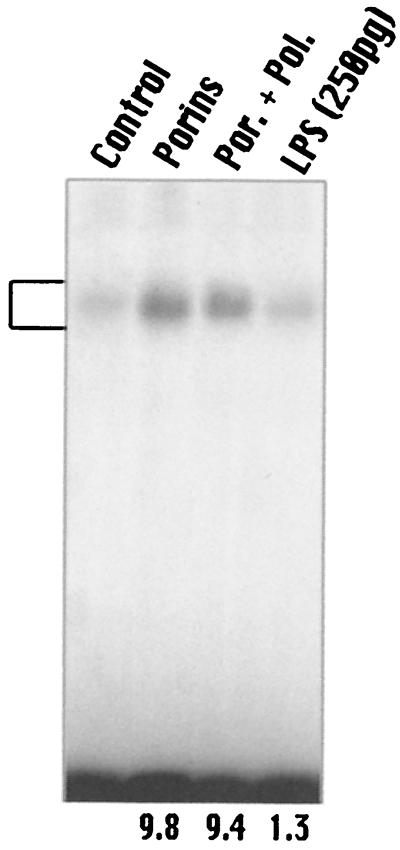
The lowest concentration of LPS that activates AP-1 and NF-κB was 100 ng/ml. In our preparation, 50 pg of LPS/μg of porins was present. Using this concentration, we were not able to observe any activation of AP-1 (Fig. 7) or NF-κB (data not shown). Porins added with polymyxin B at a ratio of 1:100 also induced activation of AP-1. Polymyxin B binds to lipid A with high affinity and inhibits the biological activity of LPS (41). These data indicate that porins and not traces of LPS activate AP-1 in our assays.
FIG. 7.
EMSA with AP-1 consensus oligonucleotide. U937 cells (3 × 106 cells/ml) were stimulated for 60 min with porins (5 μg/ml) or porins plus polymyxin B or LPS (250 pg/ml). Control, control nuclear extract (5 μg); Porins, porin-induced nuclear extract (5 μg); Por. + Pol., nuclear extract (5 μg) from cells treated with porins and the polymyxin B mixture; LPS, nuclear extract (5 μg) from cells treated with 250 pg of LPS. The bracket at the left indicates control and induced complexes. Fold increases in AP-1 activation are shown below each lane for each blot.
Dose response of AP-1 and NF-κB activation by porins.
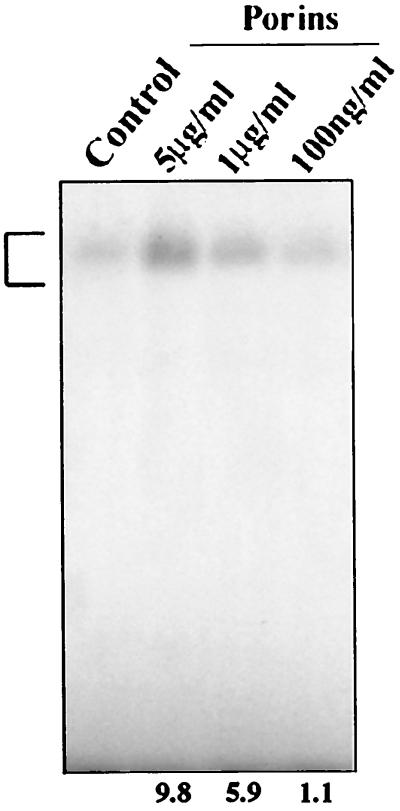
Concentrations of porins below 100 ng/ml failed to activate AP-1 and NF-κB by 60 min of stimulation. Activation of AP-1 was clearly detected at 1 μg/ml and was maximal at 5 μg/ml (Fig. 8). Accordingly, we chose to perform further cell stimulation experiments using a porin concentration of 5 μg/ml. Porin activation of NF-κB showed the same dose-response curve (data not shown).
FIG. 8.
EMSA with AP-1 consensus oligonucleotide. U937 cells (3 × 106 cells/ml) were stimulated for 60 min with different porin concentrations. Control, 5 μg of untreated nuclear extract; Porins, 5 μg of nuclear extract induced with porins at 5 μg/ml, 1 μg/ml, and 100 ng/ml. The bracket at the left indicates control and induced complexes. Fold increases in AP-1 activation are shown below each lane for each blot.
