Abstract
The neurite outgrowth inhibitor protein Nogo is one of 300 proteins that contain a reticulon homology domain, which is responsible for their association with the endoplasmic reticulum. Here we have found that the Nogo-B spliceform becomes phosphorylated at Ser107 in response to lipopolysaccharide in RAW264 macrophages or anisomycin in HeLa cells. The phosphorylation is prevented by SB 203580, an inhibitor of SAPK2a (stress-activated protein kinase 2a)/p38α and SAPK2b/p38β, and does not occur in embryonic fibroblasts generated from SAPK2a/p38α-deficient mice. Nogo-B is phosphorylated at Ser107 in vitro by MAPKAP-K2 [MAPK (mitogen-activated protein kinase)-activated protein kinase-2] or MAPKAP-K3, but not by other protein kinases that are known to be activated by SAPK2a/p38α. The anisomycin-induced phosphorylation of Ser107 in HeLa cells can be prevented by ‘knockdown’ of MAPKAP-K2 using siRNA (small interfering RNA). Taken together, our results identify Nogo-B as a new physiological substrate of MAPKAP-K2.
Keywords: MAPK (mitogen-activated protein kinase)-activated protein kinase-K2 (MAPKAP-K2), microtubule, Nogo (neurite outgrowth inhibitor protein), p38 (p38 MAPK)
Abbreviations: CNS, central nervous system; ER, endoplasmic reticulum; GFP, green fluorescent protein; GST, glutathione S-transferase; hnRNP A0, heterogeneous nuclear ribonucleoprotein A0; HSP27, heat-shock protein of 27 kDa; 5-LO, 5-lipoxygenase; LPS, lipopolysaccharide; MAG, myelin-associated glycoprotein; MAPK, mitogen-activated protein kinase; MAPKAP, MAPK-activated protein kinase; MBP, maltose-binding protein; MEF, mouse embryonic fibroblast; MKK6, MAPK kinase 6; MNK1, MAPK integrating kinase-1; MSK, mitogen- and stress-activated protein kinase; NgR, Nogo (neurite outgrowth inhibitor protein) receptor; OMgp, oligodendrocyte myelin glycoprotein; ORF, open reading frame; PDI, protein disulphide-isomerase; PKA, cyclic AMP-dependent protein kinase; SAPK, stress-activated protein kinase; siRNA, small interfering RNA
INTRODUCTION
The family of reticulon proteins encompass more than 300 members that share homology within a C-terminal RHD (reticulon-homology domain) that is involved in their association with the ER (endoplasmic reticulum). The Nogo (neurite outgrowth inhibitor protein) reticulon gene encodes three spliceforms, Nogo-A, Nogo-B and Nogo-C [1,2], which were originally identified as genes encoding the bovine neurite growth inhibitor bNI-220. Nogo-A (and to a lesser extent Nogo-B and Nogo-C), in conjunction with two other inhibitory factors, termed MAG (myelin-associated glycoprotein) and OMgp (oligodendrocyte myelin glycoprotein) was originally suggested to be involved in the inhibition of axonal regeneration when found associated with myelin in the CNS (central nervous system) [1,3,4]. Consistent with such a role, young mice lacking both Nogo-A and Nogo-B were reported to show enhanced axonal regeneration in the CNS [5]. The inhibitory effects on axonal regeneration were attributed to two regions in Nogo-A, one within the N-terminal extension specific to Nogo-A and the other located between the two ER transmembrane domains, which are common to all three Nogo isoforms [1,6]. The latter region, comprising 66 amino acid residues (Nogo-66), is thought to be intraluminal and/or extracellular and, together with MAG and OMgp, to bind to a NgR (Nogo receptor) located on another neuronal cell [7]. It was suggested that the interaction of Nogo-66 with the NgR was responsible for inducing the inhibition of axonal regeneration.
Although young mice lacking both Nogo-A and Nogo-B show enhanced axonal regeneration, this phenotype was lost as the animals aged [5]. Moreover, mice lacking Nogo-A alone, or all three spliceforms, failed to show a significant enhancement of axonal regeneration in two other studies [8,9]. Furthermore, mice lacking the NgR did not show enhanced axonal regeneration [10]. Thus exact roles of the Nogo family in axonal regeneration are unclear. On the other hand, Nogo-B (also called ASY, RTN4-B1, RTN-Xs and Foocen-M) [1,11,12] has been proposed to play different roles, for example in remodelling of the vasculature [13]. It was also implicated in the regulation of apoptosis [11], but more recent work appears to exclude this possibility [14].
In the present study we have unexpectedly discovered that Nogo-B is phosphorylated via the SAPK2 (stress-activated protein kinase 2)/p38 pathway, which plays a key role in triggering the production of pro-inflammatory cytokines and other inflammatory mediators, as well as in the cellular response to environmental stress.
MATERIALS AND METHODS
Materials
Ion-exchange and gel-filtration columns were obtained from Amersham Pharmacia Biotech (Little Chalfont, Bucks., U.K.), human fibronectin was from Sigma (Poole, Dorset, U.K.), Cytochalasin D and nocodazole {methyl[5-(2-thienylcarbonyl)-1H-benzimidazol-2-yl]carbamate} were from Calbiochem (Nottingham, U.K.) and Silencer siRNA (small interfering RNA) Construction Kit was from Ambion (Abingdon, Oxon, U.K.). Peptides were synthesized by Dr Graham Bloomberg (Molecular Recognition Centre, School of Medical Sciences, University of Bristol, U.K.). The sources of all other materials are detailed elsewhere [15].
Cell lysis
The cells were washed twice in ice-cold PBS and lysed in buffer A [50 mM Tris/HCl (pH 7.4)/150 mM KCl/0.1 mM EDTA/1% (w/v) Nonidet P40/4 mM dithiothreitol/20 mM NaF/40 mM sodium β-glycerophosphate/2 mM sodium orthovanadate/1 mM Pefabloc/complete EDTA-free protease inhibitors]. The lysates were centrifuged for 10 min at 13000 g and the supernatants removed, and used immediately or frozen in liquid N2 and stored at −80 °C.
