Abstract
Steroid hormones are important endocrine signalling molecules controlling reproduction, development, metabolism, salt balance and specialized cellular responses, such as inflammation and immunity. They are lipophilic in character and act by binding to intracellular receptor proteins. These receptors function as ligand-activated transcription factors, switching on or off networks of genes in response to a specific hormone signal. The receptor proteins have a conserved domain organization, comprising a C-terminal LBD (ligand-binding domain), a hinge region, a central DBD (DNA-binding domain) and a highly variable NTD (N-terminal domain). The NTD is structurally flexible and contains surfaces for both activation and repression of gene transcription, and the strength of the transactivation response has been correlated with protein length. Recent evidence supports a structural and functional model for the NTD that involves induced folding, possibly involving α-helix structure, in response to protein–protein interactions and structure-stabilizing solutes.
Keywords: AF1 transactivation domain, allosteric regulation, protein–nucleic acid interaction, protein–protein interaction, post-translational modification, secondary structure, steroid receptor
Abbreviations: AF, activation function; AR, androgen receptor; CBP, CREB (cAMP-response-element-binding protein)-binding protein; DBD, DNA-binding domain; ERα/β, oestrogen receptor α and β respectively; ERE, oestrogen-response element; FLASH, Fas-associated huge protein; FTIR spectroscopy, Fourier-transform infrared spectroscopy; GR, glucocorticoid receptor; GRIP1, glucocorticoid receptor-interacting protein 1; GTF, general transcription factor; HCA, hydrophobic cluster analysis; IF, inhibitory function; LBD, ligand-binding domain; MAPK, mitogen-activated protein kinase; MR, mineralocorticoid receptor; NCoR, nuclear receptor co-repressor; NTD, N-terminal domain; PIAS, protein inhibitor of activated STAT; PIC, pre-initiation complex; PKB, protein kinase B; PR, progesterone receptor; PSA, prostate-specific antigen; SC, synergy control; SHR, steroid hormone receptor; SMRT, silencing mediator of retinoid and thyroid hormone receptor; SRC-1, steroid receptor co-activator 1; TBP, TATA-binding protein; TFE, trifluoroethanol; TIF2, transcription intermediary factor 2; TMAO, trimethylamine-N-oxide
INTRODUCTION
It is 25 years since the cDNAs for the glucocorticoid [1,2] and oestrogen [3] receptors were isolated. This led to the cloning of cDNAs for other steroid receptors, as well as receptors for non-steroid ligands and a large number of orphans, and resulted in the creation of the superfamily of nuclear receptors (reviewed in [4–6]). The availability of steroid receptor cDNAs also opened up new opportunities for investigating receptor protein structure and mechanisms of action, and the genetic dissection of hormone physiology.
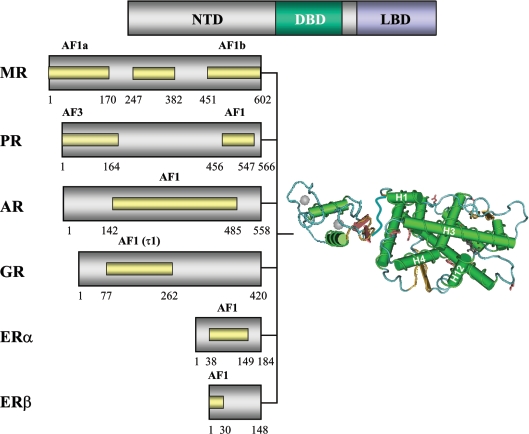
Steroid hormones and non-steroid ligands are important endocrine-signalling molecules, which control reproduction, development, metabolism, salt balance and specialized cellular responses, such as inflammation and immunity. Although the known hormones and ligands for nuclear receptors are structurally and/or chemically distinct, they share a general lipophilic character, and strikingly the individual receptor proteins show some remarkable conservation in terms of domain organization and structure. SHRs (steroid hormone receptors) are intracellular proteins, which have a well-defined domain organization consisting of a C-terminal LBD (ligand-binding domain), linked via a hinge region to a DBD (DNA-binding domain), which is followed by a variable NTD (N-terminal domain; Figure 1). The LBD binds both agonists and antagonists, and retains variable amino acid identity between receptors (approx. 20–50%). The LBD for all steroid receptors is folded into ‘12’ α-helices arranged to form three separate helical sheets, with helices 3, 4 and 12 integral to ligand binding (Figure 1). In addition, a glutamic acid residue in helix 12 and a lysine residue in helix 3, together with a hydrophobic pocket on the surface of the LBD made up of residues from helices 3, 4 and 5, are important for protein–protein interactions and represent a ligand-dependent transactivation domain, termed AF2 (activation function 2) [7]. High-resolution structures are available for both agonist- [8–13] and, in some cases, antagonist- [8,13,14] bound SHR-LBD (Figure 1).
Figure 1. Domain structure of SHRs.
The overall organization of the NTD, DBD and LBD is shown in schematic representation at the top of the Figure. The variable lengths of the different SHR-NTDs are illustrated below, together with representative structures for the DBD and hormone-bound LBD (note that the hinge region connecting the DBD and LBD has not been determined structurally). The position of regions important for transactivation (AF1 and 3) that have been mapped to sequences in the NTD are shown as yellow shaded bars.
The core DBD is a highly conserved, defining feature of this family of transcription factors, and is characterized by eight conserved cysteine residues that co-ordinate two zinc ions. The DBD folds to adopt a globular conformation consisting of two perpendicular α-helices, with residues important for DNA recognition and binding forming part of the recognition helix (Figure 1) [15–18]. Steroid receptors bind to palindromic-like sequences, consisting of a 6 bp half-site separated by 3 bp, as homodimers. A subfamily of SHRs, consisting of the androgen, glucocorticoid, mineralocorticoid and progesterone receptors, bind to the half-site sequence AGAACA, whereas the oestrogen receptor recognizes the sequence AGGTCA [19]. Although the LBD and DBD of SHRs share significant homology in both primary amino acid sequence and tertiary structure, the NTD shows little or no sequence homology between the different families of receptor, and structural information is limited. However, the NTD has been found to be important for both activation and repression of steroid-regulated genes. This review will focus on recent advances in our understanding of the structure–function relationships of the SHR-NTDs.
MAPPING OF FUNCTIONAL DOMAINS WITHIN THE SHR-NTD
Deletion of the LBD of SHRs results in a protein that is now constitutively active in reporter gene assays. This led to the identification of a hormone-independent AF1 (activation function 1) in the NTD. This contrasts with the AF2 activity in the LBD, which is dependent upon the binding of ligand.
Androgen receptor (AR; NR3C4)
In contrast with other members of the SHR subfamily, deletion of the AR-NTD results in a transcriptionally weak protein, providing evidence for the main transactivation function being located within the NTD [20,21]. The AR-AF1 is modular in nature, and regions important for transactivation have been mapped by deletion analysis [20,21], the use of fusion proteins [21] and by point mutations [22]. These studies identified amino acid residues 101–370 and 360–485 as being critical for receptor-dependent transactivation, and these have been termed TAU1 and TAU5 [21]. A fusion protein containing amino acids 142–485 (human AR), consisting of most of TAU1 and all of TAU5, retained approx. 70% the activity of the full-length NTD when measured in yeast cells (Figure 1) [23].
Comparison of the amino acid sequence of the AR-NTD from different species has highlighted three regions of conservation: the first 25 amino acids, amino acids 224–258 (within the AF1 domain) and the region immediately adjacent to the DBD (amino acids 500–541) [24–26]. The highly conserved sequence in the AF1 domain has some limited sequence identity with the GR (glucocorticoid receptor) amino acids 66–83 [26] and the highly conserved hydrophobic amino acids have been shown to be important for activity and protein–protein interactions [25]. Naturally occurring mutations in this sequence have also been identified in an animal prostate cancer model [26,27].
The first 20 amino acids of the AR-NTD contain an FxxLF motif (where x is any amino acid) that is important for interactions with the AR-LBD (reviewed in [28,29]). This is a key interaction for receptor function, which stabilizes bound hormone and is important for AR-dependent gene regulation, since mutations that disrupt this interaction impair receptor activity [30–32]. Interestingly, while other SHRs interact with co-activator proteins via LxxLL motifs, which bind in the hydrophobic groove of AF2, the AR-LBD preferentially binds the AR-NTD and co-activators with more bulky hydrophobic residues in the sequence F/WxxLF/W/Y [33,34], which may explain the relatively weak activity observed for AR-AF2. Recently, the structural basis for this preferred binding of the FxxLF motif has been reported [35–37]. X-ray crystallography studies showed that the AR-NTD FxxLF motif forms a ‘charge clamp’ with Glu-897 in helix 12 and Lys-720 at the end of helix 3, and the hydrophobic residues fit better into the surface pocket on the LBD [36,37]. In contrast, an LxxLL motif peptide fails to make hydrogen-bond contacts with the glutamic acid residue in helix 12, and makes fewer hydrophobic contacts with the surface of the LBD [36,37].
Glucocorticoid receptor (GR; NR3C1)
The AF1 domain of the human GR, originally termed τ1, was mapped by deletion analysis to amino acids 77–262 (Figure 1) [38], and was subsequently shown to function in yeast cells as a LexA fusion protein [39], and in yeast or HeLa nuclear extracts in vitro [40–42]. A 58-amino-acid core domain (amino acids 187–244) was mapped by further deletions, which retained 60–70% the activity of full-length AF1 [43]. Comparison of the sequence of the GR from different mammalian species and Xenopus reveals good amino acid conservation for the AF1 domain (32% identity) and the sequences immediately N-terminal of the DBD, amino acids 275–419 (54% identity). However, when fish sequences are included, there is less evidence of amino acid conservation.
Extensive mutagenesis studies revealed an important role for hydrophobic amino acids in AF1 core activity [44,45]. Mutations that reduced the overall acidity of the AF1 domain led to progressive impairment of transactivation [46]. It was speculated that these residues were important structurally, and defined the solvent-exposed surface of the transactivation domain. It is significant, therefore, that mutating key glutamic acid, phenylalanine or tryptophan residues in the corresponding enh2 domain (amino acids 108–317) of the rat GR impaired transactivation activity, but not the ability to repress transcription through protein–protein interactions with the transcription factor AP1 [47]. Such studies emphasize a role for different regions of the GR-NTD in gene regulation, which are likely to be mediated via protein–protein interactions (see below) [48]. More recently, using transactivation domain mutations, Yamamoto and colleagues [49] showed differential requirements for AF1 and AF2 in target gene expression. These studies emphasize a further level of selectivity in SHR signalling, after hormone and DNA binding that operates at the level of assembly of transcriptionally competent protein complexes on target gene promoters.
Mineralocorticoid receptor (MR; NR3C2)
Initial studies by Govindin and Warriar [50] identified amino acids 328–382, in the middle of the MR-NTD, as being important for transactivation function (Figure 1). However, more recent studies have mapped two distinct regions as being critical: AF1a was defined as the first 170 amino acids, and AF1b as the 147 amino acids adjacent to the DBD [51] (Figure 1). The mapping of quite distinct regions of the protein may suggest cell and/or promoter-selective activity for the MR-NTD transactivation function. Significantly, these three regions of the MR-NTD show a high degree of amino acid conservation between the mammalian MR sequences and a number of fish species: amino acids 1–170 (AF1a), 25% identity; amino acids 244–300, 54% identity; and amino acids 459–566, including AF1b, 46% identity.
Oestrogen receptor-α and -β (ERα and ERβ; NR3A1 and 2 respectively)
The ERα AF1 domain is located between amino acids 51 and 149 [52–54] (Figure 1). However, different regions of the protein were shown to have distinct cell-type and promoter selectivity [52]. More recently, the first 40 amino acids, immediately N-terminal of AF1, have been reported to be involved in receptor-dependent transcription and interactions with the ER-LBD [55]. In 1997, Gustafsson and co-workers [56] identified and cloned a second ER, termed ERβ, which has a distinct NTD. In contrast with ERα, the NTD of ERβ seems relatively weak at transactivation, but the first 31 amino acids appear to be critical for AF1 activity [54] (Figure 1).
Comparison of the ERα sequence from species as diverse as mammals, birds and Xenopus reveals high amino acid conservation within the first 39 residues (46% identity) and, overall, approx. 29% identity for the AF1 domain. This level of conservation is lost when the sequences from several fish species are analysed.
Progesterone receptor (PR; NR3C3)
There are two forms of the PR: PR-A and PR-B, which differ by the presence of an N-terminal 165-amino-acid extension (reviewed in [57]). The AF1 domain has been mapped to 91 amino acids preceding the DBD [58,59] (Figure 1). Interestingly, the autonomous function of this domain required the PR-DBD, suggesting intra-domain communication [57]. A second transactivation domain, termed AF3, is present in the PR-B isoform, and consists of the first 165 amino acids [59] (Figure 1). Comparison of the PR sequences from mammalian species and chicken indicated small regions of high conservation: amino acids 302–326, 71% identity; amino acids 357–390, 39% identity; and a region including AF1 (amino acids 459–566), 46% identity. Strikingly, the sequences corresponding to AF3, that are unique to PR-B, show only 12% amino acid identity under these conditions.
