Abstract
RPA (replication protein A) is an essential factor for DNA DSB (double-strand break) repair and cell cycle checkpoint activation. The 32 kDa subunit of RPA undergoes hyperphosphorylation in response to cellular genotoxic insults. However, the potential involvement of hyperphosphorylated RPA in DSB repair and checkpoint activation remains unclear. Using co-immunoprecipitation assays, we showed that cellular interaction of RPA with two DSB repair factors, Rad51 and Rad52, was predominantly mediated by the hyperphosphorylated species of RPA in cells after UV and camptothecin treatment. Moreover, Rad51 and Rad52 displayed higher affinity for the hyperphosphorylated RPA than native RPA in an in vitro binding assay. Checkpoint kinase ATR (ataxia telangiectasia mutated and Rad3-related) also interacted more efficiently with the hyperphosphorylated RPA than with native RPA following DNA damage. Consistently, immunofluorescence microscopy demonstrated that the hyperphosphorylated RPA was able to co-localize with Rad52 and ATR to form significant nuclear foci in cells. Our results suggest that hyperphosphorylated RPA is preferentially localized to DSB repair and the DNA damage checkpoint complexes in response to DNA damage.
Keywords: checkpoint activation, double strand break repair, preferential localization, Rad51, replication protein A, RPA hyperphosphorylation
Abbreviations: ATM, ataxia telangiectasia mutated; ATR, ATM and Rad3-related; ATRIP, ATR interacting protein; CIAP, calf intestinal alkaline phosphatase; CPT, camptothecin; DNA-PK, DNA-dependent protein kinase; DSB, double-strand break; DTT, dithiothreitol; FBS, fetal bovine serum; HR, homologous recombination; HU, hydroxyurea; IP, immunoprecipitation; NER, nucleotide excision repair; PIKK, phosphatidylinositol 3-kinase-related kinase; RPA, replication protein A; ssDNA, single-stranded DNA; SV40, simian virus 40
INTRODUCTION
RPA (replication protein A) is a eukaryotic ssDNA (single-stranded DNA)-binding protein composed of three tightly associated subunits of approx. 70, 32 and 14 kDa (referred to hereafter as RPA70, RPA32, RPA14 respectively). RPA was originally identified as a factor indispensable for in vitro SV40 (simian virus 40) DNA replication. Subsequently, it was found that RPA takes part in almost all aspects of cellular DNA metabolism, including DNA replication, recombination and repair, and plays essential roles in these pathways [1,2]. Recently, RPA has been implicated in regulating DNA damage checkpoint activation [3,4]. It is believed that RPA participates in such diverse pathways through its ability to interact with numerous protein partners and to undergo extensive post-translational modifications [1,2,5].
The involvement of RPA in human NER (nucleotide excision repair) reactions has been well documented (reviewed in [6–8]). The heterotrimeric RPA forms a complex with XPA (xeroderma pigmentosum A) protein and functions at the early stage of human NER. In the HR (homologous recombination) pathway of DSB (double strand break) repair, RPA has been shown to interact with two Rad52 epistasis group proteins, Rad51 and Rad52, and to modulate their activities [9–11]. RPA promotes Rad51 presynaptic filament assembly by reducing secondary structure in the long length of ssDNA regions, but also suppresses the assembly by occluding the DNA [12]. The inhibitory effect of RPA, nevertheless, can be overcome significantly in the presence of Rad52 because Rad52 recognizes RPA-bound ssDNA, and this activity of Rad52 allows the Rad51–Rad52 complex to gain access to ssDNA already covered with RPA [12,13]. The role of RPA in the NHEJ (non-homologous end joining) pathway of DSB repair is not well defined, although it has been observed that RPA interacts with DNA-PK (DNA-dependent protein kinase) and co-localizes with the MRN (Mre11/Rad50/Nbs1) complex [14,15].
A recent advancement towards our understanding of RPA functions is the finding of RPA involvement in the initiation of DNA damage checkpoint signalling, both in yeast and mammalian cells [3,4,16–18]. Binding of RPA to ssDNA generated during repair processes or as a result of replication fork stalling serves as a common intermediate structure for the assembly of two independent checkpoint apparatuses, Rad17-Rfc2-5/Rad9-Rad1-Hus1 and ATR [ATM (ataxia telangiectasia mutated) and Rad3-related]-ATRIP (ATR interacting protein) complexes, at the site of DNA damage [3,4,18]. RPA-mediated localization of ATR to damage sites elicits a variety of cellular responses to DNA damage, e.g., phosphorylation of a key downstream kinase Chk1 by ATR [3], which eventually leads to cell-cycle arrest.
While the 32 kDa subunit of RPA is phosphorylated in a cell cycle-dependent manner [1,2], hyperphosphorylation of RPA occurs in response to a variety of DNA damage agents such as UV or ionizing irradiation treatments [1,2,5]. Although the involvement of RPA in most DNA metabolic pathways has been described in some detail, the exact role of RPA hyperphosphorylation in cellular DNA damage responses is still poorly understood. It has been reported that cellular extracts prepared from UV-treated cells displayed a reduction in the ability to support DNA replication in the SV40 in vitro replication system [2,19]. Recently, it was found that RPA32 mutants that mimic the hyperphosphorylation were unable to localize to replication centres in cells, but were competent to associate with DNA damage foci [20]. In addition, RPA hyperphosphorylation has been suggested to have no effect on NER activity in vitro with crude cellular extracts or a purified reconstituted system [21,22].