Purification of Nogo-B from RAW and HeLa cells
RAW264 cells were stimulated for 45 min with LPS (lipopoly-saccharide; 50 ng/ml) or HeLa cells were exposed for 15 min to anisomycin (10 μg/ml). After cell lysis, 100 mg of extract protein [approx. (100–200)×107 cells] was desalted on a HiPrep 26/10 Desalting Column in buffer B [50 mM β-glycerophosphate (pH 7.4)/5% (v/v) glycerol/0.1% (v/v) 2-mercaptoethanol/0.03% (w/v) Brij 35/0.1 mM EGTA]. The desalted extracts were applied to a 10 ml column of Source Q (HR 10/10) and developed with a 200 ml linear salt gradient to 0.6 M NaCl in buffer B; fractions (2 ml each) were collected at a flow rate of 1 ml/min. The 43 kDa band recognized by the phospho-specific ‘5-LO’ (human 5-lipoxygenase) antibody was eluted at 0.45 M NaCl. These fractions were pooled, concentrated, and applied to a 24 ml column of Superose 6 (HR10/30) equilibrated in buffer C [20 mM piperazine (pH 5.6)/5% (v/v) glycerol/0.1% (v/v) 2-mercaptoethanol/0.03% (w/v) Brij 35/0.1 mM EGTA]; fractions (0.8 ml each) were collected. The 43 kDa protein was eluted at a position equivalent to a globular protein of molecular mass 600 kDa. The fractions containing this protein were pooled and applied to a 0.24 ml column of Mini Q (PC 3.2/3) equilibrated in buffer C. The column was developed with a 10 ml linear salt gradient from 0.2 to 0.3 M NaCl; fractions (0.05 ml each) were collected at a flow rate of 0.05 ml/min. The fractions containing the 43 kDa protein were pooled and analysed as described in the Results section.
MS
Tryptic peptides were analysed on a Perseptive Biosystems (Framingham, MA, U.S.A.) Elite STR MALDI-TOF (matrix-assisted laser-desorption–time-of-flight) mass spectrometer with saturated α-cyanocinnamic acid as the matrix. The mass spectrum was acquired in the reflector mode and was internally mass-calibrated. The tryptic peptide ions obtained were scanned against the Swiss-Prot and GenPep (Genbank®) databases using the MS-FIT program of the proteomics tool ProteinProspector. Where there was an ambiguity in the identity of a protein sample, liquid chromatography–tandem MS was performed. The tryptic digest was injected on to a 0.075 mm×100 mm PepMap C18 capillary column, equilibrated with 0.1% formic acid in water, attached to a LC-Packings Ultimate HPLC system [Dionex (U.K.) Ltd., Camberley, Surrey, U.K.]. The column was developed with a discontinuous acetonitrile gradient at 0.2 μl/min, and the column was interfaced to a Q-TOF2 mass spectrometer (Micromass, Wythenshaw, Manchester, U.K.). The peptide ions generated by the electrospray interface were fragmented automatically using machine-defined collision voltages. The resultant peak lists were searched using the Sonar search engine (Genomic Solutions, Ann Arbor, MI, U.S.A.) against the NCBInr database.
Plasmids
The first 586 bp of the DNA encoding human Nogo-B were amplified from EST (expressed sequence tag) KIAA0886 (kindly donated by the Kazusa DNA Research Institute, Chiba, Japan) using oligonucleotides 5′Bam-HA-Nogo-B (GGATCCGCCACCATGTACCCATACGATGTGCCAGATTACGCCGAAGACCTGGACCAGTCTCCTCTGGTC) and MP015 (TAATGTCTCTCCAGTACAGGAGGTCAACAACCACTGAGCCCGAGGAGCCCC). The 3′ end of the ORF (open reading frame) (560–1122 bp) was amplified using MP014 (TTGTTGACCTCCTGTACTGGA) and 3′BamH1-Nogo-B (GGATCCTCATTCAGCTTTGCGCTTCAATCCAGGGAT) oligonucleotides. All reactions were carried out using the GC Rich PCR System (Roche Molecular Biochemicals, Lewes, East Sussex, U.K.). A portion (10 ng) of each PCR product was mixed and used as a template for PCR with oligonucleotides 5′BamH1-HA-Nogo-B and 3′BamH1-Nogo-B. The resulting PCR fragment was cloned into pCR2.1 (Invitrogen) and sequenced to produce pCR2.1 HA-Nogo-B. pCR2.1 was digested with BamH1 and ligated into the same site in pGEX6P-1 to produce pGEX-Nogo-B. A mutant in which Ser107 was changed to alanine (pGEX-Nogo-B[S107A]) was produced in a similar manner. The 5′ end of the ORF was amplified using oligonucleotides 5′Bam-HA-Nogo-B and MP136 (GTCGACGACACCGGGCTCGGGTCCCAAGCCGGCTGCCGCTCCGGGGC), whereas the 3′ end was amplified using MP135 (GCCCCGGAGCGGCAGCCGGCTTGGGACCCGAGCCCGGTGTCGTCGAC) and 3′BamH1-Nogo-B. A portion (10 ng) of each overlapping fragment was then used in a second round of PCR using the 5′ and 3′ oligonucleotides. The resulting fragment was cloned into pCR2.1, sequenced, then subcloned into pGEX6P-1 for expression in Escherichia coli as described above, giving rise to pGEX Nogo-B[S107A].
Proteins
All proteins were expressed in E. coli strain BL21. Nogo-B was expressed as a GST (glutathione S-transferase)-fusion protein and the GST moiety cleaved with the PreScission™ Protease (Amersham). SAPK2a/p38α was expressed as an inactive GST-fusion protein and maximally activated with a MBP (maltose-binding protein) fusion of a constitutively active mutant of MKK6 [mitogen-activated protein kinase (MAPK) kinase 6] in which Ser210 and Thr214 were mutated to aspartic acid (‘D’). The [D210,D214]MBP–MKK6 was then removed by passage through amylose resin. GST–MAPKAP-K2 (GST–MAPK-activated protein kinase-2), GST-MAPKAP-K3, His6–MSK1 (hexahistidine–mitogen- and stress-activated protein kinase 1) and GST–MNK1 (GST–MAPK integrating kinase-1) were activated with GST–SAPK2a/p38α and the activating enzyme inhibited by inclusion of 10 μM SB 203580 in subsequent experiments. One unit of protein kinase activity was that amount which catalysed the incorporation of 1 nmol of phosphate into the standard substrate in 1 min. Full details of the expression, activation and assay protocols are given elsewhere [16,17].
Antibodies
An antibody was raised against the phosphopeptide CSLERQLS*LEQEVQ (where the phosphorylated serine residue is represented by ‘S*’), corresponding to residues 266–278 of 5-LO plus an N-terminal cysteine residue. The sequence is identical in the murine enzyme. The peptide was conjugated separately to both BSA and keyhole-limpet haemocyanin via the N-terminal cysteine residue before injection into sheep at Diagnostics Scotland (Edinburgh, Scotland, U.K.). The antisera were affinity purified on antigen-Sepharose [15] and used at 0.1–1.0 μg/ml in the presence of 10 μg/ml of the unphosphorylated peptide antigen. A further antibody that recognizes all forms of Nogo-B was raised by injecting a GST-fusion protein of Nogo-B into sheep and affinity-purifying the antisera followed by passage through a GST column to remove anti-GST antibodies. The characterization of antibodies that immunoprecipitate MAPKAP-K2 or MAPKAP-K3 specifically have been described previously [18]. Two antibodies raised against an N-terminal and a C-terminal sequence of human 5-LO respectively were purchased from Research Diagnostics Inc. (Flanders, NJ, U.S.A.). Rabbit anti-sheep IgG and goat anti-rabbit IgG, both conjugated to horseradish peroxidase, were obtained from Perbio Science Ltd. (Tattenhall, Cheshire, U.K.). Alexa Fluor 594 goat anti-mouse antibody, Alexa Fluor 488 donkey anti-sheep antibody, Alexa Fluor 488 chicken anti-mouse antibody were from Molecular Probes (Leiden, The Netherlands). YL 1/2 antibody (for microtubule staining) was from Sera Lab (Crawley Down, Sussex, U.K.). Anti-PDI [anti-(protein disulphide-isomerase)] antibody (for ER staining) was from Stressgen Biotechnologies (York, U.K.). Fluorescein-conjugated anti-rat IgG was from Lorne Diagnostics (Reading, Berks., U.K.).