Identification of protein subunits of AP-1 activated by porins.
The transcription factor AP-1 is composed of multiple protein complexes formed between the protein products of the proto-oncogenes c-fos and c-jun and their related gene family members. The fos family consists of c-fos, the gene for fos-related antigen-1 (fra-1), fra-2, and fosB and its naturally truncated form fosB2; the jun family consists of c-jun, junB, and junD. The AP-1 family of transcription factors consists of homodimers and heterodimers of these subunits (33).
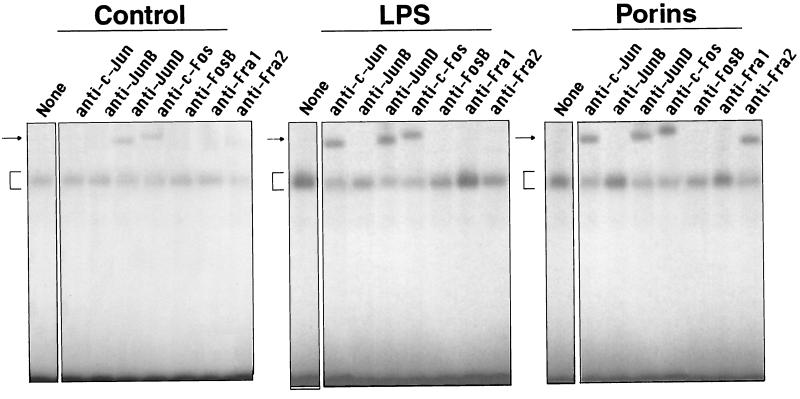
To further characterize the predominant AP-1 complex formed by 60 min of porin stimulation, supershift experiments using specific antisera were performed. Antibodies against c-Jun, JunD, Fra-2, or c-Fos specifically shifted and decreased the formation of the complex, while antibodies against JunB, Fra1, or FosB did not have any effect. These data suggest that this activated AP-1 complex is composed mainly of cJun, JunD, c-Fos, and Fra-2. In contrast, in U937 cells stimulated with 1 μg of LPS per ml, anti-c-Jun, anti-JunD, and anti-c-Fos antibodies altered the binding activity, while in the control of unstimulated cells, only anti-JunD and anti-c-Fos generated two complexes with their proteins. The effect of anti-JunD and anti-c-Fos antibodies in complex migration was modest in cells stimulated with LPS or porins compared to control cells (Fig. 9).
FIG. 9.
Supershift with AP-1 consensus oligonucleotide. U937 cells (3 × 106 cells/ml) were stimulated for 60 min with porins (5 μg/ml) or LPS (1 μg/ml). Control, control nuclear extract (5 μg); LPS, LPS-induced nuclear extract (5 μg); Porins, porin-induced nuclear extract (5 μg). The specific antibodies used are indicated above the lanes. The arrows at the left indicate the specific supershifted complex, and the brackets indicate control and induced complexes.
Implication of Raf-1-MEK1-MEK2-MAPK in AP-1 and NF-κB activation triggered by porins.