The damage-induced hyperphosphorylation of RPA32 is believed to be carried out by members of PIKK (phosphatidylinositol 3-kinase-related kinase) kinase family, including DNA-PK, ATM and ATR, although the relative contribution of these kinases to RPA hyperphosphorylation has not been defined [5]. Interestingly, each of these kinases is either involved in DSB repair (DNA-PK) or in the DNA damage checkpoint network (ATR, ATM), suggesting that RPA is an initiator as well as an effector for these two pathways. In response to DNA damage, a co-localization of RPA with various proteins involved in DSB repair and checkpoint signalling, such as ATR–ATRIP complex, ATM, BRCA1 and Rad52, has been observed [10,18,23,24]. However, the involvement of hyperphosphorylated RPA in DSB repair and checkpoint pathways as yet has not been investigated. The present study reports that the cellular interactions of two DSB repair factors Rad51 and Rad52 with RPA were predominantly mediated by the hyperphosphorylated form of RPA after UV irradiation and CPT (camptothecin) treatment. The checkpoint kinase ATR also displayed a slightly stronger interaction with the hyperphosphorylated RPA after DNA damage. The immunofluorescent analysis further demonstrated that the hyperphosphorylated RPA was able to significantly co-localize with Rad52 and ATR to form nuclear foci in cells. Our results suggest that following DNA damage, hyperphosphorylated RPA is preferentially located to DSB repair and checkpoint complexes over native RPA.
MATERIALS AND METHODS
Cell culture and treatments
Human lung adenocarcinoma cells A549 were obtained from American Type Culture Collection (A.T.C.C., Manassas, VA, U.S.A.) and maintained at 37 °C in 5% CO2 in Dulbecco's Modified Eagle's medium (Invitrogen) supplemented with 10% (v/v) FBS (fetal bovine serum; Invitrogen) and 1% penicillin/streptomycin (Invitrogen). For UV exposure, the growth medium was removed and cells were washed once with PBS, and then irradiated with various doses using a UV crosslinker (Stratagene, La Jolla, CA, U.S.A.) at a dose rate of 0.5 J·m−2·s−1. After UV exposure, the original growth medium was added back and the cells were incubated further for 2 h at 37 °C before harvesting. For drug treatments, all chemicals used in this study were purchased from Sigma, unless otherwise stated.
Preparations of whole cell extracts and nuclear extracts
To prepare whole cell extracts, cells were washed once with PBS, then scraped and collected in ice-cold PBS. After a brief centrifugation (15000 g, 20 s), cells were resuspended in solution A [50 mM Tris/HCl, pH 7.8, 400 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 0.34 M sucrose, 10% glycerol, 1 mM Na3VO4, 10 mM NaF, 10 mM β-glycerophosphate, 1 mM PMSF and a protease inhibitor cocktail (Roche, Indianapolis, IN, U.S.A.)] and left on ice for 30 min for lysis. Lysates were cleared by centrifugation (15000 g, 30 min at 4 °C) and the protein concentration was determined by the Bradford assay (Bio-Rad Laboratories, Richmond, VA, U.S.A.). For the preparation of nuclear extracts, cells were first lysed in buffer B (10 mM Hepes, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 0.1% Triton X-100, 1 mM Na3VO4, 10 mM NaF and 10 mM β-glycerophosphate, 1 mM PMSF and a protease inhibitor cocktail) on ice for 5 min. Nuclei were separated from cytoplasmic proteins by low-speed centrifugation (1300 g for 4 min). Isolated nuclei were washed once with solution B and then further lysed in solution A as above.
Western blotting
Cell lysates and immunoprecipitates were separated on 8 or 12% (for RPA32) SDS/polyacrylamide gels and transferred on to PVDF membrane (Amersham Biosciences, Piscataway, NJ, U.S.A.). The membranes were blocked for 1 h at room temperature (24 °C) with TBST (25 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.05% Tween 20) containing 5% (w/v) powdered milk and then probed using the following primary antibodies: anti-RPA32 (1:1000; Kamiya Biomedical, Seattle, WA, U.S.A.), anti-RPA70, anti-Rad51, anti-Rad52, anti-phospho-Chk1-317 (1:500, Santa Cruz Biotechnology, CA, U.S.A.) or anti-ATR (1:2000, Oncogene Science, Uniondale, NY, U.S.A.). The membranes were then incubated with horseradish peroxidase-linked secondary anti-mouse antibodies (Amersham) and bound antibodies were visualized using the ECL® chemiluminescent method (Amersham).
Co-IP (where IP stands for immunoprecipitation) assays
Whole cell extracts prepared from 1×107 cells were used for each co-IP reaction. Cell lysates were diluted with dilution buffer (50 mM Tris/HCl, pH 7.8, 1 mM EDTA, 10% glycerol, protease and phosphatase inhibitor as above), and incubated with 4 μg of rabbit anti-Rad51, anti-Rad52 and anti-ATR antibodies respectively, for 10–14 h at 4 °C with end-over-end mixing. Then, 50 μl of Protein A/G–agarose beads (Amersham) were added and the reaction mixtures mixed further for 1 h at 4 °C. The immunoprecipitates were separated from the supernatant by centrifugation and washed with PBS containing 0.05% Nonidet P-40. Proteins were extracted from the agarose beads by boiling in 1×SDS gel loading buffer and resolved by SDS/PAGE [12% (w/v) polyacrylamide gels].
RPA purification and in vitro phosphorylation by DNA-PK
Recombinant human RPA complex was expressed in Escherichia coli BL21 (DE3)-RP (Stratagene) cells and purified as described previously [25]. Recombinant RPA was phosphorylated in vitro by DNA-PK kinase (Promega, Madison, WI, U.S.A.) as described previously [26,27]. SDS/PAGE analysis showed that at least 90% RPA were hyperphosphorylated in the reaction. The protein was then purified to remove the DNA-PK kinase and calf thymus DNA in the reaction buffer. The phosphorylation reaction products were loaded on to an HR 10/30 Superdex 200 column with an AKTA purifier system (Amersham Biosciences, Uppsala, Sweden). The column was pre-equilibrated with high-salt FPLC running buffer [40 mM Hepes-KOH, pH 7.5, 2 M NaCl, 10 mM MgCl2, 10 μM ZnCl2 and 1 mM DTT (dithiothreitol)] and run in the same buffer at 4 °C. The fractions containing phosphorylated RPA were pooled and dialysed against RPA storage buffer (40 mM Hepes-KOH, pH 7.5, 50 mM NaCl, 10 mM MgCl2, 10 μM ZnCl2, 1 mM DTT and 50% glycerol). The protein concentration was determined by the Bradford assay.