siRNA construction and transfection
siRNAs for human MAPKAP-K2 (ACCACCAGCCACAACUCUU) and human MAPKAP-K3 (GGUGGUGAGUUGUUCAGCA) were prepared using the Silencer siRNA Construction Kit [Ambion (Europe) Ltd., Huntingdon, Cambs., U.K.] according to the manufacturer's instructions. Experiments were performed using a strain of easily transfectable HeLa cells, generously provided by Professor Jacques Pouyssegur (Institute of Signaling, Developmental Biology and Cancer Research, CNRS UMR 6543, Nice, France). These cells were transfected twice at 24 h intervals with a 60 nM concentration of the specified siRNA using Oligofectamine™ reagent (Invitrogen) and used 24 h following the second transfection [19]. For transfection of GFP (green fluorescent protein) and GFP-Nogo-B, DNA was transfected 16 h before the assay using FuGENE 6 at a ratio of 3 μl of FuGENE 6/1μg of DNA (Roche).
Immunofluorescence
HeLa cells stained for microtubules were fixed for 10 min at ambient temperature by the addition of 90% (v/v) methanol that had been precooled to −20 °C [20]. Cells were permeabilized with 0.1% (w/v) saponin in PBS, pH 7.4, and stained with the antibodies mentioned in the relevant Figure legends. The cells were mounted using ProLong antifade kit (Molecular Probes Europe BV, Leiden, The Netherlands), imaged on a Leica microscope and the data analysed using the Openlab software (Improvision Ltd, University of Warwick Science Park, Coventry, U.K.).
Cell culture, immunoprecipitation and immunoblotting
The murine RAW264 macrophage cell line, transformed mouse embryonic fibroblasts (MEFs) from wild-type and SAPK2a/p38α-deficient mice [21] and HeLa cells were prepared, maintained and lysed as described in [15]. Immunoblotting was also carried out as described previously using the ECL® (enhanced chemiluminescence; Amersham Pharmacia Biotech) detection system. Details of the immunoprecipitation procedure are given in the appropriate Figure legends.
RESULTS
A 5-LO phospho-specific antibody also recognizes a 43 kDa protein in LPS-stimulated RAW264 cells
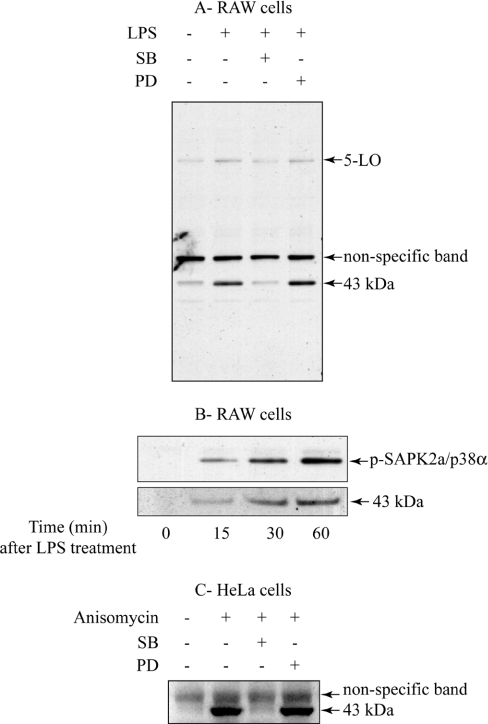
5-LO, a key enzyme involved in the production of leukotrienes from arachidonic acid [22], was reported to be phosphorylated in vitro by MAPKAP-K2, a protein kinase that is activated by SAPK2a/p38α [23]. Those authors pointed out that Ser272 of 5-LO lies within an optimal consensus sequence for phosphorylation by MAPKAP-K2, which is Hyd-Xaa-Arg-Xaa-Xaa-Ser-Hyd, where Hyd is a bulky hydrophobic residue [24]. However, they did not establish that this residue becomes phosphorylated in cells in response to agonists that activate the SAPK2a/p38α pathway. To investigate whether 5-LO is a physiological substrate for MAPKAP-K2, we raised a phospho-specific antibody against a synthetic phosphopeptide corresponding to the sequence surrounding Ser272 of human and murine 5-LO. When RAW264 cell extracts were probed with this antibody, it weakly recognized a 78 kDa (Figure 1A) band that co-migrated with 5-LO, as judged by immunoblotting with two 5-LO-specific antibodies (results not shown). The phosphorylation of this protein increased slightly in response to LPS, which appeared to be decreased by prior treatment of the cells with SB 203580, a specific inhibitor of SAPK2/p38 [25–27], but not PD 184352 (Figure 1A) [17,28]. PD 184352 completely prevents the LPS-induced activation of the classical MAPK pathway in RAW cells at the concentration used [15].
Figure 1. An unknown 43 kDa band is phosphorylated by the SAPK2/p38 pathway.
(A) RAW cells were left untreated or pre-treated for 15 min with 10 μM SB 203580 (‘SB’) or 10 μM PD 184352 (‘PD’) and then stimulated for 45 min with 50 ng/ml LPS. Lysates were separated by SDS/PAGE, transferred to nitrocellulose and immunoblotted with the 5-LO phospho-specific antibody. (B) RAW cells were left untreated or incubated with 50 ng/ml LPS for the times indicated. Lysates were denatured in SDS, followed by SDS/PAGE and immunoblotting with the 5-LO phospho-specific antibody or an antibody that recognizes phosphorylated SAPK2a/p38α (pSAPK2a/p38α). (C) HeLa cells were left untreated or pretreated for 15 min with 10 μM SB 203580 (‘SB’) or 10 μM PD 184352 (‘PD’) and then stimulated for 15 min with 10 μg/ml anisomycin. Lysates were processed as described in (A). Similar results were obtained in at least six independent experiments.
Interestingly, the 5-LO-phospho-specific antibody recognized an additional 43 kDa band much more strongly than 5-LO. The phosphorylation of this protein increased strikingly in response to LPS (Figure 1B), and was suppressed by SB 203580, but not by PD 184352 (Figure 1A). The same protein became phosphorylated in HeLa cells exposed to the protein-synthesis inhibitor anisomycin, another activator of the SAPK2a/p38α pathway (Figure 1C). The 43 kDa protein was not recognized by either of the two 5-LO antibodies, one of which was raised against an N-terminal peptide and the other against a C-terminal peptide (results not shown). These results suggested that the 43 kDa band was not a proteolytic fragment of 5-LO, but a novel physiological substrate of the SAPK2a/p38α pathway.
