Abstract
Trypanothione plays a pivotal role in defence against chemical and oxidant stress, thiol redox homoeostasis, ribonucleotide metabolism and drug resistance in parasitic kinetoplastids. In Trypanosoma brucei, trypanothione is synthesized from glutathione and spermidine by a single enzyme, TryS (trypanothione synthetase), with glutathionylspermidine as an intermediate. To examine the physiological roles of trypanothione, tetracycline-inducible RNA interference was used to reduce expression of TRYS. Following induction, TryS protein was reduced >10-fold and growth rate was reduced 2-fold, with concurrent 5–10-fold decreases in glutathionylspermidine and trypanothione and an up to 14-fold increase in free glutathione content. Polyamine levels were not significantly different from non-induced controls, and neither was the intracellular thiol redox potential, indicating that these factors are not responsible for the growth defect. Compensatory changes in other pathway enzymes were associated with prolonged suppression of TryS: an increase in trypanothione reductase and γ-glutamylcysteine synthetase, and a transient decrease in ornithine decarboxylase. Depleted trypanothione levels were associated with increases in sensitivity to arsenical, antimonial and nitro drugs, implicating trypanothione metabolism in their mode of action. Escape mutants arose after 2 weeks of induction, with all parameters, including growth, returning to normal. Selective inhibitors of TryS are required to fully validate this novel drug target.
Keywords: chemotherapy, glutathione, RNA interference, trypanosome, trypanothione
Abbreviations: ALAT, alanine aminotransferase; γ-GCS, γ-glutamylcysteine synthetase; GspdSH, glutathionylspermidine; GspS, glutathionyl-spermidine synthetase; ODC, ornithine decarboxylase; RNAi, RNA interference; TryR, trypanothione reductase; TryS, trypanothione synthetase; T(S)2/T(SH)2, trypanothione disulphide/dihydrotrypanothione couple
INTRODUCTION
Trypanosomatid species causing human disease infect an estimated 15 to 20 million people worldwide, and cause 100000–150000 deaths per annum, mainly in developing countries [1]. Current therapy for African trypanosomiasis, leishmaniasis and Chagas' disease is inadequate, and therefore the chemical or genetic validation of novel chemotherapeutic targets is an essential prerequisite for the discovery of new drugs. Trypanothione [N1,N8-bis(glutathionyl)spermidine] represents one attractive area of metabolism for chemotherapeutic intervention [2].
In the kinetoplastid parasites, trypanothione and TryR (trypanothione reductase) replace the glutathione reductase and thioredoxin reductase systems found in other cells [3]. TryR is pivotal in the redox-dependent function of trypanothione, since the dithiol form [4] provides reducing equivalents for the various trypanothione- and glutathione-dependent peroxidases, ascorbate peroxidase and ribonucleotide reductase [5–8]. TryR is essential for parasite survival and virulence [9–11] and has pronounced differences in substrate specificity compared with human glutathione reductase [4]. Trypanothione, and TryR are implicated in the mode of action of antimonials and arsenicals [12–14]. The recently characterized trypanothione S-transferase [15] and glyoxalase I and II enzymes involved in the detoxification of methylglyoxal [16,17] are also trypanothione-dependent in these parasites.
Since TryR is essential for trypanosomatid growth and survival, its substrate trypanothione should also be essential [2]. The insect trypanosomatid Crithidia fasciculata has two ATP-dependent enzymes involved in the biosynthesis of trypanothione from glutathione and spermidine [18–20, but see 20a]. GspS (glutathionylspermidine synthetase; EC 6.3.1.8) forms both the N1 and N8-isomers of glutathionylspermidine, and TryS (trypanothione synthetase; EC 6.3.1.9) catalyses the addition of the second glutathione moiety to glutathionylspermidine. However, in the pathogenic trypanosomatids Trypanosoma brucei, Trypanosoma cruzi and Leishmania major, a single TryS enzyme is able to catalyse both biosynthetic steps [21–24]. No gene encoding GspS has so far been found in the genus Trypanosoma [23], but L. major appears to harbour a GSPS pseudogene [22].
A recent report using RNAi (RNA interference) methods to knock down TryS expression has demonstrated impaired cell viability and hypersensitivity to peroxides, validating TryS as a drug target [25]. Here, we confirm and extend these findings with a detailed analysis of the metabolic and enzymatic changes in polyamine and glutathione biosynthesis. We also examine the sensitivity to trypanocidal agents arising from trypanothione knockdown, and provide a rationale for the previously observed synergism between some of these drugs.
EXPERIMENTAL
Trypanosome culture
The transgenic procyclic cell line (29-13) [26] co-expressing integrated genes for T7 RNA polymerase and tetracycline repressor was cultured in SDM-79 medium [27] with 10% foetal calf serum and haemin (10 mg·l−1) at 28 °C. Cultures were initiated at 1×106 cells·ml−1 and passaged every 3 to 4 days into fresh medium with appropriate antibiotics for selection (G418 for T7 RNA polymerase, 15 μg·ml−1; hygromycin for tetracycline repressor, 50 μg·ml−1; phleomycin for plasmid insert, 2.5 μg·ml−1). Cell densities were measured using a haemocytometer and plotted as a product of total cell density and dilution.
Generation of RNAi construct
The pZJM vector [28] was utilized to construct the TRYS RNAi plasmid. A 750 bp gene fragment of TRYS (nt 1–750) was amplified by PCR (30 cycles of 94 °C, 15 s; 55 °C, 30 s; 68 °C, 60 s) using 20 ng of pET15b-TbTRYS plasmid DNA [23] as the template. The PCR primers contained XhoI (CTCGAGatgacgaagtcggcac) and HindIII (AAGCTTtcgcctccttaccaaa) linkers (where the restriction sites are shown in uppercase letters). The TRYS gene fragment was then ligated into XhoI/HindIII-digested pZJM, replacing the α-tubulin stuffer region. Correct insertion was verified by DNA sequencing. The pZJM-TRYS plasmid was then transformed into JM109 cells and clones were obtained for plasmid amplification and purified by DNA maxi-prep kit (Qiagen). The plasmid was linearized with NotI before transfection into 29-13 procyclic parent cell lines.