Immunofluorescence
Cells were grown on 18 mm coverslips (Fisher Scientific, Fair Lawn, NJ, U.S.A.) overnight before treatment. Cells were treated with 20 J/m2 of UV irradiation followed by a 2 h recovery. After treatment, cells were washed with PBS, and then extracted with PBS containing 0.5% Nonidet P40 for 5 min on ice and fixed with 100% methanol at −20 °C for 10 min. Cells were then blocked for 30 min in PBS containing 15% FBS. Primary antibody dilutions used were as follows: rabbit anti-phospho- RPA32 Ser4/Ser8 1:2000 (Bethyl Laboratories, Montgomery, TX, U.S.A.), mouse anti-RPA32 1:1000 (Kamiya Biomedical), rabbit anti-Rad52 1:500 (Santa Cruz Biotechnology), mouse anti-Rad52 1:250 (Abcam, Cambridge, U.K.), rabbit anti-ATR 1:2000 (Oncogene) and mouse anti-ATR 1:1000 (GeneTex, San Antonio, TX, U.S.A.). Secondary antibody dilutions were as follows: anti-rabbit Alexa Fluor 488 1:250 and anti-mouse Alexa Fluor 568 1:250 (Molecular Probes). Images were captured with a Nikon inverted fluorescent microscope with attached CCD camera at ×100 magnification and processed using Photoshop 6.0 (Adobe) software. The co-localization was quantified by counting the number of co-localized foci in each of randomly selected cells.
RESULTS
Hyperphosphorylation of RPA in response to DNA damage
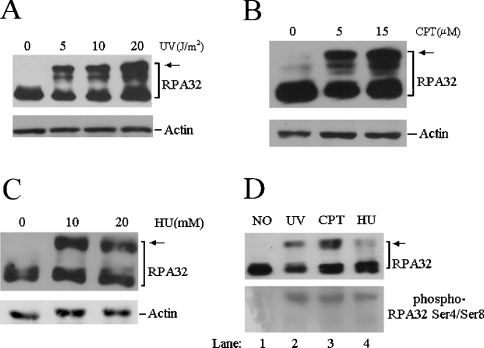
To induce the cellular hyperphosphorylation of RPA, two different types of genotoxic agents were employed for cell treatment. Cultured cells were irradiated with varying doses of UV and the whole cell lysates prepared from the irradiated cells were then subjected to Western blotting probed with anti-RPA32 antibody. Similar to previous observations, the treatment resulted in significant hyperphosphorylation of the RPA32 subunit, as indicated by the appearance of additional slower-migrating bands on SDS/PAGE (Figure 1A, [28,29]). Treatment with CPT, which induces DSBs through inhibition of topoisomerase, or with HU (hydroxyurea) that inhibits ribonucleotide reductase and leads to nucleotide depletion, also resulted in a substantial hyperphosphorylation of the RPA32 subunit (Figures 1B and 1C). The hyperphosphorylated RPA32 were predominantly located within the nucleus, while cytoplasmic fractions contained little, if any, hyperphosphorylated forms (results not shown). In addition, the hyperphosphorylation of RPA could be detected by an antibody that specifically recognized the phosphorylated Ser4 and Ser8 of RPA32 (Figure 1D, lanes 2–4 [14]).
Figure 1. Induction of RPA hyperphosphorylation by genotoxic agents in human cells.
(A) Exponentially growing A549 cells were treated with indicated doses of UV followed by a 2 h recovery. Total cellular lysates were prepared and then subjected to Western blotting with anti-RPA32 and anti-actin antibodies respectively. (B, C) A549 cells were treated with indicated doses of CPT (B) or HU (C) for 3 h. Total cell lysates were prepared for Western blotting with anti-RPA32 and anti-actin antibodies respectively. (D) A549 cells were treated with 10 J/m2 UV, 2 μM CPT and 1 mM HU (lanes 2–4) respectively, as indicated above. Whole cell lysates were prepared and probed with anti-RPA32 (upper panel) and anti-phospho-RPA32 Ser4/Ser8 (lower panel) antibodies respectively. The arrows indicate the hyperphosphorylated RPA.
Interaction of hyperphosphorylated RPA with DSB repair factors
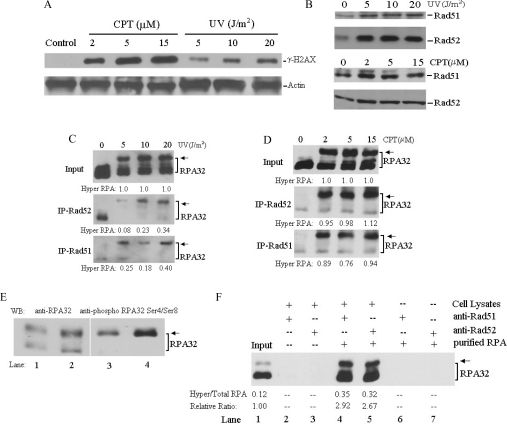
We next examined the involvement of hyperphosphorylated RPA in the DSB repair pathway. The induction of DSBs in cells after UV irradiation and CPT treatment was determined by assessing the level of phosphorylated histone H2AX (γH2AX), a marker for DSB formation (Figure 2A and [30,31]). Treatment with CPT appeared to induce DSBs more efficiently than the moderate doses of UV irradiation (Figure 2A). UV irradiation and CPT treatments also resulted in significant nuclear accumulation of Rad51 and Rad52 (Figure 2B), two essential components for HR, indicating the proficiency of DSB repair in the cell nucleus. Therefore, co-IP assays were performed to examine the interaction of RPA with Rad51 and Rad52 upon DNA damage. Strikingly, the interaction of RPA with both Rad51 and Rad52 in cells was predominantly mediated by the hyperphosphorylated species of RPA (Figures 2C and 2D). Moreover, more hyperphosphorylated RPA was immunoprecipitated from cells treated with CPT than UV irradiation (Figures 2C and 2D). Pretreatment of the cellular lysates with DNaseI or ethidium bromide before IP did not alter the interaction patterns of RPA with Rad51 and Rad52 (results not shown), suggesting that the preferential interaction of DSB repair factors with hyperphosphorylated RPA was mediated by protein–protein interactions. We also determined if this interaction was due to the direct binding of hyperphosphorylated RPA to Rad51/Rad52. For this purpose, purified RPA was hyperphosphorylated in vitro by DNA-PK. As shown