Identification of the 43 kDa band as Nogo-B
We purified the 43 kDa band from LPS-stimulated RAW cells and anisomycin-treated HeLa cells using the phospho-specific antibody to monitor its elution position after ion-exchange chromatography and gel-filtration (see the Materials and methods section). Both preparations contained two protein-staining bands with molecular masses close to 43 kDa, the lower co-migrating with the immunoreactive species (results not shown). The upper band was identified as actin (results not shown), whereas the lower band was a Nogo spliceform (Table 1). On the basis of the molecular mass of the protein (43 kDa), it would appear that this species is Nogo-B and not the far larger (140 kDa) Nogo-A. The Nogo-C spliceform does not contain the peptides corresponding to residues 105–110 or 92–104 that were detected by MS (Table 1).
Table 1. Identification of the 43 kDa band as Nogo-B.
The mass of tryptic peptides were scanned against the Swiss-Prot, Genpep and NCBInr databases as previously described [15].
| Masses | Residue no. | ||||
|---|---|---|---|---|---|
| Source of purified protein | Submitted | Matched | Start | End | Peptide sequence |
| Murine RAW cells | 802.3858 | 802.3848 | 105 | 110 | QPSWER |
| 944.4238 | 944.4226 | 235 | 242 | SDEGHPFR | |
| 1450.7661 | 1450.7694 | 168 | 180 | GSGSVVVDLLYWR | |
| 1490.7506 | 1490.7603 | 259 | 272 | YSNSALGHVNSTIK | |
| Human HeLa cells | 944.4354 | 944.4226 | 248 | 255 | SDEGHPFR |
| 1271.7121 | 1271.7112 | 92 | 104 | GPLPAAPPVAPER | |
| 1537.7695 | 1537.8015 | 180 | 193 | GSSGSVVVDLLYWR | |
| 1607.8362 | 1607.8264 | 339 | 352 | HQAQIDHYLGLANK | |
| 1807.9317 | 1807.9329 | 256 | 271 | AYLESEVAISEELVQK | |
| 3271.6215 | 3271.6938 | 57 | 91 | KPAAGLSAAPVPTAPAAGAPLMDFGNDFFVPPAPR | |
Nogo-B is phosphorylated by MAPKAP kinases in vitro
We found that SAPK2a/p38α did not phosphorylate Nogo-B in vitro, indicating that a distinct SAPK2a/p38α-activated protein kinase was responsible for the phosphorylation of Nogo-B in cells. We therefore phosphorylated Nogo-B with protein kinases known to be activated by SAPK2a/p38α, namely MAPKAP-K2, MAPKAP-K3, MSK1 and MNK1 (reviewed in [29,30]). Under these conditions, MAPKAP-K2 phosphorylated Nogo-B to 0.85 mol of phosphate/mol of protein, and MAPKAP-K3 to 0.5 mol of phosphate/mol of protein. By contrast, there was only slight phosphorylation by MSK1 and no phosphorylation by MNK1 (results not shown).
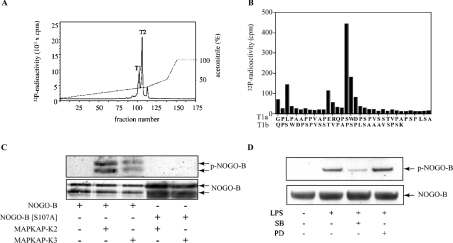
Nogo-B phosphorylated by MAPKAP-K2 in vitro was digested with trypsin and chromatographed on a C18 column, which resolved two major 32P-labelled peaks, termed T1 and T2 (Figure 2A). Edman sequencing showed that T1 comprised a major phosphopeptide T1a, starting at residue 92, and a minor phosphopeptide, T1b, starting at residue 105. A small amount of 32P radioactivity was released after the third cycle of Edman degradation and a much larger amount after the sixteenth cycle (Figure 2B). The results demonstrate that there is a single site of phosphorylation at Ser107, T1a arising from incomplete tryptic cleavage of the Arg–Gln bond between residues 104 and 105. Peak T2 was almost exclusively peptide T1a, the site of phosphorylation again being Ser107 (results not shown).
Figure 2. Nogo-B is phosphorylated at Ser107 by MAPKAP-K2.
(A) Nogo-B (1 μM) was phosphorylated for 60 min with 10 units/ml active MAPKAP-K2, 10 mM MgCl2 and 0.1 mM [γ-32P]ATP and subjected to SDS/PAGE. The band corresponding to Nogo-B was excised, digested with trypsin and the resulting peptides separated by chromatography on a C18 column equilibrated in 0.1% (v/v) trifluoroacetic acid. 32P radioactivity is shown by the continuous line and the acetonitrile gradient by the broken line. (B) The peak T1 in (A) was shown by Edman sequencing to be a mixture of two peptides. The major peptide T1a started at residue 92 and the minor peptide T1b at residue 105. Solid-phase sequencing [40] identified Ser107 as the site of phosphorylation in both peptides. (C) Bacterially expressed Nogo-B or Nogo-B[S107A] were left unphosphorylated (−) or phosphorylated for 60 min with 1 unit/ml MAPKAP-K2 or MAPKAP-K3 (+) and Mg-[γ32-P]ATP. The proteins were denatured in SDS, subjected to SDS/PAGE and immunoblotted with the 5-LO phospho-specific antibody (‘p-Nogo-B’) as described in Figure 1 or with an antibody that recognizes all forms of Nogo-B (‘Nogo-B’). In contrast with the endogenous Nogo-B in cells, bacterially expressed Nogo-B migrates as two major bands, the lower generated by proteolysis near the C-terminus. (D) RAW cells were left untreated or incubated for 15 min with 5 μM SB 203580 or 10 μM PD 184352 and then stimulated for 45 min with 50 ng/ml LPS. Nogo-B was immunoprecipitated from the lysates, using 1 μg of anti-Nogo-B per mg of cell lysate, then denatured in SDS, followed by immunoblotting with the 5-LO phospho-specific antibody (‘p-Nogo-B’) as described in Figure 1 or with an antibody that recognizes all forms of Nogo-B (‘Nogo-B’). Similar results were obtained in two independent experiments.
In order to check that no other phosphorylation site had been missed, for example as a result of another 32P peptide precipitating on the C18 column, we mutated Ser107 to alanine and expressed the mutant Nogo-B in E. coli. In contrast with wild-type Nogo-B, Nogo-B[S107A] was not phosphorylated by MAPKAP-K2 (results not shown), confirming Ser107 as the only site of phosphorylation for MAPKAP-K2 in vitro.