Transfection of the parent cell line
Trypanosomes were electroporated as described previously [26] with some modifications. Briefly, cells were grown to mid-exponential phase [(2–5)×106 cells·ml−1], washed with Cytomix buffer at room temperature (≈22 °C) and resuspended at a density of (2–3)×107 cells per 0.45 ml of Cytomix. Cells were placed in 4 mm gap cuvettes (BTX) with 10–12 μg per 10 μl of linearized pZJM-TRYS and electroporated at 1.7 kV with 3×100 μs pulses using a BTX 630 electroporator. Each electroporation was immediately transferred to 24 ml of pre-warmed medium (without antibiotics) and incubated overnight for recovery. The following day, aliquots of cells (1 ml) were plated out into 12-well plates, and selection was applied by the addition of 2.5 μg·ml−1 phleomycin. As controls, cells were electroporated either with linearized pZJM or without DNA. RNAi of TRYS was induced by the daily addition of tetracycline (1 μg·ml−1, prepared fresh every 7 days) to cultures.
Western blotting
Approx. (1–2)×108 cells were pelleted at 800 g for 10 min at room temperature, washed with 1 ml of Dulbecco's PBS (Invitrogen) and lysed in 0.1–0.2 ml of lysis buffer [20 mM Tris/HCl (pH 7.5)/5 mM MgCl2/10 mM EDTA/0.5% (v/v) Triton X-100/0.25 μg·ml−1 RNase/0.5·μg ml−1 DNase/1 mM PMSF/50 mM tris-carboxyethyl phosphine/1 mM benzamidine, with one tablet of EDTA-free protease inhibitor cocktail (Roche) per 25 ml of lysis buffer]. Lysates were inactivated by three cycles of freezing in liquid nitrogen and thawing in a 37 °C water bath. Cell debris was cleared by centrifugation (13000 g for 10 min at 4 °C). Protein content was measured using Coomassie Blue (Bio-Rad) with BSA as the standard. Lysates were boiled in SDS/PAGE loading buffer and 30 μg of protein from each extract was separated on 4–12% NuPAGE Bistris gels (Invitrogen). Proteins were transferred to a PVDF membrane and blocked with 5% (w/v) milk in PBS/0.1% (v/v) Tween 20 before incubation with primary antibodies. Rat polyclonal antisera were obtained in-house against recombinant T. brucei TryS and T. cruzi TryR (cross-reactive with T. brucei TryR). Rabbit polyclonal antisera raised against recombinant T. brucei ODC (ornithine decarboxylase) and T. brucei γ-GCS (γ-glutamylcysteine synthetase) were provided by Dr Margaret Phillips and Dr Tu Huynh (UT Southwestern Medical Center at Dallas, TX, U.S.A.) [29]. Antisera were diluted 1:500 (T. brucei TryS and T. cruzi TryR) or 1:5000 (T. brucei ODC and T. brucei γ-GCS) in 1% (w/v) milk in PBS/0.1% (v/v) Tween 20 before use. Antigen recognition was visualized by horseradish peroxidase detection reagents (ECL® Western blotting kit, Amersham Biosciences) with either anti-rat IgG–horse-radish peroxidase conjugate (Dako) or anti-rabbit IgG–horseradish peroxidase conjugate (Upstate) diluted 1:5000. The same blots were re-probed with antibodies after stripping using a Western blot recycling kit (Auto Biogen Clear). Protein band intensities were determined by Labworks software (UVP).
Enzyme analysis
Cells (approx. 5×109 cells·ml−1 of lysis buffer) were freeze-thawed as described above, sonicated (3×1 min pulses with cooling) and debris was removed by centrifugation (13000 g for 10 min at 4 °C). Dialysed cell-free extracts were then assayed for TryR and ALAT (alanine aminotransferase) spectrophotometrically, as described previously [10,30].
Metabolite analysis
The disulphide content of extracts was measured using a continuous recycling assay with Ellman's reagent with either TryR (5 m-units·ml−1) or glutathione reductase (1.5 m-units·ml−1) [31,32]. For disulphide quantification, 109 cells were pelleted (1600 g at 4 °C for 10 min), resuspended in a total volume of 0.05 ml of 40 mM Hepps, pH 8.0/4 mM diethylenetriaminepenta-acetate buffer and treated with excess N-ethylmaleimide (final concentration of 6 mM) at 70 °C for 3 min to alkylate free thiols. Protein was removed by treatment with an equal volume of 20% (w/v) trichloroacetic acid (in 10 mM HCl) at 4 °C for 30 min, followed by centrifugation to remove precipitate. Excess N-ethylmaleimide was removed by extraction 10 times with water-saturated ethyl acetate, followed by gently bubbling nitrogen through the sample. The reaction rates for trypanothione and glutathione disulphides in this recycling assay were established to be linear from 0.1–2.0 μM. Reaction rates of cell extracts were assayed, and the disulphide content was calculated from standard curves. Cell samples spiked with trypanothione disulphide were used to verify recovery.
Free thiols were analysed following pre-column derivatization with monobromobimane, as described previously [33]. To measure the total thiol content (disulphides and free thiol), cells were suspended in buffer with 1 mM tris-carboxyethyl phosphine and then derivatized with monobromobimane. Acid-hydrolysed polyamines were analysed following pre-column derivatization with dansyl chloride, as described previously [34]. Cell extracts were analysed for amino-positive metabolites, also as described previously [35].
Measurement of redox potential
Total thiol and disulphide content measurements obtained above were used to calculate redox potentials for TRYS RNAi and control trypanosomes. Redox potentials for the trypanothione disulphide/dihydrotrypanothione couple [T(S)2/T(SH)2; −242 mV at 25 °C) and the glutathione disulphide/glutathione couple (GSSG/2GSH; −240 mV at 25 °C) were calculated assuming an intracellular pH of 7.0 and using the appropriate Nernst equations [13,36]. Cell volumes used in the calculations were measured using a Scharfe Systems CASY1 cell counter, yielding 3.45±0.19 and 3.31±0.11 μl·(108 cells)−1 for TRYS RNAi-induced and control procyclic trypanosomes respectively. As a control, S427 bloodstream trypomastigotes were measured using the same method, and the value obtained [5.89 μl·(108 cells)−1] was in good agreement with that obtained by the inulin exclusion method [5.8 μl·(108 cells)−1] [37].