in Figure 2(E), the in vitro hyperphosphorylated RPA (lanes 2 and 4) had the same migration pattern on SDS/PAGE as the endogenous phosphorylated RPA prepared from UV-treated cells (lanes 1 and 3 and [28]), and could be detected by the anti-phosphoRPA32 Ser4/Ser8 antibody (lane 4). This indicated that the in vitro phosphorylated RPA could be used as an equivalent to endogenous phosphorylated RPA. After IP of cell lysates with anti-Rad51 or anti-Rad52 antibodies, the immunoprecipitates were washed with the buffer containing a high concentration of salt (0.6 M NaCl) to remove the proteins (including endogenous RPA) associated with the immunoprecipitated Rad51 or Rad52 (Figure 2F, lanes 2 and 3). Then the in vitro hyperphosphorylated RPA was supplied to allow for interaction with the endogenous Rad51 or Rad52 in the immunoprecipitates in the normal buffer. As shown in Figure 2(F), both Rad51 and Rad52 did directly interact with RPA with relatively higher affinities to hyperphosphorylated RPA than to unphosphorylated RPA (2.92- and 2.67-fold respectively, lanes 4 and 5). It appears, however, that these moderately increased affinities in vitro may not fully account for the prominent interactions of Rad51 and Rad52 with hyperphosphorylated RPA in cells.
Figure 2. Co-IP of Rad51 and Rad52 with hyperphosphorylated RPA after DNA damage.
(A) Exponentially growing cells were treated with increasing doses of UV irradiation or CPT as above, and then total cell lysates were prepared and probed with anti-γH2AX and anti-actin antibodies respectively. (B) Cells were treated with the indicated doses of UV irradiation followed by a 2 h recovery or treated with the indicated doses of CPT for 3 h. Nuclear extracts were prepared and analysed by Western blotting with anti-Rad52 or anti-Rad51 antibodies. (C, D) Cells were treated with indicated doses of UV or CPT and then total cell lysates were prepared for co-IP assays with anti-Rad51 and anti-Rad52 antibodies. Proteins from the immunoprecipitates were detected by Western blotting using anti-RPA32 antibody. As control, 10% of the total volumes of the whole cell lysates used for the IP were also included (Input, upper panel). The hyperphosphorylated RPA immunoprecipitated with Rad51 and Rad52 was quantified by densitometry and further normalized to the inputs that were designated as of value 1. (E) Total cell lysates prepared from 20 J/m2 UV irradiated cells (lanes 1 and 3), and 4 pmol in vitro phosphorylated RPA (mixed with 1 pmol native RPA as an internal control) (lanes 2 and 4) were probed by Western blotting with anti-RPA32 (lanes 1 and 2) and anti-phospho-RPA32 Ser4/Ser8 (lanes 3 and 4) antibodies respectively. (F) Total cell lysates prepared from untreated cells were incubated with Rad51 or Rad52 antibodies. The immunoprecipitates were washed three times with PBS containing 0.05% Nonidet P-40, and further incubated in a high concentration of salt buffer (15 mM Tris/HCl, pH 7.5, 600 mM NaCl and 0.1% NP-40) for 30 min at 4 °C to remove endogenously bound proteins (lanes 2 and 3). Then the purified RPA containing a 1:7 mix of in vitro phosphorylated/native protein (lane 1) was added and incubated in RPA binding buffer for 4–6 h. The IPs were washed free of unbound proteins before the bound proteins were detected by Western blotting with RPA32 antibody (lanes 4 and 5). Anti-Rad51, anti-Rad52 or protein A/G beads did not bind non-specifically to the purified RPA (lanes 6 and 7).
Co-localization of hyperphosphorylated RPA with Rad52 on DNA damage
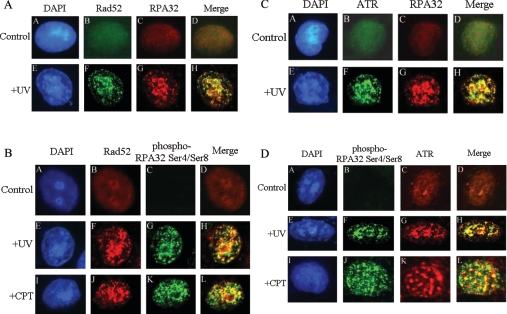
In order to explore the role of co-localization of RPA-Rad52 in DNA damage, immunofluorescence experiments were carried out. In mock-treated cells, RPA and Rad52 appeared to be homogeneously distributed throughout the nucleus (Figure 3A, subpanels B and C). After 20 J/m2 UV treatment, there was a clear redistribution of RPA and Rad52 that formed discrete nuclear foci and, as expected, co-localized mostly with each other (Figure 3A subpanels F–H). These results indicate that RPA (including both native and phosphorylated forms) interacted efficiently with Rad52 at the DSB sites to support DSB repair. To probe the co-localization of Rad52 specifically with hyperphosphorylated RPA, immunofluorescence assays with the antibody uniquely recognizing the phosphorylated RPA32 were performed. Under mock-treatment conditions, no staining with the phospho-specific RPA antibody was observed (Figure 3B, subpanel C). Following treatment of cells with UV or CPT, hyperphosphorylated-RPA aggregated into nuclear foci that also showed efficient co-localization with Rad52 foci (Figure 3B, subpanels F–H and J–L). These results are in full agreement with those obtained from our IP assays, and together suggest that hyperphosphorylated RPA could efficiently participate in the DSB repair pathway.
Figure 3. Co-localization of RPA with Rad52 and ATR after DNA damage.