The 5′-LO phospho-specific antibody recognizes Nogo-B phosphorylated at Ser107
The amino acid sequence surrounding Ser107 (indicated by ‘S*’) of human Nogo-B (PVAPERQPS*WDPSPV) resembles that surrounding Ser272 of human 5′-LO (SLRERQLS*LEQEVQ), suggesting that Ser107 might be the residue in Nogo-B that becomes phosphorylated in LPS-stimulated RAW264 cells and is recognized by the 5-LO phospho-specific antibody. This was established by incubating wild-type Nogo-B and Nogo-B[S107A] with MAPKAP-K2 or MAPKAP-K3 in vitro and then subjecting each protein to immunoblotting using the 5′-LO phospho-specific antibody (Figure 2C).
To establish that the 43 kDa band detected in extracts from LPS-stimulated RAW264 cells really was Nogo-B, we repeated the experiment reported in Figure 1, except that Nogo-B was first immunoprecipitated from the lysates before immunoblotting with the 5-LO phospho-specific antibody. The same result was obtained, establishing that the 43 kDa protein is indeed Nogo-B (Figure 2D).
Phosphorylation of Nogo-B in transformed MEFs from wild-type and SAPK2a/p38α-deficient mice
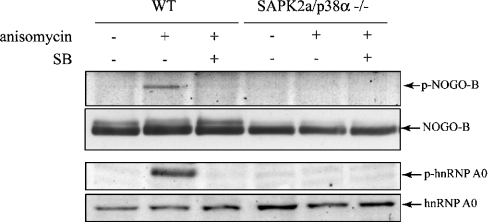
To check that the suppression of Nogo-B phosphorylation by SB 203580 resulted from the inhibition of SAPK2a/p38α and not from the inhibition of the closely related SAPK2b/p38β or the non-specific inhibition of another protein kinase, we also examined whether Nogo-B was phosphorylated in SAPK2a/p38α-deficient MEFs. In MEFs from wild-type mice, Nogo-B became phosphorylated at Ser107 in response to anisomycin, and this was prevented by SB 203580, but not by PD 184352. No phosphorylation of Nogo-B at Ser107 occurred in the SAPK2a/p38α-deficient MEFs (Figure 3) [31]. Similar results were obtained in control experiments using a phosphospecific antibody that recognizes hnRNP A0 (heterogeneous nuclear ribonucleoprotein A0) phosphorylated at Ser84 (Figure 3), an authentic physiological substrate for MAPKAP-K2 [15]. This is consistent with the finding that, in these MEFs, no MAPKAP-K2 activity is discernible, suggesting that SAPK2b/p38β cannot compensate for the absence of SAPK2a/p38α in activating this enzyme in fibroblasts [31].
Figure 3. Nogo-B is phosphorylated at Ser107 in cells from wild-type mice, but not in mice deficient in SAPK2a/p38α.
Wild-type (WT) and SAPK2a/p38α−/− MEFs were left untreated (−) or preincubated for 15 min with 5 μM SB 203580 (‘SB’; +) and then incubated for 30 min without (−) or with (+) 10 μg/ml anisomycin. Nogo-B was immunoprecipitated from the lysates and denatured in SDS. After separation by SDS/PAGE and transfer to nitrocellulose, the membrane was probed with the 5-LO phospho-specific antibody (‘p-Nogo-B’) as described in Figure 1 or with an antibody that recognizes all forms of Nogo-B (‘Nogo-B’). Another aliquot of the cell lysate was immunoblotted (without immunoprecipitation) with a phospho-specific antibody that recognizes hnRNP A0 phosphorylated at Ser84 (‘p-hnRNP A0’) or with an antibody that recognizes all forms of hnRNP A0 (‘hnRNP A0’). Similar results were obtained in two independent experiments.
The phosphorylation of Nogo-B is abolished by siRNA against MAPKAP-K2 and MAPKAP-K3
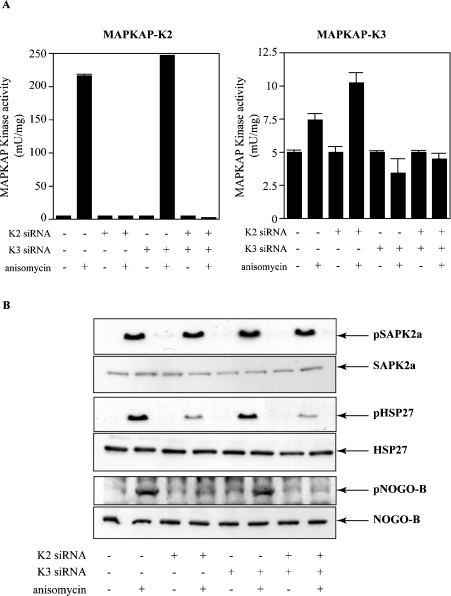
We made use of siRNA technology to investigate which protein kinases activated by SAPK2a/p38α were responsible for phosphorylating Nogo-B in cells. After treating HeLa cells with siRNAs against MAPKAP-K2 and MAPKAP-K3 alone or in combination, we immunoprecipitated each protein kinase from the extracts of anisomycin-stimulated cells and assayed their activity (Figure 4A). These results demonstrated that siRNA against MAPKAP-K2, but not siRNA against MAPKAP-K3, effectively blocked MAPKAP-K2 activity, whereas siRNA against MAPKAP-K3, but not siRNA against MAPKAP-K2, effectively blocked MAPKAP-K3 activity. It should be noted that the MAPKAP-K2 activity in these cell extracts greatly exceeded that of MAPKAP-K3 (note the 20-fold difference in scale of the ordinates in the left- and right-hand panels of Figure 4A).
Figure 4. Knock-down of MAPKAP-K2 and MAPKAP-K3 using siRNA prevents the phosphorylation of Nogo-B at Ser107.
HeLa cells were either mock-transfected (−) or transfected with siRNA against MAPKAP-K2, siRNA against MAPKAP-K3 or siRNA against both MAPKAP-K2 and MAPKAP-K3 (+) as indicated. (A) The cells were then left untreated or exposed for 30 min to 10 μg/ml anisomycin. MAPKAP-K2 (‘K2’) or MAPKAP-K3 (‘K3’) were immunoprecipitated from HeLa cell extracts as described [18] and assayed. Activity is shown in units/mg of cell extract. (B) The HeLa cell extracts from (A) (30 μg of protein) were immunoblotted with antibodies that recognize phosphorylated SAPK2a/p38α (‘pSAPK2a’), all forms of SAPK2a/p38α (‘SAPK2a’), HSP27 phosphorylated at Ser15 (‘pHSP27’), all forms of HSP27 (HSP27) or with the 5-LO phospho-specific antibody (‘p-Nogo-B’) and antibodies that recognize all forms of Nogo-B (‘Nogo-B’). Similar results were obtained in three independent experiments.