The two isoforms of the PR (A and B), which have AF1 (NTD) and AF2 (LBD) in common, show different patterns of transcriptional activity, with the B-form being in general the stronger activator (reviewed in [57]). Although this may reflect the presence of an additional activation surface, AF3, in PR-B, a selective and ‘transferable IF (inhibitory function)’ has also been mapped to the 291 amino acids upstream of AF1 (residues 165–455) [60]. Huse et al. [61] similarly noted a ‘negative modulation domain’ within 120 amino acids of the PR-NTD (amino acids 230–350). The IF was found to suppress the activity of AF1 and AF2, but interestingly not AF3. Furthermore, the inhibitory activity could be transferred to ERα, reducing ER-dependent transactivation when inserted upstream of the NTD [60]. The mechanism for this selective inhibition is not clear, but does not appear to require additional factors, and may involve intramolecular communication with AF1 or AF2 or the blocking of key protein–protein interactions with these domains [60]. More recently, Wang and Simons [62] identified amino acids 468–508, within the AF1 domain, as being necessary for co-repressor binding (Table 1, and see below). Significantly, this sequence could substitute for a corresponding activity in the GR-AF1 domain (amino acids 154–236 of the rat GR), despite showing no significant sequence homology [62]. This would argue for a structural basis for this inhibitory activity, but since these sequences are necessary for the binding of the co-repressors NcoR (nuclear receptor co-repressor) and SMRT (silencing mediator of retinoid and thyroid hormone receptor), the mechanism is likely to be distinct from the IF activity identified by Horwitz and co-workers [60].
Table 1. Selective protein–protein interactions involving the SHR-NTD.
Abbreviations: aa, amino acids. FL, interaction with full-length receptor, although binding site/region not delineated; –, no interaction observed; ?, studied in reporter gene assays, but receptor binding site not delineated; ns, not studied.
| Protein | AR | GR | ERα/β | MR | PR | References |
|---|---|---|---|---|---|---|
| GTFs | ||||||
| TFIID/TBP | aa 142–485 | aa 77–262; | aa 1–185 (α) | ns | ns | [42,45,110–112,139] |
| aa 187–244 | ||||||
| TFIIF | aa 142–485 | ns | ns | ns | ns | [23,111,137] |
| TFIIH | aa 38–561 | ns | ns | ns | ns | [114] |
| pTEFb | aa 38–643 | ns | ns | ns | ns | [115] |
| ELL | ? | FL | ns | aa 163–437; | ns | [116] |
| aa 445–602 | ||||||
| Co-activators | ||||||
| ARA24 | aa 11–208 | ns | ns | ns | ns | [158] |
| ARA160 | aa 38–643 | ? | ns | ns | ? | [159] |
| ART27 | aa 153–336†; | aa 107–237† | ? | ns | ns | [160] |
| aa 336–500† | ||||||
| CBP/p300 | aa 1–503; | aa 187–244 | aa 1–184 (α); | aa 1–169 | ? | [45,51,161–165] |
| aa 297–640†; | aa 56–72 (α); | |||||
| aa 142–485‡ | aa 62–72 (β) | |||||
| p68 | ? | ns | aa 1–180 (α) | ? | ns | [85] |
| RHA | ns | ns | ns | aa 1–169 | ns | [164] |
| SAGA complex* (ada2) | ns | aa 77–262; | ns | ns | ns | [45,166] |
| aa 187–244 | ||||||
| SRC-1 | aa 1–555; | – | ? | ? | aa 1–535; | [23, 32,121–125] |
| aa 142–485; | aa 163–534 | |||||
| aa 360–494 | ||||||
| SRC-2 | aa 1–555 | ? | aa 38–116 (α) | ? | ? | [51,32,121,123] |
| SRC-3 | aa 1–660 | ns | aa 1–184 (α) | ns | ? | [121,126] |
| SWI/SNF complex* (?) | ns | aa 77–262; | – | ns | ns | [167] |
| TRAP/DRIP complex* (DRIP150) | – | aa 107–237† | ns | ns | ns | [168] |
| Co-repressors | ||||||
| AES | aa 1–599 | ns | – | ns | ns | [169] |
| DAXX | aa 1–560 | aa 1–450 | ns | aa 170–433 | ns | [129,170] |
| FLASH | ? | aa 1–450 | ns | aa 170–433 | ? | [129–131] |
| HEY-1 | aa 1–556 | – | – | ns | – | [128] |
| NcoR | aa 1–500; | aa 1–551; | ? | ns | aa 468–508 | [62,171] |
| aa 171–505 | aa 154–236; | |||||
| aa 206–236 | ||||||
| SMRT | aa 1–682; | aa 154–236; | ? | ns | aa 468–508 | [62,172] |
| aa 171–328 | aa 206–236 | |||||
| Co-regulators | ||||||
| BAG-1 | aa 360–554 | ? | ? | ns | ns | [132,173] |
| CHIP | aa 220–270; | ? | ns | ns | ns | [26] |
| PIAS1/xα/Miz1 | aa 544–634/DBD | FL | FL (α/β) | aa 1–602 | FL | [87,91,174] |
| Prp6p (ANT-1) | aa 1–153 | aa 14–438 | – | ns | ns | [175] |
| TSG101 | aa 18–500† | aa 107–237† | ns | ns | ns | [168] |
*Multi-subunit complexes. SHR-NTD binding partner indicated in brackets, where known.
†Amino acids for the rat AR and GR.
‡D. N. Lavery and I. J. McEwan, unpublished work.
The AF1 domain or related SHR-NTD activities have been shown to function in a variety of mammalian cell types and in the budding yeast Saccharomyces cerevisiae. Although this suggests a potential conservation in transactivation activity between SHRs, there is also considerable evidence available that the transactivation activity of individual NTDs can exhibit cell and/or promoter selectivity. When taken together, these studies illustrate that different surfaces, even within a single SHR-NTD, can be involved in activation and/or repression of transcription. Furthermore, the strength of the respective SHR-NTDs for activating transcription is also variable. Wilson and co-workers [36], using Gal4–DBD fusion proteins, recently demonstrated a nice correlation between AF1 activity (and, inversely, AF2 activity) and the length of the NTD, with the longer AR-, PR- and GR-NTDs showing the highest activity.
POST-TRANSLATIONAL MODIFICATIONS
SHRs are also targets for post-translational modifications, and the NTD is the site of both phosphorylation and sumoylation (reviewed in [63–65]), whereas lysine residues targeted for acetylation or ubiquitination are found in the hinge region between the DBD and LBD ([65], and references therein). The roles of these modifications are beginning to be elucidated, but the possible effects on structure are less clear. What has also emerged in recent years is that post-translational modifications may be interdependent and not necessarily ‘stand-alone’ events (see [66,67]). The five classes of SHR are phosphoproteins, with key phosphorylation sites mapped to serine, threonine and tyrosine residues. However, what are less clear are the consequences of phosphorylation for receptor action.
In the case of the AR, the majority of phosphorylated sites have been identified in the NTD. Initial studies reported phosphorylation of the AR on serine residues 81 and 94 [68], and more recently, Gioeli et al. [69] identified an increase in phosphorylation of serines 16, 81, 256, 308, 424 and 650 in response to hormone treatment of LNCaP cells. However, the kinase enzymes responsible for phosphorylating the AR at these sites are less well defined. Activation of the MAPK (mitogen-activated protein kinase) and Akt/PKB (protein kinase B) pathways has also been implicated in AR function. Chang and co-workers [70] correlated phosphorylation of Ser-514, possibly by MAPK, with an increase in the PSA (prostate-specific antigen) gene transcription. A similar increase in PSA expression, coupled with an increase in cell survival, was observed by Wen et al. [71] after phosphorylation of Ser-213 and Ser-791 by Akt/PKB. In contrast, Lin et al. observed a decrease in AR-dependent gene expression upon phosphorylation of these two residues by Akt [72]. Consistent with Akt/PKB having a positive role, Manin et al. [73] observed that the phosphoinositide 3-kinase/Akt pathway was involved in an up-regulation of AR expression levels. However, that study did not determine whether this was due to a direct phosphorylation of AR protein.
The GR is phosphorylated in both mammalian and yeast cells in a similar manner, and as with the AR studies described above, phosphorylation enhanced, repressed or had little effect on GR-dependent gene expression [46,74,75]. The human and rat GRs are phosphorylated at Ser-203 and Ser-211 (numbering for human receptor) by cyclin-dependent kinase in yeast and mammalian cells [75], whereas Thr-171 and Thr-246 are phosphorylated in the rat GR by glycogen synthetase 3-kinase and the MAPK, JNK (c-Jun N-terminal kinase), respectively [76]. Phosphorylation of Ser-203 and Ser-211 has been shown to be hormone-dependent, and using phosphospecific antibodies, Garabedian and co-workers [77] were able to show that the GR was cytoplasmic when phosphorylated on Ser-203, and both cytoplasmic and nuclear when Ser-211 was modified. This supported the hypothesis that phosphorylation of specific sites altered the intracellular location of the GR, and presumably its function. The MR has also been shown to be phosphorylated, but the domain or residues modified have not been identified [78,79].
A number of potential phosphorylation sites exist within the PR-NTD. The AF3 region, which is unique to the PR-B isoform, is phosphorylated, but mutating phosphorylated residues had little effect on transactivation activity of the full-length receptor or the isolated transactivation domain [80]. In contrast, mutating Ser-190 or a cluster of residues adjacent to the AF1 and DBD did reduce transactivation of the PR-A isoform in a promoter- and cell-context-dependent manner [80], whereas Ser-294 was reported to be a substrate for MAPK, which then targeted the PR for degradation via the proteasomal pathway [81].
Both ERα and β have been shown to be phosphorylated on serine or threonine residues in the NTD. Phosphorylation of Ser-87 (human ERβ) and the corresponding residues in ERα, Ser-104 and Ser-106, together with Ser-118, was shown to be important for ligand-independent activation of transcription and recruitment of the co-activator protein SRC-1 (steroid receptor co-activator 1) [82–84]. Similarly, binding of p68/p72 has also been shown to be phosphorylation-dependent [85], as was the interaction between ERα and the orphan receptor COUP-TF1 (chicken ovalbumin upstream promoter-transcription factor 1) [86]. Ser-87 (ERβ) and Thr-311 (ERα) have both been shown to be substrates for MAPK enzymes [82], and Ser-118 (ERα) is a target for the GTF (general transcription factor) TFIIH/cyclin-dependent kinase 7 activity [83]. Thus several residues in the ER-NTD can serve as targets for cross-talk with other signalling pathways, and lead to modulation of protein–protein interactions.
SUMO-1 (small ubiquitin-related modifier-1) is part of a family of small peptides that are covalently linked to a range of proteins and is the most recent post-translational modification to be described for SHRs. The E2-ligase, ubc9, and members of the PIAS [protein inhibitor of activated STAT (signal transducers and activators of transcription)] family of E3-ligase enzymes have been shown to interact with the six principal SHRs, and to modulate receptor-dependent activation (Table 1) [87]. Interestingly, all the SHR-NTDs with the exception of ERα and β have one or more consensus motif, ψKxE (where ψ represents a hydrophobic residue), which contains an acceptor lysine (K) residue [88]. Sumoylation of the AR, GR and MR leads to repression of transcription, which appears to be promoter-specific and may also depend upon cell type [87–92]. Similarly, the IF activity identified in the PR-NTD has been correlated with sumoylation of the receptor, since mutation of an acceptor lysine residue (Lys-388) abolishes trans-repression [93]. However, the picture is more complicated since the different PIAS family members have been found to activate as well as repress SHR-dependent transcription. This may be a direct effect of the PIAS proteins [87], or result from sumoylation of co-activator proteins, such as members of the p160 family [94].
The consensus sumoylation motif has previously been described as a SC (synergy control) sequence [95]. Synergism or co-operativity between SHR dimers [96] and/or other transcription factors [97] can involve the NTD, and is likely to play an important role in determining the magnitude of the receptor-dependent transcription response at different promoters. Deletion of the SC sequence resulted in increased activity from multiple hormone response elements [95], and the data from the above studies support the model that this sequence functions, at least in part, to ‘dampen down’ transactivation activity via modification of the receptor protein by SUMO-1.
Post-translational modification of the SHR-NTD can result in changes in intracellular localizations, turnover and protein–protein interactions. The continuing challenge is to determine the functional and/or structural significance of individual modifications, and to determine whether such modifications may actually act co-operatively to regulate receptor function.
PROTEIN–PROTEIN INTERACTIONS WITH THE SHR-NTD
SHRs function in the main as ligand-activated transcription factors that are targeted to regulated genes via DNA-response element recognition and binding. The DNA-bound SHR may then activate transcription though modifying the underlying chromatin organization and by recruitment of the transcription machinery (reviewed in [98–100]). Alternatively, the DNA-bound receptor may repress transcription through so-called negative DNA-response elements (reviewed in [101]). These actions involve the assembly of multiprotein complexes through receptor–protein interactions, which are discussed below. In addition, certain SHRs, most notably the GR, can regulate gene expression in the absence of specific DNA-binding through protein–protein interactions with other transcription factors and inhibiting their transactivation function [101,102].
The basal transcription machinery
The RNA polymerase II enzyme comprises 10 or 11 subunits, and is highly conserved in all eukaryotes [103,104]. Transcription initiation involves the assembly of a PIC (pre-initiation complex) containing the multi-subunit RNA polymerase II enzyme and up to six GTFs [103,104]. A number of the subunits making up the PIC exhibit DNA-binding activity, most notably the TBP (TATA-binding protein), which is part of the TFIID complex. The assembly of the PIC can follow a stepwise progression in vitro, but most likely involves recruitment of pre-existing protein complexes, for example TFIID [TBP+TAFs (TBP-associated factors)], holo-RNA polymerase II (polymerase enzyme+mediator complex+GTFs) and the multi-subunit TFIIH factor (see [99,105–109]). TFIIH has a number of defined enzymatic activities that play an important role in initiation and promoter escape by the RNA polymerase enzyme, including a helicase activity required to unwind the DNA to create an open complex and a kinase that phosphorylates the C-terminal domain of the large subunit of RNA polymerase II [104].