Cells were mock-treated, treated with 20 J/m2 UV irradiation followed by 2 h recovery or treated with 10 μM CPT for 3 h. After extraction of cytoplasmic proteins, cells were fixed and incubated with primary and secondary antibodies and visualized by fluorescence microscopy. (A) Cells were stained with rabbit anti-Rad52 antibody (green, subpanels B and F) and mouse anti-RPA32 antibody (red, subpanels C and G). Subpanels D and H are the merged images of the anti-Rad52 and anti-RPA32 stained cells. Subpanels A and E are the DAPI (4′,6-diamidino-2-phenylindole)-stained nuclei. (B) Cells were stained with mouse anti-Rad52 antibody (red, subpanels B, F and J) and rabbit anti-phospho-RPA32 Ser4/Ser8 antibody (green, subpanels C, G and K). Subpanels D, H and L are the merged images of the anti-Rad52 and anti-phospho-RPA32 Ser4/Ser8 stained cells. (C) Cells were stained with rabbit anti-ATR antibody (green, subpanels B and F) and mouse anti-RPA32 antibody (red, subpanels C and G). Subpanels D and H are the merged images of the anti-ATR and anti-RPA32 stained cells. Subpanels A and E are the DAPI stained nuclei. (D) Cells were stained with rabbit anti-phospho-RPA32 Ser4/Ser8 antibody (green, subpanels B, F and J) and mouse anti-ATR antibody (red, subpanels C, G and K). Subpanels D, H and L are the merged images of the anti-ATR and anti-phospho-RPA32 Ser4/Ser8 stained cells.
Participation of hyperphosphorylated RPA in checkpoint pathways
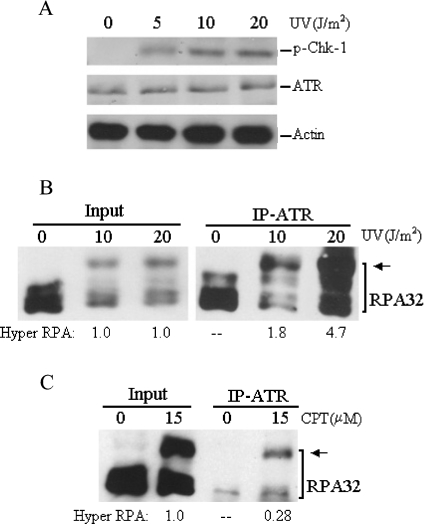
As shown in Figure 4(A), the UV irradiation led to the phosphorylation of Chk1 protein kinase [3], a key substrate of ATR–ATRIP, indicating that the checkpoint was specifically activated in these cells. ATR has been reported to interact with RPA through its interacting partner ATRIP [20]. The interactions of RPA and hyperphosphorylated RPA with ATR in response to DNA damage were therefore examined. As revealed in Figures 4(B) and 4(C), the ATR did interact with the hyperphosphorylated RPA following UV and CPT treatments and, in comparison with unphosphorylated RPA, more hyperphosphorylated RPA was co-immunoprecipitated with ATR antibody (from at least three independent experiments). Furthermore, hyperphosphorylated RPA appeared to be immunoprecipitated by ATR more efficiently from cell lysates prepared from UV irradiated cells than from CPT-treated cells (Figures 4B and 4C), probably reflecting the fact that the CPT-induced DSBs may activate an alternative checkpoint, e.g. the ATM kinase pathway which is RPA-independent [32].
Figure 4. Co-IP of ATR with hyperphosphorylated RPA after DNA damage.
(A) Cells were treated with indicated doses of UV irradiation, and then whole cell lysates were prepared and probed with anti-phospho-Chk1-317, anti-ATR and anti-β-actin antibodies respectively. (B) and (C) Cells were treated with indicated doses of UV or CPT, and then whole cell lysates were prepared for co-IP assays with ATR antibody. Proteins from the immunoprecipitates were detected by Western blotting using anti-RPA32 antibody. The arrows indicate the hyperphosphorylated RPA. The hyperphosphorylated RPA immunoprecipitated with ATR was quantified by densitometry and normalized to the inputs that were designated as of value 1.
Immunofluorescence experiments were also performed to examine the co-formation of nuclear foci of ATR and RPA in cells. In mock-treated cells, immunostaining with ATR antibody revealed a few diffusive nuclear spots (Figure 3C, subpanel B; and Figure 3D, subpanel C), which is consistent with the proposed role of ATR in monitoring genome integrity during normal cell-cycle progression [18]. On exposure to UV, a clear redistribution of ATR in nucleus occurred and some brighter nuclear foci were formed (Figure 3C, subpanel F; and Figure 3D, subpanel G). When overlapped, ATR and RPA foci displayed obvious co-localization as expected (Figure 3C, subpanel H). Then the antibody that recognized only the phosphorylated RPA but not the native RPA was applied for immunofluorescent staining. As shown in Figure 3D (subpanel H), after UV irradiation, ATR co-localized with the hyperphosphorylated RPA foci with an efficiency comparable with that of ATR with the total RPA (compare Figure 3C, subpanel H with Figure 3D, subpanel H). Some extent of co-localization of ATR with hyperphosphorylated RPA was also observed after treatment with CPT (Figure 3D, subpanel L). Altogether these data supported the conclusion that hyperphosphorylated RPA was involved in the ATR-dependent checkpoint signalling process in response to DNA damage.