Drug susceptibility in vitro
Cell density was determined using Alamar Blue [38]. Control experiments showed that the fluorescence intensity was proportional to cell numbers between 5×103 and 6×106 procyclics per well for incubation times of 1.5–5 h. Procyclic trypanosomes (stably transfected with RNAi vector pZJM-TRYS) were induced by the addition of 1 μg·ml−1 tetracycline for 3 days before drug sensitivity assays were conducted. Serial doubling dilutions of drugs (20–30 dilutions from 0.2 mM or 2 mM stocks) in a total volume of 0.05 ml were made in flat-bottomed 96-well plates, to which 1×105 cells in 0.05 ml medium were added. Plates were incubated for 72 h, with the daily addition of 1 μg·ml−1 tetracycline to induce RNAi against TRYS. Alamar Blue solution (0.01 ml; Serotec Ltd) was added to each well, and plates were incubated for a further 3 h before fluorescence was measured with a Bio-Tek fluorescence plate reader (FLX-800) set at 528 nm excitation and 590 nm emission wavelengths. Non-induced pZJM-TRYS cells were assayed similarly; wells without drugs and medium-only wells served as controls. IC50 values were calculated using GraFit software (Erithacus Software Ltd) with a non-linear, four-parameter robust curve fit.
RESULTS
Induction of TRYS RNAi in procyclic trypanosomes
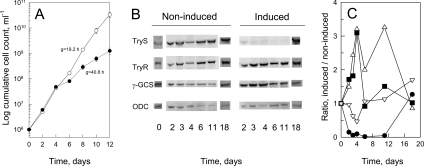
Transgenic procyclics were transformed with pZJM-TRYS and selected for resistance to phleomycin. Stable cell lines with the tetracycline-inducible RNAi construct integrated into the ribosomal locus were obtained 3 to 4 weeks after transfection. Three independent cell lines were examined for their growth phenotypes in response to induction of RNAi with tetracycline (cell line A is shown in Figure 1A). Cells grew exponentially at the same rate until day 4, after which the generation time (‘g’ in Figure 1A) significantly increased in RNAi-induced cells 2.1-, 1.6- and 1.3-fold for cell lines A, B and C respectively. Since cell line A gave the most pronounced growth reduction upon RNAi induction, it was selected for analysis of its biochemical phenotype.
Figure 1. Effect of TRYS RNAi on trypanosome growth and enzymatic phenotype.
(A) Growth of cell line A: RNAi-induced cells (●), non-induced cells (○); ‘g’ indicates the generation time. The results are means±S.D. for three separate cultures. (B) Western blot analysis of RNAi-induced and non-induced trypanosomes. The blot is representative of two separate experiments. (C) Ratio of induced:non-induced band densitometry measurements: TryS (●), TryR (■), γ-GCS (△), ODC (▽). Densitometry measurements were obtained from (B).
Effect of TRYS RNAi on enzymatic phenotype
Owing to the high endogenous ATPase activity in whole cell extracts, TryS enzymatic activity was too low to be measured by spectrophotometric methods. Accordingly, Western blot analysis (Figure 1B) and densitometry (Figure 1C) were used to monitor the effect of RNAi on TryS and other enzymes involved in trypanothione metabolism. Following induction, the amount of TryS decreased 10-fold by day 3, and remained the same until day 11; it subsequently returned to basal levels by day 18 (Figure 1B, and 1C, filled-in circles). This decrease in TryS levels correlates with the onset of decreased growth rate (Figure 1A). The same blots were stripped and re-probed with antibodies raised against ODC, TryR and γ-GCS (Figure 1B and 1C). Following induction, the ratio of induced over non-induced ODC transiently decreased 2-fold compared with the control, subsequently returning to basal levels by day 6. TryR expression is the inverse of ODC, increasing 3-fold by day 4 before returning to basal level by day 6, whereas γ-GCS expression is the inverse of TryS, remaining elevated 2–3-fold until at least day 11, before returning to basal levels by day 18. To confirm that changes in protein levels measured by Western blotting reflect changes in enzyme activity, TryR and ALAT activities were measured in a subsequent experiment (Table 1). Basal TryR activity (0.044±0.003 units·mg−1) was found to be comparable with that described in a previous paper [39], and the subsequent change in TryR activity (2-fold by day 3) followed that obtained by Western analysis. Enzyme activity of ALAT, an unrelated housekeeping enzyme, remains essentially constant throughout the time course (Table 1). These results indicate that changes in Western blots serve as a suitable estimate of enzyme activity.
Table 1. Effect of TryS RNAi on TryR and ALAT activities.
Enzyme activities were measured spectrophometrically as described in the Experimental section. Results are the means±S.D. for three separate cultures.
| TryR (units·mg−1) | ALAT (units·mg−1) | |||||
|---|---|---|---|---|---|---|
| Day | Induced (I) | Non-induced (N) | Ratio (I/N) | Induced (I) | Non-induced (N) | Ratio (I/N) |
| 0 | 0.044±0.003 | 0.044±0.003 | 1.0 | 0.17±0.02 | 0.17±0.02 | 1.0 |
| 3 | 0.087±0.007 | 0.042±0.026 | 2.1 | 0.23±0.01 | 0.24±0.02 | 1.0 |
| 6 | 0.051±0.007 | 0.040±0.010 | 1.3 | 0.16±0.01 | 0.19±0.02 | 0.8 |
| 8 | 0.075±0.004 | 0.044±0.017 | 1.7 | 0.15±0.01 | 0.18±0.01 | 0.8 |
Effect of TRYS RNAi on metabolic phenotype
Metabolic control theory has demonstrated that changes in enzyme activity do not necessarily reflect changes in intermediate levels or metabolic flux through a pathway [40]. Therefore we determined the thiol and polyamine content of induced cells during maximum TryS depletion (day 8) and compared them with non-induced cells (Table 2). Free glutathione, the substrate for TryS, is markedly increased 5-fold in induced cells and, conversely, the products of the reaction, glutathionylspermidine and trypanothione, are significantly reduced 4.5-fold and 7.2-fold respectively. The overall total intracellular glutathione (free and conjugated to spermidine) is slightly increased in induced cells. Free spermidine levels are also slightly increased, but this difference is not significant. Therefore the depletion of TryS is associated with increases in substrates and significant decreases in the products of the TryS reaction.