Multiple interactions between SHRs and members of the basal transcription machinery have been reported over the past 10 years (Table 1), and include interactions with TFIID/TBP [42,45,110–113], TFIIF [23,111] and TFIIH [114], and the elongation factors pTEFb [115] and ELL [116]. It seems likely that such interactions play a part in recruitment of the GTFs and RNA polymerase II, resulting in the assembly of the PIC and/or regulating the early steps of promoter clearance and transcription elongation.
Co-activators: faithful interactions or promiscuous partners?
Evidence for the existence of co-activator proteins originally came from studies involving the phenomenon of squelching [117–119]. However, perhaps what was less expected was the huge number of proteins that would be identified that could bind to and/or modulate SHR activity in vivo. In order to make sense of the myriad of interactions involving the SHR-NTD, it is helpful to group the different target proteins: (1) those that directly regulate the receptor transactivation function, including components of the general transcription machinery and co-activators that may act as a ‘bridge’ between the DNA-bound receptors and the transcriptional machinery, or those that harbour specific enzyme activities, such as histone acetyltransferases or methyltransferases; (2) co-repressors, proteins that mediate receptor-dependent repression of transcription, which may block the transactivation domain or recruit complexes with histone deacetylase activity; and (3) co-regulatory proteins that act indirectly on receptor-dependent transactivation activity by regulating receptor stability or intracellular localization; again, such proteins may have modifying enzymatic activity, such as sumoylation or phosphorylation. Table 1 lists representatives from each of these three groupings that have been reported to bind to the NTD of one or more SHR.
A significant proportion of research effort has been focused on the identification and characterization of co-activator proteins. Co-activators can be defined as proteins that interact directly with transactivation domains, and are recruited to promoter and enhancer DNA sequences. Such proteins should harbour enzymatic activity and/or interact with components of the general transcription machinery, and function to enhance the level of transcription of receptor-target genes. As co-activators were identified and characterized, the idea of receptor-specificity was an attractive possibility with respect to possible mechanism(s) for gene regulation. However, as shown in Table 1, whereas some co-activators appear restricted to one or two SHRs, e.g. ARA24, ARA160 and DRIP150, others, such as CBP [CREB (cAMP-response-element-binding protein)-binding protein] and the SRC proteins, are more promiscuous in their binding profiles. The SRC or p160 family of co-activators represents a particularly interesting group. Members of the p160 co-activator family [SRC-1, SRC-2/TIF2 (transcription intermediary factor 2)/GRIP1 (glucocorticoid receptor-interacting protein 1) and SRC-3/ACTR (activator for thyroid hormone and retinoid receptors)/RAC3/AIB1] were originally identified as LBD-AF2-binding proteins via so-called NR-box (LxxLL) motifs [7,120]. Subsequently, they have been shown to interact with SHR-NTD sequences and, at least to some degree, increase receptor transactivation function [32,121–126]. However, the level of co-activation for different receptors can vary, and may also be dependent on the cellular background. Interactions with the p160 proteins have also been shown to be important for synergism between the SHR-AF1 and -AF2 domains [55,127], and for ligand-independent activity, possibly resulting from phosphorylation of the NTD [84].
In addition to the abundance of enzymatic activities exhibited by co-activators, or co-regulatory proteins in general, are the concepts of redundancy and the abundance of multi-subunit complexes that share subunits, but show distinct patterns of recruitment and/or activity. This was most elegantly demonstrated by the recent Herculean analysis of the transcription events at the oestrogen-regulated pS2 promoter [108]. Gannon and co-workers, using ChIP (chromatin immunoprecipitation) and ‘Re-ChIP’ assays, demonstrated the importance of ER binding to the promoter for the recruitment of different multi-subunit complexes in order to initiate cyclic rounds of transcription initiation of the target gene. These workers found evidence for redundancy and selectivity in the ER interactions, and could distinguish several distinct protein complexes needed for transactivation, including p68/TBP, SRC-1 or SRC-3/CBP or Tip60/and/or methylase transferases, SWI/SNF and TRAP (Mediator) [108]. The recruitment of different protein complexes has also been shown for the AR [106,107] and the GR and PR [109].
In summary, SHR co-activators can function at several steps during transcription, from recruitment of chromatin-remodelling complexes (SWI/SNF) and histone-modifying complexes [e.g. SAGA (Spt-Ada-Gcn5 acetyltransferase) and CBP–p/CAF (CREB-binding protein-associated factor)] to the regulation of transcription initiation and elongation by the polymerase II holo-enzyme. Initially, it was presumed that co-activators made specific interactions with specific receptors. Indeed, the transactivation potential of many co-activators differs, depending on cell type and receptor employed. Although the majority of co-activators identified to date have been shown to interact with more than one SHR (Table 1), there are likely to be subtle levels of control depending on the levels of co-activator, post-translational modifications and the architecture of target gene promoters. Thus further studies of co-activators and SHR relationships will need to address these issues in order to confidently dissect the molecular and functional interactions.
Co-repressors: more than a road block?
Studies relating to SHR co-repressors have been somewhat overshadowed by the wealth of information gathered relating to co-activators. However, the co-repressors themselves provide an interesting insight into transcriptional repression and the balance between switching genes ‘on’ and ‘off’.
Table 1 illustrates a selection of co-repressors that bind to SHR-NTDs. There generally appear to be fewer AF1-interacting co-repressors in comparison with co-activators. This may reflect a bias in scientific focus, which has generally been directed towards co-activators and ligand (AF2)-dependent gene regulation. Alternatively, the cell may not require a large array of co-repressors to switch ‘off’ transcription; indeed, a handful of co-repressors may be sufficient to accurately fulfil the task in hand.
Co-repressors respond to both inter- and intra-cellular signals when switching ‘off’ transcription. In response to specific cues, co-repressors will bind sites on SHRs, which are distinct but may overlap with the transactivation functions AF1 or AF2, and recruit histone deacetylase complexes, effectively re-condensing the chromatin and packing away target genes. In addition, co-repressors may also effect receptor turnover by recruiting the 26 S proteasome subunit or modulate cellular signalling cascades (reviewed in [101]). Interestingly, co-repressors may physically alter the flexible N-terminal transactivation domain and block the recruitment of co-activators engaging in transcription [62]. For example, elegant AF1 domain-swapping experiments on the GR and PR reveal that the co-repressors NCoR and SMRT bind to both the N- and C-termini, highlighting the importance of intramolecular interactions [62]. In addition, irrespective of the different affinities of co-repressor–GR/PR binding, a TIF2 (SRC-2) peptide was able to compete for AF1 binding. It has already been shown that N/C intra-/inter-domain bridging is important for a fully functioning SHR [28,29], so it may be plausible to presume that an additional mechanism of co-repressor action is to inhibit this interaction and prevent recruitment of co-activators like the p160 family of proteins, thus down-regulating transcription of target genes.
Selective interactions between co-repressor and specific SHRs may complement the actions of the more ‘general’ co-repressors (NcoR, SMRT and Daxx). For example, the basic helix–loop–helix orange protein, HEY-1, appears to be specific for the AR [128]. Discovered through a yeast-two-hybrid screen, HEY-1 was shown to interact with residues in the AF1 domain. Furthermore, the authors noted that HEY-1, albeit weakly, interacts with the GR, PR and ERα, but functions as a specific co-repressor for AR in reporter gene assays [128]. A second co-repressor, FLASH (Fas-associated huge protein), has been shown to interact with both MR and GR AF-1 domains [129]. Although that study indicates that FLASH up-regulates MR- and GR-dependent transcription, two further studies by Chrousos and co-workers [130,131] indicate that FLASH is able to repress the actions of the GR, PR and AR, and has no effect on the ER. Such differences are intriguing, and again may reflect differences in cellular environments and the balance between endogenous co-activator and co-repressor proteins.
Taken together, it seems that co-repressors may act in several ways. There may be specific interactions between co-repressors and selective SHRs that may depend on cell context and type. In addition, more general forms of co-repression may involve abrogation of intramolecular bridging or competition with co-activators for similar binding sites or regions.
Co-regulator proteins
Co-regulatory proteins represent an interesting set of factors that can affect SHR-dependent transactivation and includes molecular chaperones and a number of proteins with specific enzymatic activities, for example, acetyltransferases and ubiquitin and SUMO-conjugating enzymes (Table 1). Chaperones such as the BAG-1 family of proteins have been shown to increase transcription of the AR, presumably by recruiting a complex of co-activators or through active cycling of the receptor protein [132]. In stark contrast to the AR, BAG-1 proteins have been shown to inhibit GR-dependent transactivation by binding to the hinge region of the receptor [133].
The importance of the SHR-NTD for receptor-dependent gene regulation is underpinned by the large amount of data amassed on the binding of GTFs, co-activators, co-repressors and co-regulator proteins. The interplay between transactivation and transrepression is likely to be dependent on both receptor type and discrete, but possibly overlapping, amino acid sequences. The differential binding of co-regulators has been implicated in both gene- and cell-specific expression, and taken in context suggests that a large and varied response to stimuli can be achieved in vivo.
STRUCTURAL ANALYSIS OF THE AF1 TRANSACTIVATION DOMAIN
As indicated earlier there is little, if any, sequence homology between the different SHR-NTDs. However, analysis of the amino acid composition of these domains suggests some shared characteristics. The NTDs of SHRs have a high proportion of proline and/or serine residues (greater than 10% of the total amino acids). The AF1 domains of the AR and ERα show a similar high content of these residues, and for all the SHRs, the regions mapped as being important for transactivation are rich in the amino acids glycine and/or leucine relative to the corresponding NTD. With the exception of the PR-AF1 domain, the mapped transactivation domains all have acidic isoelectric points (pI 4.4–5.0), which is higher than the archetypal acidic activator VP16/Vma65 (pI 3.4), and suggests they may not necessarily fit the paradigm of acidic activators. The PR-AF1 domain stands out, with a pI of 7.6 and less than 5% acidic residues.
In contrast with the DBD and the LBD, there are no high-resolution structures available to date for the NTD of any member of the nuclear receptor superfamily. In the case of the SHR, the lengths of the NTDs are likely to contribute some practical challenges in terms of structure determination, and the situation is complicated further by the lack of amino acid sequence identity and the functional properties of the AF1 domains (as discussed below). However, in the absence of high-resolution structures, significant structural information has been obtained from CD, NMR, fluorescence and Fourier transform infrared (FTIR) spectroscopy, together with secondary structure prediction algorithms and biochemical analysis.
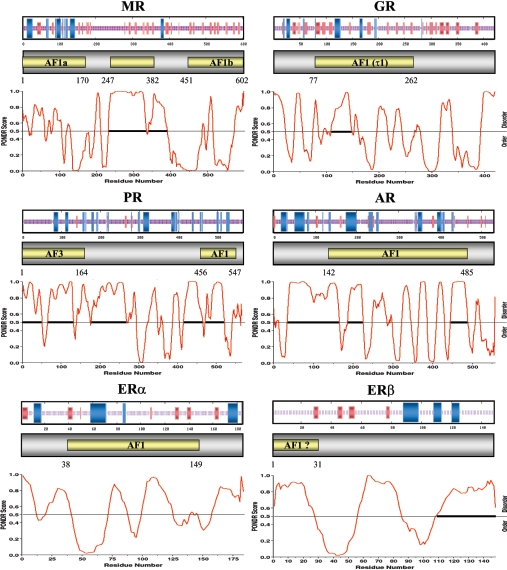
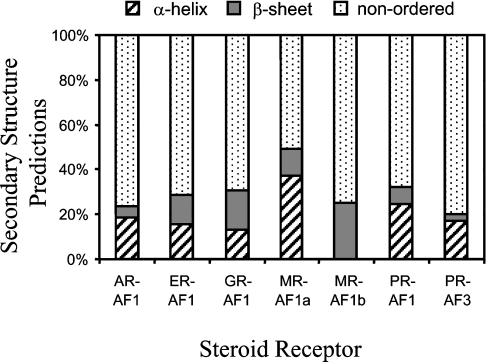
Secondary structure and regions of natural disordered structure can be predicted on the basis of amino acid sequence and composition. For each SHR-NTD, the distribution of secondary structure types (α-helix, β-strand and coil) is shown in relation to the mapped transactivation domains (Figure 2). It can be seen that the MR-AF1a, GR-AF1, AR-AF1 and ERα-AF1 domains consist of potentially a mixture of α-helix, β-strand and coil conformations. In contrast, PR-AF1 and AF3 are likely to be predominantly α-helix, whereas MR-AF1b and ERβ are predicted to be exclusively β-strand (Figure 2). In the case of the AR-AF1, this predictive analysis has proved informative when combined with mutagenesis to disrupt suspected helical regions [22,134]. Figure 3 shows the relative proportion of α-helix, β-strand and non-ordered/other structure for the different transactivation domains. With the exception of the MR-AF1a/b domains, the mapped transactivation domains are predicted to have similar levels of α-helix (15–20%) and generally less than 10% β-strand. In contrast, the MR-AF1a and -AF1b domains have 40 and 0% α-helix respectively, and MR-AF1b is predicted to be predominantly β-strand (greater than 20%; Figure 3).
Figure 2. Secondary and natural disordered structure predictions.
At the top of the panel representing each SHR-NTD is a consensus secondary structure prediction. The blue and red lines/bars represent α-helix and β-strand respectively [157]. Below is shown the PONDR® (www.pondr.com) plots indicating likely regions of natural disordered structure (scores above the 0.5 threshold); the longer the region the more accurate the prediction. The solid horizontal lines represent regions of highest probability, with 40 or more consecutive residues predicted to be disordered.
Figure 3. Proportions of predicted secondary structure in SHR-NTD transactivation functions.
The percentage contents of α-helix, β-strand and non-ordered/other structure are plotted for each SHR-AF1 and PR-AF3 domain.