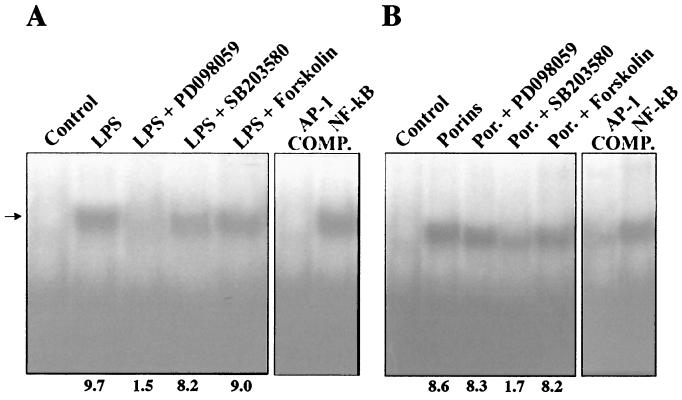
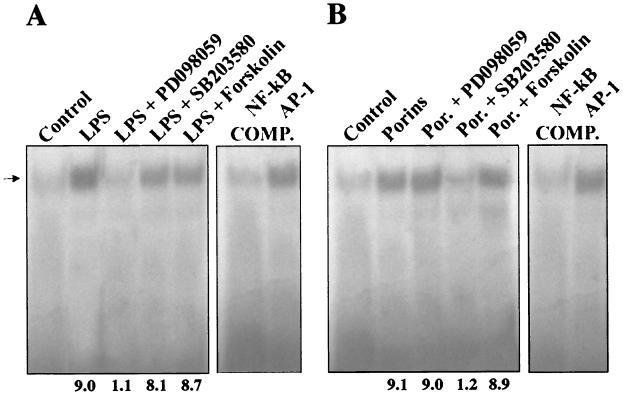
To further investigate the role of MAPK intermediates in the signal transduction pathway resulting in AP-1 and NF-κB activation, the effects of specific inhibitors on the activation of these DNA-binding factors were analyzed. When nuclear extracts were prepared from cells treated with porins (5 μg/ml) for 60 min and pretreated for 60 min with PD-098059 (New England Biolabs, Inc.) (100 μM for 1 h) (1), a highly selective inhibitor of MEK1 activator and the MAPK cascade (1, 11), no significant decrease in the shift of the labeled probe was observed compared to the AP-1 (Fig. 10) and NF-κB (Fig. 11) transduction detected in cells treated only with porins. From nuclear extracts of cells treated with LPS (1 μg/ml) and pretreated with PD-098059, an evident decrease in the shift of the labeled probe was observed compared to controls.
We next tested the effect of SB203580 (Calbiochem-Novabiochem GmbH, Schwalbach, Germany), a specific inhibitor of the p38 pathway (8). Stimulation of U937 cells pretreated with SB203580 (10 μM for 60 min) with porins (5 μg/ml) resulted in a significant decrease in the AP-1 and NF-κB DNA-binding activity compared with controls stimulated with porins and not treated with SB203580. When cells pretreated with SB203580 were stimulated with LPS (1 μg/ml), no significant difference in AP-1 and NF-κB DNA-binding activity compared with cells treated only with LPS was observed.
Further, we tested the effect of forskolin (Calbiochem-Novabiochem GmbH), an inhibitor at the level of Raf-1 kinase (48). Nuclear extract prepared from U937 cells pretreated with forskolin (20 μg/ml for 10 min) and stimulated with porins (5 μg/ml) or LPS (1 μg/ml) did not show any modification in AP-1 and NF-κB translocation with respect to the controls stimulated with porins or LPS and not treated with forskolin.
At these concentrations the inhibitors did not cause cellular damage as verified in preliminary tests measuring leakage of LDH activity from the cells into the supernatant.
DISCUSSION
It is well established that AP-1 activation, resulting from the engagement of several different surface receptors with their extracellular ligands, usually involves the Raf, MEK, and MAPK intermediates (9, 40). In contrast, only limited observations suggest that the signaling pathway leading to NF-κB activation may be linked to this well-defined Raf-MEK-MAPK pathway.
Recently, Raf-1 was found to induce the dissociation of cytoplasmic NF-κB-IκB complexes (37), suggesting that a Raf-1-dependent pathway may be involved in NF-κB activation. However, it is known that PKC-ζ triggers the activation of a number of kinases, suggesting that MEK and ERK may also participate in NF-κB activation by enhancing an AP-1-NF-κB cross-coupling mechanism (5). Activation of the ERK pathway is one of the early biochemical events that follow LPS treatment of macrophages or their infection by virulent and attenuated Salmonella strains (29, 45), as tested in various cell types, and LPS can induce activation of JNKs (25) and p38 (26).
From our results, MAPK activation of cells treated with porins begins a few minutes after stimulation, reaches the highest level by 20 min, and goes back to the background level by 60 min. Moreover, the activation is dose dependent; porin concentrations of as low as 500 ng/ml (0.02 μM) still induce activation, while concentrations of 20 μg/ml (0.8 μM) do not further increase the phosphorylation of the Raf-1-MEK1-MEK2-MAPK pathways and of the transcription factors AP-1 and NF-κB. The minimum LPS concentration which was able to induce Raf-1-MEK1-MEK2-MAPK phosphorylation was 100 ng/ml. Therefore, the amount of LPS present in our preparation was insignificant in this experimental model. Outer membrane blebbing or immune- or antibiotic-mediated bacterial lysis in sites of Salmonella infection is sufficient to generate the concentrations of porins and LPS used in these experiments (59). Gram-negative bacterial cells contain about 105 molecules of porins (molecular mass, about 36 kDa) per cell and about 3.4 × 106 molecules of LPS (molecular mass, about 4.5 kDa) per cell (42); therefore, 106 to 109 bacterial cells are enough for porin concentrations of 500 ng/ml to 20 μg/ml (0.02 to 0.8 μM) or LPS concentrations of 100 ng/ml to 10 μg/ml (0.05 to 5 μM) to be reached.