The next step was to verify that these siRNAs could block the phosphorylation of known substrates without affecting other pathways. Anisomycin treatment of HeLa cells triggered the phosphorylation of HSP27 (heat-shock protein of 27 kDa) at Ser15, a downstream target of MAPKAP-K2, which was greatly decreased in cells transfected with MAPKAP-K2 siRNA alone or in combination with the MAPKAP-K3 siRNA (Figure 4B). Similar results were obtained when the membranes were immunoblotted with a phospho-specific antibody that recognizes hnRNP A0 phosphorylated at Ser84 (results not shown). In contrast, the anisomycin-stimulated SB 203580-sensitive phosphorylation of CREB (cyclic AMP-response element binding protein), which is mediated by MSK1/MSK2 [32], was unaffected by siRNA knockdown of MAPKAP-K2 and/or MAPKAP-K3. Moreover, the anisomycin-induced phosphorylation of SAPK2a/p38α (Figure 4B) and JNK (c-Jun N-terminal kinase) (results not shown) were also unaffected, indicating that the siRNAs had not affected the activation of SAPK2a/p38α or another downstream element of the pathway.
Interestingly, the siRNA against MAPKAP-K2 greatly decreased the anisomycin-stimulated phosphorylation of Nogo-B at Ser107 (Figure 4B), consistent with the much higher level of MAPKAP-K2 relative to MAPKAP-K3.
Nogo-B is associated with microtubules in cells
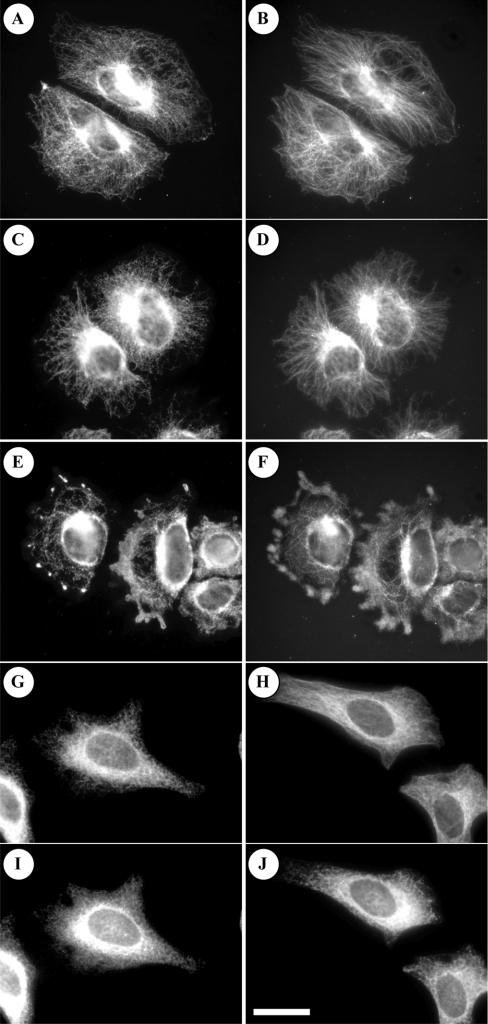
Nogo-B, like other members of the Nogo family, associates with the ER (see the Introduction). To visualize this, we stained HeLa cells with an antibody raised against the full-length Nogo-B protein. This revealed that endogenous Nogo-B forms a fibrillar pattern (Figure 5A), strikingly similar to the arrangement of α-tubulin in the same cells (Figure 5B). Disrupting the actin cytoskeleton with cytochalasin D did not destroy the fibrillar pattern of either Nogo-B or tubulin (Figures 5C and 5D), but incubation with nocodazole to collapse the microtubules did disrupt the staining pattern of either protein (Figures 5E and 5F). This could be rescued by washing away the nocodazole and letting the microtubules repolymerize (results not shown). A similar distribution of Nogo-B was observed when a GFP–Nogo-B fusion protein was transfected into the cells (results not shown), indicating that the observed localization of Nogo-B was not due to lack of specificity of the anti-Nogo-B antibody. The localization of both Nogo-B (Figure 5G) and microtubules (Figure 5H) is also consistent with that of the ER, as illustrated by staining with an antibody that recognizes PDI and calreticulin markers for the ER (Figures 5I and 5J).
Figure 5. Nogo-B co-localizes with microtubules.
HeLa cells were plated on Lab-Tek chamber slides (Nunc, Roskilde, Denmark) coated with fibronectin (5 μg/ml), left to adhere for 1 h and then incubated for 30 min without (A, B, G, H, I and J) or with 10 μM cytochalasin D (C and D) or 5 μM nocodazole (E and F). Nogo-B was visualized using the antibody that recognizes all forms of Nogo-B equally well coupled to anti-sheep IgG (A, C, E and G), whereas microtubules were visualized with an anti-(tyrosinated α-tubulin) antibody (YL-1/2) coupled to anti-rat IgG (B, D, F and H). The ER was visualized with an antibody that recognizes PDI and calreticulin coupled to anti-mouse IgG (I and J). The Figure shows that Nogo-B co-localizes with microtubules. Similar results were obtained in four (A and B) or two (C–J) independent experiments.
DISCUSSION
In the present study we identified Nogo-B as a new physiological substrate for MAPKAP-K2. Interestingly, the phosphorylation occurred at Ser107, which does not lie in the conventional MAPKAP-K2 consensus sequence. Studies with synthetic peptide substrates have revealed that the optimal consensus sequence for MAPKAP-K2 is Hyd-Xaa-Arg-Xaa-Xaa-Ser-Hyd, where Hyd is a bulky hydrophobic residue (typically leucine or phenylalanine). In Nogo-B the large hydrophobic residue five residues N-terminal to Ser107 (termed the ‘n−5’ position) is replaced by proline. A short peptide containing proline at n−5 was phosphorylated far less efficiently by MAPKAP-K2 in previously described work [24]. Nevertheless, the Nogo-B protein is phosphorylated by MAPKAP-K2 at a similar rate to HSP27 in vitro (results not shown), indicating that another residue(s) in the protein is able to compensate for the lack of leucine or phenylalanine at n−5. Thus physiological substrates of protein kinases cannot always be identified simply by searching databases with optimal consensus sequences for phosphorylation. Similar observations have been made for other protein kinases. For example TAB1, a regulatory subunit of TAK1 (transforming-growth-factor-β-activated kinase 1), is phosphorylated by the proline-directed protein kinase SAPK2a/p38α at Ser423 in vivo, even though this residue is followed not by proline but by alanine [21]. Similarly, PKA (cyclic AMP-dependent protein kinase) phosphorylates inhibitor-1 (an inhibitor of protein phosphatase-1) at Thr46 in vivo, and at similar rates to other physiological substrates, even though the substitution by threonine of serine prevents the phosphorylation of small synthetic peptides by PKA [33].