Table 2. Effect of TryS RNAi on trypanosome thiol and polyamine content.
Total glutathione was calculated as [free glutathione+glutathionylspermidine+2×trypanothione]; free spermidine was calculated as [total spermidine−(trypanothione+glutathionylspermidine)]. Measurements were obtained from day 8 cultures following RNAi induction. Results are the means±S.D. for at least three separate cultures. Student's t test analysis was performed to examine statistically significant differences between non-induced and induced cells (*P<0.005; †P<0.05). Other values were not significantly different (P>0.05).
| Metabolite content [nmol·(108 cells)−1] | |||
|---|---|---|---|
| Metabolite | Non-induced (N) | Induced (I) | Ratio (I/N) |
| Thiol | |||
| Free glutathione | 1.20±0.34 | 5.80±0.37* | 4.83 |
| Glutathionylspermidine | 0.22±0.03 | 0.05±0.01* | 0.22 |
| Trypanothione | 1.69±0.12 | 0.23±0.03* | 0.14 |
| Total glutathione | 4.80±0.59 | 6.34±0.55† | 1.31 |
| Polyamine | |||
| Putrescine | 12.43±1.18 | 11.31±1.47 | 0.91 |
| Total spermidine | 7.57±1.00 | 6.50±1.06 | 0.86 |
| Free spermidine | 5.66±1.14 | 6.21±1.09 | 1.10 |
The effects of TRYS knockdown on other thiol, polyamine and amino acid metabolites were also analysed in this experiment. Ovothiol A content remained unaltered [0.32±0.03 nmol·(108 cells)−1 for non-induced cells; 0.45±0.08 nmol·(108 cells)−1 for induced cells]. However, upon HPLC analysis of monobromobimane-derivatized thiols, two unidentified fluorescent peaks present in non-induced cells were abolished following induction. The peak areas amounted to 0.1 nmol·(108 cells)−1 when calculated with the fluorescence response factor of glutathionylspermidine-bimane. As derivatization of cells with monobromobimane is conducted under linear reagent conditions, this is not due to formation of mono-derivatized trypanothione species. Neither do these peaks co-elute with the bimane conjugates of glutathionylspermine, bis(glutathionyl)spermine, cysteinylglycine, γ-glutamylcysteine or ovothiol C [33]. Both peaks were unchanged following reduction with tris-carboxyethyl phosphine before derivatization, but were abolished by alkylation with N-ethylmaleimide. Therefore they appear to be trypanothione-dependent, thiol-group-containing metabolites. However, these thiols were not detectable in the bloodstream form of the parasite. Further experiments are under way to determine the chemical nature of these metabolites.
Trace amounts of cadaverine [0.5 nmol·(108 cells)−1] were detected in both non-induced and induced cells, but no aminopropylcadaverine or homotrypanothione was observed. The amino acid profiles obtained from non-induced and induced trypanosomes were virtually identical, apart from a reduction in lysine in induced cells. No significant changes in S-adenosylmethionine or decarboxylated S-adenosylmethionine content were noted.
Effect of TRYS RNAi on thiol redox
Since trypanothione and glutathione content is markedly changed following RNAi induction of TryS, we investigated whether this would affect the intracellular thiol redox balance. Absolute amounts of thiols and disulphides and cell volumes were determined in both non-induced and induced cells during maximum TryS depletion from day 7 cultures (Table 3). RNAi induction did not cause perturbation in cell volume. The measurements also revealed that trypanothione and glutathione disulphide content of both induced and non-induced cells are less than 0.5% of the total thiol pool, indicating that TryR activity and thiol–disulphide exchange remained efficient in induced cells despite a 9.6-fold reduction in trypanothione content. The half-cell reduction potentials for the T(S)2/T(SH)2 and GSSG/2GSH couples did not change significantly following RNAi induction (Table 3), suggesting that the intracellular thiol redox balance remains unaffected. Similar to Leishmania donovani promastigotes and amastigotes [13], the reduction potential of the trypanothione couple was lower than the glutathione couple, indicating the lack of equilibrium between the two systems.
Table 3. Effect of TryS RNAi on trypanosome redox metabolism.
Total thiols, disulphides, intracellular concentrations and thiol redox potentials were determined as described in the Experimental section. Measurements were obtained from day 7 cultures following RNAi induction. Results are the means±S.D. for at least three separate cultures. The redox potentials between non-induced and induced cells were not significantly different (Student's t test; P>0.05).