Interaction of hyperphosphorylated RPA with Rad51, Rad52 and ATR after a high dose of UV irradiation
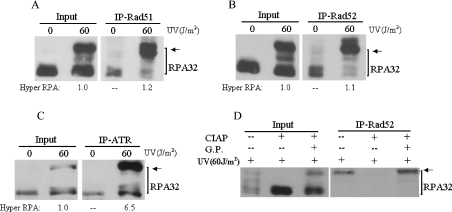
In general, high-doses of DNA damage agents are required to induce extensive DSBs and a strong checkpoint response. To further address the hypothesis that hyperphosphorylated RPA is preferentially localized to DSB repair and checkpoint complexes, we irradiated cells with 60 J/m2 of UV, a dose that has been widely used to activate DNA damage checkpoint pathways and induce extensive DSBs in cells. The cells were then harvested 2 h after treatment, when the early cellular DNA damage responses were highly active, but long before the cells started the process of apoptosis. Remarkably, following 60 J/m2 of UV irradiation, significantly more hyperphosphorylated RPA was immunoprecipitated by DSB repair factors Rad51 and Rad52 (Figures 5A and 5B). The interaction of ATR with hyperphosphorylated RPA was also slightly enhanced by a high dose of UV treatment (compare Figure 5C with Figure 4B). Treatment of cell lysates with DNase I or ethidium bromide before the IP had little effect on the hyperphosphorylated RPA interactions with ATR, Rad51 and Rad52 (results not shown). Altogether, these results suggest that the interactions of hyperphosphorylated RPA with Rad51, Rad52 and ATR are dependent on DSB generation and/or checkpoint activation and therefore further support a preferential participation of hyperphosphorylated RPA in DSB repair and checkpoint pathways.
Figure 5. Interactions of Rad51, Rad52 and ATR with hyperphosphorylated RPA after 60 J/m2 UV irradiation of cells.
Cells were treated with 60 J/m2 UV irradiation followed by 2 h recovery and total cellular lysates were prepared for co-IP assays with anti-Rad51 (A), anti-Rad52 (B) or anti-ATR (C) antibodies. Proteins from the immunoprecipitates were detected by Western blotting using anti-RPA32 antibody. Arrows indicate the hyperphosphorylated RPA. The hyperphosphorylated RPA immunoprecipitated with ATR was quantified by densitometry and normalized to the inputs that were designated to be of value 1. (D) Whole cell lysates were prepared from 60 J/m2 UV irradiated cells. Before co-IP assays, the lysates were treated with 200 units of CIAP for 1 h at 37 °C in the absence (−/+) or presence (+/+) of 50 mM glycerophosphate (G.P.), or mock treated (–/–). Treated cell lysates were then subjected to co-IP with anti-Rad52 antibody. The bound proteins were then detected by Western blotting using anti-RPA32 antibody.
Since Rad51, Rad52 and ATR appeared to interact primarily with hyperphosphorylated species of RPA in cells (either directly or indirectly), we attempted to verify if phosphorylation was required for the interactions. To this end, whole cell lysates from 60 J/m2 UV-treated cells were pre-incubated with CIAP (calf intestinal alkaline phosphatase) before the co-IP assays. As shown in Figure 5(D), the CIAP treatment abolished RPA interaction with DSB repair factors Rad52 and Rad51 (results not shown). However, the presence of the CIAP inhibitor, glycerophosphate, preserved the interaction (Figure 5D), indicating that the disruption of Rad52–RPA interaction was indeed the result of de-phosphorylation of proteins (hyperphosphorylated RPA, other cellular phospho-proteins or both). In addition, treatment of the cells with 60 J/m2 UV irradiation in the presence of wortmannin, an inhibitor of PIKK family kinases which also inhibits RPA hyperphosphorylation [33], abrogated co-IP of ATR with RPA (results not shown). Taken together, these results indicate that the cellular protein phosphorylation may be essential for the efficient interaction of RPA with DSB repair factors Rad51 and Rad52 and checkpoint kinase ATR, although the exact proteins involved (other than RPA) remain to be identified.
DISCUSSION
The damage-induced phosphorylations of RPA primarily target the N-terminus of the 32 kDa subunit of RPA (RPA32), an unstructured domain containing multiple serine and threonine residues. Phosphorylations or mutations that add negative charges to this domain have been suggested to cause a conformational change in the RPA trimeric complex that may regulate RPA interactions with DNA and protein partners [2,5,34,35]. It has been shown that phosphorylation caused the decrease in dsDNA binding and helix destabilization activity of RPA [34], and altered RPA interactions with numerous proteins involved in DNA replication, repair and checkpoint response, including T antigen, DNA polymerase α, p53, ATM and DNA-PK [15,36,37]. These observations illustrated the modulation of RPA functions through phosphorylation in response to DNA damage.
Although the involvement of RPA in DSB repair and ATR-dependent checkpoint pathways has been well demonstrated, the potential participation of hyperphosphorylated RPA in these processes has not previously been investigated. In the present study, we showed that interaction of RPA with DSB repair factors Rad51 and Rad52, two essential components in HR pathway of DSB repair, was mostly mediated by the hyperphosphorylated form of RPA in cells. Interestingly, the extent of the cellular interaction between Rad51/Rad52 and hyperphosphorylated RPA was dependent on the type of DNA damaging agent (Figures 2C and 2D). The CPT treatment which directly leads to the formation of DSBs triggered a significant interaction, while the lower doses of UV irradiation (5–20 J/m2) induced relatively less interaction (Figures 2C and 2D). However, the interaction was apparently enhanced when the UV dose was further increased to 60 J/m2 (Figures 5A and 5B). In contrast, hyperphosphorylated RPA interacted with ATR more efficiently upon UV irradiation than after CPT treatment. This is in line with the facts that UV induces less DSBs than CPT, and that ATR mainly responds to UV-induced DNA damage or alike [8]. Altogether, these results suggest that the quantitative localization of hyperphosphorylated RPA to DSB repair and checkpoint pathways relies on the amount of DSB generated and the degree of checkpoint activation after DNA damage. The participation of hyperphosphorylated RPA in these processes was confirmed by immunofluorescent analysis showing the co-localization of hyperphosphorylated RPA with Rad52 and ATR in cells. Moreover, the MRN complex, which functions in the NHEJ pathway of DSB repair and as a damage sensor upstream of ATM activation, and the Rad9–Rad1–Hus1 checkpoint complex, which is recruited to DNA damage sites independently of ATR, also have been reported to interact and co-localize with hyperphosphorylated RPA after UV treatment of cells [14,38], further supporting our hypothesis.