Moreover, additional experiments to detect JNK and p38 activation were done with macrophages from LPS-hyporesponsive C3H/HeJ mice. We found that porins stimulated phosphorylation of cell enzymes from the C3H/HeJ macrophages, whereas LPS failed to elicit any response different from that of controls. Porins treated with polymyxin B also activate AP-1.
Using several inhibitors, we confirmed that the activation of AP-1 and NF-κB following S. enterica serovar Typhimurium porin treatment of U937 cells may involve the Raf-1-MEK1-MEK2-MAPK signaling pathway. We found that activation of transcription factors after treatment with porins was slightly modulated by treatment with PD-098059 and was markedly affected by treatment with SB203580. It has been demonstrated that the activation of ERK2 and, to a lesser extent, of ERK1 is reduced by PD-098059 (1). Our results showed that PD-098059 acts mainly in LPS-treated cells; therefore, activation induced by porins appeared to be less affected by PD-098059 than that induced by LPS. SB203580, although only partially blocking phosphorylation due to LPS, is able to block more strongly phosphorylation due to porins. These results indicate that in U937 cells, the p38 pathway is mainly involved in AP-1 and NF-κB activation by porins, while in AP-1 and NF-κB activation by LPS, ERK1/2 and, to a minor extent, p38 pathways are involved. Since MAPKs are located downstream of Raf-1 in the classical MAPK signaling pathway, our data also suggest that a Raf-1 signal may regulate AP-1 and NF-κB activation. Forskolin, an inhibitor of Raf-1 activation (48), did not block stimulation by porins or LPS, suggesting that a Raf-1-independent pathway may also be involved in AP-1 and NF-κB activation by porins or LPS.
Activation of AP-1 and NF-κB by endotoxin has been previously described to be dependent only on LPS stimulation (22, 31). We now show that AP-1 and NF-κB are also activated by porins, the pore-forming proteins of the gram-negative outer membrane. Our results show that the porins of S. enterica serovar Typhimurium activate AP-1 and NF-κB in U937 cells at concentrations of 0.1 to 5 μg/ml by 30 to 60 min of stimulation. The expression of Fos and Jun proteins is temporally coordinated in response to various stimuli, with the result that the AP-1 composition may change in the cell as a function of time and stimulus (6, 7, 14, 46). The binding affinity for a given target DNA sequence is determined by the different AP-1 dimer combinations and the context of the surrounding sequences (24).
Antibody EMSA using antibodies to specific transcription factor protein subunits showed that in U937 cell controls the AP-1 complex contains JunD and c-Fos heterodimers and probably no other homodimers or heterodimers. In U937 cells treated with porins, different complexes including c-Jun and Fra-2 subunits appeared, while in cells treated with LPS, c-Jun was also present. fra-2 is the only member of the fos gene family which is present at high levels in adult tissues, while generally every member of the jun family is moderately expressed in the same tissues (14). Whereas LPS leads to AP-1 complexes containing JunD, c-Fos and c-Jun stimulation by porins induces AP-1 complexes containing Fra-2 in addition to the other subunits.