| Physiological measurement | Non-induced | Induced |
|---|---|---|
| Intracellular concentration (μM) | ||
| [T(SH)2+T(S)2] | 480.3±45.5 | 50.3±10.8 |
| T(S)2 | 1.61±0.42 | 0.11±0.03 |
| [GSH+GSSG] | 251.0±13.1 | 797.0±117.9 |
| GSSG | 0.73±0.04 | 0.78±0.25 |
| Disulphide as a percentage of total thiol | ||
| T(S)2 | 0.34 | 0.22 |
| GSSG | 0.29 | 0.09 |
| Redox potential (E; in mV) | ||
| T(S)2/T(SH)2 | −315.3±4.2 | −320.7±2.7 |
| GSSG/2GSH | −208.5±1.9 | −237.9±3.1 |
TRYS RNAi escape mutants
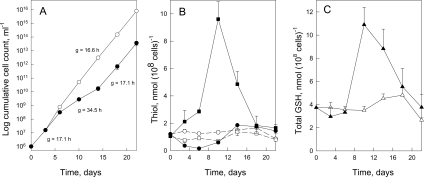
The increase in TryS expression by day 18 of RNAi induction (Figure 1B) prompted us to continue growth of cells under tetracycline induction over a longer time period (22 days; Figure 2A). The generation time of induced cells between days 6 and 14 doubled compared with that of non-induced and day 0 to day 6 induced cells. After 14 days, induced cells reverted back to initial growth rates. This suggests that escape mutants had overcome the knockdown effect of TRYS RNAi, a phenomenon that has been observed with other RNAi experiments in African trypanosomes (examples are quoted in [41]). This interpretation was verified by analysis of thiol content over the whole time course (Figure 2B). In induced cells, between day 0 and day 6, trypanothione decreases 8.3-fold and glutathionylspermidine 5.3-fold (results not shown). This 8-fold depletion of trypanothione over the first 6 days of induction can be accounted for by the 300-fold increase in cell number, which equates to just over eight cell doublings. Therefore virtually all trypanothione biosynthesis is halted immediately after TryS knockdown. Concurrently, the free glutathione content increases 3.2-fold by day 6, reaching a maximum of 13.5-fold by day 10. At this time point, the total glutathione content of induced cells increases nearly 4-fold over basal levels (Figure 2C). Based on a cell volume of 3.45 μl·(108 cells)−1 for induced trypanosomes, this amounts to an intracellular concentration of 3.2 mM, similar to that found in some Leishmania spp. promastigotes [33]. By the time trypanosomes resume initial growth rates (days 14–22), trypanothione and free glutathione content also revert back to basal levels, suggesting that all trypanothione-related enzyme activities have returned back to basal expression.
Figure 2. Growth of procyclic TRYS RNAi revertant trypanosomes and changes in thiol content.
(A) Growth of cell line A: RNAi-induced cells (●), non-induced cells (○); ‘g’ indicates the generation time. (B) Free glutathione (■) and trypanothione (●) content of RNAi-induced cells; free glutathione (□) and trypanothione (○) content of non-induced cells are also shown. (C) Total glutathione content of RNAi-induced cells (▲) and non-induced cells (△). Total glutathione was calculated as [glutathione+glutathionylspermidine+2×trypanothione]. All results are the means±S.D. for separate cultures.
Effect of TRYS RNAi on procyclic susceptibility to trypanocides
Since a number of trypanocides exert their effect by inhibiting and/or perturbing trypanothione metabolism [14], we investigated the susceptibility of TryS knockdown procyclics to a selection of these compounds (Table 4). RNAi-induced procyclics showed increased sensitivity to the arsenical, melarsen oxide, and the trivalent antimonial, triostam (Scheme 1), which affect trypanothione metabolism [12,13]. In addition, RNAi-induced procyclics became more susceptible to the redox cycling nitroheterocyclic compounds megazol and nifurtimox. In contrast, pentamidine, which does not affect trypanothione metabolism [14], did not increase the sensitivity of RNAi-induced cells. Sensitivity to buthionine sulphoximine, an inhibitor of T. brucei γ-GCS [42], was marginally decreased in induced cells, which is consistent with elevated levels of glutathione and γ-GCS expression.
Table 4. Susceptibility of procyclic trypanosomes to trypanocidal compounds.
Trypanosomes were RNAi-induced for 3 days before trypanocidal compounds were added. Student's t test analysis was performed to examine statistically significant differences between non-induced and induced cells (*P<0.001; †P<0.01). Other values were not significantly different (P>0.05). Results are the means±S.D. for at least 3 separate experiments.
| IC50 (nM) | |||
|---|---|---|---|
| Compound | Non-induced (N) | Induced (I) | Ratio (N/I) |
| Melarsen oxide | 405±27 | 135±1* | 3.0 |
| Triostam | 314±22 | 129±7* | 2.4 |
| Megazol | 201±46 | 61±5† | 3.3 |
| Nifurtimox | 3605±408 | 1664±21* | 2.2 |
| Buthionine sulphoximine | 67700±140 | 72600±100 | 0.9 |
| Pentamidine | 391±4 | 401±17 | 1.0 |
Scheme 1. Biosynthetic pathway of trypanothione and chemotherapeutic intervention points.
SOD, superoxide dismutase (all other abbreviations for enzymes are explained in the main text); metabolites: AdoSMe, methylthioadenosine; dAdoMet, S-adenosylmethionine; γ-GluCys, γ-glutamylcysteine; GspdSH, glutathionylspermidine; Put, putrescine; Spd, spermidine; T(SH)2, dihydrotrypanothione; T(S)2, trypanothione disulphide; drugs: BSO, buthionine sulphoximine; DFMO, difluoromethylornithine; RAs=O, arsenicals; RNO2, nitro compounds (nifurtimox, megazol); Sb3+, trivalent antimony.
DISCUSSION
The RNAi studies presented here indicate that TryS and trypanothione are required for normal cell viability and growth in procyclic forms of T. brucei. The profound depletion of glutathionylspermidine and trypanothione, together with the pronounced increase in glutathione content, is entirely consistent with the substrate specificity and enzymatic properties of TryS [23]. In addition, the absence of any accumulation of glutathionylspermidine supports our previous suggestion that a functional GspS is absent in T. brucei; it also indicates that no alternative biosynthetic routes to trypanothione are present (Scheme 1). Interestingly, perturbation of trypanothione biosynthesis is associated with compensatory changes in other enzymes in the pathway. First, polyamine biosynthesis is initially down-regulated by a decrease in ODC expression; this would account for the fact that spermidine levels do not rise markedly as a consequence of decreased incorporation into glutathionylspermidine and trypanothione. Secondly, trypanothione depletion up-regulates TryR expression; since the intracellular concentration of trypanothione disulphide is less than one-tenth the Km value for TryR, this compensation serves to maintain a favourable intracellular redox potential. Thirdly, γ-GCS expression is elevated so that free glutathione increases up to 14-fold over basal levels, presumably to maintain thiol buffer capacity. In general, regulation of gene expression occurs mainly at the post-transcriptional level [43], and it would be interesting to determine the mechanism by which this is achieved for the above enzymes. An eventual escape from the inhibitory effects of RNAi and the restoration of normal growth, mRNA, protein and metabolite levels (examples quoted in [41]) appear to be indicative of the essential nature of an enzyme in African trypanosomes. Our results therefore strongly support the conclusion of Comini et al. [25] that trypanothione is essential for growth and survival in the African trypanosome. These authors obtained more dramatic growth impairment upon TryS RNAi; this may be due to the improved host cell line (carrying double tetracycline repressor genes) and improved vector (targeted to a different integration site within the genome) used in their experiments. However, the slower and partial growth reduction obtained in our experiments enabled us to study the enzymatic and metabolic phenotype of TryS depletion in much more detail than would have been possible with their model.