Two possible scenarios can be considered regarding the relationship between the hyperphosphorylation of RPA and its preferential localization to DSB repair and DNA damage checkpoint complexes in cells. First, upon DNA damage, RPA is subject to hyperphosphorylation, which then stimulates RPA localization to DSB repair or checkpoint complexes. The phosphorylation-mediated enhancement of the interaction of RPA with the proteins specifically involved in DSB repair and checkpoint activation (e.g. Rad51 and Rad52, Figure 2) may modulate these preferential localizations. In the second scenario, RPA is efficiently hyperphosphorylated by the PIKK family of kinases after it is recruited to DSB repair and checkpoint pathways. This hyperphosphorylation in turn stabilizes and facilitates the involvement of RPA in DSB repair and checkpoint pathways. It has been suggested that cellular RPA hyperphosphorylation depends on active DNA replication [28,29], which is necessary for the activation of the ATR-dependent DNA damage checkpoint and conversion of the unrepaired UV lesions or intermediates into DSBs [39,40]. In addition, efficient in vitro phosphorylation of RPA by PIKK kinases requires RPA binding to ssDNA and the optimal stimulation was observed when ssDNA longer than 30 nt was applied [41,42]. It is well known that in the processes of DSB repair and checkpoint activation, longer ssDNA intermediates are generated. Moreover, the kinases responsible for RPA hyperphosphorylation (DNA-PK, ATR and ATM) are involved in DSB repair and checkpoint activation [5]. All these data imply that RPA participating in DSB repair and checkpoint activation pathways is preferentially hyperphosphorylated. Upon phosphorylation, RPA may undergo structural changes and alter its binding affinities to protein partners (possibly also to DNA), which then modulate cellular functions of RPA in response to DNA damage.
Acknowledgments
This study was supported by NCI grant CA86927 (to Y.Z.). We thank Drs Antonio E. Rusinol and K. Singh (East Tennessee State University, Johnson City, TN, U.S.A.) for their assistance in using the immunofluorescence technique. We also thank Dr P. Musich (East Tennessee State University, Johnson City, TN, U.S.A.) for his critical reading of this paper.
References
- 1.Wold M. S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 2.Iftode C., Daniely Y., Borowiec J. A. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- 3.Zou L., Elledge S. J. Sensing DNA damage through ATRIP recognition of RPAssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 4.Zou L., Liu D., Elledge S. J. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binz S. K., Sheehan A. M., Wold M. S. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst) 2004;3:1015–1024. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 6.Thoma B. S., Vasquez K. M. Critical DNA damage recognition functions of XPChHR23B and XPA-RPA in nucleotide excision repair. Mol. Carcinog. 2003;38:1–13. doi: 10.1002/mc.10143. [DOI] [PubMed] [Google Scholar]
- 7.Costa R. M., Chigancas V., Galhardo R., da S., Carvalho H., Menck C. F. The eukaryotic nucleotide excision repair pathway. Biochimie. 2003;85:1083–1099. doi: 10.1016/j.biochi.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 8.Sancar A., Lindsey-Boltz L. A., Unsal-Kaccmaz K., Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 9.Golub E. I., Gupta R. C., Haaf T., Wold M. S., Radding C. M. Interaction of human rad51 recombination protein with single-stranded DNA binding protein, RPA. Nucleic Acids Res. 1998;26:5388–5393. doi: 10.1093/nar/26.23.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park M. S., Ludwig D. L., Stigger E., Lee S. H. Physical interaction between human Rad52 and RPA is required for homologous recombination in mammalian cells. J. Biol. Chem. 1996;271:18996–19000. doi: 10.1074/jbc.271.31.18996. [DOI] [PubMed] [Google Scholar]
- 11.Stauffer M. E., Chazin W. J. Physical interaction between replication protein A and Rad51 promotes exchange on single-stranded DNA. J. Biol. Chem. 2004;279:25638–25645. doi: 10.1074/jbc.M400029200. [DOI] [PubMed] [Google Scholar]
- 12.Sung P., Krejci L., Van Komen S., Sehorn M. G. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 13.Sugiyama T., Kowalczykowski S. C. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 2002;277:31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- 14.Robison J. G., Elliott J., Dixon K., Oakley G. G. Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J. Biol. Chem. 2004;279:34802–34810. doi: 10.1074/jbc.M404750200. [DOI] [PubMed] [Google Scholar]
- 15.Shao R. G., Cao C. X., Zhang H., Kohn K. W., Wold M. S., Pommier Y. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA: DNA-PK complexes. EMBO J. 1999;18:1397–1406. doi: 10.1093/emboj/18.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bomgarden R. D., Yean D., Yee M. C., Cimprich K. A. A novel protein activity mediates DNA binding of an ATR-ATRIP complex. J. Biol. Chem. 2004;279:13346–13353. doi: 10.1074/jbc.M311098200. [DOI] [PubMed] [Google Scholar]
- 17.Dodson G. E., Shi Y., Tibbetts R. S. DNA replication defects, spontaneous DNA damage, and ATM-dependent checkpoint activation in replication protein A-deficient cells. J. Biol. Chem. 2004;279:34010–34014. doi: 10.1074/jbc.C400242200. [DOI] [PubMed] [Google Scholar]
- 18.Dart D. A., Adams K. E., Akerman I., Lakin N. D. Recruitment of the cell cycle checkpoint kinase ATR to chromatin during S-phase. J. Biol. Chem. 2004;279:16433–16440. doi: 10.1074/jbc.M314212200. [DOI] [PubMed] [Google Scholar]