It is becoming increasingly apparent that regions or even whole domains of naturally disordered structure can play an important role in protein function (reviewed in [135,136]). The presence of a high proportion of proline, serine and glycine residues in the NTDs of steroid receptors conforms to the amino acid compositional bias associated with a signature for intrinsic disorder [136]. The trained neural network program, PONDR®, uses primary amino acid sequences to predict the occurrence of such regions. Figure 2 shows the PONDR® score plot indicating regions of disorder (threshold 0.5 or above) for each of the six principal SHR-NTDs. Regions of 40 or more consecutive residues represent the highest probability of naturally disordered structure (Figure 2, solid bar). There is a good inverse relationship between predicted secondary structure elements and regions of disordered structure. It is also striking that regions of highest probability for disordered structure generally overlap with regions important for transactivation (Figure 2).
The actual secondary structure content has been determined for the AR-AF1 [134,137], ERα/β-NTD [112] and GR-AF1 and AF1 core [138,139] by CD and FTIR spectroscopy. The far-UV CD spectrums for all five polypeptides in aqueous solution are characterized by large minima at 190 nm, which is characteristic of proteins with little stable secondary structure content. The experimentally measured α-helical content of AR-AF1 and GR-AF1 was 13–16% and 27% respectively, with the non-ordered structure of 24–36% for the AR and 39% for GR [134,137,139]. Both polypeptides adopt a more α-helical conformation, at the expense of non-ordered or possibly β-structure, when placed in a hydrophobic environment or in the presence of the natural osmolyte TMAO (trimethylamine-N-oxide) [134,137–140]. Folding of the GR-AF1 and AR-AF1 domains, in the presence of TMAO, to a more structured conformation was also followed by measuring the steady-state fluorescence emission spectrum for aromatic amino acids and by partial proteolysis [113,134,140]. Under these conditions, the AF1 domain adopts a more protease-resistant conformation, with the tryptophan residues being less solvent-exposed. Three induced α-helical regions within the GR-AF1 core were identified by NMR spectroscopy in the presence of the hydrophobic solvent TFE (trifluoroethanol) [138]. The ability to form α-helix structure was correlated with transactivation potential of the GR through the introduction of proline mutations to disrupt the helical segments and alter the secondary structure of the receptor polypeptide [138,141]. Similarly, introduction of mutations into the AR-AF1 region to disrupt putative helical regions led to alterations in the partial proteolysis profile, consistent with a loss of structure and supporting the existence of α-helical segments in the polypeptide [134]. A common feature of transactivation domains, apart from the general lack of stable structure, has been their modular nature. This was illustrated elegantly for the GR-AF1 core domain by analysing the activity of individual and multiple combinations of helices 1 and 2 [142]. These studies demonstrated that, although helix 1 and 2 were able to bind to target proteins (Ada2 and CBP), they were not capable of activating transcription alone. However, multiple copies of either helix or, even better, combinations of the two were as active as the original AF1 core polypeptide [142]. These findings emphasize the importance of multiple protein–protein interactions and the structural architecture of the transactivation domain for function. Taken together, the biophysical and biochemical analysis of the AR- and GR-TAD conformation revealed structurally flexible polypeptides that could fold into more compact conformations in the presence of structure-stabilizing solutes (TFE and TMAO); this folded structure may involve a coil-to-helix transition.
Although the structural predictions for the PR-NTD are not significantly different from those of the other SHRs (Figures 2 and 3), this domain is thought to have significant structure, as revealed by limited proteolysis, which was stabilized further by the presence of the DBD [143,144]. Physicochemical properties suggested that the NTDs of both PR-A and PR-B were monomeric in solution, with a non-globular, extended conformation, and that the AF3 region of the B-isoform, in contrast with AF1, showed a more extended conformation [143,144].
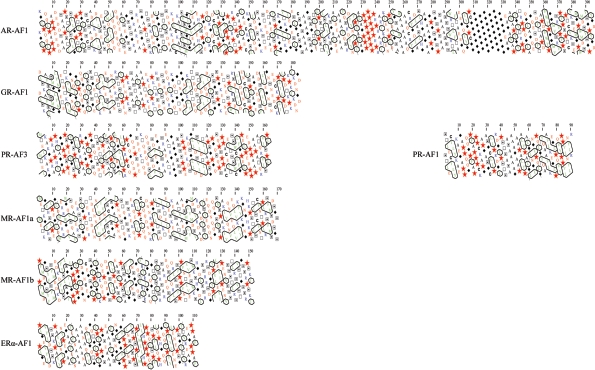
Another procedure that has proved informative, in the absence of high-resolution structures and sequence similarity, is HCA (hydrophobic cluster analysis), again based on examination of the primary amino acid sequence [145,146]. This technique is based on pattern recognition, and has proved useful in revealing structural relationships in the absence of any obvious sequence homology, and is therefore ideal for analysing SHR-NTDs. The amino acid sequence is duplicated and represented on a flattened α-helical structure (without making assumptions about the underlying structure), and hydrophobic residues are circled, revealing patches of hydrophobicity, which can be related to the folded conformation. Figure 4 shows the HCA plots for the transactivation domains of six SHR-NTDs. Similarities in the HCA profiles, even in the absence of clear sequence alignments, has helped predict that proteins may share similar folded conformation [145,146]. From the plots shown, there are clear patches of hydrophobic residues (Val, Ile, Leu, Phe, Met, Trp and Tyr) and clusters of both proline residues (stars), which break possible secondary structures, and glycine residues (diamonds), which confer flexibility in the polypeptide chain. However, there is no obvious similarity in the patterns of hydrophobic residues among the different transactivation domains (Figure 4). This is in striking contrast with a similar analysis of the AR-, GR- and PR-DBD/LBD sequences, which show high similarity between the HCA plots (I. J. McEwan, unpublished work) and which would be expected from the crystal structures of these SHR-LBDs. Thus the analysis fails to reveal any likely shared conformational folding between the different SHR transactivation domains, but may prove useful in studying the folding of the individual domains. This might suggest that, although the SHR-NTD share the general properties of being structurally flexible, with little stable secondary structure and the propensity to adopt α-helix conformation, the overall folding of these domains may result in distinct conformations, which in turn may depend on cellular and target gene environment.
Figure 4. HCA of the SHR-transactivation domains (AF1 and AF3).
HCA is based on pattern recognition and allows for the identification of potentially shared structural folds. The amino acid sequence of the individual transactivation regions has been duplicated and represented on a two-dimensional α-helix [145,146]. Hydrophobic residues (in green) are enclosed by black lines, and proline (red stars), glycine (filled black diamonds), serine ( ), threonine (□) and cysteine (C) residues are highlighted.
), threonine (□) and cysteine (C) residues are highlighted.
Allosteric regulation: role of DNA-response element binding
There is increasing evidence to support the view that DNA-response elements can play a more active role in transcription factor action than merely tethering the protein to the DNA (reviewed in [147]). Thus DNA binding may cause structural changes within the SHR-NTD, and reciprocal intramolecular-domain communications may influence response element recognition and/or binding affinity.
Evidence for DNA binding affecting the structure of the NTD of steroid receptors comes again from spectroscopy analysis and sensitivity to limited proteolysis. Changes in the structure of the GR-NTD were observed by both near-UV CD and fluorescence spectroscopy, with the changes possibly involving β-sheet secondary structure and folding of the domain [148], whereas Greenfield et al. [149] observed an increase in thermal stability and α-helical content upon binding of the full-length ERα to DNA. Given the known structural content of the DBD and LBD, these changes in conformation are most likely to reflect folding of the NTD and/or the hinge region. Evidence for intradomain communication within ERα was originally suggested from studies using domain-specific antibodies [150], and was supported further by the observation of altered protease sensitivity for both ERα and β when bound to different EREs (oestrogen-response elements) [151–153]. In the case of ERβ, binding to different EREs led to distinct patterns of recruitment of members of the p160 co-activator family and strongly supported a link between protein–protein interactions and SHR-NTD conformation [153]. More recently, binding to specific DNA sequences was shown to alter the steady state fluorescence emission spectrum for a construct of the AR-NTD-DBD and alter the pattern of proteolysis for the receptor fragment [154] or the full-length receptor [155]. In addition, it was found that the presence of the AR-NTD reduced the affinity of the DBD for binding to both selective and non-selective DNA-response elements [154]. Interestingly, although the presence of the DBD stabilized the hinge and NTD of the PR, there was no additional affect observed in the presence of a specific DNA-response element [143]. Taken together, these studies highlight the possibilities for domain communication within the steroid receptor molecule, and the potential reciprocal effects on protein folding and DNA binding.
Allosteric regulation: role of target protein binding
All the structural analyses to date on the isolated SHR-NTD/AF1 indicate that these regions of the protein are likely to lack stable structure, but adopt a more folded conformation under certain conditions. This does not exclude the possibility that, in the context of the full-length receptor, the structure of the NTD may be more ordered and stabilized by the presence of the DBD and/or LBD. However, in recent years there has been increasing interest in proteins or protein domains that appear natively unstructured, but are nonetheless functionally important [135,136]. The proportion of naturally disordered proteins in eukaryotes has been estimated to be as high as 35–51% [135]. A number of functions have been proposed for intrinsically unstructured regions that seem particularly relevant to a discussion of SHR-NTDs, given their role in multiple protein–protein interactions and gene regulation. The first is in molecular recognition, where structural flexibility: (a) allows high specificity, without the need for high affinity; (b) allows the ability to make multiple different interactions; and (c) results in a relatively very large interaction surface as the protein folds around the binding partner [135,136]. Secondly, natural disorder in SHR-NTDs may help to facilitate molecular assembly by allowing proteins to form loose groupings, which then undergo a folding step to form the functional complex [135]. Thirdly, sites of post-translational modification are often found in unstructured regions, which allow access to the amino acid side chain by the modifying enzyme [135].
Kumar, Thompson and co-workers have shown that folding of the GR-AF1 domain with TMAO resulted in enhanced binding of the targets TBP, CBP and the p160 co-activator, SRC-2 (GRIP1) [113]. These authors have proceeded to show that the binding of TBP to GR-AF1 resulted in a coil-to-α-helix transition and alteration in the chemical shifts of glycine residues present in the GR-AF1 core region [139]. TBP binding also results in a structural change in the ERα-NTD, consistent with an increase in α-helix/β-sheet secondary structure [112].
The interaction of the AR-AF1 domain with the large subunit of the GTF, TFIIF, also induces folding and an increase in α-helix structure similar to that seen with TMAO [134,137]. Furthermore, folding of the AF1 domain by either TMAO or TFIIF binding resulted in enhanced binding of the co-activator protein SRC-1a [137]. This is a significant finding, since it suggests, together with the data for GR-AF1, that induced folding of the AF1 domain leads to the creation of surfaces for further protein–protein interactions, and may illustrate a mechanism for regulating assembly of different transcriptionally competent complexes. The model would be that some interactions induce folding (Figure 5), whereas other interactions would require a more ordered binding surface. Alternatively, as suggested above, there could be concomitant folding of the receptor transactivation domain with the different target proteins, resulting in the assembly of a multi-subunit, transcriptionally competent complex. These models will need to be tested in order to fully understand SHR-NTD structure and function.
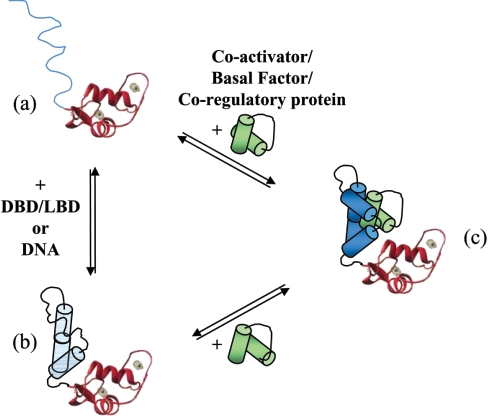
Figure 5. Model illustrating the putative conformers adopted by SHR-NTD.
(a–c) The NTD is represented by line or helical structures (shown in blue) linked to the folded globular structure of the DBD. The binding of the target protein is shown in green. The NTD has regions of significant intrinsic disorder (a), which may be stabilized by other receptor domains, or on binding DNA (b). The NTD can be thought of as a number of conformers with a flexible structure. The binding of a target protein (in green) induces and/or stabilizes the conformation of the SHR-NTD (c).
STRUCTURAL BASIS FOR STEROID RECEPTOR ACTION
Thus, in spite of the lack of crystal structure data, a large body of evidence has been gathered underlining an induced structure model for the SHR-NTD/AF1 upon DNA and/or protein binding. Figure 5 illustrates this model, and indicates that different conformers of the SHR-NTD/AF1 may exist in equilibrium (parts a and b), with regions of partial structure stabilized by intramolecular interactions with the DBD (shown) and/or the LBD (not shown). Upon specific protein–protein interaction, there is induction of folding and/or further stabilization of structural elements within the NTD (Figure 5c), which is important in the assembly of a transcriptionally competent receptor complex. Such a model is attractive, as it allows for specificity and multiple target protein binding to the SHR-NTD. Interestingly, the few NTD-target protein interactions that have been analysed kinetically suggest that protein–protein-binding affinities exist in the micromolar range ([112,132], and D. N. Lavery and I. J. MacEwan, unpublished work). Thus induced folding and the creation of new binding sites within the NTD suggest a way of regulating the different interactions required to assemble transcriptionally competent complexes. It is also possible to include in this model of induced transactivation domain folding distinct roles that charged and hydrophobic amino acids may play. Wright and co-workers have argued, on the basis of kinetic evidence, for a fast step involving long-range charged interactions, followed by a slow folding step involving the burying of hydrophobic residues at the binding surface [156]. In addition to the kinetic data, support for this model of AF1 structure and function comes from mutational analysis, highlighting the importance of charge [46,48], hydrophobicity [25,44,48] and structural flexibility [25].