What factors could be responsible for the decreased rate of proliferation in these parasites? In mammalian cells, decreases in ODC activity, polyamine biosynthesis and/or polyamine transport reduce cell proliferation [44]. However, since putrescine and spermidine levels are not significantly different from non-induced controls in African trypanosomes, these factors clearly cannot be responsible for the growth defect. Likewise, an increase in glutathione thiol-redox potential is implicated in the transition from a proliferative to non-proliferative or apoptotic state in mammalian cells [36]. Again, the absence of significant changes in either redox potential for trypanothione or glutathione indicates that this cannot be responsible here. Deoxyribonucleotide synthesis in T. brucei is also trypanothione-dependent [8]. Although millimolar concentrations of trypanothione can act as a direct reductant, at physiological concentrations of trypanothione (480 μM; Table 3) the reaction is accelerated by tryparedoxin [45]. The Km of tryparedoxin for trypanothione in this reaction is not known precisely, but is approx. 150 μM for the tryparedoxin-catalysed reduction of GSSG by trypanothione [45]. This would translate to a 3-fold decrease in the rate of reduction of tryparedoxin when trypanothione is reduced to 50 μM following RNAi (Table 3). Whether alterations in ribonucleotide reductase activity, methylglyoxal metabolism [17] or some other effect is responsible for the growth defect deserves further investigation.
RNAi-induced procyclics are significantly more susceptible to trypanocides (e.g. melarsen oxide and triostam) shown to act on trypanothione metabolism (Scheme 1) than those that are not (pentamidine) [14]. Procyclics are also more sensitive to the redox-cycling agents, nifurtimox and megazol, which decrease trypanothione content in T. cruzi epimastigotes [46]. This agrees with the observation that TryS knockdown causes an increased sensitivity to peroxides [25]. It is notable that the ODC inhibitor difluoromethylornithine has pronounced synergistic effects with melarsoprol, nifurtimox or antimony tartrate in T. brucei [47]. Difluoromethylornithine depletes both polyamine and trypanothione content, which is followed initially by cytostasis and ultimately by cell death [48,49]. However, our results show that enhanced susceptibility to these trypanocides is due to interaction with trypanothione metabolism, since polyamine content is essentially unchanged by TRYS RNAi. In the short term, these findings provide a clear rationale for combination chemotherapy for African sleeping sickness to reduce drug costs by either shortening the duration, or reducing the dose of these expensive (e.g. difluoromethylornithine) or toxic (e.g. nifurtimox or melarsoprol) drugs. In the longer term, our results indicate that TryS, which has no counterpart in mammalian cells, is an attractive novel target for drug discovery.
Acknowledgments
We thank Professor George Cross and Dr Simone Leal (Rockefeller University, New York, U.S.A.) for the provision of transgenic cell lines, Professor Paul Englund (John Hopkins University School of Medicine, Baltimore, MD, U.S.A.) for the RNAi vector, Professor Margaret Phillips and Dr Tu Huynh (University of Texas Southwestern Medical Center at Dallas, TX, U.S.A.) for γ-GCS and ODC antibodies, and Dr Bernard Blessington (Bradford University, Yorks., U.K.) for decarboxylated S-adenosylmethionine standard. This study was supported by the Wellcome Trust.
References
- 1.World Health Organization. The World Health Report 2002: Reducing risks, promoting healthy life. Geneva: World Health Organization; 2002. Statistical Annex; pp. 169–232. [Google Scholar]
- 2.Fairlamb A. H. Target discovery and validation with special reference to trypanothione. In: Fairlamb A. H., Ridley R. G., Vial H. J., editors. Drugs Against Parasitic Diseases: R&D Methodologies and Issues. Geneva: TDR Publications, World Health Organization; 2003. pp. 107–118. [Google Scholar]
- 3.Fairlamb A. H., Cerami A. Metabolism and functions of trypanothione in the Kinetoplastida. Annu. Rev. Microbiol. 1992;46:695–729. doi: 10.1146/annurev.mi.46.100192.003403. [DOI] [PubMed] [Google Scholar]
- 4.Shames S. L., Fairlamb A. H., Cerami A., Walsh C. T. Purification and characterization of trypanothione reductase from Crithidia fasciculata, a newly discovered member of the family of disulphide-containing flavoprotein reductases. Biochemistry. 1986;25:3519–3526. doi: 10.1021/bi00360a007. [DOI] [PubMed] [Google Scholar]
- 5.Henderson G. B., Fairlamb A. H., Cerami A. Trypanothione dependent peroxide metabolism in Crithidia fasciculata and Trypanosoma brucei. Mol. Biochem. Parasitol. 1987;24:39–45. doi: 10.1016/0166-6851(87)90113-7. [DOI] [PubMed] [Google Scholar]
- 6.Nogoceke E., Gommel D. U., Kiess M., Kalisz H. M., Flohé L. A unique cascade of oxidoreductases catalyses trypanothione-mediated peroxide metabolism in Crithidia fasciculata. Biol. Chem. 1997;378:827–836. doi: 10.1515/bchm.1997.378.8.827. [DOI] [PubMed] [Google Scholar]