In the present study we found that Nogo-B was associated with microtubules (Figure 5), an observation consistent with the reported association of the ER with microtubules [34] and the localization of Nogo spliceforms to the ER (see the Introduction). A small proportion of each Nogo spliceform is also reported to be localized at the plasma membrane, where Nogo-66 is thought to be extracellular and to interact with the NgR (see the Introduction). Another model in which the region N-terminal to the Nogo-66 sequence (equivalent to residues 1–184 of Nogo-B) is extracellular has been proposed [35]. However, our finding that Ser107 is located in this region makes this less likely, because Ser107 must presumably be intracellular at the time it is phosphorylated by MAPKAP-K2. Thus our data support a model in which Nogo-B is mainly associated with the ER. Its phosphorylation is likely to occur at this location, since the extent of phosphorylation of Nogo-B by MAPKAP-K2 in cells is substantial (greater than 50%), as judged by an upward bandshift that is prevented by SB 203580 (results not shown); moreover, no discernible change in the staining pattern of Nogo-B is detected following stimulation with agonists that activate MAPKAP-K2 (results not shown).
The Nogo family of proteins has generated considerable interest in recent years, but whether the major role of Nogo is to regulate axonal regeneration, remodel the vasculature or some other process is still unclear. In the present paper we have introduced an additional level of complexity into this story by identifying Nogo-B as a new physiological substrate for MAPKAP-K2 that is likely to be involved in regulating one or more of its biological functions. Interestingly Ser107 is conserved in Nogo-A but not in Nogo-C.
The identification of Nogo-B as a substrate of MAPKAP-K2 extends the list of well-authenticated substrates for this protein kinase, which include HSP27 [36], hnRNP A0 [15], CAPZIP (CapZ-interacting protein) [37] and LSP1 (leucocyte-specific protein 1) [38]. The phosphorylation of three of these substrates appears to regulate actin dynamics and, hence, processes such as cell motility (discussed in [37]). By contrast, the phosphorylation of hnRNP A0 appears to enhance its interaction with the AU-rich elements that confer instability to the mRNAs encoding several pro-inflammatory cytokines [15] and may participate in the MAPKAP-K2-mediated increase in the production of these substances [39]. However, how phosphorylation modifies the function of Nogo-B (and perhaps Nogo-A) and mediates one or more MAPKAP-K2-dependent processes is unknown. The answer to this question will have to await more definitive information about the biological roles of this intriguing protein.
Acknowledgments
We thank the Protein Production and Antibody Purification teams, Division of Signal Transduction Therapy, University of Dundee (co-ordinated by Hilary McLauchlan and James Hastie) for expression and purification of proteins and affinity purification of antibodies, the Sequencing Service, School of Life Sciences, University of Dundee (www.dnaseq.co.uk), for DNA sequencing, and Leanne Brown for tissue-culture support. S.R. acknowledges receipt of a Senior Postdoctoral Fellowship from the Canadian Institute of Health Research. We are grateful to the UK Medical Research Council, The Royal Society, AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Merck and Co., Merck KGaA and Pfizer for financial support.
References
- 1.Chen M. S., Huber A. B., van der Haar M. E., Frank M., Schnell L., Spillmann A. A., Christ F., Schwab M. E. Nogo-A is a myelin-associated neurite outgrowth inhibitor and an antigen for monoclonal antibody IN-1. Nature (London) 2000;403:434–439. doi: 10.1038/35000219. [DOI] [PubMed] [Google Scholar]
- 2.Fournier A. E., Strittmatter S. M. Repulsive factors and axon regeneration in the CNS. Curr. Opin. Neurobiol. 2001;11:89–94. doi: 10.1016/s0959-4388(00)00178-1. [DOI] [PubMed] [Google Scholar]
- 3.McKerracher L., Winton M. J. Nogo on the go. Neuron. 2002;36:345–348. doi: 10.1016/s0896-6273(02)01018-8. [DOI] [PubMed] [Google Scholar]
- 4.Spillmann A. A., Bandtlow C. E., Lottspeich F., Keller F., Schwab M. E. Identification and characterization of a bovine neurite growth inhibitor (bNI-220) J. Biol. Chem. 1998;273:19283–19293. doi: 10.1074/jbc.273.30.19283. [DOI] [PubMed] [Google Scholar]
- 5.Kim J. E., Li S., GrandPre T., Qiu D., Strittmatter S. M. Axon regeneration in young adult mice lacking Nogo-A/B. Neuron. 2003;38:187–199. doi: 10.1016/s0896-6273(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 6.GrandPre T., Nakamura F., Vartanian T., Strittmatter S. M. Identification of the Nogo inhibitor of axon regeneration as a Reticulon protein. Nature (London) 2000;403:439–444. doi: 10.1038/35000226. [DOI] [PubMed] [Google Scholar]
- 7.Fournier A. E., GrandPre T., Strittmatter S. M. Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature (London) 2001;409:341–346. doi: 10.1038/35053072. [DOI] [PubMed] [Google Scholar]
- 8.Simonen M., Pedersen V., Weinmann O., Schnell L., Buss A., Ledermann B., Christ F., Sansig G., van der Putten H., Schwab M. E. Systemic deletion of the myelin-associated outgrowth inhibitor Nogo-A improves regenerative and plastic responses after spinal cord injury. Neuron. 2003;38:201–211. doi: 10.1016/s0896-6273(03)00226-5. [DOI] [PubMed] [Google Scholar]
- 9.Zheng B., Ho C., Li S., Keirstead H., Steward O., Tessier-Lavigne M. Lack of enhanced spinal regeneration in Nogo-deficient mice. Neuron. 2003;38:213–224. doi: 10.1016/s0896-6273(03)00225-3. [DOI] [PubMed] [Google Scholar]
- 10.Zheng B., Atwal J., Ho C., Case L., He X. L., Garcia K. C., Steward O., Tessier-Lavigne M. Genetic deletion of the Nogo receptor does not reduce neurite inhibition in vitro or promote corticospinal tract regeneration in vivo. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1205–1210. doi: 10.1073/pnas.0409026102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q., Qi B., Oka K., Shimakage M., Yoshioka N., Inoue H., Hakura A., Kodama K., Stanbridge E. J., Yutsudo M. Link of a new type of apoptosis-inducing gene ASY/Nogo-B to human cancer. Oncogene. 2001;20:3929–3936. doi: 10.1038/sj.onc.1204536. [DOI] [PubMed] [Google Scholar]
- 12.Tagami S., Eguchi Y., Kinoshita M., Takeda M., Tsujimoto Y. A novel protein, RTN-XS, interacts with both Bcl-XL and Bcl-2 on endoplasmic reticulum and reduces their anti-apoptotic activity. Oncogene. 2000;19:5736–5746. doi: 10.1038/sj.onc.1203948. [DOI] [PubMed] [Google Scholar]