- 19.Carty M. P., Zernik-Kobak M., McGrath S., Dixon K. UV light-induced DNA synthesis arrest in HeLa cells is associated with changes in phosphorylation of human singlestranded DNA-binding protein. EMBO J. 1994;13:2114–2123. doi: 10.1002/j.1460-2075.1994.tb06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vassin V. M., Wold M. S., Borowiec J. A. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol. Cell. Biol. 2004;24:1930–1943. doi: 10.1128/MCB.24.5.1930-1943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan Z.-Q., Park C. H., Amin A. A., Hurwitz J., Sancar A. Phosphorylated and unphosphorylated forms of human single-stranded DNA-binding protein are equally active in simian virus 40 DNA replication and in nucleotide excision repair. Proc. Natl. Acad. Sci. U.S.A. 1995;92:4636–4640. doi: 10.1073/pnas.92.10.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ariza R. R., Keyse S. M., Moggs J. G., Wood R. D. Reversible protein phosphorylation modulates nucleotide excision repair of damaged DNA by human cell extracts. Nucleic Acids Res. 1996;24:433–440. doi: 10.1093/nar/24.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plug A. W., Peters A. H., Xu Y., Keegan K. S., Hoekstra M. F., Baltimore D., de Boer P., Ashley T. ATM and RPA in meiotic chromosome synapsis and recombination. Nat. Genet. 1997;17:457–461. doi: 10.1038/ng1297-457. [DOI] [PubMed] [Google Scholar]
- 24.Choudhary S. K., Li R. BRCA1 modulates ionizing radiation-induced nuclear focus formation by the replication protein A p34 subunit. J. Cell Biochem. 2002;84:666–674. doi: 10.1002/jcb.10081. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z. G., Liu Y., Mao L. Y., Zhang J. T., Zou Y. Dimerization of human XPA and formation of XPA2-RPA protein complex. Biochemistry. 2002;41:13012–13020. doi: 10.1021/bi026064z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niu H., Erdjument-Bromage H., Pan Z-Q., Lee S. H., Tempst P., Hurwitz J. Mapping of amino acid residues in the p34 subunit of human single-stranded DNA-binding protein phosphorylated by DNA-dependent protein kinase and Cdc2 kinase in vitro. J. Biol. Chem. 1997;272:12634–12641. doi: 10.1074/jbc.272.19.12634. [DOI] [PubMed] [Google Scholar]
- 27.Zernik-Kobak M., Vasunia K., Connelly M., Anderson C. W., Dixon K. Sites of UV-induced phosphorylation of the p34 subunit of replication protein A from HeLa cells. J. Biol. Chem. 1997;272:23896–23904. doi: 10.1074/jbc.272.38.23896. [DOI] [PubMed] [Google Scholar]
- 28.Oakley G. G., Loberg L. I., Yao J., Risinger M. A., Yunker R. L., Zernik-Kobak M., Khanna K. K., Lavin M. F., Carty M. P., Dixon K. UV-induced hyperphosphorylation of replication protein A depends on DNA replication and expression of ATM protein. Mol. Biol. Cell. 2001;12:1199–1213. doi: 10.1091/mbc.12.5.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodrigo G., Roumagnac S., Wold M. S., Salles B., Calsou P. DNA replication but not nucleotide excision repair is required for UVC-induced replication protein A phosphorylation in mammalian cells. Mol. Cell. Biol. 2000;20:2696–2705. doi: 10.1128/mcb.20.8.2696-2705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward I. M., Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- 31.Redon C., Pilch D., Rogakou E., Sedelnikova O., Newrock K., Bonner W. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 2002;12:162–169. doi: 10.1016/s0959-437x(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 32.Bartek J., Lukas C., Lukas J. Checking on DNA damage in S phase. Nat. Rev. Mol. Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 33.Sarkaria J. N., Tibbetts R. S., Busby E. C., Kennedy A. P., Hill D. E., Abraham R. T. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 1998;58:4375–4382. [PubMed] [Google Scholar]
- 34.Binz S. K., Lao Y., Lowry D. F., Wold M. S. The phosphorylation domain of the 32-kDa subunit of replication protein A (RPA) modulates RPA-DNA interactions. Evidence for an intersubunit interaction. J. Biol. Chem. 2003;278:35584–35591. doi: 10.1074/jbc.M305388200. [DOI] [PubMed] [Google Scholar]
- 35.Georgaki A., Hübscher U. DNA unwinding by replication protein A is a property of the 70 kDa subunit and is facilitated by phosphorylation of the 32 kDa subunit. Nucleic Acids Res. 1993;21:3659–3665. doi: 10.1093/nar/21.16.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oakley G. G., Patrick S. M., Yao J., Carty M. P., Turchi J. J., Dixon K. RPA phosphorylation in mitosis alters DNA binding and protein-protein interactions. Biochemistry. 2003;42:3255–3264. doi: 10.1021/bi026377u. [DOI] [PubMed] [Google Scholar]
- 37.Abramova N. A., Russell J., Botchan M., Li R. Interaction between replication protein A and p53 is disrupted after UV damage in a DNA repair-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 1997;94:7186–7191. doi: 10.1073/pnas.94.14.7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu X., Shell S. M., Zou Y. Interaction and co-localization of Rad9/Rad1/Hus1 checkpoint complex with replication protein A in human cells. Oncogene. 2005;24:4728–4735. doi: 10.1038/sj.onc.1208674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lupardus P. J., Byun T., Yee M. C., Hekmat-Nejad M., Cimprich K. A. A requirement for replication in activation of the ATR-dependent DNA damage checkpoint. Genes Dev. 2002;16:2327–2332. doi: 10.1101/gad.1013502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dunkern T. R., Kaina B. Cell proliferation and DNA breaks are involved in ultraviolet light-induced apoptosis in nucleotide excision repair-deficient Chinese hamster cells. Mol. Biol. Cell. 2002;13:348–361. doi: 10.1091/mbc.01-05-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blackwell L. J., Borowiec J. A., Masrangelo I. A. Single-stranded-DNA binding alters human replication protein A structure and facilitates interaction with DNA-dependent protein kinase. Mol. Cell. Biol. 1996;16:4798–4807. doi: 10.1128/mcb.16.9.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartrand A. J., Iyasu D., Brush G. S. DNA stimulates Mec1-mediated phosphorylation of replication protein A. J. Biol. Chem. 2004;279:26762–26767. doi: 10.1074/jbc.M312353200. [DOI] [PubMed] [Google Scholar]