The formation of a different complex represent a further difference between stimulation with LPS and stimulation with porins, to be added to past observations that cytokine release after stimulation with porins begins after 120 min and continues for 5 to 6 h, whereas cytokine release following LPS stimulation begins after 30 min and decreases at 120 min (19). In LPS-stimulated macrophages, MAPK participates in the control of the production of inflammatory mediators such as phosphatidylglycerols, leukotrienes, and platelet-activating factor (27, 47); the data reported here show that Raf-1-MEK1-MEK2-MAPK activation is also part of the response of macrophages to porin signal transduction. Porins, with respect to LPS, activate mainly but not exclusively the p38 pathway. The stimulation of different signal transmission pathways leads to a sum of effects (49); therefore, different bacterial components, such as LPS or porins, induce increased expression and release of cytokines through different pathways. Regulation at the transcriptional and posttranscriptional levels, or regulation of transcriptional factors and the subunit compositions of transcriptional factors, may affect the adaptive mechanism to rapidly modulate the production of biologically active proteins. Even though they show similar biological effects, LPS and porins differentially regulate the expression and release of cytokines and other biologically active molecules at several levels. The different potential associations between Fos and Jun protein subunits resulting from stimulation by different surface components of gram-negative bacteria could provide new molecular insights into the pathophysiological events of each component during bacterial infection. The engagement of different pathways during signal transmission, due to different bacterial components, makes the use of molecular inhibitors as therapeutic agents much more complex.
FIG. 10.
EMSA with AP-1 consensus oligonucleotide. U937 cells (3 × 106 cells/ml) were stimulated for 60 min with porins (5 μg/ml) or LPS (1 μg/ml) pretreated with inhibitors. (A) Control, control nuclear extract (5 μg); LPS, LPS-induced nuclear extract (5 μg); LPS + PD-098059, LPS-induced nuclear extract (5 μg) pretreated with PD-098059; LPS + SB202190, LPS-induced nuclear extract (5 μg) pretreated with SB203580; LPS + Forskolin, LPS-induced nuclear extract (5 μg) pretreated with forskolin; COMP., binding reactions with unlabeled excesses of AP-1 and NF-κB consensus oligonucleotides. The arrow to the left indicates control and induced complexes. (B) The same experiment using porins instead of LPS. Fold increases in AP-1 activation are shown below each lane for each blot.
FIG. 11.
EMSA with NF-κB consensus oligonucleotide. U937 cells (3 × 106 cells/ml) were stimulated for 60 min with porins (5 μg/ml) or LPS (1 μg/ml) pretreated with inhibitors. (A) Control, control nuclear extract (5 μg); LPS, LPS-induced nuclear extract (5 μg); LPS + PD-098059, LPS-induced nuclear extract (5 μg) pretreated with PD-098059; LPS + SB202190, LPS-induced nuclear extract (5 μg) pretreated with SB202190; LPS + Forskolin, LPS-induced nuclear extract (5 μg) pretreated with forskolin; COMP., binding reactions with unlabeled excesses of NF-κB and AP-1 consensus oligonucleotides. The arrow to the left indicates control and induced complexes. (B) The same experiment using porins instead of LPS. Fold increases in NF-κB activation are shown below each lane for each blot.
Editor: A. D. O’Brien
REFERENCES
- 1.Alessi, D. R., A. Cuenda, P. Cohen, D. T. Dudley, and A. R. Saltiel. 1995. PD-098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J. Biol. Chem. 270:27489–27494. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angel, P., and M. Karin. 1991. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta 1072:129–157. [DOI] [PubMed] [Google Scholar]
- 4.Arditi, M., J. Zhou, M. Torres, D. L. Durden, M. Stins, and K. S. Kim. 1995. Lipopolysaccharide stimulates the tyrosine phosphorylation of mitogen-activated protein kinases p44, p42, and p41 in vascular endothelial cells in a soluble CD14-dependent manner. Role of protein tyrosine phosphorylation in lipopolysaccharide-induced stimulation of endothelial cells. J. Immunol. 155:3994–4003. [PubMed] [Google Scholar]