- 13.Acevedo L., Yu J., Erdjument-Bromage H., Miao R. Q., Kim J. E., Fulton D., Tempst P., Strittmatter S. M., Sessa W. C. A new role for Nogo as a regulator of vascular remodeling. Nat. Med. 2004;10:382–388. doi: 10.1038/nm1020. [DOI] [PubMed] [Google Scholar]
- 14.Oertle T., Merkler D., Schwab M. E. Do cancer cells die because of Nogo-B? Oncogene. 2003;22:1390–1399. doi: 10.1038/sj.onc.1206278. [DOI] [PubMed] [Google Scholar]
- 15.Rousseau S., Morrice N., Peggie M., Campbell D. G., Gaestel M., Cohen P. Inhibition of SAPK2a/p38 prevents hnRNP A0 phosphorylation by MAPKAP-K2 and its interaction with cytokine mRNAs. EMBO J. 2002;21:6505–6514. doi: 10.1093/emboj/cdf639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bain J., McLauchlan H., Elliott M., Cohen P. The specificities of protein kinase inhibitors: an update. Biochem. J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies S. P., Reddy H., Caivano M., Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clifton A. D., Young P. R., Cohen P. A comparison of the substrate specificity of MAPKAP kinase-2 and MAPKAP kinase-3 and their activation by cytokines and cellular stress. FEBS Lett. 1996;392:209–214. doi: 10.1016/0014-5793(96)00816-2. [DOI] [PubMed] [Google Scholar]
- 19.Berra E., Benizri E., Ginouves A., Volmat V., Roux D., Pouyssegur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prescott A. R., Vestberg M., Warn R. M. Microtubules rich in modified α-tubulin characterize the tail processes of motile fibroblasts. J. Cell Sci. 1989;94:227–236. doi: 10.1242/jcs.94.2.227. [DOI] [PubMed] [Google Scholar]
- 21.Cheung P. C., Campbell D. G., Nebreda A. R., Cohen P. Feedback control of the protein kinase TAK1 by SAPK2a/p38alpha. EMBO J. 2003;22:5793–5805. doi: 10.1093/emboj/cdg552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radmark O. Basel: Birkhauser; 1999. 5-Lipoxygenase. [Google Scholar]
- 23.Werz O., Klemm J., Samuelsson B., Radmark O. 5-Lipoxygenase is phosphorylated by p38 kinase-dependent MAPKAP kinases. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5261–5266. doi: 10.1073/pnas.050588997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stokoe D., Caudwell B., Cohen P. T., Cohen P. The substrate specificity and structure of mitogen-activated protein (MAP) kinase-activated protein kinase-2. Biochem. J. 1993;296:843–849. doi: 10.1042/bj2960843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beyaert R., Cuenda A., Vanden Berghe W., Plaisance S., Lee J. C., Haegeman G., Cohen P., Fiers W. The p38/RK mitogen-activated protein kinase pathway regulates interleukin-6 synthesis response to tumor necrosis factor. EMBO J. 1996;15:1914–1923. [PMC free article] [PubMed] [Google Scholar]
- 26.Cuenda A., Rouse J., Doza Y. N., Meier R., Cohen P., Gallagher T. F., Young P. R., Lee J. C. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 27.Lee J. C., Laydon J. T., McDonnell P. C., Gallagher T. F., Kumar S., Green D., McNulty D., Blumenthal M. J., Heys J. R., Landvatter S. W., et al. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature (London) 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 28.Sebolt-Leopold J. S., Dudley D. T., Herrera R., Van Becelaere K., Wiland A., Gowan R. C., Tecle H., Barrett S. D., Bridges A., Przybranowski S., Leopold W. R., Saltiel A. R. Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo. Nat. Med. 1999;5:810–816. doi: 10.1038/10533. [DOI] [PubMed] [Google Scholar]
- 29.Arthur J. S. C., Cohen P. Sigma-RBI Handbook. 4th edn. Gillingham, Dorset: Sigma–Aldrich; 2001. MAPK-activated protein kinases; pp. 174–175. [Google Scholar]
- 30.Cohen P. The search for physiological substrates of MAP and SAP kinases in mammalian cells. Trends Cell Biol. 1997;7:353–360. doi: 10.1016/S0962-8924(97)01105-7. [DOI] [PubMed] [Google Scholar]
- 31.Adams R. H., Porras A., Alonso G., Jones M., Vintersten K., Panelli S., Valladares A., Perez L., Klein R., Nebreda A. R. Essential role of p38α MAP kinase in placental but not embryonic cardiovascular development. Mol. Cell. 2000;6:109–116. [PubMed] [Google Scholar]
- 32.Wiggin G. R., Soloaga A., Foster J. M., Murray-Tait V., Cohen P., Arthur J. S. MSK1 and MSK2 are required for the mitogen- and stress-induced phosphorylation of CREB and ATF1 in fibroblasts. Mol. Cell Biol. 2002;22:2871–2881. doi: 10.1128/MCB.22.8.2871-2881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen P. The role of cyclic AMP-dependent protein kinase in the regulation of glycogen metabolism in mammalian skeletal muscle. Curr. Top. Cell Regul. 1978;14:117–196. doi: 10.1016/b978-0-12-152814-0.50008-3. [DOI] [PubMed] [Google Scholar]
- 34.Klopfenstein D. R., Kappeler F., Hauri H. P. A novel direct interaction of endoplasmic reticulum with microtubules. EMBO J. 1998;17:6168–6177. doi: 10.1093/emboj/17.21.6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dodd D. A., Niederoest B., Bloechlinger S., Dupuis L., Loeffler J. P., Schwab M. E. Nogo-A, -B, and -C are found on the cell surface and interact together in many different cell types. J. Biol. Chem. 2005;280:12494–12502. doi: 10.1074/jbc.M411827200. [DOI] [PubMed] [Google Scholar]
- 36.Stokoe D., Engel K., Campbell D. G., Cohen P., Gaestel M. Identification of MAPKAP kinase 2 as a major enzyme responsible for the phosphorylation of the small mammalian heat shock proteins. FEBS Lett. 1992;313:307–313. doi: 10.1016/0014-5793(92)81216-9. [DOI] [PubMed] [Google Scholar]
- 37.Eyers C. E., McNeill H., Knebel A., Morrice N., Arthur S. J., Cuenda A., Cohen P. The phosphorylation of CapZ-interacting protein (CapZIP) by stress-activated protein kinases triggers its dissociation from CapZ. Biochem. J. 2005;389:127–135. doi: 10.1042/BJ20050387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang C. K., Zhan L., Ai Y., Jongstra J. LSP1 is the major substrate for mitogen-activated protein kinase-activated protein kinase 2 in human neutrophils. J. Biol. Chem. 1997;272:17–19. doi: 10.1074/jbc.272.1.17. [DOI] [PubMed] [Google Scholar]
- 39.Kotlyarov A., Neininger A., Schubert C., Eckert R., Birchmeier C., Volk H. D., Gaestel M. MAPKAP kinase 2 is essential for LPS-induced TNF-α biosynthesis. Nat. Cell Biol. 1999;1:94–97. doi: 10.1038/10061. [DOI] [PubMed] [Google Scholar]
- 40.Stokoe D., Campbell D. G., Nakielny S., Hidaka H., Leevers S. J., Marshall C., Cohen P. MAPKAP kinase-2; a novel protein kinase activated by mitogen-activated protein kinase. EMBO J. 1992;11:3985–3994. doi: 10.1002/j.1460-2075.1992.tb05492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]