Steroid receptors are targeted to specific DNA sequences usually located in the promoter and/or enhancers of target genes. The influence of the steroid receptor is likely to be at one or more of the different steps of the transcription cycle. For example, the DNA-bound receptor may recruit chromatin-remodelling complexes (SWI/SNF) or histone-modifying enzymes (histone acetyltransferases, methyltransferases and histone kinases) in order to open up the chromatin structure to other transcription factors and/or the transcription machinery. However, SHRs can also recruit the transcription machinery either directly through interactions with GTFs or indirectly via co-activator proteins. Subsequent to transcription initiation by RNA polymerase II, steroid receptors may act to regulate transcription elongation or mRNA processing. The different actions of steroid receptors on target gene regulation will depend upon the nature of the DNA-response element and the binding of different combinations of target proteins, and the characteristics of these interactions. Taken together, some or all of these macromolecular interactions are likely to influence the structure of the AF1 domain.
CONCLUSIONS AND FUTURE PERSPECTIVES
The last 25 years have seen tremendous progress in our understanding of the structure–function relationships of members of the nuclear receptor superfamily. This has included the isolation of receptor cDNAs, the solving at atomic resolution of the structure of the isolated DBD and LBD, the identification of binding partners, and a clearer appreciation of the role of mutations in nuclear receptors, resulting in a wide range of pathological states. The NTD is variable in terms of amino acid sequence and length, but has been shown to be important for SHR-dependent gene regulation, and is a major site for post-translational modifications. Thus, in the next quarter century, there will continue to be both challenges to, and advances in, our understanding of the structure and function of this domain. In particular, it will be important to: (i) identify gene targets and regulatory networks for each SHR, which are dependent upon the NTD; (ii) study the role of different post-translational modifications for SHR action, and identify the enzymes responsible and potential interdependence of post-translational events for receptor function; and (iii) arrive at a better structural understanding of the SHR-AF1/NTD. Ultimately, this will depend upon the solving of a high-resolution structure (perhaps of the ER with the smallest NTD or one of the non-steroid receptor members of the superfamily). In the meantime, better quantification (binding kinetics, affinity and stoichiometry) of individual receptor–protein interactions would provide a better picture of the molecular details underpinning receptor-dependent transactivation.
As befits such a large family of proteins involved in development, metabolic regulation and reproduction, there is intensive research into all aspects of SHR structure and function. In the future, it is assured that there will be further dramatic developments as researchers strive to understand the biology and physiology of SHRs, the structural and biochemical basis for receptor action and the role genetic alterations in receptor signalling play in disease.
Acknowledgments
D.N.L. is supported by a PhD studentship funded by the AICR (Association of International Cancer Research). Work in the I.J.M. laboratory is supported by the AICR and Biotechnology and Biological Sciences Research Council (U.K.). We are also grateful to colleagues in the field for fruitful and stimulating conversations on the questions addressed in this review.
References
- 1.Hollenberg S. M., Weinberger C., Ong E. S., Cerelli G., Oro A., Lebo R., Thompson E. B., Rosenfeld M. G., Evans R. M. Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature(London) 1985;318:635–641. doi: 10.1038/318635a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miesfeld R., Godowski P. J., Maler B. A., Yamamoto K. R. Glucocorticoid receptor mutants that define a small region sufficient for enhancer activation. Science. 1987;236:423–427. doi: 10.1126/science.3563519. [DOI] [PubMed] [Google Scholar]
- 3.Green S., Walter P., Kumar V., Krust A., Bornert J. M., Argos P., Chambon P. Human oestrogen receptor cDNA: sequence, expression and homology to v-erb-A. Nature (London) 1986;320:134–139. doi: 10.1038/320134a0. [DOI] [PubMed] [Google Scholar]
- 4.Escriva H., Bertrand S., Laudet V. The evolution of the nuclear receptor superfamily. Essays Biochem. 2004;40:11–26. doi: 10.1042/bse0400011. [DOI] [PubMed] [Google Scholar]
- 5.Thornton J. W. Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5671–5676. doi: 10.1073/pnas.091553298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thornton J. W., Need E., Crews D. Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science. 2003;301:1714–1717. doi: 10.1126/science.1086185. [DOI] [PubMed] [Google Scholar]
- 7.Darimont B. D., Wagner R. L., Apriletti J. W., Stallcup M. R., Kushner P. J., Baxter J. D., Fletterick R. J., Yamamoto K. R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brzozowski A. M., Pike A. C., Dauter Z., Hubbard R. E., Bonn T., Engstrom O., Ohman L., Greene G. L., Gustafsson J. A., Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature (London) 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 9.Fagart J., Wurtz J. M., Souque A., Hellal-Levy C., Moras D., Rafestin-Oblin M. E. Antagonism in the human mineralocorticoid receptor. EMBO J. 1998;17:3317–3325. doi: 10.1093/emboj/17.12.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanenbaum D. M., Wang Y., Williams S. P., Sigler P. B. Crystallographic comparison of the estrogen and progesterone receptor's ligand binding domains. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5998–6003. doi: 10.1073/pnas.95.11.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams S. P., Sigler P. B. Atomic structure of progesterone complexed with its receptor. Nature (London) 1998;393:392–396. doi: 10.1038/30775. [DOI] [PubMed] [Google Scholar]
- 12.Matias P. M., Donner P., Coelho R., Thomaz M., Peixoto C., Macedo S., Otto N., Joschko S., Scholz P., Wegg A., et al. Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations. J. Biol. Chem. 2000;275:26164–26171. doi: 10.1074/jbc.M004571200. [DOI] [PubMed] [Google Scholar]
- 13.Kauppi B., Jakob C., Farnegardh M., Yang J., Ahola H., Alarcon M., Calles K., Engstrom O., Harlan J., Muchmore S., et al. The three-dimensional structures of antagonistic and agonistic forms of the glucocorticoid receptor ligand-binding domain: RU-486 induces a transconformation that leads to active antagonism. J. Biol. Chem. 2003;278:22748–22754. doi: 10.1074/jbc.M212711200. [DOI] [PubMed] [Google Scholar]
- 14.Pike A. C., Brzozowski A. M., Hubbard R. E., Bonn T., Thorsell A. G., Engstrom O., Ljunggren J., Gustafsson J. A., Carlquist M. Structure of the ligand-binding domain of oestrogen receptor beta in the presence of a partial agonist and a full antagonist. EMBO J. 1999;18:4608–4618. doi: 10.1093/emboj/18.17.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hard T., Kellenbach E., Boelens R., Maler B. A., Dahlman K., Freedman L. P., Carlstedt-Duke J., Yamamoto K. R., Gustafsson J. A., Kaptein R. Solution structure of the glucocorticoid receptor DNA-binding domain. Science. 1990;249:157–160. doi: 10.1126/science.2115209. [DOI] [PubMed] [Google Scholar]
- 16.Schwabe J. W., Neuhaus D., Rhodes D. Solution structure of the DNA-binding domain of the oestrogen receptor. Nature (London) 1990;348:458–461. doi: 10.1038/348458a0. [DOI] [PubMed] [Google Scholar]
- 17.Schwabe J. W., Chapman L., Finch J. T., Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- 18.Shaffer P. L., Jivan A., Dollins D. E., Claessens F., Gewirth D. T. Structural basis of androgen receptor binding to selective androgen response elements. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4758–4763. doi: 10.1073/pnas.0401123101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Claessens F., Gewirth D. T. DNA recognition by nuclear receptors. Essays Biochem. 2004;40:59–72. doi: 10.1042/bse0400059. [DOI] [PubMed] [Google Scholar]
- 20.Simental J. A., Sar M., Lane M. V., French F. S., Wilson E. M. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J. Biol. Chem. 1991;266:510–518. [PubMed] [Google Scholar]
- 21.Jenster G., van der Korput H. A., Trapman J., Brinkmann A. O. Identification of two transcription activation units in the N-terminal domain of the human androgen receptor. J. Biol. Chem. 1995;270:7341–7346. doi: 10.1074/jbc.270.13.7341. [DOI] [PubMed] [Google Scholar]
- 22.Chamberlain N. L., Whitacre D. C., Miesfeld R. L. Delineation of two distinct type 1 activation functions in the androgen receptor amino-terminal domain. J. Biol. Chem. 1996;271:26772–26778. doi: 10.1074/jbc.271.43.26772. [DOI] [PubMed] [Google Scholar]
- 23.Reid J., Murray I., Watt K., Betney R., McEwan I. J. The androgen receptor interacts with multiple regions of the large subunit of general transcription factor TFIIF. J. Biol. Chem. 2002;277:41247–41253. doi: 10.1074/jbc.M205220200. [DOI] [PubMed] [Google Scholar]
- 24.Choong C. S., Kemppainen J. A., Wilson E. M. Evolution of the primate androgen receptor: a structural basis for disease. J. Mol. Evol. 1998;47:334–342. doi: 10.1007/pl00006391. [DOI] [PubMed] [Google Scholar]
- 25.Betney R., McEwan I. J. Role of conserved hydrophobic amino acids in androgen receptor AF-1 function. J. Mol. Endocrinol. 2003;31:427–439. doi: 10.1677/jme.0.0310427. [DOI] [PubMed] [Google Scholar]
- 26.He B., Bai S., Hnat A. T., Kalman R. I., Minges J. T., Patterson C., Wilson E. M. An androgen receptor NH2-terminal conserved motif interacts with the COOH terminus of the Hsp70-interacting protein (CHIP) J. Biol. Chem. 2004;279:30643–30653. doi: 10.1074/jbc.M403117200. [DOI] [PubMed] [Google Scholar]
- 27.Han G., Buchanan G., Ittmann M., Harris J. M., Yu X., Demayo F. J., Tilley W., Greenberg N. M. Mutation of the androgen receptor causes oncogenic transformation of the prostate. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1151–1156. doi: 10.1073/pnas.0408925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He B., Wilson E. M. The NH2-terminal and carboxyl-terminal interaction in the human androgen receptor. Mol. Genet. Metab. 2002;75:293–298. doi: 10.1016/S1096-7192(02)00009-4. [DOI] [PubMed] [Google Scholar]
- 29.Steketee K., Berrevoets C. A., Dubbink H. J., Doesburg P., Hersmus R., Brinkmann A. O., Trapman J. Amino acids 3–13 and amino acids in and flanking the 23FxxLF27 motif modulate the interaction between the N-terminal and ligand-binding domain of the androgen receptor. Eur. J. Biochem. 2002;269:5780–5791. doi: 10.1046/j.1432-1033.2002.03276.x. [DOI] [PubMed] [Google Scholar]
- 30.Langley E., Zhou Z. X., Wilson E. M. Evidence for an anti-parallel orientation of the ligand-activated human androgen receptor dimer. J. Biol. Chem. 1995;270:29983–29990. doi: 10.1074/jbc.270.50.29983. [DOI] [PubMed] [Google Scholar]
- 31.Langley E., Kemppainen J. A., Wilson E. M. Intermolecular NH2-/carboxyl-terminal interactions in androgen receptor dimerization revealed by mutations that cause androgen insensitivity. J. Biol. Chem. 1998;273:92–101. doi: 10.1074/jbc.273.1.92. [DOI] [PubMed] [Google Scholar]
- 32.He B., Kemppainen J. A., Voegel J. J., Gronemeyer H., Wilson E. M. Activation function 2 in the human androgen receptor ligand binding domain mediates interdomain communication with the NH2-terminal domain. J. Biol. Chem. 1999;274:37219–37225. doi: 10.1074/jbc.274.52.37219. [DOI] [PubMed] [Google Scholar]
- 33.He B., Kemppainen J. A., Wilson E. M. FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J. Biol. Chem. 2000;275:22986–22994. doi: 10.1074/jbc.M002807200. [DOI] [PubMed] [Google Scholar]
- 34.Hsu C. L., Chen Y. L., Yeh S., Ting H. J., Hu Y. C., Lin H., Wang X., Chang C. The use of phage display technique for the isolation of androgen receptor interacting peptides with (F/W)XXL(F/W) and FXXLY new signature motifs. J. Biol. Chem. 2003;278:23691–23698. doi: 10.1074/jbc.M211908200. [DOI] [PubMed] [Google Scholar]
- 35.Dubbink H. J., Hersmus R., Verma C. S., van der Korput H. A., Berrevoets C. A., van Tol J., Ziel-van der Made A. C., Brinkmann A. O., Pike A. C., Trapman J. Distinct recognition modes of FXXLF and LXXLL motifs by the androgen receptor. Mol. Endocrinol. 2004;18:2132–2150. doi: 10.1210/me.2003-0375. [DOI] [PubMed] [Google Scholar]
- 36.He B., Gampe R. T., Jr, Kole A. J., Hnat A. T., Stanley T. B., An G., Stewart E. L., Kalman R. I., Minges J. T., Wilson E. M. Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance. Mol. Cell. 2004;16:425–438. doi: 10.1016/j.molcel.2004.09.036. [DOI] [PubMed] [Google Scholar]