- 5.Berra, E., T. Diaz-Meco, J. Lonzano, S. Frutos, M. M. Municio, P. Sànchez, L. Sanz, and J. Moscat. 1995. Evidence for a role of MEK and MAPK during signal transduction by protein kinase Cζ. EMBO J. 14:6157–6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, D. R., P. C. Ferreria, R. Gentz, B. R. Franza, Jr., and T. Curran. 1989. The product of a fos-related gene, fra-1, binds cooperatively to the AP-1 site with Jun : scription factor AP-1 is comprised of multiple protein complexes. Genes Dev. 3:173–184. [DOI] [PubMed] [Google Scholar]
- 7.Cook, S. J., N. Aziz, and M. McMahon. 1999. The repertoire of Fos and Jun proteins expressed during the G1 phase of the cell cycle is determined by the duration of mitogen-activated protein kinase activation. Mol. Cell. Biol. 19:330–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuenda, A., J. Rouse, Y. N. Doza, R. Meier, P. Cohen, T. F. Gallagher, P. R. Yang, and J. C. Lee. 1995. SB203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stress and interleukin-1. FEBS Lett. 364:229–233. [DOI] [PubMed] [Google Scholar]
- 9.Dèrijard, B., M. Hibi, I. H. Wu, T. Barrett, B. Su, T. Deng, M. Karin, and R. J. Davis. 1994. JNK1:a protein otein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76:1025–1037. [DOI] [PubMed] [Google Scholar]
- 10.Dofferhoff, A. S., J. H. Nijland, H. G. de Vries-Hospers, P. O. Mulder, J. Weits, and V. J. Bom. 1991. Effects of different types and combinations of antimicrobial agents on endotoxin release from gram-negative bacteria: in vitro and in vivo study. Scand. J. Infect. Dis. 23:745–754. [DOI] [PubMed] [Google Scholar]
- 11.Dudley, D. T. 1995. A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proc. Natl. Acad. Sci. USA 92:7686–7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans, M. E., and M. Pollack. 1993. Effect of antibiotic class and concentration on the release of lipopolysaccharide from Escherichia coli. J. Infect. Dis. 167:1336–1343. [DOI] [PubMed] [Google Scholar]
- 13.Evans, T. J., E. Strivens, A. Carpenter, and J. Cohen. 1993. Differences in cytokine response and induction of nitric oxide synthase in endotoxin-resistant and endotoxin-sensitive mice after intravenous Gram-negative infection. J. Immunol. 150:5033–5040. [PubMed] [Google Scholar]
- 14.Foletta, V. C. 1996. Transcription factor AP-1 and the role of Fra-2. Immunol. Cell. Biol. 74:121–133. [DOI] [PubMed] [Google Scholar]
- 15.Galanos, C., O. Luderitz, and O. Westphal. 1969. A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 9:245–249. [DOI] [PubMed] [Google Scholar]
- 16.Galdiero, F., M. A. Tufano, M. Galdiero, S. Masiello, and M. Di Rosa. 1990. Inflammatory effects of Salmonella typhimurium porins. Infect. Immun. 58:3183–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galdiero, F., G. Cipollaro de l’Ero, N. Benedetto, M. Galdiero, and M. A. Tufano. 1993. Release of cytokines by Salmonella typhimurium porins. Infect. Immun. 61:155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galdiero, F., M. A. Tufano, L. Sommese, A. Folgore, and F. Tedesco,. 1984. Activation of the complement system by porins extracted from Salmonella typhimurium. Infect. Immun. 46:559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galdiero, M., G. Cipollaro de l’Ero, G. Donnarumma, A. Marcatili, and F. Galdiero. 1995. Interleukin-1 and interleukin-6 gene expression in human monocytes stimulated with Salmonella typhimurium porins. Immunology 86:612–619. [PMC free article] [PubMed] [Google Scholar]
- 20.Galdiero, M., M. D’Amico, F. Gorga, C. Di Filippo, M. D’Isanto, M. Vitiello, A. Longanella, and A. Tortora. 2001. Haemophilus influenzae porin contributes to signaling of the inflammatory cascade in rat brain. Infect. Immun. 69:221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldfeld, A. E., C. Doyle, and T. Maniatis. 1990. Human tumor necrosis factor α gene regulation by virus and lipopolysaccharide. Proc. Natl. Acad. Sci. USA 87:9769–9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groupp, E. R., and M. Donovan-Peluso. 1996. Lipopolysaccharide induction of THP-1 cells activates binding of c-Jun, Ets, and Egr-1 to the tissue factor promoter. J. Biol. Chem. 24:12423–12430. [DOI] [PubMed] [Google Scholar]