- 7.Wilkinson S. R., Obado S. O., Mauricio I. L., Kelly J. M. Trypanosoma cruzi expresses a plant-like ascorbate-dependent hemoperoxidase localized to the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13453–13458. doi: 10.1073/pnas.202422899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dormeyer M., Reckenfelderbaumer N., Ludemann H., Krauth-Siegel R. L. Trypanothione-dependent synthesis of deoxyribonucleotides by Trypanosoma brucei ribonucleotide reductase. J. Biol. Chem. 2001;276:10602–10606. doi: 10.1074/jbc.M010352200. [DOI] [PubMed] [Google Scholar]
- 9.Dumas C., Ouellette M., Tovar J., Cunningham M. L., Fairlamb A. H., Tamar S., Olivier M., Papadopoulou B. Disruption of the trypanothione reductase gene of Leishmania decreases its ability to survive oxidative stress in macrophages. EMBO J. 1997;16:2590–2598. doi: 10.1093/emboj/16.10.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tovar J., Cunningham M. L., Smith A. C., Croft S. L., Fairlamb A. H. Down-regulation of Leishmania donovani trypanothione reductase by heterologous expression of a trans-dominant mutant homologue: effect on parasite intracellular survival. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5311–5316. doi: 10.1073/pnas.95.9.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krieger S., Schwarz W., Ariyanayagam M. R., Fairlamb A. H., Krauth-Siegel R. L., Clayton C. Trypanosomes lacking trypanothione reductase are avirulent and show increased sensitivity to oxidative stress. Mol. Microbiol. 2000;35:542–552. doi: 10.1046/j.1365-2958.2000.01721.x. [DOI] [PubMed] [Google Scholar]
- 12.Fairlamb A. H., Henderson G. B., Cerami A. Trypanothione is the primary target for arsenical drugs against African trypanosomes. Proc. Natl. Acad. Sci. U.S.A. 1989;86:2607–2611. doi: 10.1073/pnas.86.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wyllie S., Cunningham M. L., Fairlamb A. H. Dual action of antimonial drugs on thiol redox metabolism in the human pathogen Leishmania donovani. J. Biol. Chem. 2004;279:39925–39932. doi: 10.1074/jbc.M405635200. [DOI] [PubMed] [Google Scholar]
- 14.Fries D. S., Fairlamb A. H. Antiprotozoal Agents. In: Abraham D. J., editor. Burger's Medicinal Chemistry and Drug Discovery, vol. 5: Chemotherapeutic Agents. New York: Wiley and Sons; 2003. pp. 1033–1087. [Google Scholar]
- 15.Vickers T. J., Fairlamb A. H. Trypanothione S-transferase activity in a trypanosomatid ribosomal elongation factor 1B. J. Biol. Chem. 2004;279:27246–27256. doi: 10.1074/jbc.M311039200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vickers T. J., Greig N., Fairlamb A. H. A trypanothione-dependent glyoxalase I with a prokaryotic ancestry in Leishmania major. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13186–13191. doi: 10.1073/pnas.0402918101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irsch T., Krauth-Siegel R. L. Glyoxalase II of African trypanosomes is trypanothione-dependent. J. Biol. Chem. 2004;279:22209–22217. doi: 10.1074/jbc.M401240200. [DOI] [PubMed] [Google Scholar]
- 18.Smith K., Nadeau K., Walsh C., Fairlamb A. H. Purification of glutathionylspermidine and trypanothione synthetases from Crithidia fasciculata. Protein Sci. 1992;1:874–883. doi: 10.1002/pro.5560010705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tetaud E., Manai F., Barrett M. P., Nadeau K., Walsh C. T., Fairlamb A. H. Cloning and characterization of the two enzymes responsible for trypanothione biosynthesis in Crithidia fasciculata. J. Biol. Chem. 1998;273:19383–19390. doi: 10.1074/jbc.273.31.19383. [DOI] [PubMed] [Google Scholar]
- 20.Comini M., Menge U., Flohé L. Erratum [Koenig, K., Menge, U., Kiess, M., Wray, V. and Flohé, L. (1997) Convenient isolation and kinetic mechanism of glutathionylspermidine synthetase from Crithidia fasciculata. J. Biol. Chem. 272, 11908–11915] J. Biol. Chem. 2005;280:7407. doi: 10.1074/jbc.272.18.11908. [DOI] [PubMed] [Google Scholar]
- 20a.Comini M., Menge U., Wissing J., Flohé L. Trypanothione synthesis is Crithidia revisited. J. Biol. Chem. 2005;280:6850–6860. doi: 10.1074/jbc.M404486200. [DOI] [PubMed] [Google Scholar]
- 21.Oza S. L., Tetaud E., Ariyanayagam M. R., Warnon S. S., Fairlamb A. H. A single enzyme catalyses formation of trypanothione from glutathione and spermidine in Trypanosoma cruzi. J. Biol. Chem. 2002;277:35853–35861. doi: 10.1074/jbc.M204403200. [DOI] [PubMed] [Google Scholar]
- 22.Oza S. L., Shaw M. P., Wyllie S., Fairlamb A. H. Trypanothione biosynthesis in Leishmania major. Mol. Biochem. Parasitol. 2005;139:107–116. doi: 10.1016/j.molbiopara.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Oza S. L., Ariyanayagam M. R., Aitcheson N., Fairlamb A. H. Properties of trypanothione synthetase from Trypanosoma brucei. Mol. Biochem. Parasitol. 2003;131:25–33. doi: 10.1016/s0166-6851(03)00176-2. [DOI] [PubMed] [Google Scholar]
- 24.Comini M., Menge U., Flohe L. Biosynthesis of trypanothione in Trypanosoma brucei brucei. Biol. Chem. 2003;384:653–656. doi: 10.1515/BC.2003.072. [DOI] [PubMed] [Google Scholar]
- 25.Comini M. A., Guerrero S. A., Haile S., Menge U., Lunsdorf H., Flohe L. Validation of Trypanosoma brucei trypanothione synthetase as drug target. Free Radical Biol. Med. 2004;36:1289–1302. doi: 10.1016/j.freeradbiomed.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Wirtz E., Leal S., Ochatt C., Cross G. A. M. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 27.Brun R., Schonenberger M. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- 28.Wang Z. F., Morris J. C., Drew M. E., Englund P. T. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- 29.Lueder D. V., Phillips M. A. Characterization of Trypanosoma brucei γ-glutamylcysteine synthetase, an essential enzyme in the biosynthesis of trypanothione (diglutathionylspermidine) J. Biol. Chem. 1996;271:17485–17490. doi: 10.1074/jbc.271.29.17485. [DOI] [PubMed] [Google Scholar]