- 37.Hur E., Pfaff S. J., Payne E. S., Gron H., Buehrer B. M., Fletterick R. J. Recognition and accommodation at the androgen receptor coactivator binding interface. PLoS Biol. 2004;2:E274. doi: 10.1371/journal.pbio.0020274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hollenberg S. M., Evans R. M. Multiple and cooperative trans-activation domains of the human glucocorticoid receptor. Cell. 1988;55:899–906. doi: 10.1016/0092-8674(88)90145-6. [DOI] [PubMed] [Google Scholar]
- 39.Wright A. P., McEwan I. J., Dahlman-Wright K., Gustafsson J. A. High level expression of the major transactivation domain of the human glucocorticoid receptor in yeast cells inhibits endogenous gene expression and cell growth. Mol. Endocrinol. 1991;5:1366–1372. doi: 10.1210/mend-5-10-1366. [DOI] [PubMed] [Google Scholar]
- 40.McEwan I. J., Wright A. P., Dahlman-Wright K., Carlstedt-Duke J., Gustafsson J. A. Direct interaction of the tau 1 transactivation domain of the human glucocorticoid receptor with the basal transcriptional machinery. Mol. Cell. Biol. 1993;13:399–407. doi: 10.1128/mcb.13.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McEwan I. J., Almlof T., Wikstrom A. C., Dahlman-Wright K., Wright A. P., Gustafsson J. A. The glucocorticoid receptor functions at multiple steps during transcription initiation by RNA polymerase II. J. Biol. Chem. 1994;269:25629–25636. [PubMed] [Google Scholar]
- 42.Ford J., McEwan I. J., Wright A. P., Gustafsson J. A. Involvement of the transcription factor IID protein complex in gene activation by the N-terminal transactivation domain of the glucocorticoid receptor in vitro. Mol. Endocrinol. 1997;11:1467–1475. doi: 10.1210/mend.11.10.9995. [DOI] [PubMed] [Google Scholar]
- 43.Dahlman-Wright K., Almlof T., McEwan I. J., Gustafsson J. A., Wright A. P. Delineation of a small region within the major transactivation domain of the human glucocorticoid receptor that mediates transactivation of gene expression. Proc. Natl. Acad. Sci. U.S.A. 1994;91:1619–1623. doi: 10.1073/pnas.91.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Almlof T., Gustafsson J. A., Wright A. P. Role of hydrophobic amino acid clusters in the transactivation activity of the human glucocorticoid receptor. Mol. Cell. Biol. 1997;17:934–945. doi: 10.1128/mcb.17.2.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Almlof T., Wallberg A. E., Gustafsson J. A., Wright A. P. Role of important hydrophobic amino acids in the interaction between the glucocorticoid receptor tau 1-core activation domain and target factors. Biochemistry. 1998;37:9586–9594. doi: 10.1021/bi973029x. [DOI] [PubMed] [Google Scholar]
- 46.Almlof T., Wright A. P., Gustafsson J. A. Role of acidic and phosphorylated residues in gene activation by the glucocorticoid receptor. J. Biol. Chem. 1995;270:17535–17540. doi: 10.1074/jbc.270.29.17535. [DOI] [PubMed] [Google Scholar]
- 47.Iniguez-Lluhi J. A., Lou D. Y., Yamamoto K. R. Three amino acid substitutions selectively disrupt the activation but not the repression function of the glucocorticoid receptor N terminus. J. Biol. Chem. 1997;272:4149–4156. doi: 10.1074/jbc.272.7.4149. [DOI] [PubMed] [Google Scholar]
- 48.Rogatsky I., Hittelman A. B., Pearce D., Garabedian M. J. Distinct glucocorticoid receptor transcriptional regulatory surfaces mediate the cytotoxic and cytostatic effects of glucocorticoids. Mol. Cell. Biol. 1999;19:5036–5049. doi: 10.1128/mcb.19.7.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogatsky I., Wang J. C., Derynck M. K., Nonaka D. F., Khodabakhsh D. B., Haqq C. M., Darimont B. D., Garabedian M. J., Yamamoto K. R. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13845–13850. doi: 10.1073/pnas.2336092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Govindan M. V., Warriar N. Reconstitution of the N-terminal transcription activation function of human mineralocorticoid receptor in a defective human glucocorticoid receptor. J. Biol. Chem. 1998;273:24439–24447. doi: 10.1074/jbc.273.38.24439. [DOI] [PubMed] [Google Scholar]
- 51.Fuse H., Kitagawa H., Kato S. Characterization of transactivational property and coactivator mediation of rat mineralocorticoid receptor activation function-1 (AF-1) Mol. Endocrinol. 2000;14:889–899. doi: 10.1210/mend.14.6.0467. [DOI] [PubMed] [Google Scholar]
- 52.Metzger D., Ali S., Bornert J. M., Chambon P. Characterization of the amino-terminal transcriptional activation function of the human estrogen receptor in animal and yeast cells. J. Biol. Chem. 1995;270:9535–9542. doi: 10.1074/jbc.270.16.9535. [DOI] [PubMed] [Google Scholar]
- 53.McInerney E. M., Katzenellenbogen B. S. Different regions in activation function-1 of the human estrogen receptor required for antiestrogen- and estradiol-dependent transcription activation. J. Biol. Chem. 1996;271:24172–24178. doi: 10.1074/jbc.271.39.24172. [DOI] [PubMed] [Google Scholar]
- 54.Delaunay F., Pettersson K., Tujague M., Gustafsson J. A. Functional differences between the amino-terminal domains of estrogen receptors alpha and beta. Mol. Pharmacol. 2000;58:584–590. doi: 10.1124/mol.58.3.584. [DOI] [PubMed] [Google Scholar]
- 55.Metivier R., Penot G., Flouriot G., Pakdel F. Synergism between ERalpha transactivation function 1 (AF-1) and AF-2 mediated by steroid receptor coactivator protein-1: requirement for the AF-1 alpha-helical core and for a direct interaction between the N- and C-terminal domains. Mol. Endocrinol. 2001;15:1953–1970. doi: 10.1210/mend.15.11.0727. [DOI] [PubMed] [Google Scholar]
- 56.Kuiper G. G., Enmark E., Pelto-Huikko M., Nilsson S., Gustafsson J. A. Cloning of a novel receptor expressed in rat prostate and ovary. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takimoto G. S., Tung L., Abdel-Hafiz H., Abel M. G., Sartorius C. A., Richer J. K., Jacobsen B. M., Bain D. L., Horwitz K. B. Functional properties of the N-terminal region of progesterone receptors and their mechanistic relationship to structure. J. Steroid Biochem. Mol. Biol. 2003;85:209–219. doi: 10.1016/s0960-0760(03)00197-3. [DOI] [PubMed] [Google Scholar]
- 58.Meyer M. E., Quirin-Stricker C., Lerouge T., Bocquel M. T., Gronemeyer H. A limiting factor mediates the differential activation of promoters by the human progesterone receptor isoforms. J. Biol. Chem. 1992;267:10882–10887. [PubMed] [Google Scholar]
- 59.Tung L., Shen T., Abel M. G., Powell R. L., Takimoto G. S., Sartorius C. A., Horwitz K. B. Mapping the unique activation function 3 in the progesterone B-receptor upstream segment. Two LXXLL motifs and a tryptophan residue are required for activity. J. Biol. Chem. 2001;276:39843–39851. doi: 10.1074/jbc.M106843200. [DOI] [PubMed] [Google Scholar]
- 60.Hovland A. R., Powell R. L., Takimoto G. S., Tung L., Horwitz K. B. An N-terminal inhibitory function, IF, suppresses transcription by the A-isoform but not the B-isoform of human progesterone receptors. J. Biol. Chem. 1998;273:5455–5460. doi: 10.1074/jbc.273.10.5455. [DOI] [PubMed] [Google Scholar]
- 61.Huse B., Verca S. B., Matthey P., Rusconi S. Definition of a negative modulation domain in the human progesterone receptor. Mol. Endocrinol. 1998;12:1334–1342. doi: 10.1210/mend.12.9.0164. [DOI] [PubMed] [Google Scholar]
- 62.Wang D., Simons S. S., Jr Corepressor binding to progesterone and glucocorticoid receptors involves the AF-1 domain and is inhibited by molybdate. Mol. Endocrinol. 2005;19:1483–1500. doi: 10.1210/me.2005-0012. [DOI] [PubMed] [Google Scholar]
- 63.Weigel N. L. Steroid hormone receptors and their regulation by phosphorylation. Biochem. J. 1996;319:657–667. doi: 10.1042/bj3190657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lange C. A. Making sense of cross-talk between steroid hormone receptors and intracellular signaling pathways: Who will have the last word? Mol. Endocrinol. 2004;18:269–278. doi: 10.1210/me.2003-0331. [DOI] [PubMed] [Google Scholar]
- 65.McEwan I. J. Sex, drugs and gene expression: Signalling by members of the nuclear receptor superfamily. Essays Biochem. 2004;40:1–10. doi: 10.1042/bse0400001. [DOI] [PubMed] [Google Scholar]
- 66.Lin H. K., Wang L., Hu Y. C., Altuwaijri S., Chang C. Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by akt require Mdm2 E3 ligase. EMBO J. 2002;21:4037–4048. doi: 10.1093/emboj/cdf406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fu M., Wang C., Wang J., Zhang X., Sakamaki T., Yeung Y. G., Chang C., Hopp T., Fuqua S. A., Jaffray E., et al. Androgen receptor acetylation governs transactivation and MEKK1-induced apoptosis without affecting in vitro sumoylation and trans-repression function. Mol. Cell. Biol. 2002;22:3373–3388. doi: 10.1128/MCB.22.10.3373-3388.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jenster G., de Ruiter P. E., van der Korput H. A., Kuiper G. G., Trapman J., Brinkmann A. O. Changes in the abundance of androgen receptor isotypes: effects of ligand treatment, glutamine-stretch variation, and mutation of putative phosphorylation sites. Biochemistry. 1994;33:14064–14072. doi: 10.1021/bi00251a015. [DOI] [PubMed] [Google Scholar]
- 69.Gioeli D., Ficarro S. B., Kwiek J. J., Aaronson D., Hancock M., Catling A. D., White F. M., Christian R. E., Settlage R. E., Shabanowitz J., et al. Androgen receptor phosphorylation. Regulation and identification of the phosphorylation sites. J. Biol. Chem. 2002;277:29304–29314. doi: 10.1074/jbc.M204131200. [DOI] [PubMed] [Google Scholar]
- 70.Yeh S., Lin H. K., Kang H. Y., Thin T. H., Lin M. F., Chang C. From HER2/Neu signal cascade to androgen receptor and its coactivators: a novel pathway by induction of androgen target genes through MAP kinase in prostate cancer cells. Proc. Natl. Acad. Sci. U.S.A. 1999;96:5458–5463. doi: 10.1073/pnas.96.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wen Y., Hu M. C., Makino K., Spohn B., Bartholomeusz G., Yan D. H., Hung M. C. HER-2/neu promotes androgen-independent survival and growth of prostate cancer cells through the akt pathway. Cancer Res. 2000;60:6841–6845. [PubMed] [Google Scholar]
- 72.Lin H. K., Yeh S., Kang H. Y., Chang C. Akt suppresses androgen-induced apoptosis by phosphorylating and inhibiting androgen receptor. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7200–7205. doi: 10.1073/pnas.121173298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Manin M., Baron S., Goossens K., Beaudoin C., Jean C., Veyssiere G., Verhoeven G., Morel L. Androgen receptor expression is regulated by the phosphoinositide 3-kinase/Akt pathway in normal and tumoral epithelial cells. Biochem. J. 2002;366:729–736. doi: 10.1042/BJ20020585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bodwell J. E., Orti E., Coull J. M., Pappin D. J., Smith L. I., Swift F. Identification of phosphorylated sites in the mouse glucocorticoid receptor. J. Biol. Chem. 1991;266:7549–7555. [PubMed] [Google Scholar]
- 75.Krstic M. D., Rogatsky I., Yamamoto K. R., Garabedian M. J. Mitogen-activated and cyclin-dependent protein kinases selectively and differentially modulate transcriptional enhancement by the glucocorticoid receptor. Mol. Cell. Biol. 1997;17:3947–3954. doi: 10.1128/mcb.17.7.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rogatsky I., Waase C. L., Garabedian M. J. Phosphorylation and inhibition of rat glucocorticoid receptor transcriptional activation by glycogen synthase kinase-3 (GSK-3). Species-specific differences between human and rat glucocorticoid receptor signaling as revealed through GSK-3 phosphorylation. J. Biol. Chem. 1998;273:14315–14321. doi: 10.1074/jbc.273.23.14315. [DOI] [PubMed] [Google Scholar]
- 77.Wang Z., Frederick J., Garabedian M. J. Deciphering the phosphorylation ‘code’ of the glucocorticoid receptor in vivo. J. Biol. Chem. 2002;277:26573–26580. doi: 10.1074/jbc.M110530200. [DOI] [PubMed] [Google Scholar]
- 78.Alnemri E. S., Maksymowych A. B., Robertson N. M., Litwack G. Overexpression and characterization of the human mineralocorticoid receptor. J. Biol. Chem. 1991;266:18072–18081. [PubMed] [Google Scholar]
- 79.Galigniana M. D. Native rat kidney mineralocorticoid receptor is a phosphoprotein whose transformation to a DNA-binding form is induced by phosphatases. Biochem. J. 1998;333:555–563. doi: 10.1042/bj3330555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Takimoto G. S., Hovland A. R., Tasset D. M., Melville M. Y., Tung L., Horwitz K. B. Role of phosphorylation on DNA binding and transcriptional functions of human progesterone receptors. J. Biol. Chem. 1996;271:13308–13316. doi: 10.1074/jbc.271.23.13308. [DOI] [PubMed] [Google Scholar]