- 23.Gupta, S., D. Kumar, H. Vohra, and N. Kumar. 1999. Involvement of signal transduction pathways in Salmonella typhimurium porin activated gut macrophages. Mol. Cell. Biochem. 194:235–243. [DOI] [PubMed] [Google Scholar]
- 24.Halazonetis, T. D., K. Georgeopoulus, M. E. Greenberg, and P. Leder. 1988. c-Jun dimerizes with itself and with c-Fos forming complexes with different binding affinities. Cell 55:917–924. [DOI] [PubMed] [Google Scholar]
- 25.Hambleton, J., S. L. Weistein, L. Lem, and A. L. DeFranco. 1996. Activation of c-Jun N-terminal kinase in bacterial lipopolysaccharide-stimulated macrophages. Proc. Natl. Acad. Sci. USA 93:2774–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han, J., J. D. Lee, L. Bibbs, and R. J. Ulevitch. 1994. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 265:808–811. [DOI] [PubMed] [Google Scholar]
- 27.Hanahan, D. J. 1986. Platelet-activating factor: biologically active phosphoglyceride. Annu. Rev. Biochem. 55:483–509. [DOI] [PubMed] [Google Scholar]
- 28.Henderson, B., S. Poole, and M. Wilson. 1996. Bacterial modulins: l class of virulence factors which cause host tissue pathology by inducing cytokine synthesis. Microbiol. Rev. 60:316–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hobbie, S., L. M. Chen, R. J. Davis, and J. E. Galan. 1997. Involvement of mitogen-activated protein kinase pathways in the nuclear responses and cytokine production induced by Salmonella typhimurium in cultured intestinal epithelial cells. J. Immunol. 159:550–559. [PubMed] [Google Scholar]
- 30.Iovane, G., P. Pagnini, M. Galdiero, G. Cipollaro de L’Ero, M. Vitiello, M. D’Isanto, and A. Marcatili. 1998. Role of Pasteurella multocida porin on cytokine expression and release by murine splenocytes. Vet. Immunol. Immunopathol. 66:391–404. [DOI] [PubMed] [Google Scholar]
- 31.Ishikawa, Y., N. Mukaida, K. Kuno, N. Rice, S. Okamoto, and K. Matsushima. 1995. Establishment of lipopolysaccharide-dependent nuclear factor κB activation in a cell-free system. J. Biol. Chem. 270:4158–4164. [PubMed] [Google Scholar]
- 32.Karin, M. 1996. The regulation of AP-1 activity by mitogen-activated protein kinases. Philos. Trans. R. Soc. London B 351:127–134. [DOI] [PubMed] [Google Scholar]
- 33.Karin, M., Z. G. Liu, and E. Zandi. 1997. AP-1 function and regulation. Curr. Opin. Cell Biol. 9:240–246. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature 277:680–685. [DOI] [PubMed] [Google Scholar]
- 35.Leeson, M. C., Y. Fujihara, and D. C. Morrison. 1994. Evidence for lipopolysaccharide as the predominant proinflammatory mediator in supernatants of antibiotic-treated bacteria. Infect. Immun. 62:4975–4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, J. J., C. Westergaard, P. Ghosh, and N. H. Colburn. 1997. Inhibitors of both nuclear factor-kappa B and activator protein-1 activation block the neoplastic transformation response. Cancer Res. 57:3569–3576. [PubMed] [Google Scholar]
- 37.Li, S., and J. M. Sedivy. 1993. Raf-1 protein kinase activates the NF-κB transcription factor by dissociating the cytoplasmic NF-κB-IκB complex. Proc. Natl. Acad. Sci. USA 90:9247–9251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265–275. [PubMed] [Google Scholar]
- 39.Mahon, T. M., and L. A. J. O’Neill. 1995. Studies into the effect of the tyrosine kinase inhibitor herbimycin A on NF-κB activation in T lymphocytes. J. Biol. Chem. 270:28557–28564. [DOI] [PubMed] [Google Scholar]
- 40.Marais, R., J. Wynne, and R. Treisman. 1993. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell 73:381–393. [DOI] [PubMed] [Google Scholar]
- 41.Morrison, D. C., and D. M. Jacobs. 1976. Binding of polymyxin B to the lipid A portion of bacterial lipopolysaccharides. Immunochemistry 13:813–818. [DOI] [PubMed] [Google Scholar]
- 42.Nikaido, H., and M. Vaara. 1987. Outer membrane, p.7–22. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, D.C.