- 30.Tovar J., Fairlamb A. H. Extrachromosomal, homologous expression of trypanothione reductase and its complementary mRNA in Trypanosoma cruzi. Nucleic Acids Res. 1996;24:2942–2949. doi: 10.1093/nar/24.15.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamilton C. J., Saravanamuthu A., Eggleston I. M., Fairlamb A. H. Ellman's-reagent-mediated regeneration of trypanothione in situ: substrate-economical microplate and time-dependent inhibition assays for trypanothione reductase. Biochem. J. 2003;369:529–537. doi: 10.1042/BJ20021298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cribb A. E., Leeder J. S., Spielberg S. P. Use of a microplate reader in an assay of glutathione reductase using 5,5′-dithiobis(2-nitrobenzoic acid) Anal. Biochem. 1989;183:195–196. doi: 10.1016/0003-2697(89)90188-7. [DOI] [PubMed] [Google Scholar]
- 33.Ariyanayagam M. R., Fairlamb A. H. Ovothiol and trypanothione as antioxidants in trypanosomatids. Mol. Biochem. Parasitol. 2001;115:189–198. doi: 10.1016/s0166-6851(01)00285-7. [DOI] [PubMed] [Google Scholar]
- 34.Hunter K. J. A dansyl chloride-HPLC method for the determination of polyamines. In: Morgan D., editor. Polyamine Protocols. Totowa, NJ: Humana Press; 1998. pp. 119–123. [DOI] [PubMed] [Google Scholar]
- 35.Hunter K. J., Fairlamb A. H. The determination of polyamines and amino acids by a fluorescamine-HPLC method. In: Morgan D., editor. Polyamine Protocols. Totowa, NJ: Humana Press; 1998. pp. 125–130. [DOI] [PubMed] [Google Scholar]
- 36.Schafer F. Q., Buettner G. R. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radical Biol. Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 37.Opperdoes F. R., Baudhuin P., Coppens I., de Roe C., Edwards S. W., Weijers P. J., Misset O. Purification, morphometric analysis, and characterization of the glycosomes (microbodies) of the protozoan hemoflagellate Trypanosoma brucei. J. Cell Biol. 1984;98:1178–1184. doi: 10.1083/jcb.98.4.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raz B., Iten M., Grether-Buhler Y., Kaminsky R., Brun R. The Alamar Blue® assay to determine drug sensitivity of African trypanosomes (T. b. rhodesiense and T. b. gambiense) in vitro. Acta Trop. 1997;68:139–147. doi: 10.1016/s0001-706x(97)00079-x. [DOI] [PubMed] [Google Scholar]
- 39.Fairlamb A. H., Carter N. S., Cunningham M., Smith K. Characterisation of melarsen-resistant Trypanosoma brucei brucei with respect to cross-resistance to other drugs and trypanothione metabolism. Mol. Biochem. Parasitol. 1992;53:213–222. doi: 10.1016/0166-6851(92)90023-d. [DOI] [PubMed] [Google Scholar]
- 40.Fell D. London: Portland Press; 1996. Understanding the Control of Metabolism. [Google Scholar]
- 41.Schlecker T., Schmidt A., Dirdjaja N., Voncken F., Clayton C., Krauth-Siegel R. L. Substrate specificity, localization, and essential role of the glutathione peroxidase-type tryparedoxin peroxidases in Trypanosoma brucei. J. Biol. Chem. 2005;280:14385–14394. doi: 10.1074/jbc.M413338200. [DOI] [PubMed] [Google Scholar]
- 42.Huynh T. T., Huynh V. T., Harmon M. A., Phillips M. A. Gene knockdown of gamma-glutamylcysteine synthetase by RNAi in the parasitic protozoa Trypanosoma brucei demonstrates that it is an essential enzyme. J. Biol. Chem. 2003;278:39794–39800. doi: 10.1074/jbc.M306306200. [DOI] [PubMed] [Google Scholar]
- 43.Clayton C. E. Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas T., Thomas T. J. Polyamines in cell growth and cell death: molecular mechanisms and therapeutic applications. Cell Mol. Life Sci. 2001;58:244–258. doi: 10.1007/PL00000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ludemann H., Dormeyer M., Sticherling C., Stallmann D., Follmann H., Krauth-Siegel R. L. Trypanosoma brucei tryparedoxin, a thioredoxin-like protein in African trypanosomes. FEBS Lett. 1998;431:381–385. doi: 10.1016/s0014-5793(98)00793-5. [DOI] [PubMed] [Google Scholar]
- 46.Repetto Y., Opazo E., Maya J. D., Agosin M., Morello A. Glutathione and trypanothione in several strains of Trypanosoma cruzi: effect of drugs. Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 1996;115:281–285. doi: 10.1016/0305-0491(96)00112-5. [DOI] [PubMed] [Google Scholar]
- 47.Jennings F. W. Future prospects for the chemotherapy of human trypanosomiasis 2. Combination chemotherapy and African trypanosomiasis. Trans. R. Soc. Trop. Med. Hyg. 1990;84:618–621. doi: 10.1016/0035-9203(90)90125-x. [DOI] [PubMed] [Google Scholar]
- 48.Fairlamb A. H., Henderson G. B., Bacchi C. J., Cerami A. In vivo effects of difluoromethylornithine on trypanothione and polyamine levels in bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 1987;24:185–191. doi: 10.1016/0166-6851(87)90105-8. [DOI] [PubMed] [Google Scholar]
- 49.Bellofatto V., Fairlamb A. H., Henderson G. B., Cross G. A. M. Biochemical changes associated with α-difluoromethylornithine uptake and resistance in Trypanosoma brucei. Mol. Biochem. Parasitol. 1987;25:227–238. doi: 10.1016/0166-6851(87)90086-7. [DOI] [PubMed] [Google Scholar]