- 81.Lange C. A., Shen T., Horwitz K. B. Phosphorylation of human progesterone receptors at serine-294 by mitogen-activated protein kinase signals their degradation by the 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1032–1037. doi: 10.1073/pnas.97.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tremblay A., Tremblay G. B., Labrie F., Giguere V. Ligand-independent recruitment of SRC-1 to estrogen receptor beta through phosphorylation of activation function AF-1. Mol. Cell. 1999;3:513–519. doi: 10.1016/s1097-2765(00)80479-7. [DOI] [PubMed] [Google Scholar]
- 83.Chen D., Riedl T., Washbrook E., Pace P. E., Coombes R. C., Egly J. M., Ali S. Activation of estrogen receptor alpha by S118 phosphorylation involves a ligand-dependent interaction with TFIIH and participation of CDK7. Mol. Cell. 2000;6:127–137. [PubMed] [Google Scholar]
- 84.Dutertre M., Smith C. L. Ligand-independent interactions of p160/steroid receptor coactivators and CREB-binding protein (CBP) with estrogen receptor-alpha: Regulation by phosphorylation sites in the A/B region depends on other receptor domains. Mol. Endocrinol. 2003;17:1296–1314. doi: 10.1210/me.2001-0316. [DOI] [PubMed] [Google Scholar]
- 85.Endoh H., Maruyama K., Masuhiro Y., Kobayashi Y., Goto M., Tai H., Yanagisawa J., Metzger D., Hashimoto S., Kato S. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor alpha. Mol. Cell. Biol. 1999;19:5363–5372. doi: 10.1128/mcb.19.8.5363. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 86.Metivier R., Gay F. A., Hubner M. R., Flouriot G., Salbert G., Gannon F., Kah O., Pakdel F. Formation of an hER alpha-COUP-TFI complex enhances hER alpha AF-1 through Ser118 phosphorylation by MAPK. EMBO J. 2002;21:3443–3453. doi: 10.1093/emboj/cdf344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kotaja N., Aittomaki S., Silvennoinen O., Palvimo J. J., Janne O. A. ARIP3 (androgen receptor-interacting protein 3) and other PIAS (protein inhibitor of activated STAT) proteins differ in their ability to modulate steroid receptor-dependent transcriptional activation. Mol. Endocrinol. 2000;14:1986–2000. doi: 10.1210/mend.14.12.0569. [DOI] [PubMed] [Google Scholar]
- 88.Poukka H., Karvonen U., Janne O. A., Palvimo J. J. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1) Proc. Natl. Acad. Sci. U.S.A. 2000;97:14145–14150. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nishida T., Yasuda H. PIAS1 and PIASxalpha function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J. Biol. Chem. 2002;277:41311–41317. doi: 10.1074/jbc.M206741200. [DOI] [PubMed] [Google Scholar]
- 90.Tian S., Poukka H., Palvimo J. J., Janne O. A. Small ubiquitin-related modifier-1 (SUMO-1) modification of the glucocorticoid receptor. Biochem. J. 2002;367:907–911. doi: 10.1042/BJ20021085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tallec L. P., Kirsh O., Lecomte M. C., Viengchareun S., Zennaro M. C., Dejean A., Lombes M. Protein inhibitor of activated signal transducer and activator of transcription 1 interacts with the N-terminal domain of mineralocorticoid receptor and represses its transcriptional activity: Implication of small ubiquitin-related modifier 1 modification. Mol. Endocrinol. 2003;17:2529–2542. doi: 10.1210/me.2003-0299. [DOI] [PubMed] [Google Scholar]
- 92.Callewaert L., Verrijdt G., Haelens A., Claessens F. Differential effect of small ubiquitin-like modifier (SUMO)-ylation of the androgen receptor in the control of cooperativity on selective versus canonical response elements. Mol. Endocrinol. 2004;18:1438–1449. doi: 10.1210/me.2003-0313. [DOI] [PubMed] [Google Scholar]
- 93.Abdel-Hafiz H., Takimoto G. S., Tung L., Horwitz K. B. The inhibitory function in human progesterone receptor N termini binds SUMO-1 protein to regulate autoinhibition and transrepression. J. Biol. Chem. 2002;277:33950–33956. doi: 10.1074/jbc.M204573200. [DOI] [PubMed] [Google Scholar]
- 94.Chauchereau A., Amazit L., Quesne M., Guiochon-Mantel A., Milgrom E. Sumoylation of the progesterone receptor and of the steroid receptor coactivator SRC-1. J. Biol. Chem. 2003;278:12335–12343. doi: 10.1074/jbc.M207148200. [DOI] [PubMed] [Google Scholar]
- 95.Iniguez-Lluhi J. A., Pearce D. A common motif within the negative regulatory regions of multiple factors inhibits their transcriptional synergy. Mol. Cell. Biol. 2000;20:6040–6050. doi: 10.1128/mcb.20.16.6040-6050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wright A. P., Gustafsson J. A. Mechanism of synergistic transcriptional transactivation by the human glucocorticoid receptor. Proc. Natl. Acad. Sci. U.S.A. 1991;88:8283–8287. doi: 10.1073/pnas.88.19.8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schule R., Muller M., Kaltschmidt C., Renkawitz R. Many transcription factors interact synergistically with steroid receptors. Science. 1988;242:1418–1420. doi: 10.1126/science.3201230. [DOI] [PubMed] [Google Scholar]
- 98.Belandia B., Parker M. G. Nuclear receptors: A rendezvous for chromatin remodeling factors. Cell. 2003;114:277–280. doi: 10.1016/s0092-8674(03)00599-3. [DOI] [PubMed] [Google Scholar]
- 99.Acevedo M. L., Kraus W. L. Transcriptional activation by nuclear receptors. Essays Biochem. 2004;40:73–88. doi: 10.1042/bse0400073. [DOI] [PubMed] [Google Scholar]
- 100.Kinyamu H. K., Archer T. K. Modifying chromatin to permit steroid hormone receptor-dependent transcription. Biochim. Biophys. Acta. 2004;1677:30–45. doi: 10.1016/j.bbaexp.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 101.Moehren U., Eckey M., Baniahmad A. Gene repression by nuclear hormone receptors. Essays Biochem. 2004;40:89–104. doi: 10.1042/bse0400089. [DOI] [PubMed] [Google Scholar]
- 102.Cato A. C., Wade E. Molecular mechanisms of anti-inflammatory action of glucocorticoids. Bioessays. 1996;18:371–378. doi: 10.1002/bies.950180507. [DOI] [PubMed] [Google Scholar]
- 103.Butler J. E., Kadonaga J. T. The RNA polymerase II core promoter: A key component in the regulation of gene expression. Genes Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- 104.Woychik N. A., Hampsey M. The RNA polymerase II machinery: Structure illuminates function. Cell. 2002;108:453–463. doi: 10.1016/s0092-8674(02)00646-3. [DOI] [PubMed] [Google Scholar]
- 105.Shang Y., Hu X., DiRenzo J., Lazar M. A., Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 106.Shang Y., Myers M., Brown M. Formation of the androgen receptor transcription complex. Mol. Cell. 2002;9:601–610. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 107.Huang Z. Q., Li J., Sachs L. M., Cole P. A., Wong J. A role for cofactor-cofactor and cofactor-histone interactions in targeting p300, SWI/SNF and mediator for transcription. EMBO J. 2003;22:2146–2155. doi: 10.1093/emboj/cdg219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Metivier R., Penot G., Hubner M. R., Reid G., Brand H., Kos M., Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 109.Li X., Wong J., Tsai S. Y., Tsai M. J., O'Malley B. W. Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification. Mol. Cell. Biol. 2003;23:3763–3773. doi: 10.1128/MCB.23.11.3763-3773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sadovsky Y., Webb P., Lopez G., Baxter J. D., Fitzpatrick P. M., Gizang-Ginsberg E., Cavailles V., Parker M. G., Kushner P. J. Transcriptional activators differ in their responses to overexpression of TATA-box-binding protein. Mol. Cell. Biol. 1995;15:1554–1563. doi: 10.1128/mcb.15.3.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McEwan I. J., Gustafsson J. Interaction of the human androgen receptor transactivation function with the general transcription factor TFIIF. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8485–8490. doi: 10.1073/pnas.94.16.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Warnmark A., Wikstrom A., Wright A. P., Gustafsson J. A., Hard T. The N-terminal regions of estrogen receptor alpha and beta are unstructured in vitro and show different TBP binding properties. J. Biol. Chem. 2001;276:45939–45944. doi: 10.1074/jbc.M107875200. [DOI] [PubMed] [Google Scholar]
- 113.Kumar R., Lee J. C., Bolen D. W., Thompson E. B. The conformation of the glucocorticoid receptor af1/tau1 domain induced by osmolyte binds co-regulatory proteins. J. Biol. Chem. 2001;276:18146–18152. doi: 10.1074/jbc.M100825200. [DOI] [PubMed] [Google Scholar]
- 114.Lee D. K., Duan H. O., Chang C. From androgen receptor to the general transcription factor TFIIH. Identification of cdk activating kinase (CAK) as an androgen receptor NH2-terminal associated coactivator. J. Biol. Chem. 2000;275:9308–9313. doi: 10.1074/jbc.275.13.9308. [DOI] [PubMed] [Google Scholar]
- 115.Lee D. K., Duan H. O., Chang C. Androgen receptor interacts with the positive elongation factor P-TEFb and enhances the efficiency of transcriptional elongation. J. Biol. Chem. 2001;276:9978–9984. doi: 10.1074/jbc.M002285200. [DOI] [PubMed] [Google Scholar]
- 116.Pascual-Le Tallec L., Simone F., Viengchareun S., Meduri G., Thirman M. J., Lombes M. The elongation factor ELL (eleven-nineteen lysine-rich leukemia) is a selective coregulator for steroid receptor functions. Mol. Endocrinol. 2005;19:1158–1169. doi: 10.1210/me.2004-0331. [DOI] [PubMed] [Google Scholar]
- 117.Tora L., White J., Brou C., Tasset D., Webster N., Scheer E., Chambon P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989;59:477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- 118.Berger S. L., Cress W. D., Cress A., Triezenberg S. J., Guarente L. Selective inhibition of activated but not basal transcription by the acidic activation domain of VP16: Evidence for transcriptional adaptors. Cell. 1990;61:1199–1208. doi: 10.1016/0092-8674(90)90684-7. [DOI] [PubMed] [Google Scholar]
- 119.Tasset D., Tora L., Fromental C., Scheer E., Chambon P. Distinct classes of transcriptional activating domains function by different mechanisms. Cell. 1990;62:1177–1187. doi: 10.1016/0092-8674(90)90394-t. [DOI] [PubMed] [Google Scholar]
- 120.Heery D. M., Kalkhoven E., Hoare S., Parker M. G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 121.Webb P., Nguyen P., Shinsako J., Anderson C., Feng W., Nguyen M. P., Chen D., Huang S. M., Subramanian S., McKinerney E., Katzenellenbogen B. S., Stallcup M. R., Kushner P. J. Estrogen receptor activation function 1 works by binding p160 coactivator proteins. Mol. Endocrinol. 1998;12:1605–1618. doi: 10.1210/mend.12.10.0185. [DOI] [PubMed] [Google Scholar]
- 122.Onate S. A., Boonyaratanakornkit V., Spencer T. E., Tsai S. Y., Tsai M. J., Edwards D. P., O'Malley B. W. The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J. Biol. Chem. 1998;273:12101–12108. doi: 10.1074/jbc.273.20.12101. [DOI] [PubMed] [Google Scholar]
- 123.Ma H., Hong H., Huang S. M., Irvine R. A., Webb P., Kushner P. J., Coetzee G. A., Stallcup M. R. Multiple signal input and output domains of the 160-kilodalton nuclear receptor coactivator proteins. Mol. Cell. Biol. 1999;19:6164–6173. doi: 10.1128/mcb.19.9.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Alen P., Claessens F., Verhoeven G., Rombauts W., Peeters B. The androgen receptor amino-terminal domain plays a key role in p160 coactivator-stimulated gene transcription. Mol. Cell. Biol. 1999;19:6085–6097. doi: 10.1128/mcb.19.9.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bevan C. L., Hoare S., Claessens F., Heery D. M., Parker M. G. The AF1 and AF2 domains of the androgen receptor interact with distinct regions of SRC1. Mol. Cell. Biol. 1999;19:8383–8392. doi: 10.1128/mcb.19.12.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tan J. A., Hall S. H., Petrusz P., French F. S. Thyroid receptor activator molecule, TRAM-1, is an androgen receptor coactivator. Endocrinology. 2000;141:3440–3450. doi: 10.1210/endo.141.9.7680. [DOI] [PubMed] [Google Scholar]
- 127.Tetel M. J., Giangrande P. H., Leonhardt S. A., McDonnell D. P., Edwards D. P. Hormone-dependent interaction between the amino- and carboxyl-terminal domains of progesterone receptor in vitro and in vivo. Mol. Endocrinol. 1999;13:910–924. doi: 10.1210/mend.13.6.0300. [DOI] [PubMed] [Google Scholar]
- 128.Belandia B., Powell S. M., Garcia-Pedrero J. M., Walker M. M., Bevan C. L., Parker M. G. Hey1, a mediator of notch signaling, is an androgen receptor corepressor. Mol. Cell. Biol. 2005;25:1425–1436. doi: 10.1128/MCB.25.4.1425-1436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Obradovic D., Tirard M., Nemethy Z., Hirsch O., Gronemeyer H., Almeida O. F. DAXX, FLASH, and FAF-1 modulate mineralocorticoid and glucocorticoid receptor-mediated transcription in hippocampal cells – toward a basis for the opposite actions elicited by two nuclear receptors? Mol. Pharmacol. 2004;65:761–769. doi: 10.1124/mol.65.3.761. [DOI] [PubMed] [Google Scholar]
- 130.Kino T., Chrousos G. P. Tumor necrosis factor alpha receptor- and fas-associated FLASH inhibit transcriptional activity of the glucocorticoid receptor by binding to and interfering with its interaction with p160 type nuclear receptor coactivators. J. Biol. Chem. 2003;278:3023–3029. doi: 10.1074/jbc.M209234200. [DOI] [PubMed] [Google Scholar]