- 43.Nurminen, M., K. Lounatmaa, M. Sarvas, P. H. Makela, and T. Nakae. 1976. Bacteriophage-resistant mutants of Salmonella typhimurium deficient in two major outer membrane proteins. J. Bacteriol. 127:941–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nurminen, M. 1985. Isolation of porin trimers. Enterobacterial surface antigens: methods for molecular characterization. FEMS Symp. 25:294. [Google Scholar]
- 45.Procyk, K. S., P. Kovarik, A. von Gabain, and M. Baccarini. 1999. Salmonella typhimurium and lipopolysaccharide stimulate extracellularly regulated kinase activation in macrophages by a mechanism involving phosphatidylinositol 3-kinase and phospholipase D as novel intermediates. Infect. Immun. 67:1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryseck, R. P., and R. Bravo. 1991. Jun, JunB and JunD differ in their binding affinities to AP-1 and CRE consensus sequences; effect of Fos proteins. Oncogene 6:533–542. [PubMed] [Google Scholar]
- 47.Samuelsson, B., S. E. Dahlen, J. A. Lindgren, C. A. Rouzer, and C. N. Serhan. 1987. Leukotrienes and lipoxins: structures, biosynthesis, and biological effects. Science 237:1171–1176. [DOI] [PubMed] [Google Scholar]
- 48.Scamon, K. B., and J. W. Daly. 1986. Forskolin : biological and chemical properties. Adv. Cyclic Nucleotide Protein Phosphoregulation Res. 20:1–50. [PubMed] [Google Scholar]
- 49.Schaeffer, H. J., and M. J. Weber. 1999. Mitogen activated protein kinases : specific messages from ubiquitous messengers. Mol. Cell. Biol. 19:2435–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siebenlist, U., G. Franzoso, and K. Brown. 1994. Structure, regulation and function of NF-κB. Annu. Rev. Cell Biol. 10:405–455. [DOI] [PubMed] [Google Scholar]
- 51.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver-stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115–119. [DOI] [PubMed] [Google Scholar]
- 52.Turner, M., D. Chantry, G. Buchan, K. Barrett, and M. Feldmann. 1989. Regulation of expression of human IL-1α, IL-1β genes. J. Immunol. 143:3556–3561. [PubMed] [Google Scholar]
- 53.Vandel, L., C. M. Pfarr, S. Huguier, L. Loiseau, A. Sergeant, and M. Castellazzi. 1995. Increased transforming activity of JunB and JunD by introduction of a heterologous homodimerisation domain. Oncogene 10:495–507. [PubMed] [Google Scholar]
- 54.Van der Bruggen, T., S. Nijenhuis, E. van Raaij, J. Verhoef, and B. S. van Asbeck. 1999. Lipopolysaccharide-induced tumor necrosis factor alpha production by human monocytes involves the Raf-1/MEK1-MEK2/ERK1-ERK2 pathway. Infect. Immun. 67:3824–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whitmarsh, A. J., and R. J. Davis. 1996. Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J. Mol. Med. 74:589–607. [DOI] [PubMed] [Google Scholar]
- 56.Willis, S. A., and P. D. Nisen. 1996. Differential induction of the mitogen-activated protein kinase pathway by bacterial lipopolysaccharide in cultured monocytes and astrocytes. Biochem. J. 313:519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yagisawa, M., K. Saeki, E. Okuma, T. Kitamura, S. Kitagawa, H. Hirai, Y. Yazaki, F. Takaku, and A. You. 1999. Signal transduction pathways in normal human monocytes stimulated by cytokines and mediators: comparative study with normal human neutrophils or transformed cells and the putative roles in functionality and cell biology. Exp. Hematol. 27:1063–1076. [DOI] [PubMed] [Google Scholar]
- 58.Yin, E. T., C. Galanos, S. Kinsky, R. A. Bradshaw, S. Wessler, O. Luderity, and M. F. Sarmiento. 1972. Picogram-sensitive assay for endotoxin: gelation of Limulus polyphemus blood cell lysate induced by purified lipopolysaccharide and lipid A from Gram-negative bacteria. Biochim. Biophys. Acta 261:284–289. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, H., D. W. Nielsen, J. W. Peterson, and G. R. Klimpel. 1998. Lipoprotein release by bacteria: potential factor in bacterial pathogenesis. Infect. Immun. 66:5196–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]