- 131.Kino T., Ichijo T., Chrousos G. P. FLASH interacts with p160 coactivator subtypes and differentially suppresses transcriptional activity of steroid hormone receptors. J. Steroid Biochem. Mol. Biol. 2004;92:357–363. doi: 10.1016/j.jsbmb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 132.Shatkina L., Mink S., Rogatsch H., Klocker H., Langer G., Nestl A., Cato A. C. The cochaperone bag-1L enhances androgen receptor action via interaction with the NH2-terminal region of the receptor. Mol. Cell. Biol. 2003;23:7189–7197. doi: 10.1128/MCB.23.20.7189-7197.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schneikert J., Hubner S., Langer G., Petri T., Jaattela M., Reed J., Cato A. C. Hsp70-RAP46 interaction in downregulation of DNA binding by glucocorticoid receptor. EMBO J. 2000;19:6508–6516. doi: 10.1093/emboj/19.23.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Reid J., Kelly S. M., Watt K., Price N. C., McEwan I. J. Conformational analysis of the androgen receptor amino-terminal domain involved in transactivation. Influence of structure-stabilizing solutes and protein-protein interactions. J. Biol. Chem. 2002;277:20079–20086. doi: 10.1074/jbc.M201003200. [DOI] [PubMed] [Google Scholar]
- 135.Dunker A. K., Brown C. J., Lawson J. D., Iakoucheva L. M., Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 136.Dyson H. J., Wright P. E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 137.Kumar R., Betney R., Li J., Thompson E. B., McEwan I. J. Induced alpha-helix structure in AF1 of the androgen receptor upon binding transcription factor TFIIF. Biochemistry. 2004;43:3008–3013. doi: 10.1021/bi035934p. [DOI] [PubMed] [Google Scholar]
- 138.Dahlman-Wright K., Baumann H., McEwan I. J., Almlof T., Wright A. P., Gustafsson J. A., Hard T. Structural characterization of a minimal functional transactivation domain from the human glucocorticoid receptor. Proc. Natl. Acad. Sci. U.S.A. 1995;92:1699–1703. doi: 10.1073/pnas.92.5.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kumar R., Volk D. E., Li J., Lee J. C., Gorenstein D. G., Thompson E. B. TATA box binding protein induces structure in the recombinant glucocorticoid receptor AF1 domain. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16425–16430. doi: 10.1073/pnas.0407160101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Baskakov I. V., Kumar R., Srinivasan G., Ji Y. S., Bolen D. W., Thompson E. B. Trimethylamine N-oxide-induced cooperative folding of an intrinsically unfolded transcription-activating fragment of human glucocorticoid receptor. J. Biol. Chem. 1999;274:10693–10696. doi: 10.1074/jbc.274.16.10693. [DOI] [PubMed] [Google Scholar]
- 141.Dahlman-Wright K., McEwan I. J. Structural studies of mutant glucocorticoid receptor transactivation domains establish a link between transactivation activity in vivo and alpha-helix-forming potential in vitro. Biochemistry. 1996;35:1323–1327. doi: 10.1021/bi952409k. [DOI] [PubMed] [Google Scholar]
- 142.Warnmark A., Gustafsson J. A., Wright A. P. Architectural principles for the structure and function of the glucocorticoid receptor tau 1 core activation domain. J. Biol. Chem. 2000;275:15014–15018. doi: 10.1074/jbc.M001007200. [DOI] [PubMed] [Google Scholar]
- 143.Bain D. L., Franden M. A., McManaman J. L., Takimoto G. S., Horwitz K. B. The N-terminal region of the human progesterone A-receptor. Structural analysis and the influence of the DNA binding domain. J. Biol. Chem. 2000;275:7313–7320. doi: 10.1074/jbc.275.10.7313. [DOI] [PubMed] [Google Scholar]
- 144.Bain D. L., Franden M. A., McManaman J. L., Takimoto G. S., Horwitz K. B. The N-terminal region of human progesterone B-receptors: biophysical and biochemical comparison to A-receptors. J. Biol. Chem. 2001;276:23825–23831. doi: 10.1074/jbc.M102611200. [DOI] [PubMed] [Google Scholar]
- 145.Gaboriaud C., Uze G., Lutfalla G., Mogensen K. Hydrophobic cluster analysis reveals duplication in the external structure of human alpha-interferon receptor and homology with gamma-interferon receptor external domain. FEBS Lett. 1990;269:1–3. doi: 10.1016/0014-5793(90)81103-u. [DOI] [PubMed] [Google Scholar]
- 146.Callebaut I., Labesse G., Durand P., Poupon A., Canard L., Chomilier J., Henrissat B., Mornon J. P. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell Mol. Life Sci. 1997;53:621–645. doi: 10.1007/s000180050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lefstin J. A., Yamamoto K. R. Allosteric effects of DNA on transcriptional regulators. Nature (London) 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- 148.Kumar R., Baskakov I. V., Srinivasan G., Bolen D. W., Lee J. C., Thompson E. B. Interdomain signaling in a two-domain fragment of the human glucocorticoid receptor. J. Biol. Chem. 1999;274:24737–24741. doi: 10.1074/jbc.274.35.24737. [DOI] [PubMed] [Google Scholar]
- 149.Greenfield N., Vijayanathan V., Thomas T. J., Gallo M. A., Thomas T. Increase in the stability and helical content of estrogen receptor alpha in the presence of the estrogen response element: analysis by circular dichroism spectroscopy. Biochemistry. 2001;40:6646–6652. doi: 10.1021/bi002846l. [DOI] [PubMed] [Google Scholar]
- 150.Traish A. M., Pavao M. Binding of site-directed monoclonal antibodies to an epitope located in the A/B region (amino acids 140–154) of human estrogen receptor-induced conformational changes in an epitope in the DNA-binding domain. Steroids. 1996;61:549–556. doi: 10.1016/s0039-128x(96)00109-2. [DOI] [PubMed] [Google Scholar]
- 151.Wood J. R., Greene G. L., Nardulli A. M. Estrogen response elements function as allosteric modulators of estrogen receptor conformation. Mol. Cell. Biol. 1998;18:1927–1934. doi: 10.1128/mcb.18.4.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Klinge C. M., Jernigan S. C., Smith S. L., Tyulmenkov V. V., Kulakosky P. C. Estrogen response element sequence impacts the conformation and transcriptional activity of estrogen receptor alpha. Mol. Cell. Endocrinol. 2001;174:151–166. doi: 10.1016/s0303-7207(01)00382-3. [DOI] [PubMed] [Google Scholar]
- 153.Loven M. A., Wood J. R., Nardulli A. M. Interaction of estrogen receptors alpha and beta with estrogen response elements. Mol. Cell. Endocrinol. 2001;181:151–163. doi: 10.1016/s0303-7207(01)00491-9. [DOI] [PubMed] [Google Scholar]
- 154.Brodie J., McEwan I. J. Intra-domain communication between the N-terminal and DNA-binding domains of the androgen receptor: modulation of androgen response element DNA binding. J. Mol. Endocrinol. 2005;34:603–615. doi: 10.1677/jme.1.01723. [DOI] [PubMed] [Google Scholar]
- 155.Geserick C., Meyer H. A., Barbulescu K., Haendler B. Differential modulation of androgen receptor action by deoxyribonucleic acid response elements. Mol. Endocrinol. 2003;17:1738–1750. doi: 10.1210/me.2002-0379. [DOI] [PubMed] [Google Scholar]
- 156.Hermann S., Berndt K. D., Wright A. P. How transcriptional activators bind target proteins. J. Biol. Chem. 2001;276:40127–40132. doi: 10.1074/jbc.M103793200. [DOI] [PubMed] [Google Scholar]
- 157.Combet C., Blanchet C., Geourjon C., Deleage G. NPS@: network protein sequence analysis. Trends Biochem. Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 158.Hsiao P. W., Lin D. L., Nakao R., Chang C. The linkage of Kennedy's neuron disease to ARA24, the first identified androgen receptor polyglutamine region-associated coactivator. J. Biol. Chem. 1999;274:20229–20234. doi: 10.1074/jbc.274.29.20229. [DOI] [PubMed] [Google Scholar]
- 159.Hsiao P. W., Chang C. Isolation and characterization of ARA160 as the first androgen receptor N-terminal-associated coactivator in human prostate cells. J. Biol. Chem. 1999;274:22373–22379. doi: 10.1074/jbc.274.32.22373. [DOI] [PubMed] [Google Scholar]
- 160.Markus S. M., Taneja S. S., Logan S. K., Li W., Ha S., Hittelman A. B., Rogatsky I., Garabedian M. J. Identification and characterization of ART-27, a novel coactivator for the androgen receptor N terminus. Mol. Biol. Cell. 2002;13:670–682. doi: 10.1091/mbc.01-10-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ikonen T., Palvimo J. J., Janne O. A. Interaction between the amino- and carboxyl-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J. Biol. Chem. 1997;272:29821–29828. doi: 10.1074/jbc.272.47.29821. [DOI] [PubMed] [Google Scholar]
- 162.Aarnisalo P., Palvimo J. J., Janne O. A. CREB-binding protein in androgen receptor-mediated signaling. Proc. Natl. Acad. Sci. U.S.A. 1998;95:2122–2127. doi: 10.1073/pnas.95.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fronsdal K., Engedal N., Slagsvold T., Saatcioglu F. CREB binding protein is a coactivator for the androgen receptor and mediates cross-talk with AP-1. J. Biol. Chem. 1998;273:31853–31859. doi: 10.1074/jbc.273.48.31853. [DOI] [PubMed] [Google Scholar]
- 164.Kitagawa H., Yanagisawa J., Fuse H., Ogawa S., Yogiashi Y., Okuno A., Nagasawa H., Nakajima T., Matsumoto T., Kato S. Ligand-selective potentiation of rat mineralocorticoid receptor activation function 1 by a CBP-containing histone acetyltransferase complex. Mol. Cell. Biol. 2002;22:3698–3706. doi: 10.1128/MCB.22.11.3698-3706.2002. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 165.Kobayashi Y., Kitamoto T., Masuhiro Y., Watanabe M., Kase T., Metzger D., Yanagisawa J., Kato S. p300 mediates functional synergism between AF-1 and AF-2 of estrogen receptor alpha and beta by interacting directly with the N-terminal A/B domains. J. Biol. Chem. 2000;275:15645–15651. doi: 10.1074/jbc.M000042200. [DOI] [PubMed] [Google Scholar]
- 166.Henriksson A., Almlof T., Ford J., McEwan I. J., Gustafsson J. A., Wright A. P. Role of the ada adaptor complex in gene activation by the glucocorticoid receptor. Mol. Cell. Biol. 1997;17:3065–3073. doi: 10.1128/mcb.17.6.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wallberg A. E., Neely K. E., Hassan A. H., Gustafsson J. A., Workman J. L., Wright A. P. Recruitment of the SWI-SNF chromatin remodeling complex as a mechanism of gene activation by the glucocorticoid receptor tau1 activation domain. Mol. Cell. Biol. 2000;20:2004–2013. doi: 10.1128/mcb.20.6.2004-2013.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hittelman A. B., Burakov D., Iniguez-Lluhi J. A., Freedman L. P., Garabedian M. J. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 1999;18:5380–5388. doi: 10.1093/emboj/18.19.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yu X., Li P., Roeder R. G., Wang Z. Inhibition of androgen receptor-mediated transcription by amino-terminal enhancer of split. Mol. Cell. Biol. 2001;21:4614–4625. doi: 10.1128/MCB.21.14.4614-4625.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lin D. Y., Fang H. I., Ma A. H., Huang Y. S., Pu Y. S., Jenster G., Kung H. J., Shih H. M. Negative modulation of androgen receptor transcriptional activity by daxx. Mol. Cell. Biol. 2004;24:10529–10541. doi: 10.1128/MCB.24.24.10529-10541.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hodgson M. C., Astapova I., Cheng S., Lee L. J., Verhoeven M. C., Choi E., Balk S. P., Hollenberg A. N. The androgen receptor recruits nuclear receptor CoRepressor (N-CoR) in the presence of mifepristone via its N and C termini revealing a novel molecular mechanism for androgen receptor antagonists. J. Biol. Chem. 2005;280:6511–6519. doi: 10.1074/jbc.M408972200. [DOI] [PubMed] [Google Scholar]
- 172.Dotzlaw H., Moehren U., Mink S., Cato A. C., Iniguez Lluhi J. A., Baniahmad A. The amino terminus of the human AR is target for corepressor action and antihormone agonism. Mol. Endocrinol. 2002;16:661–673. doi: 10.1210/mend.16.4.0798. [DOI] [PubMed] [Google Scholar]
- 173.Cutress R. I., Townsend P. A., Sharp A., Maison A., Wood L., Lee R., Brimmell M., Mullee M. A., Johnson P. W., Royle G. T., Bateman A. C., Packham G. The nuclear BAG-1 isoform, BAG-1L, enhances oestrogen-dependent transcription. Oncogene. 2003;22:4973–4982. doi: 10.1038/sj.onc.1206688. [DOI] [PubMed] [Google Scholar]
- 174.Tan J. A., Hall S. H., Hamil K. G., Grossman G., Petrusz P., French F. S. Protein inhibitors of activated STAT resemble scaffold attachment factors and function as interacting nuclear receptor coregulators. J. Biol. Chem. 2002;277:16993–17001. doi: 10.1074/jbc.M109217200. [DOI] [PubMed] [Google Scholar]
- 175.Zhao Y., Goto K., Saitoh M., Yanase T., Nomura M., Okabe T., Takayanagi R., Nawata H. Activation function-1 domain of androgen receptor contributes to the interaction between subnuclear splicing factor compartment and nuclear receptor compartment. Identification of the p102 U5 small nuclear ribonucleoprotein particlebinding protein as a coactivator for the receptor. J. Biol. Chem. 2002;277:30031–30039. doi: 10.1074/jbc.M203811200. [DOI] [PubMed] [Google Scholar]